Abstract
Directed evolution is a useful method for the discovery of nucleic acids, peptides or proteins that have desired binding abilities or functions. Because of the abundance and importance of glycosylation in nature, directed evolution of glycopeptides and glycoproteins is also highly desirable. However, common directed evolution platforms such as phage-, yeast-, or mammalian-cell display are limited for these applications by several factors. Glycan structure at each glycosylation site is not genetically encoded, and yeast and mammalian cells produce a heterogenous mixture of glycoforms at each site on the protein. Although yeast, mammalian and Escherichia coli cells can be engineered to produce a homogenous glycoform at all glycosylation sites, there are just a few specific glycan structures that can readily be accessed in this manner. Recently, we reported a novel system for the directed evolution of glycopeptide libraries, which could in principle be decorated with any desired glycan. Our method combines in vitro peptide selection by mRNA display with unnatural amino acid incorporation and chemical attachment of synthetic oligosaccharides. Here, we provide an updated and optimized protocol for this method, which is designed to create glycopeptide mRNA display libraries containing ~1013 sequences and select them for target binding. The target described here is the HIV broadly-neutralizing monoclonal antibody 2G12; 2G12 binds to cluster of high-mannose oligosaccharides on the HIV envelope glycoprotein gp120 and glycopeptides that mimic this epitope may be useful in HIV vaccine applications. This method is expected to be readily applicable for other types of glycans and targets of interest in glycobiology.
Keywords: glycopeptide/glycoprotein, multivalency, HIV, CuAAC, click chemistry, broadly neutralizing antibodies, 2G12, mRNA display, directed evolution, selection
1. Introduction
Directed evolution is a method to engineer biological molecules such as proteins/peptides and nucleic acids by mimicking natural selection in the laboratory. It is useful for generating biomolecules with novel function or properties, and can also be used to generate data on structure-activity relationships. For directed evolution of peptides, a number of systems have been developed (Lane & Seelig, 2014; Packer & Liu, 2015). However, for glycopeptides or glycoproteins, only a few in vitro directed evolution methods have been reported (Arai, Tsutsumi, & Mihara, 2013; Horiya, Bailey, Temme, Guillen Schlippe, & Krauss, 2014a; Ng et al., 2015); phage- and cell-surface display methods have also been reported, but in these methods, control of glycosylation is limited (Çelik, Fisher, Guarino, Mansell, & DeLisa, 2010; Grimm, Battles, & Ackerman, 2015; Steichen et al., 2016). Yeast and mammalian cells normally produce a heterogenous mixture of glycoforms that is not genetically encoded, and wild-type (non-engineered) E. coli do not glycosylate proteins. Although all of these cell types can be engineered to produce a homogenous glycoform at all sites, only a few specific glycan structures can readily be accessed this way. Here, we describe a detailed method for in vitro directed evolution of glycopeptides using mRNA display (Horiya et al., 2014a), which is one of the most powerful methods of in vitro peptide selection, combined with chemical glycosylation, enabling library decoration with a glycan of potentially any homogenous structure. The strengths of our method compared with other in vitro techniques (Arai et al., 2013; Ng et al., 2015) are the large library diversity (~1013 sequences) and the fact that glycan incorporation can occur at multiple sites anywhere in the peptide. Multivalency is a particularly useful attribute to engineer into protein-binding glycopeptides because typical monovalent carbohydrate-protein interactions are weak (mM to μM KD) and require multivalent enhancement to achieve strong (nM) avidity (Kiessling, Young, Gruber, & Mortell, 2008).
We developed the in vitro glycopeptide selection system for the purpose of reverse engineering glycopeptide HIV vaccine candidates from known broadly neutralizing HIV antibodies (bnAb) (Bailey, Nguyen, Horiya, & Krauss, 2016; Horiya, MacPherson, & Krauss, 2014b). BnAbs are monoclonal antibodies that have been isolated from HIV-positive individuals and discovered to neutralize a broad spectrum of HIV strains (Burton & Hangartner, 2016). Many bnAbs such as 2G12 (Binley et al., 2004; Trkola et al., 1996) and a more recent large family known as the “PGT” antibodies (Walker et al., 2011) bind to epitopes comprising three or four glycans and usually some polypeptide motifs on the HIV envelope protein gp120 (Calarese et al., 2003; Garces et al., 2014; Julien et al., 2013; Pejchal et al., 2011; Scanlan et al., 2002). Our in vitro selection approach yields glycopeptides in which the glycans are optimally presented on the peptide scaffold to afford tight binding to the bnAb. As possible mimics of bnAb epitopes on gp120, these glycopeptides are being tested as immunogens for their ability to “re-elicit” bnAbs in vivo.
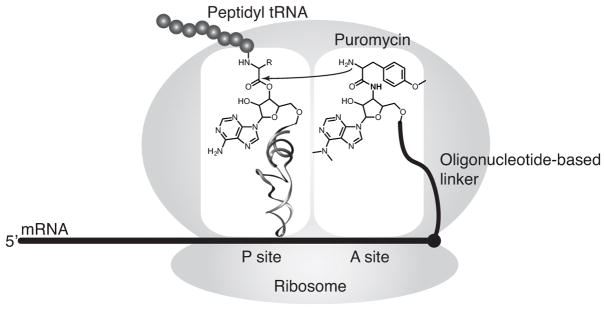
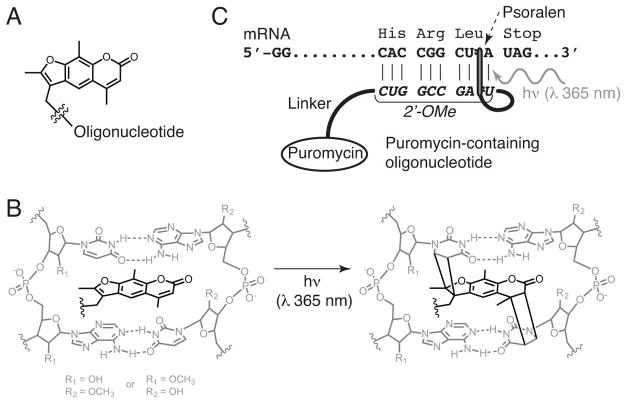
Our method is based on mRNA display, a technique that covalently links library peptides to their encoding mRNAs via a puromycin-functionalized oligonucleotide linker (Nemoto, Miyamoto-Sato, Husimi, & Yanagawa, 1997; Roberts & Szostak, 1997). In this method, mRNAs encoding the peptide library are conjugated to puromycin near their 3′-termini, then subjected to in vitro translation (Figure 1). As the ribosome reaches the end of the open reading frame (ORF), the puromycin moiety, which is an analogue of the 3′-end of tyrosyl-tRNA, enters the A-site of ribosome and is covalently joined to the C-terminus of the nascent peptide through a stable amide bond. In the resulting mRNA-peptide fusions, the mRNA part functions as the “genotype”, and the displayed peptide as the “phenotype”; these fusions are amenable to selections for a function of interest, which is typically binding to a certain target. Using a large, yet practical volume of in vitro translation mixture (hundred(s) of μL to 10 mL), mRNA display enables selections with large libraries containing 1012 to 1013 sequences.
Figure 1.
Puromycin-mediated formation of mRNA-peptide fusions in the ribosome.
mRNA display was originally developed to evolve peptides/proteins composed of natural amino acids. More recently, mRNA display has been combined with technologies enabling incorporation of unnatural amino acids in in vitro translation (Guillen Schlippe, Hartman, Josephson, & Szostak, 2012; Josephson, Ricardo, & Szostak, 2014; Kawakami, Ogawa, Hatta, Goshima, & Natsume, 2016; Passioura & Suga, 2017; Yamagishi et al., 2011). Evolved unnatural peptides, with altered functional group diversity, may exhibit enhanced stability and function. Critical for the method described here, functional groups can be incorporated that enable chemical conjugation of moieties far too large to be incorporated in translation. In our case the chemical conjugation method utilized is copper(I)-catalyzed azide alkyne cycloaddition (CuAAC), the most common “click” reaction (Kolb, Finn, & Sharpless, 2001; Rostovtsev, Green, Fokin, & Sharpless, 2002).
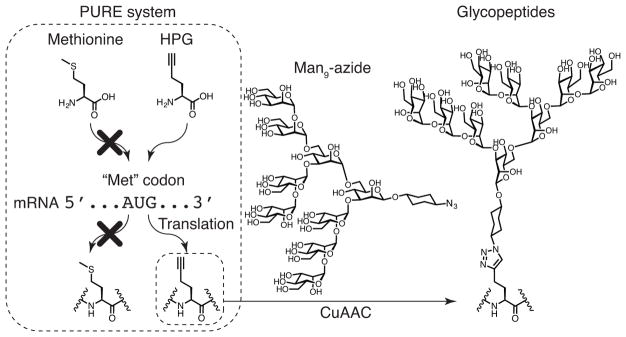
For incorporation of a “clickable” alkynyl amino acid, we have found it most convenient to replace methionine with homopropargylglycine (HPG) (van Hest, Kiick, & Tirrell, 2000), using PURE system translation (Josephson, Hartman, & Szostak, 2005) (Figure 2). The PURE system is a translation system that is reconstituted from purified components and, unlike cellular lysates and extracts, is highly customizable. For this application, the most essential advantage of the PURE system is that the natural amino acids present in cellular lysates and extracts can be excluded and replaced by unnatural amino acids with very little possibility of competition. In our system, HPG is thus incorporated into peptides at every AUG codon instead of methionine, and the alkyne groups of the incorporated HPG can be used for conjugation with synthetic azide-modified sugars by CuAAC to make “genetically encoded” glycopeptides.
Figure 2.
The customized PURE system, in which methionine is omitted and HPG is added, decodes AUG codons to HPG rather than methionine, producing alkynyl peptides. The alkynyl groups on peptides are conjugated with Man9-azide sugars through CuAAC click chemistry to form glycopeptides.
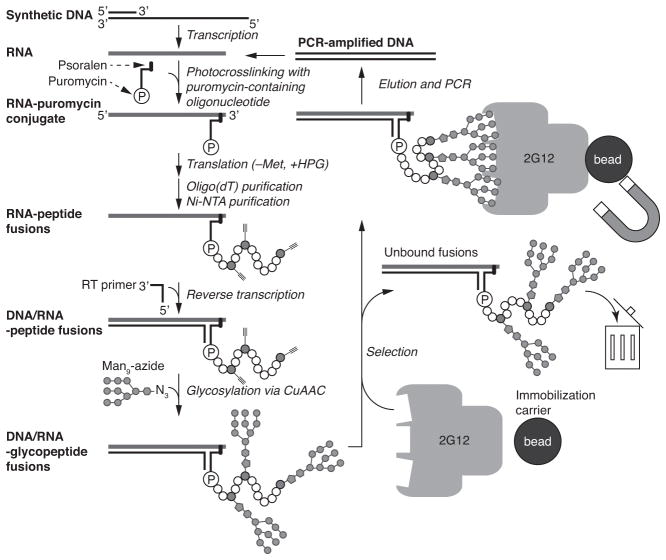
Here, we present a detailed protocol for directed evolution of glycopeptides using mRNA display, using 2G12 as a model target. The procedure is summarized in Figure 3: starting from a synthetic DNA library, RNA is transcribed and attached to puromycin. The RNA is then translated in a customized PURE system that contains HPG but not methionine, to form mRNA-alkynyl-peptide fusions. After purification steps, cDNA is synthesized to form cDNA/mRNA-peptide fusions by reverse transcription. Next, synthetic azide-modified sugars are conjugated by CuAAC. These cDNA/mRNA-glycopeptide fusions are selected for binding to the target, and the cDNA of bound fusions is amplified by PCR to yield DNA for the next round of selection. This selection cycle is repeated until the library is sufficiently enriched in target-binding glycopeptides, and then their sequences are determined. Using this method, we have successfully created libraries of ~1013 sequences, selected for binding to mAb 2G12 and identified multiple glycopeptides with subnanomolar to low nanomolar KDs for 2G12.
Figure 3.
Cycles of glycopeptide mRNA display and in vitro selection.
2. Overview of the PURE system
The PURE system is a cell-free translation system created by Ueda and colleagues (Shimizu et al., 2001). The PURE system reconstitutes the E. coli translation machinery, including purified, fully recombinant aminoacyl tRNA synthetases (AARSs) and translation factors, energy system, ribosomes, pooled tRNAs and amino acids. The translation components are customizable, which allows us to utilize a system in which methionine is omitted and HPG is added.
Currently, the PURE system is commercially available from New England Biolabs (NEB), with amino acids and tRNAs provided in separate tubes, if desired. If the amino acids are replaced with a customized mixture; in this case where methionine is replaced with HPG, the commercial PURE system can in principle be used. However, mainly for cost considerations, we have prepared our own PURE system; for the protocol shown here, > 5 mL of the PURE system are used throughout selection, equivalent to > 20 packs of NEB kit (PURExpress® Δ (aa, tRNA) Kit $327/250 μL). Moreover, although we have successfully utilized NEB PURE system in translation of many test RNA sequences, in one case, we experienced isolation of fragments of a test peptide when using NEB PURE system with NEB ribosomes. This is possibly due to the presence of small amounts of proteases or RNasesin the NEB ribosomes, which are purified from an E. coli BL21 derivative using a sucrose cushion method. In our hands, the fragmentation was reduced when using our own ribosomes, purified from the A19 strain in-house by hydrophobic chromatography or only a sucrose cushion method (data not shown). If the NEB PURE system is used, replacement of the commercially-supplied ribosomes with A19 strain ribosomes may be considered; in this case, PURE system Δ (ribosomes, aa, tRNA) can be obtained by special order.
Although several protocols describing detailed preparations of the PURE system are available (Z. Ma & Hartman, 2012; Shimizu & Ueda, 2010), the PURE system that we produce and utilize in our group possesses a unique feature besides the HPG introduction. We have included a protease inhibitor cocktail, which further reduces the minor residual fragmentation observed with A19 ribosomes. Our detailed protocol for the preparation of the PURE system is described in the appendix. Most components, e. g., ribosomes and His6-tagged forms of methionyl-tRNA formyltransferase (MTF), 9 translation factors (IF1, IF2, IF3, EF-G, EF-Tu, EF-Ts, RF1, RF3, RRF), and 20 AARSs are expressed in E. coli. Other PURE system components (amino acids, energy system, etc.) are assembled from commercially available products. Production of all components takes weeks to a few months, depending on how many proteins can be expressed and purified simultaneously. However, most of these are obtained in sufficient quantity for multiple selections. Following the procedures in the appendix for in-house preparation of proteins, yields sufficient for > 200 mL of PURE system translation have been obtained, for most proteins. The exception is EF-Tu, which is the most consumed protein in our system; in this case, the amount obtained is sufficient for ~60 mL of translation. The ribosome preparation protocol affords amounts sufficient for ~15 mL of the translation reaction.
3. Quality control/evaluation of target protein
3.1. Confirmation of target activity
Prior to selection, we recommend that the activity of the target protein be confirmed. When binding is considered to be an acceptable readout of protein function, numerous biophysical methods can be used to confirm that the target protein interacts with a positive control ligand, for instance Isothermal Titration Calorimetry (ITC), Surface Plasmon Resonance (SPR), Biolayer Interferometry (BLI) or Fluorescence Polarization (FP) (Du et al., 2016). In our previous selection for 2G12, the BLItz system (Pall ForteBio) with His6-gp120 immobilized on Ni-NTA biosensors was used to confirm strong binding by 2G12 to gp120 (Horiya et al., 2014a). The BLItz system allows for analysis with small amounts of samples, but is similar to Octet or Biacore systems. An ELISA measurement is a simple alternative that is also practical for antibody targets.
3.2. Confirmation of target capture method
The method for solid-phase capture of the target protein needs to be considered. 2G12, the target discussed herein, is a monoclonal antibody, and can therefore be captured on immobilized protein A or protein G. Magnetic beads are utilized as the solid support because they are easy to separate from solution. We recommend calibrating the amount of beads required to capture all of the target in solution by the method below. In our case, 0.12 mg of Dynabeads protein G (Invitrogen) or 0.24 mg of Dynabeads protein A (Invitrogen) were required to capture in 10–100 nM 2G12 in 20 μL selection buffer (1x solution: 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 % (v/v) Triton X-100; store at −20 °C as 5x solution).
Resuspend Dynabeads in the commercial vial and transfer 4 or 8 μL (0.12 or 0.24 mg) to 1.5 mL microcentrifuge tubes.
Add 80 or 160 μL of selection buffer and resuspend the beads. Briefly spin down the tubes with a mini-centrifuge, place the tubes on the magnetic rack for 1 minute, and remove the supernatant. Repeat this washing step for a total of three times.
Add 20 μL of 10–100 nM 2G12 to the beads and tumble at room temperature for 1 hour.
Briefly spin down the tubes with a mini-centrifuge and place the tubes on the magnetic rack for 1 minute.
Remove the supernatant and mix with 20 μL 2x Laemmli Sample Buffer (Bio-Rad). Add 40 μL of 1x Laemmli Sample Buffer to the beads.
Heat samples at 95 °C for 5 minutes and cool down to room temperature.
Analyze the samples by SDS-PAGE, staining with Pierce Silver Stain Kit (Thermo Scientific) to confirm that no 2G12 is present in supernatant.
If antibodies are observed in the supernatant fraction, increase the bead amount and repeat this experiment to determine the bead amount sufficient to capture all 2G12.
3.3. Choosing a method for the retrieval of cDNA from selected fusions
The method for recovery of selected cDNA/mRNA-peptide fusions or cDNA alone also needs to be considered. Although a PCR reaction directly from library fusions while they are still bound to the target and beads may be possible (Seelig, 2011), this may increase undesired sequences, such as binders to the solid support, capture protein or other non-specific binders. Ideally, conditions should be found to elute only those fusions that are bound to the target protein (Keefe, 2001), prior to amplification. In the case of 2G12, we tested the conditions to inactivate 2G12 binding activity to gp120 using the BLItz instrument in advance. Incubation at 70 °C for 20 minutes (or lower temperature in the presence of DTT) irreversibly inactivates 2G12 in the buffer used in the selection (Horiya et al., 2014a). On the other hand, when 2G12 was captured on protein A or G magnetic beads and heated at 70 °C for 20 minutes, then chilled on ice for 5 minutes and incubated at room temperature at 10 minutes, none or little of 2G12 is present in the supernatant, suggesting that the capture of 2G12 on protein A or G is barely affected by this heating and chilling step. In addition, the cDNA/mRNA duplex of the library fusions shown here can be denatured at 95 °C but not at 70 °C (Horiya et al., 2014a). Therefore, heat denaturation at 70 °C followed by chilling elutes intact fusions that bind to 2G12’s gp120-binding site, along with their cDNA, while likely minimizing the non-specific elution cDNA from fusions that remain bound to off-target sites on 2G12, the capture protein or beads. Conversely, higher temperatures that denature the cDNA/RNA duplex may again increase the elution of the undesired sequences.
4. Glycopeptide mRNA display selection procedures
4.1. Design and preparation of library DNA
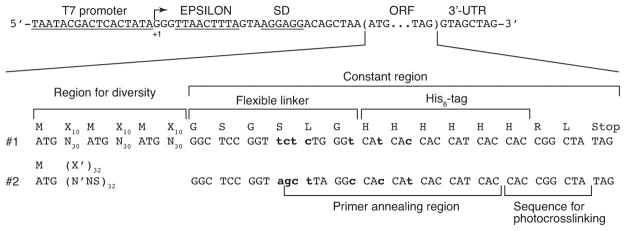
The selection procedure begins with the library DNA preparation. DNA libraries we have used start with a T7 promotor, EPSILON enhancer (Forster, Weissbach, & Blacklow, 2001) and Shine-Dalgarno sequence, followed by the start codon of the library peptide ORF; usually, the peptide sequence includes affinity tags (e.g. His6-tag). The DNA libraries we have used previously are shown in Figure 4. DNA libraries can be created in various ways. One of the simplest methods is to use only synthetic DNA with neither polymerase extension nor PCR amplification, in which the antisense strand of the library is annealed with an oligonucleotide containing the T7 promotor sequence. However, in this method, the length of the library DNA is typically confined to ~200 nt due to limitations of oligonucleotide synthesis. We have successfully obtained two different 198-mer DNA libraries that contain 90 or 96 random nucleotides (equivalent to 30 or 32 random amino acid residues) by ordering from commercial suppliers. When the required DNA sequence is longer than can be synthesized, mutual priming extension by polymerase or PCR-based methods is also possible. With regard to random codons, the frequency of AUG codons needs to be considered because AUG-codons encode potential glycosylation sites. The frequency of AUG in typical random amino codons NNS (S is G or C) or NNK (K is G or U) is 1/32. For the two libraries presented in this example, the AUG frequency in random codons was adjusted according to how many constant AUG codons were also present. In Library #1 (“Fixed”), constant AUG codons were present at amino acid positions 1, 12, and 23, so NNN codons (1/64 AUG) were used for all random positions. In Library #2 (“Variable”) only one constant AUG codon was present (for the Start signal), so the AUG content of random codons was increased to 1/20 to ensure many library peptides containing 3–4 HPGs. This increase was accomplished with biased (N′NS) codons, in which the fraction of A at the first nucleotide (N′) of each codon is changed from 1/4 to 2/5 (2/5 x 1/4 x 1/2 = 1/20). With respect to the DNA sequences for constant regions, we utilize different codon combinations to encode the His6-tag region in each library; the His6 codons are part of the library primer for reverse transcription and PCR, so this practice minimizes the possibility of cross-contamination or mix-up between libraries.
Figure 4.
Library sequences that have been used in this method. The sequences are shown as a sense strand. From 5′ to 3′, the libraries comprise a T7 promotor, the 5′ untranslated region (UTR) starting with three G’s and containing the EPSILON enhancer and the Shine-Dalgarno sequence, ORF and the 3′-UTR. Two library sequences (#1 and #2) are shown. The DNA sequences, but not peptide, of the constant regions of the two libraries are different (as shown in bold lowercase) so that each library has a specific primer for reverse transcription and PCR. These primer differences are designed to prevent the libraries from cross-contamination and/or mix-up. N is an equimolar mixture of A, C, G and T. N′ is a mixture of 40 % A and 20 % each of C, G and T. S is an equimolar mixture of C and G. NNN codons encode AUG codons with a probability of 0.016 per position. N′NS codons encode AUG codons with a probability of 0.05 per position. Random amino acids encoded by NNN and N′NS are shown as X and X′, respectively. In the formation of mRNA-peptide fusions, leucine at the C-terminus is not decoded and arginine prior to it is linked to puromycin (Josephson et al., 2005).
4.1.1. Purification of the library DNA
The protocol below describes PAGE purification of 198-mer DNA (antisense strand of Library #1 or #2) synthesized on a 1 μmol scale. The DNA can be obtained from W.M. Keck Biotechnology Resource Laboratory, Yale University or a comparable facility. The scale of the DNA synthesis may be as low as 200 nmol. In our experience, when half of the DNA synthesized on a 200 nmol scale was purified by PAGE, it yielded an amount sufficient or nearly sufficient for selection (~300–780 pmol). The volumes of the solutions to dissolve the DNA and/or percentage of the gel shown below can vary depending on the length and yields of the DNA.
-
Dissolve the unpurified or desalted DNA pellet in 1 mL TE buffer.
TE buffer: 10 mM Tris-HCl, 1 mM EDTA, pH 8.0 (make fresh). Mix 250 μL of this solution with the same volume of 8 M urea. Store the remaining DNA solution at −20 °C.
Using SequaGel UreaGel System (National Diagnostics) and Vertical Electrophoresis System, C.B.S. Lite Slab Gel Kit, 16.5cm(w) x 14.5cm(h) (C.B.S. Scientific), prepare a 4 or 5 % denaturing gel of 1.5 mm thickness with 6 wells.
Using 1x TBE buffer (National Diagnostics) as the running buffer, pre-run the gel for ~30 minutes at 18–22 W.
Heat the DNA solution at 70 °C for 5 minutes.
Flush out the urea accumulated in the wells, and immediately but gently load 100 μL DNA solution per each of 5 wells. Load 10 μL of DNA gel loading dye that contains bromophenol blue, xylene cyanol and glycerol into another well.
-
Run the gel at 18–22 W until the xylene cyanol reaches approximately 2/3 from the top the gel. This may take 40–60 minutes.
Note: Occasionally we have experienced the glass plates cracking as a result of the heat produced during electrophoresis at 22 W. Thus, we recommend caution in this step.
-
Remove the gel from the glass plates and cover it with plastic wrap. Next, place onto a TLC plate that is covered with plastic wrap, and irradiate with 254 nm UV using a compact UV lamp (UVP LLC, Model UVG-11) to visualize DNA by UV shadowing. Promptly mark positions of appropriate DNA bands on plastic wrap using a fine permanent marker.
Note: Usually bands of interest are visualized as the darkest and topmost among all the bands. Other bands are likely due to incomplete DNA synthesis and may appear as a smear in the lane. The duration of UV irradiation should be minimized, both for DNA stability and safety. For safety, the UV lamp should always be directed downward and away from the eyes or skin.
Guided by the markings made above, use a new, clean razor blade to cut through the plastic wrap and excise gel pieces containing the DNA band of interest.
Set up the Elutrap Electroelution System (Whatman) with 0.5X TBE buffer, according to manufacturer instructions. Place the gel pieces in the elution chamber and elute DNA by running at 200–300 V for 2–3 hours into the trap.
Stop the run, quickly switch the cathode and anode, run for additional 15 seconds, and stop the run again. Immediately collect the eluent in the trap.
Add 1/9 volume of 3 M KOAc (pH 5.5) (Ambion) and 3 volumes of ethanol to the eluent for ethanol precipitation.
Chill the solution in a −20 °C freezer for 1 hour to overnight.
Centrifuge at 16,000–21,000g at 4 °C for 10 minutes. Remove supernatant.
Briefly rinse the pellet with ice cold 70 % (v/v) ethanol and centrifuge at 16,000–21,000g at 4 °C for 5 minutes. Remove supernatant.
Dry the pellet and dissolve the DNA in water. Check the concentration by UV spectroscopy.
4.1.2. Optional procedure: TA cloning of DNA libraries and sequencing
As an optional step, DNA libraries prior to selection can be cloned into plasmids by TA cloning methods as is done following selection. Though not essential, this practice can provide useful control data, establishing whether there is sequence bias and providing arbitrary non-selected sequences for optimization of translations and click reactions. In our work with long peptide libraries, only ~20 % of clones examined in this manner were free of premature termination codons, frameshift or unreadable sequences. Even among sequences lacking such defects, not all will be translated equally well.
4.2. Library RNA preparation by T7 transcription
The DNA library purified as above is next transcribed to RNA using T7 RNA polymerase. We describe a method of library RNA preparation using synthetic ssDNA and a kit for T7 transcription. Although T7 RNA polymerase or transcription kits are available from various suppliers, His6-tagged T7 RNA polymerase can be expressed and purified in a similar manner as the PURE system proteins, described in the appendix. We therefore provide an alternative protocol using T7 RNA polymerase prepared in-house, with PCR-amplified DNA as a template.
4.2.1. T7 transcription using a kit and synthetic DNA
This protocol utilizes a MEGAshortscript T7 Transcription Kit (Ambion) and PAGE-purified synthetic DNA corresponding to the antisense strand of the library.
-
Combine the following solutions on ice:
Stock Final Amount T7 10X Reaction Buffer 10X 1X 50 μL T7 ATP Solution 75 mM 7.5 mM 50 μL T7 CTP Solution 75 mM 7.5 mM 50 μL T7 GTP Solution 75 mM 7.5 mM 50 μL T7 UTP Solution 75 mM 7.5 mM 50 μL T7 FP1 (sequence is shown below) 100 μM 0.6 μM 3 μL DNA library above 0.5 μM 250 pmol T7 Enzyme Mix 50 μL Add water to 500 μL T7 FP DNA: 5′-TAATACGACTCACTATAGGGTTAACTTTAG-3′ (dissolved in water to a final concentration of 100 μM)
Incubate the reaction mixture at 37 °C for 2 hours or more (up to overnight).
Add 25 μL of Turbo DNase and incubate at 37 °C for 15 minutes.
Add 40 μL of 0.5 M EDTA (pH 8.0). Then add ~350 mg solid urea and dissolve.
Heat the solution at 70 °C for 5 minutes, then purify by 5 % denaturing PAGE as described above.
Repeat these steps until ~5 nmol of RNA is obtained. In our experience, 780 pmol of DNA was required for Library #1 and 300 pmol of DNA was required for Library #2.
4.2.2. Alternative protocol for T7 transcription
This protocol utilizes His6-tagged T7 RNA Polymerase produced in-house, which is an economical alternative to the purchase of commercial kits. The following protocol is optimized for dsDNA from PCR, though ssDNA may be used as well with added T7 primer, as above.
-
Combine the following solutions on ice.
Stock Final Amount Transcription Buffer 5X 1X 200 μL PEG-8000 400 mg/mL 50 mg/mL 125 μL MgCl2 1 M 25 mM 25 μL ATP 100 mM 4 mM 40 μL CTP 100 mM 4 mM 40 μL GTP 100 mM 4 mM 40 μL UTP 100 mM 4 mM 40 μL Inorganic pyrophosphatase 0.1 U/μL 2.5 U/mL 25 μL PCR-amplified DNA 75 nM 75 pmol T7 RNA polymerase 0.05 μg/μL 50 μg Add water to 1000 μL 5X Transcription buffer: 400 mM HEPES-KOH (pH 7.6), 200 mM DTT, 10 mM spermidine (store at −20 °C)Note: NTPs are obtained as a set of 100 mM solutions (Thermo Scientific).Note: See appendix for preparation of Inorganic pyrophosphatase.Note: T7 RNA polymerase (His6-tagged) can be prepared in a manner analogous to the PURE system translation factors (see appendix), except that it should be dialyzed against 50 % (v/v) glycerol-containing storage buffer and stored at −20 °C. Incubate at 37 °C for 2 hours or more (up to overnight).
Add 25 μL of Turbo DNase (Ambion) and incubate at 37 °C for 15 minutes.
Add 75 μL of 0.5 M EDTA (pH 8).
Precipitate RNA by ethanol precipitation as described above. The transcription reaction contains PEG-8000, which has been reported to enhance transcription yields owing to macromolecular crowding effects (Ge, Luo, & Xu, 2011). Since it can impede the RNA migration in PAGE to some extent, it should be removed prior to PAGE by alcohol precipitation.
Dissolve the pellet in 8 M urea and purify RNA by PAGE as described above.
4.3. Modification of RNA with puromycin
mRNA is modified with puromycin by crosslinking a puromycin-containing oligonucleotide near the 3′-end of the ORF (Figures 4 and 5). The crosslinking is mediated by a photochemical reaction of the psoralen moiety in the puromycin oligonucleotide, triggered by long-wavelength ultraviolet light (320–380 nm), resulting in formation of cyclobutane ring(s) between the 5,6 double bond of uracil base(s) and either the 4′,5′ or 3,4 double bond or both of the psoralen (Figure 5B). In the original mRNA display protocol, mRNA and puromycin-containing oligonucleotides were ligated by T4 DNA ligase using splint DNA, which must be removed prior to lysate-based translation to prevent degradation by RNase H (Barrick, Takahashi, Balakin, & Roberts, 2001a). The photo-crosslinking method was later developed to obviate purification of the puromycin-modified RNA (Kurz, Gu, & Lohse, 2000). The following protocol is adapted from the one described by Seelig (Seelig, 2011).
Figure 5.
Conjugation of puromycin-containing oligonucleotide to mRNA by photocrosslinking between psoralen and uracil. (A) Structure of the psoralen moiety connected to oligonucleotide containing puromycin. (B) Structure of the photo-crosslinked psoralen moiety with two uracil adducts. (C) Overview of the psoralen-mediated photo-crosslinking of puromycin-containing oligonucleotides and mRNA at the end of the open reading frame.
-
Combine the following solutions:
Stock Final Amount XL buffer 10X 1X 175 μL PAGE-purified library RNA 3 μM 5250 pmol XL-PSO 100 μM 7.5 μM 131.25 μL Add water to 1750 μL XL buffer, 10X: 200 mM HEPES-KOH, 1 M KCl, 10 mM spermidine, 10 mM EDTA, pH 7.5 (store at −20 °C).Oligonucleotide XL-PSO: (C6 psoralen) 2′-OMe(UAGCCGGUG)(dA)15(Spacer 9)2dA(dC)2Puromycin. This oligonucleotide can be obtained from the W.M. Keck Biotechnology Resource Laboratory, Yale University, or similar oligonucleotide synthesis service. When the synthesis scale is 1 μmol, dissolve the pellet of the crude oligonucleotide in 2.5 mL water, and desalt using a NAP25 column (GE Healthcare) with water by following manufacturer instructions. Measure the ABS260 and dilute the solution to ~100 μM with water. Divide into 200 μL aliquots and store at −20 °C with protection from light.
Save 5 μL of the above mixture as a negative control for PAGE in step 9.
Divide the remaining mixture into 50 μL aliquots in 0.2 mL PCR tubes.
To anneal oligonucleotides, use a thermal cycler (Bio-Rad S1000) to heat at 70 °C for 3 minutes, then slowly cool to 25 °C with a ramp of 0.1 °C/sec, incubate at 25 °C for 5 minutes, and then chill at 4 °C.
Transfer the annealed solutions into wells of an open Costar 96-well plate (100 μL solution per well) and place the plate on an ice water bath prepared in an empty pipette-tip box lid or similar bath.
Irradiate at 365 nm UV by putting a handheld UV lamp (UVP Compact UV Lamps, 4 watts, UVL-21) directly on top of the wells of the plate for 20 minutes.
Collect the solution from the wells and combine.
Save 5 μL as a control for PAGE in step 9.
Precipitate the RNA by ethanol precipitation and rinse with 70 % (v/v) ethanol. Dry pellets and dissolve them in a total of 600 μL water.
-
After ethanol precipitation, analyze by 5 % denaturing PAGE to assess the crosslinking reaction and recovery of RNA.
Note: Typically, the efficiency of the photocrosslinking should be roughly 50 %. Unless the RNA is lost in ethanol precipitation, the recovery of the RNA should be ~100 %. In this case, the concentration becomes 8.7 μM; therefore, the concentration of photocrosslinked RNA is ~4.35 μM.
4.3.1. Optional procedure: purification of puromycin modified RNA
Although the resulting RNA solution is a mixture of crosslinked RNA, non-crosslinked RNA and XL-PSO, the mixture can be directly used for translation. Optionally, crosslinked RNA can be purified by denaturing PAGE as described above. To visualize the RNA in the PAGE gel slab, the use of shortwave UV light (254 nm) should be avoided, since this can reverse the photo-crosslinking reaction. Therefore, we use RNA Staining Solution (Abnova), which stains RNA with a blue color that is visible under ambient light. Because RNA stained with this dye tends to give low yields in passive elution from gel pieces, we do electro-elution followed by ethanol precipitation and 70 % (v/v) ethanol rinse to recover RNA as described above.
We purify the crosslinked RNA in two situations. The first is when using 3H-histidine in translation. In order to make the specific (radio)activity of histidine high enough for detection, mostly radioactive histidine is used, but the commercial stock is only 17–33 μM; this stock is diluted at least 5-fold in the reaction, leading to a limited histidine concentration of ~3.4–6.6 μM. As this concentration is low, it is better to remove non-crosslinked RNA so that histidine is not depleted by incorporation into non-RNA-fused His6-tagged peptides. The second situation when non-crosslinked RNA should be removed is when using oligo (dT) magnetic beads for fusion purification, as explained in the section describing that protocol.
4.4. Translation and formation of RNA-peptide fusions
Using a commercially available PURE system and/or the in-house components described in the appendix later in this chapter, a PURE system translation can be set up according to the recipe below. A 5.2 mL translation reaction volume (for first-round library preparation) is shown here, from which we have successfully obtained libraries of ~1013 glycopeptides, using in-house components. Note that the procedure using commercially available PURE system shown below has not been tested on large scale and is provisional. Whatever the source of the PURE system, it contains many valuable components; thus we strongly recommend testing the reaction on smaller volume pilot translation.
-
Combine the following components in a 50 mL conical centrifuge tube; (a) for commercial PURE system (b) for in-house components prepared as described in appendix.
-
Using commercial PURE system: PURExpress Δ(amino acids, tRNA, ribosomes) can be obtained by special order from NEB as catalog number E3315S; combine the components as listed below. Components included in the kit are marked by asterisks. See the appendix for preparation of amino acid mixture and A19 ribosomes. To use commercial PURE system including commercial BL21 ribosomes, order PURExpress Δ(amino acids, tRNA) as NEB standard catalog item E6840S, and modify this recipe accordingly.
Stock Final Amount Solution A (minus aa and tRNA)* 5x 1x 1040 μL 19 amino acid mixture 3 mM each 300 μM each 520 μL E. coli tRNA* 10x 1x 520 μL Factor Mix* 8.3x 1x 624 μL A19 ribosome varies 1.2 μM 6240 pmol cysteine or histidine (see note) varies < 300 μM varies Add water to 4600 μL -
Using in-house PURE system: prepare components described in the appendix and combine as follows:
Stock Final Amount Translation buffer 8x 1x 650 μL DTT 500 mM 1 mM 10.4 μL Protease inhibitor cocktail 10x 1x 520 μL ATP and GTP mix 25 mM each 1 mM each 208 μL Creatine phosphate 0.5 M 20 mM 208 μL 19 amino acid mixture 3 mM each 300 μM each 520 μL E. coli total tRNA 2,000 ABS260 48 ABS260 124.8 μL 5,10-methyltetrahydrofolate 2 mg/mL 10 μg/mL 26 μL Creatine kinase 10 ABS280 0.04 ABS280 20.8 μL Nucleoside 5′-diphosphate kinase varies 0.85 U/mL 4.42 U Myokinase varies 6.8 U/mL 35.36 U Inorganic pyrophosphatase 100 U/mL 1 U/mL 52 μL Factor mix varies 1x varies 20 AARS mix 20X 1X 260 μL A19 ribosome varies 1.2 μM 6240 pmol cysteine or histidine (see note) varies < 300 μM varies Add water to 4600 μL Note: When using radioactive cysteine or histidine, dilute 35S-cysteine and 3H-histidine with the non-radioactive amino acids to obtain the desired specific activity. Never add radioactive and non-radioactive versions of an amino acid to translation separately, without pre-mixing, because it can cause inaccuracy in the radioactivity-based quantitation of the fusions. The specific activity of the amino acid should be adjusted by changing the ratio of radioactive to non-radioactive amino acid. It is preferred that the specific activity be sufficiently high for detecting radioactivity in the selected binders when only a small portion (< 2.5 %) of the sample is sacrificed for liquid scintillation counting. This should be estimated by a pilot experiment.
-
Add 600 μL of 8.7 μM RNA (an approximately 50:50 mixture of puromycin modified and unmodified RNA) to the translation mix, to a final concentration of 1 μM. In the case of puromycin modified RNA that is purified by PAGE, add the RNA and water to a final concentration of 0.5 μM in 5.2 mL.
-
Incubate at 37 °C for 1 hour.
Note: The reaction time can be shortened to 15–30 minutes. We recommend optimizing the reaction time by pilot experiments. -
Add 1560 μL (0.3 vol) of a solution of 2.05 M KCl and 172 mM Mg(OAc)2 and incubate at room temperature for 15 minutes. Store at −20 °C.
Note: The above addition of magnesium and potassium is known to enhance fusion formation in the rabbit reticulocyte lysate cell-free translation system (R. Liu, Barrick, Szostak, & Roberts, 2000). This also applies to the PURE system (data not shown).
4.5. Purification of fusions
In this section, we describe the purification of mRNA-peptide fusions prior to reverse transcription and click reaction. Since XL-PSO contains a (dA)15 stretch, oligo(dT) purification retrieves mRNA-peptide fusions, as well as XL-PSO or XL-PSO-crosslinked RNA that is not fused with peptides. The latter is removed by subsequent affinity purification using Ni-NTA agarose resins prior to reverse transcription. Alternatively, reverse transcription can be done prior to Ni-NTA affinity purification, which may help to prevent unpredictable loss of fusions due to insolubility (aggregation, sticking to surfaces, etc.) in subsequent steps by increasing the solubility of fusions with newly generated DNA strands. However, reverse transcription after Ni-NTA purification requires much less enzyme, because the RNA-peptide fusions are only a small portion (typically < 10 %) of the XL-PSO-crosslinked RNA used in translation.
4.5.1. Oligo(dT) purification
Oligo(dT)-cellulose has previously been used for purification of mRNA-peptide fusions (Cho, Keefe, Liu, Wilson, & Szostak, 2000; Guillen Schlippe et al., 2012; Horiya et al., 2014a; Keefe, 2001; Seelig, 2011). However, most commercially available oligo(dT)-cellulose products (from Ambion and Sigma) have been discontinued; to our knowledge, only New England Biolabs supplies oligo(dT)25 cellulose, and this product has afforded inferior yields in our hands (data not shown), and in other labs (Sau, Larsen, & Chaput, 2014). In-house synthesis of oligo(dT) resins is also possible (Sau et al., 2014). Below is the protocol utilizing oligo(dT) cellulose, followed by an alternative protocol that uses oligo(dT) magnetic beads.
4.5.2. Fusion purification using oligo(dT) cellulose
Here, the purification from 6.76 mL of crude RNA-peptide fusion solution (resulting from 5.2 mL translation reaction followed by the addition 1.56 mL of the potassium and magnesium solution) is described. This procedure is preferred over the procedure using oligo(dT) magnetic beads if the translation utilizes unpurified cross-linked RNA, and radioactivity-based quantification of fusions is required.
-
1
Save 20 μL from 6.76 mL crude RNA-peptide fusion solution as non-purified sample.
-
2
Weigh 130 mg of oligo(dT) cellulose (Ambion, Sigma or other sources) into a 2 mL microcentrifuge tube. Repeat this to have a total of 4 sets (100 mg per 1 mL translation reaction volume prior to magnesium/potassium addition).
-
3
Resuspend each batch of resin in 1 mL water. Spin down at 16,000–21,000g for 30 seconds and remove supernatant. Repeat this wash step.
-
4
Wash each with 1 mL oligo(dT) cellulose binding buffer. Repeat this step twice.
Oligo(dT) binding buffer: 20 mM Tris-HCl, 1 M NaCl, 10 mM EDTA, pH 8.0 (store at 4 °C), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. -
5
Transfer two batches of 130 mg resin (260 mg total) into each of two 50 mL conical centrifuge tubes, resuspending the contents of each 50 mL tube in 30.33 mL (9 volumes of crude fusion solution) of oligo(dT) binding buffer.
-
6
Add 3.37 mL crude fusion solution to each.
Note: Stalled ribosomes may remain associated with some mRNA-peptide fusions. EDTA is present in the oligo(dT) binding buffer to chelate the Mg2+ that stabilizes the ribosome, thus promoting its dissociation from mRNA-peptide fusions. -
7
Tumble in a 4 ° C room for 15 minutes.
-
8
Transfer each suspension to two Econo-Pac Chromatography Columns (Bio-rad) with tips snapped off, and drain the flow-through.
-
9
Use the flow-through fractions to rinse remaining oligo(dT) resin from the 50 mL tubes into the columns. Perform this rinse a second time and then save the flow-through.
-
10
Wash each column three times with 10 mL oligo(dT) binding buffer and three times with 10 mL oligo(dT) washing buffer. Save the washes.
Oligo(dT) washing buffer: 20 mM Tris-HCl, 300 mM NaCl, 10 mM EDTA, pH 8.0 (store at 4 °C), with 0.1 % (v/v) Tween 20 and 5 mM β-mercaptoethanol added immediately prior to use. -
11
Elute each column 8 times with 1 mL 0.1 % (v/v) Tween 20, collecting each eluent in a different tube.
-
12
Take a few μL from each fraction and measure the radioactivity by liquid scintillation counting. Confirm a gradual decrease of radioactivity throughout the washes and an increase throughout the elution. Combine the eluent fractions that contain most or all of the fusions. In addition, estimate yields of the fusions based on the liquid scintillation counting results as follows.
First, the total cpm of fusions is obtained. If a translation reaction without RNA is performed in parallel and subjected to oligo(dT) purification, this cpm value can be subtracted but is usually negligible.
Using 1 μL from the unpurified solution saved in step 1, measure radioactivity by liquid scintillation counting to calculate specific activity (cpm/pmol) of cysteine or histidine. Do this measurement multiple times and take the average.
Using those values, calculate fusion yields. 16.7 pmol are equivalent to 1013 fusion molecules.
Note 1: the average expected number of cysteine (or histidine) residue(s) per library sequence is the number of random codons times the frequency of the cysteine (or histidine) codons, plus the number of constant cysteine (or histidine) codons.
Note 2: Although we have obtained more than 1014 fusion molecules by this step, the number is gradually decreased in the subsequent purification steps. The radioactivity is frequently measured by liquid scintillation counting in all of the following steps to check the yields of the fusions.
-
13
Filter the solution using 0.22 μm Ultrafree-MC GV Centrifugal Filter Units (Millipore). Analyze the solution by SDS-PAGE together with the unpurified solution and the flow-through fraction. For the visualization of 35S-labelled fusions, the gel is fixed in a solution containing 22.5 % (v/v) ethanol and 5 % (v/v) acetic acid for 15 minutes, dried on filter paper (using a gel dryer) and exposed to a phosphorimaging screen to visualize on a phosphorimager. For the visualization of 3H-labelled fusions, gel is treated with Amersham Amplify Fluorographic Reagent (GE Healthcare, NAMP100) according to the manufacturer protocol, dried and exposed to X-ray film at −80 °C to visualize by fluorography.
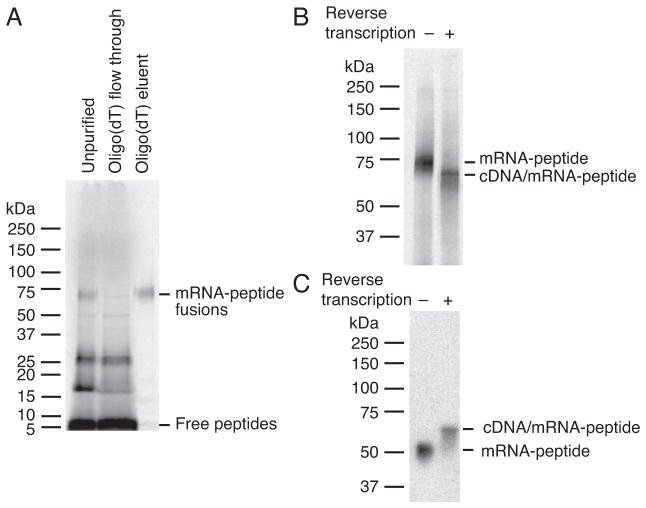
Note: Almost all of the fusions are usually recovered by oligo(dT) purification, as shown in Figure 6A.
Figure 6.
SDS-PAGE of mRNA-peptide and cDNA/mRNA-peptide fusions. The molecular weight markers in the gels were Precision Plus Protein Dual Xtra or Color Standards (Bio-Rad). (A) Analysis of the purification of the fusions using oligo(dT) cellulose. The fusions shown are from Library #2 prior to round 1 selection and labeled with 35S-cysteine. 4–20 % Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad) was used. Typically, fusion recovery is high in this purification method. (B) Analysis of the reverse transcription of the same fusions as A. 7.5 % Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad) was used. (C) Analysis of the reverse transcription of the shorter library fusions that consist of 112 nt RNA conjugated 22-mer peptide through puromycin-modified oligonucleotide (MX9GSGSLH6R-puromycin-linker-RNA, where X is a random amino acid encoded by an NNS codon). Fusions were labeled with 3H-histidine. Home-made 7.5 % polyacrylamide gel was used.
4.5.3. Alternative fusion purification using oligo(dT) magnetic beads
Our preliminary results suggest that the method shown below does not completely purify out non-fused free peptides, which are mostly translated from non-crosslinked RNA when using non-purified RNA mixture. If an accurate radioactivity-based estimation of fusion yields is required, we recommend that this protocol be utilized only with purified cross-linked RNA. The protocol shown here is for 100 μL crude RNA-peptide fusion solution (~77 μL translation reaction followed by the addition of ~23 μL of potassium and magnesium containing solution), but can be scaled as desired.
-
To equilibrate beads, add 400 μL oligo(dT) binding buffer (high EDTA) to 100 μL Oligod(T)25 Magnetic Beads (NEB) and vortex to wash. Place on a magnetic tube rack for 1 minute and discard supernatant. Repeat this wash step.
Oligo(dT) binding buffer (high EDTA): 20 mM Tris-HCl, 1 M NaCl, 50 mM EDTA, pH 8.0 (store at 4 °C), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. -
Resuspend magnetic beads in 100 μL of 2x oligo(dT) binding buffer (high EDTA) and add the resuspended magnetic beads to 100 μL translation mixture.
2x Oligo(dT) binding buffer (high EDTA): 40 mM Tris-HCl, 2 M NaCl, 100 mM EDTA, pH 8.0 (store at 4 °C), with 0.4 % (v/v) Triton X-100 and 10 mM β-mercaptoethanol added immediately prior to use.Note: This concentration of EDTA is chosen to provide a molar excess compared to Mg2+ in the fusion solution. Tumble for 30 minutes at room temperature.
Place the tube on a magnetic tube rack for 1 minute and remove supernatant.
Wash by adding 400 μL oligo(dT) binding buffer and vortexing.
Place tube on a magnetic tube rack for 1 minute and remove the wash. Repeat this wash step for a total of 4 times.
Wash by adding 400 uL oligo(dT) washing buffer and vortexing.
Place tube on a magnetic tube rack for 1 minute and remove the wash. Repeat this wash step for a total of 3 times.
-
Elute by adding 100 μL 0.1 % (v/v) Tween-20 and heating at 70 °C for 2 minutes.
Note: Heating at 70 °C may not be necessary to elute fusions.
Place tube on a magnetic tube rack for 1 minute and remove eluent to save.
Repeat this elution step for a total of three times, or until complete, which can be estimated by liquid scintillation counting of the eluent.
Combine all eluent and filter through Ultra-free 0.22 μm centrifugal filter unit (Millipore) into a 2 mL centrifuge tube to remove extra beads. Remove 30 μL to save. Total volume is ~270 μL. For translation reactions utilizing purified cross-linked RNA, radioactive fusions can now reliably be quantified by liquid scintillation counting as described above.
4.5.4. Ni-NTA agarose purification
RNA-peptide fusions are purified by capture of the peptide His6-tag on Ni-NTA agarose followed by washing away of RNA and XL-PSO, which are usually present in excess of RNA-peptide fusions. Ni-NTA-captured fusions are next eluted by a solution containing concentrated imidazole. The concentrated imidazole is removed by gel filtration to avoid its precipitation in subsequent ethanol precipitation.
-
Precipitate RNA-peptide fusions by ethanol precipitation in the presence of 1–2 μL of 5 mg/mL linear acrylamide carrier (Ambion) per one microcentrifuge tube. Dry pellets and redissolve in 1.6 mL denaturing bind buffer. Remove 1 μL to measure the radioactivity by liquid scintillation counting.
Denaturing bind buffer: 100 mM NaH2PO4, 10 mM Tris, 6 M guanidinium hydrochloride, NaOH to pH 8.0 (store at 4 °C), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. To equilibrate resin, add 100 μL Ni-NTA agarose (Qiagen) to a Poly-Prep Chromatography Column (Bio-Rad) whose bottom tip is snapped off, and add 1 mL denaturing bind buffer twice. Drain the flow through and close the bottom with a cap.
Transfer the fusion solution to the column. Rinse the tube(s) that contained the fusions with 400 μL denaturing bind buffer total and transfer to the column. Total volume of the fusion solution into the column becomes 2 mL.
Close the top of the column with lid and tumble on a tube rotator for 3 hours at 4 °C.
Open the bottom and drain the flow through. Collect the flow-through and re-apply onto the column. Repeat this once. Save the flow-through.
Wash by adding 500 μL denaturing bind buffer to the column. Repeat this wash step once.
-
Wash by adding 500 μL native wash buffer to the column. Repeat this wash step for a total of three times.
Native wash buffer: 100 mM NaH2PO4, 300 mM NaCl, NaOH to pH 8.0 (store at 4 °C), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. -
Elute by adding 100 μL native elute buffer to the column. Repeat this elution step for a total of five times.
Native elute buffer: 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, NaOH to pH 8.0 (store at 4 °C), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. Take 0.5–1 μL of each elution and measure radioactivity by liquid scintillation counting. Calculate yields of the fusions. Unlike in oligo(dT) purification, Ni-NTA agarose does not capture all fusions. When a large portion of the radioactivity (40 % or more of total radioactivity) is present in the flow through fraction, reapply the flow through onto freshly equilibrated Ni-NTA agarose and repeat these purification steps to maximize the recovery of the fusions. However, usually all fusions cannot be recovered in this step.
4.5.5. Desalting by gel filtration
In this section, concentrated imidazole is removed to avoid its precipitation in subsequent ethanol precipitation by using a pre-packed column for gel filtration with gravity. This protocol is written for the desalting of 500 μL fusion solution.
-
Equilibrate a NAP5 column (GE Healthcare) with 10 mL gel filtration buffer.
Gel filtration buffer: 10 mM Tris-HCl (pH 7.5), 1 mM EDTA (pH 8) (make fresh), with 0.2 % (v/v) Triton X-100 and 5 mM β-mercaptoethanol added immediately prior to use. -
Apply 500 μL fusion solution in native elute buffer (described above) onto the columns.
Note: Save flow through fractions.
Elute with 1 mL gel filtration buffer. Save 10 μL for analysis. Check radioactivity of fusions by liquid scintillation counting. Typically, fusions are recovered in high yield during this step.
4.6. Reverse transcription to form cDNA/RNA-peptide fusions
RNA-peptide fusions are subjected to reverse transcription reaction to convert to cDNA/RNA-peptide fusions. We recommend this reaction be done prior to the click reaction because click reaction prior to reverse transcription carries a risk of RNA degradation, leading to failure of cDNA synthesis (Horiya et al., 2014a).
Divide 990 μL fusions from the previous step equally into three 1.5 mL microcentrifuge tubes. Precipitate the fusions with ethanol in the presence of 1–2 μL of linear acrylamide carrier and wash the fusions with 70 % (v/v) ethanol as described above. Dry the pellets by opening tubes in the air at room temperature for 10–60 minutes.
-
Add the following solutions to each of the fusion pellets
Stock Final (after step 5) Amount dNTP mix 10 mM each 0.5 mM each 1.5 μL RT primer 50 μM 2.5 μM 1.5 μL 0.18 % (v/v) Triton X-100 0.18 % (v/v) 0.1 % (v/v) 16.5 μL dNTP mix (10 mM each): made from deoxynucleoside Triphosphate Set, PCR Grade (Roche)
RT primer for the Library #1: 5′-T15GTGATGGTGgTGaTGaCCcAgag-3′
RT primer for the Library #2: 5′-T15GTGATGGTGaTGgTGgCCtAagc-3′
(Nucleotides shown in bold lowercase are those that differentiate the two primers.)
Note: We have had successful reverse transcriptions with RT primer concentrations in the range of 0.1–5 μM. The number of molecules of RT primer has to be at least equivalent to, and preferably exceed, the number of the anticipated amount of fusions.
Leave on ice for ~30 minutes to dissolve fusions.
Heat at 65 °C for 5 minutes and then chill on ice for more than 5 minutes.
-
Add the following to the fusion solution:
Stock Final Amount First strand buffer 5X 1x 6 μL DTT 0.1 M 5 mM 1.5 μL SUPERase•In RNase Inhibitor (Ambion) 20 U/μL 1 U/μL 1.5 μL Superscript III RT (Invitrogen) 200 U/μL 10 U/μL 1.5 μL Note: First strand buffer and DTT are supplemented with Superscript III RT. We do not recommend diluting the reverse transcriptase.
Incubate the tube at 55 °C for 30 minutes.
-
Remove 1 μL from each tube and run SDS-PAGE to confirm the reaction.
Note: In the cases of the libraries shown in Figure 6, SDS-PAGE (typically 7.5 %) separates RNA-peptide fusions with and without cDNA. Despite the larger molecular weight of the cDNA/RNA-peptide fusions compared to RNA-peptide fusions, they migrate faster than the RNA-peptide fusions or primer annealed RNA-peptide fusions as previously reported in some cases (Figure 6B) (Barrick, Takahashi, Ren, Xia, & Roberts, 2001b; Seelig, 2011). However, we have observed this is not the case when using shorter libraries, in which cDNA/RNA-peptide fusions migrate slower than RNA-peptide fusions (Figure 6C). Thus, it is important to know the migration pattern of RNA-peptide and cDNA/RNA-peptide fusions in SDS-PAGE (or possibly by other types of PAGE) and find out the conditions to separate them beforehand. In addition, RNA-peptide fusions annealed to RT primer exhibit yet another migration pattern (data not shown), and it is informative to know that pattern as well. To test this, we recommend setting up a control reaction without reverse transcriptase.
4.7. Click glycosylation
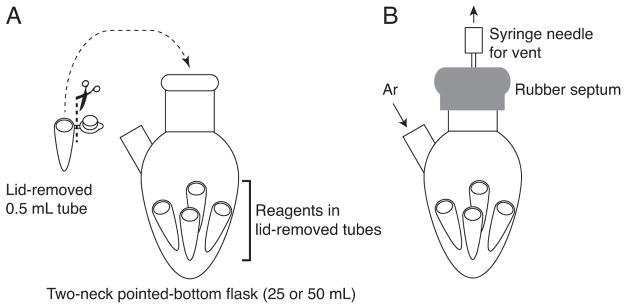
cDNA/mRNA-peptide fusions are glycosylated with synthetic azide sugars through CuAAC using Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) ligand (Hong, Presolski, Ma, & Finn, 2009). The Man9-azide sugar discussed in this protocol (Figure 2) is synthesized in our laboratory (Temme, Drzyzga, MacPherson, & Krauss, 2013), though we have seen similar results with smaller and larger oligosaccharides (data not shown). Although THPTA is also synthesized in-house, it is now commercially available from Sigma. We carry out the click reaction under inert gas (Temme & Krauss, 2015), with separate click reaction components prepared inside microcentrifuge tubes that are placed inside an argon-filled 25- or 50 mL two-neck pointed-bottom flask. The cDNA/RNA-peptide fusion solution is placed within one 0.5 mL microcentrifuge tube and other tubes contain reagents for the click reaction (Figure 7).
Figure 7.
Diagram of the setup for the click reaction under argon (Ar).
Precipitate cDNA/RNA-peptide fusions in a 0.5 mL microcentrifuge tube by ethanol precipitation followed by 70 % (v/v) ethanol rinse. Dry the pellet and dissolve the fusions in 10 μL of 40 mM HEPES-KOH (pH 7.6), 0.1 % (v/v) Triton X-100. Otherwise, precipitate in a larger tube, dissolve and transfer to a 0.5 mL microcentrifuge tube.
-
Add the following to the fusions:
Stock Amount Volume aminoguanidine 50 mM 150 nmol 3 μL water to 22.5 μL -
Prepare the solution containing copper, ligand, and Man9-azide in a 0.5 mL tube (mixture A). CuSO4 and THPTA ligand are mixed first and then Man9-azide is added to the mixture.
Stock Amount Volume CuSO4 20 mM 30 nmol 1.5 μL THPTA 40 mM 60 nmol 1.5 μL Man9-azide 50 mM 150 nmol 3 μL Note: For an efficient click reaction, Man9-azide concentrations should be on the order of mM. 3–5 mM or more is recommended.
Weigh a few mg of sodium L-ascorbate into a 0.5 mL microcentrifuge tube.
Remove the lids of the tubes and then place them into the two-neck pointed-bottom flask with argon line (Figure 7A). Cap with a septum with a vent needle (Figure 7B).
Purge the flask under argon flow for 1 hour to degas the solutions.
Under argon efflux, add mixture A to the fusion solution. Dissolve sodium L-ascorbate in degassed water to make a 100 mM solution. Then, add 1.5 μL to the tube and mix by pipetting to start the reaction.
After recapping followed by 15 minutes of argon purge, remove the vent needle to keep the system under positive argon pressure.
After 2 hours 45 minutes, stop the reaction by adding 7.5 μL of 10 mM EDTA (pH 8). Dilute to 300 μL with 0.1 % (v/v) Triton X-100.
4.7.1. Alternative protocol for click glycosylation
We also run the click reaction in the following way using the same flask set up. In this procedure, additional Man9-azide, THPTA and sodium L-ascorbate are added in the middle of the reaction. The protocol shown here is the smallest volume reaction that we have tested.
Precipitate cDNA/RNA-peptide fusions (~20 pmol or less) in a 0.5 mL microcentrifuge tube by ethanol precipitation followed by 70 % (v/v) ethanol rinse. Dry the pellet and dissolve the fusions in 2 μL of 250 mM HEPES-KOH (pH 7.6), 0.075 % (v/v) Triton X-100.
-
Add the following to the fusions:
Stock Amount Volume aminoguanidine 50 mM 25 nmol 0.5 μL subtotal 2.5 μL -
Prepare the solution containing copper, ligand, and Man9-azide in a 0.5 mL microcentrifuge tube (mixture A). CuSO4 and THPTA ligand are mixed first and then Man9-azide is added to the mixture. Prepare the supplementary solution containing THPTA and Man9-azide in another tube (mixture B).
Mixture A: Stock Final Volume CuSO4 20 mM 5 nmol 0.25 μL THPTA 20 mM 5 nmol 0.25 μL Man9-azide 50 mM 15 nmol 0.3 μL Water 1.7 μL Subtotal 2.5 μL Mixture B: Stock Final Volume THPTA 10 mM 2.5 nmol 0.25 μL Man9-azide 50 mM 7.5 nmol 0.15 μL Water 2.6 μL Subtotal 3 μL Weigh a few mg of sodium L-ascorbate into a 0.5 mL microcentrifuge tube.
Remove the lids of the tubes and then place them into the two-neck pointed-bottom flask with argon line (Figure 7A). Cap with a septum with a vent needle (Figure 7B).
Purge the flask under argon flow for 1 hour to degas the solutions.
Under argon efflux, add mixture A to the fusion solution. Dissolve sodium L-ascorbate in degassed water to make a 100 mM solution. Then, add 0.5 μL to the tube and mix by pipetting to start the reaction.
After recapping followed by 15 minutes of argon purge, remove the vent needle to keep the system under positive pressure.
After 1 hour 15 minutes, add mixture B to the fusion solution and 0.25 μL of 100 mM sodium L-ascorbate under argon efflux.
After recapping followed by 15 minutes of argon purge, remove the vent needle to keep the system under positive argon pressure.
-
After 1 hours 15 minutes, stop the reaction by adding 1.25 μL of 10 mM EDTA (pH 8.0).
Note: At this point, the volume is down to ~2.5 μL by evaporation.
Dilute to ~100 μL with 0.1 % (v/v) Triton X-100.
4.7.2. Evaluation of click reaction
The click reaction can be evaluated by SDS-PAGE of nuclease-digested fusions. Nuclease P1 hydrolyzes the 3′-phosphoester linkage between the puromycin moiety and the linker DNA in XL-PSO, and generates peptide-puromycin-PO3H2 (Hofmann, Szostak, & Seebeck, 2012; Josephson et al., 2005). The molecular weights of the peptides in these libraries are approximately 5–6 kDa and that of the Man9-azide sugar is 1.6 kDa. Although glycopeptides do not migrate identically to proteins of the same molecular weight in SDS-PAGE, the gel described below successfully resolves the glycopeptides containing different numbers of glycans so that we are able to evaluate the efficiency of the click glycosylation as well as the population of each glycosylation valency on the peptide.
Remove a part of the cDNA/RNA-glycopeptide fusions (enough to visualize, usually 0.05–1 pmol) after the click reaction and precipitate in the presence of 1 μL of linear acrylamide carrier by ethanol precipitation.
-
Add 5 μL of 200 mM ammonium acetate (pH 5.3) and 1 μL of nuclease P1 (1 unit/μL) to the pellet.
Nuclease P1 from Penicillium citrinum (Sigma): Dissolve lyophilized powder to a final concentration of 1 unit/μL in 200 mM ammonium acetate pH 5.3. Make 10 μL aliquots and store at −20 °C. Incubate at 37 °C for 1 hour.
Add 1 μL 1 M Tris-HCl (pH 8) and 7 μL 2x Laemmli Sample Buffer (Bio-Rad) with 5 % (v/v) β-mercaptoethanol.
Heat the sample at 95 °C for 5 minutes and apply to a 4–20 % Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad). Run at 300 V for 16–20 minutes.
Visualize radioactive digested (glyco)peptide fusions by phosphorimaging or fluorography as described above.
Note: Occasionally the efficiency of the click reaction may happen to be low. In that case, re-purification of fusions by Ni-NTA affinity and/or gel filtration followed by ethanol precipitation and resubjection to the click reaction may improve the result. We have also observed that click efficiency may be low due to the quality of THPTA used; in some cases, the use of HPLC-purified THPTA was found to improve the click reaction.
4.8. First round selection
cDNA/RNA-peptide fusions are now ready to be selected for the function of interest. Here is the protocol to select binders using mAb 2G12 as a target.
-
Precipitate fusions by ethanol precipitation and rinse with 70 % (v/v) ethanol as described above. Dry and redissolve the pellets of glycosylated fusions (~1013 sequences = 16.7 pmol) in 500 μL of selection buffer.
Note: The composition of the buffer is described above. Add 2G12 to a final concentration of 100 nM.
Incubate at room temperature for 1 hour.
Meanwhile, equilibrate protein G Dynabeads as follows: put 100 μL of protein G Dynabeads (Invitrogen) into a 1.5 mL microcentrifuge tube, place on a magnetic rack for 1 minute and remove supernatant. Resuspend the beads in 1 mL of selection buffer, briefly spin down the suspension using a mini-centrifuge, place on a magnetic rack for 1 minute, and remove supernatant. Repeat this step twice and resuspend in 100 μL of selection buffer.
After the mixture of fusions and 2G12 has been incubated for 1 hour, add the protein G magnetic bead suspension in 100 μL of selection buffer.
Tumble at room temperature for 20 minutes.
Place on a magnetic rack for 1 minute and remove supernatant.
Resuspend the beads in 500 μL of selection buffer, briefly spin down the suspension using a mini-centrifuge, place on a magnetic rack for 1 minute, and remove supernatant. Repeat this step twice.
Resuspend the beads in 100 μL of selection buffer and heat at 70 °C for 30 minutes to denature the paratope of 2G12 (conditions determined as described above).
Place on ice for 5 minutes and tumble at room temperature for 10 minutes.
Place on a magnetic rack for 1 minute. Remove supernatant and save as eluted fusions. To rinse the remaining fusions off the beads, resuspend the beads in 100 μL of selection buffer, briefly spin down the suspension using a mini-centrifuge, place on a magnetic rack for 1 minute and remove supernatant to save as eluted fusions. Repeat this rinse step once.
Use a part of each fraction (unbound, washes, eluents and beads) to measure radioactivity by liquid scintillation counting and calculate the amount of radioactivity in eluent fractions compared to total radioactivity.
4.9. PCR amplification of the DNA of selected fusions
The cDNA of selected fusions is then amplified by PCR. Fusions in selection buffer shown above should be diluted at least 8-fold in the PCR reaction since the buffer with less dilution inhibits the reaction. Prior to a large-scale reaction, a pilot PCR experiment is done using a small amount of the fusion solution to determine the optimum PCR conditions. In addition, a pilot PCR is set up with two different primer sets for each of two libraries when using two different libraries in parallel. This is to confirm each library is amplified only with its own primer set, indicating no cross-contamination between the two libraries. Furthermore, a PCR reaction without fusion solutions is also prepared to confirm no DNA is amplified without fusion solution.
Combine the eluted fusion solutions (~300 μL) and dilute to 320 μL with selection buffer.
-
Take two 10 μL portions of the solution for PCR reactions with two different primer sets and combine for PCR as follows:
Stock Final volume (μL) PCR Reaction Buffer with MgCl2 (Roche) 10X 1X 8 Tris-HCl (pH 8.3) 1 M 100 mM 8 dNTP mix 2.5 mM each 0.1 mM each 3.2 Library FP 100 μM 1 μM 8 Library RP 100 μM 1 μM 8 Taq DNA polymerase 5 U/μL 0.025 U/μL 0.4 Water 34.4 Selected fusions or selection buffer 10 Total 80 Library FP: 5′-TAATACGACTCACTATAGGGTTAACTTTAGTAAGGAGG-3′
Library RP for the Library #1: 5′-CTAGCTACCTATAGCCGGTGGTGATGGTGgTGaTGaCCcAgag-3′
Library RP for the Library #2: 5′-CTAGCTACCTATAGCCGGTGGTGATGGTGaTGgTGgCCtAagc-3′
(Nucleotides shown in bold lowercase are those that differentiate the two primers.)
Make 7 aliquots of 10 μL in 0.2 mL PCR tubes. Using a thermal cycler, run a PCR reaction of 7 arbitrary different cycles (e.g. 12, 14, 16, 18, 20, 22, and 24 cycles) of 94 °C for 30 seconds, 62 °C for 30 seconds, and 74 °C for 30 seconds.
-
Analyze the products by 2 % agarose gel electrophoresis to determine the optimum cycle number for PCR.
Note: We typically choose the number of cycles in which the DNA of correct size just reaches a plateau, yet the side products (bands of inappropriate size that are often observed) are minimized.
Based on the result, prepare a large volume reaction of 2.4 mL. Make 50 μL aliquots in 0.2 mL PCR tubes and run the reaction in the thermal cycler with the optimum cycle number determined in step 4. Optionally, when TA cloning of the DNA sequences is planned after the PCR, add a step of 74 °C for 30 minutes at the end of the thermal cycles to ensure that all PCR products are full length and have 3′ A-overhangs.
After the PCR reaction, combine the solutions into 4 microcentrifuge tubes and add an equal volume phenol:chloroform:isoamyl alcohol (25:24:1). Vortex the tube vigorously and spin at maximum speed for 1 minute to separate the organic and aqueous phases.
Transfer the aqueous phases to new microcentrifuge tubes and add an equal volume of chloroform. Vortex the tubes vigorously and spin at maximum speed for 1 minute to separate the organic and aqueous phases.
Precipitate DNA by ethanol precipitation followed by 70 % (v/v) ethanol rinse as described above.
Dissolve DNA in 100 μL water.
-
Estimate the concentration of the amplified DNA by running a 2 % agarose gel to compare PCR product with dsDNA controls whose concentrations are known and comparing the band intensities.
Note: Usually, the yield of PCR product is in large excess compared to the amount needed to make RNA for the next round of selection. Thus, it is easy to start over from the transcription in the case of failure of the latter step(s) in the second or subsequent rounds.
4.10. Subsequent rounds of selections
After the first round of selection is complete, repeat the selection for an additional 6–9 rounds, or possibly more, but with only 0.25–0.5 mL of translation reaction. Scale down all the other steps accordingly, except the volume of selection (the step to isolate the fusions binding to the target) and subsequent PCR. In the latter rounds of selection, increasing selection stringency (lowering concentration of target, increasing the number of washes, adding competitor, increasing the temperature, etc.) may help the enrichment of stronger binders. Regarding the stoichiometry of fusions and target, we maintain excess target compared to fusions so that stringency is controlled only by target concentrations. It is important to note that translation yield increases in later rounds, so that 10–20 pmol of library can be obtained from smaller translation volumes. When the target concentration is reduced from 100 to 10 nM, we increase the selection volume to 1700 μL (17 pmol target) if ~15 pmol fusions are present. Regarding types of selective pressure, in our experience, an increase of the temperature from room temperature to 37 °C has made a striking difference in the composition of the selected library pools (Horiya et al., 2014a), as also observed in our similar selections of glycan modified-DNA (Temme et al., 2013; Temme, MacPherson, DeCourcey, & Krauss, 2014). The effect of selection temperature on library multivalency was readily visualized by PAGE gel comparison of selected library pools at each round. We also switched protein G beads to A beads every round or every other round to avoid enrichment of binders to protein A or G. To remove binders independent of glycosylation, it is also possible to do “negative” selection, in which the fusions without glycosylation are incubated with 2G12, and the fusions in the unbound fraction are retrieved, desalted by ethanol precipitation, subjected to click glycosylation and carried on to the next “positive” selection (Horiya et al., 2014a). An additional precaution for avoiding unwanted artifacts is to pre-incubate the library solution with empty beads (protein A or G magnetic beads themselves, in this case), then to discard the beads prior to the addition of target. This “pre-clearing” step is particularly important in later rounds to remove binders to the solid phase. In order to track the enrichment of the binders in rounds 2 and later, it can be informative to subject a small portion of the glycopeptide fusions to 1st-round selection conditions in parallel with the increasingly stringent selection conditions used on the whole library. The fraction bound with the 1st-round selection conditions can be compared with that achieved in the present round, affording a readout of enrichment (Chaput & Szostak, 2004; Horiya et al., 2014a).
4.11. Determination of selected sequences
After PCR using Taq polymerase, the fragments are cloned into plasmids by TA cloning (Taq amplified) for sequencing. Usually, this is done for the PCR-amplified DNA after round 7 or later selection rounds, depending on the enrichment of the binders. Here, a protocol using TOPO TA cloning kit (Invitrogen) is shown. If screening of colonies containing plasmid with insert DNA is necessary, we recommend doing colony PCR, but not blue/white screening based on the activity of the β-galactosidase reporter on X-gal containing LB plates. This is because colony color can sometimes be pale blue even when the DNA is inserted, and screening white colonies may cause undesired bias in this step.
-
Prepare the following mixture.
Fresh PCR product 20–30 ng of the amplified DNA fragment Salt Solution 1 μL Water to a final volume of 5 μL Add 1 μL of TOPO vector, mix the reaction gently and incubate for 30 minutes at room temperature.
-
Transform 2–3 μL of the reaction into arbitrary E. coli competent cells by heat shock, add 1 mL S.O.C. medium to the cells, shake for 1 hour at 37 °C, and spread onto LB plate containing 30 μg/mL kanamycin.
S.O.C. (Super Optimal broth with Catabolite repression) medium: Autoclave a solution containing 0.5 % (w/v) yeast extract, 2 % (w/v) tryptone, 10 mM NaCl, and 2.5 mM KCl. Once the solution is cool, add freshly filtered solutions of 1 M glucose, 1 M MgSO4 and 1 M MgCl2 for final concentrations of 20 mM, 10 mM and 10 mM, respectively. Store as aliquots at −20 °C. Incubate the plates overnight at 37 °C to grow colonies.
-
Pick colonies, inoculate 1–5 mL LB containing 30 μg/mL kanamycin with each colony, isolate plasmid DNA using a Plasmid Mini Prep kit (Qiagen or Zymo Research), and determine the sequences by the Sanger method.
Note: We recommend checking the migration of the plasmid DNA in 1 % agarose gel because we sometimes observe plasmid dimers.
5. Binding assay utilizing translated glycopeptides
Table 1 shows the sequences of 12 clones within each library after round 10 selection with 2G12. Some of the sequences were observed multiple times. Interestingly, all sequences contained 3–5 glycosylation sites, similar to the numbers of 2–4 Man9(GlcNAc)2 glycans that interact with 2G12 (Calarese et al., 2003). To evaluate the selected glycopeptides’ binding affinity/avidity with 2G12, radioactive glycopeptides are prepared and apparent KD’s are determined using a radiometric assay.
Table 1.
Selected peptide sequences for 2G12 and their binding constants
Only the sequence in the random region (positions 1–33) is shown. M in bold denotes a potential Man9-glycosylation site. The observed consensus motif MxSIP(–/x)YTY(L/xW)(–/x)P, where lower case x is an unspecified amino acid, is highlighted.
The peptide sequences used in the 2G12-binding assay were followed by a linker, a His6-tag and a FLAG-tag (GSGSLGHHHHHHRDYKDDDDK). Errors reported are the standard errors of the curve fit. These data have been reported previously (Horiya et al., 2014a).
5.1. Preparation of radioactive glycopeptides
Using plasmids encoding selected sequences as templates, DNA for T7 transcription are amplified by PCR and transcribed to RNA. The RNA is translated to peptides using the PURE system in the presence of radioactive amino acids (usually histidine, unless the peptide contains multiple cysteines). The peptides are then glycosylated with Man9-azide sugars by click reaction.
-
Prepare a PCR reaction using Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific) as follows:
Stock Final volume (μL) Phusion HF buffer 5X 1X 20 dNTP mix 2.5 mM each 0.05 mM each 2 Library FP (described above) 100 μM 2 μM 2 FLAG RP 100 μM 2 μM 2 Plasmid encoding a selected sequence 10 ng/μL 0.01 ng/μL 1 Phusion Hot Start II DNA Polymerase 2 U/μL 0.02 U/μL 1 Water 72 Total 100 FLAG RP for clones from Library #1: 5′-CTAGCTACCTATTTGTCATCGTCGTCTTTATAATCCCGGTGGTGATGGTGgTGaTGaCCcAg-3′
FLAG RP for clones from Library #2: 5′-CTAGCTACCTATTTGTCATCGTCGTCTTTATAATCCCGGTGGTGATGGTGaTGgTGgCCtAa-3′
(Nucleotides shown in bold lowercase are those that differentiate the two primers.)
Note: The reverse primers are designed to attach a FLAG-tag to the peptides for possible FLAG affinity purification, which we normally do not need to do. Alternatively, Library RP as described above can be used for this PCR to make peptides without the FLAG-tag.
Run a PCR reaction of heating at 98 °C of 30 seconds followed by 25 cycles of 98 °C for 5 seconds, 66 °C for 10 seconds, and 72 °C for 10 seconds.
Purify the amplified DNA fragments by an appropriate DNA purification kit such as QIAquick PCR Purification Kit (Qiagen), E.Z.N.A. ® Cycle Pure Kit (Omega Bio-tek), or DNA Clean & Concentrator™-5 (Zymo Research). We normally use water for the elution step. Alternatively, DNA can be purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction followed by chloroform extraction and ethanol precipitation. In the case that DNA is purified using a kit, determine the concentration by UV spectroscopy. Confirm the size of the products using 2 % agarose gel electrophoresis. In the case that DNA is purified by phenol extraction followed by ethanol precipitation and 70 % (v/v) ethanol rinse, run control DNA with known concentrations in the same gel and estimate concentration of the amplified DNA fragments comparing to the controls.
Using DNA as templates, transcribe RNA by T7 transcription (typically 10 to 100 μL reaction volume) and purify by PAGE as described above. Alternatively, purify RNA using MEGAclear™ Transcription Clean-Up Kit followed by ethanol precipitation and 70 % (v/v) ethanol rinse and dissolve RNA in water.
-
Translate RNA to peptide using the PURE system (typically 25 to 50 μL) with radioactive amino acid (3H-histidine or 35S-cysteine) as described above, except that the concentration of RNA is 1 μM and incubation at 37 °C is for 2 to 4 hours. In a parallel manner, prepare non-radioactive peptides without radioactivity in the PURE system for MALDI-TOF MS in the next step.
Note: Specific (radio)activity needs to be high enough to be detected in the binding assay.
-
Purify peptides using Ni-NTA agarose as follows: add 1 volume of Ni-NTA agarose suspension and 4 volumes of Bind Buffer for peptide, and tumble at room temperature for 1 hour. Transfer the suspensions to a 0.22 μm Ultrafree-MC GV Centrifugal Filter Unit (Millipore) and rinse the tube with 4 volumes of Bind Buffer for peptide to transfer the rest of the resins to the same filter unit. Spin the filter unit at 2,000g for 30 seconds to remove the flow through fraction. Wash the resins by adding 8 volumes of Bind Buffer for peptide and spin the filter unit with the same conditions. Repeat this washing step twice. Switching to Wash Buffer for peptide, repeat this washing step 2 to 4 times, except the final centrifugation is at 14,000g for 30 seconds. Elute the peptide into a clean 1.5 mL microcentrifuge tube by the addition of 1 volume of 0.2 % (v/v) trifluoroacetic acid (TFA) to the resins in the filter unit, followed by centrifugation at 14,000g for 30 seconds. Repeat this elution step. Determine the yield by liquid scintillation counting as described as above. In addition, using non-radioactive peptides, mix 1.5 μL of peptide solution with the same volume of 10 mg/mL α-Cyano-4-hydroxycinnamic acid matrix in 50 % acetonitrile and 0.05 % TFA solution. Spot 1–1.5 μL on a sample plate and measure the mass by MALDI-TOF MS, following manufacturer instructions, to confirm the mass of peptides.
Bind Buffer for peptide: 50 mM Tris-HCl, 300 mM NaCl, pH 7.8 (store at 4 °C), with 5 mM β-mercaptoethanol added immediately prior to use.Wash Buffer for peptide: 50 mM Tris-HCl, pH 7.8 (store at 4 °C), with 5 mM β-mercaptoethanol added immediately prior to use.Note: If radioactive materials are permitted in the MALDI-TOF-MS instrument, radioactive peptides also can be used for MALDI-TOF-MS; however, it may be hard to detect the expected peaks because the low amino acid concentration needed for high specific activity can result in low peptide yield. Add 1/100 volume of 10 % (v/v) Triton X-100 to the peptide solution and dialyze against 0.1 % (v/v) Triton X-100 containing water using a Slide-A-Lyzer™ MINI device (0.1 mL capacity, 3.5 MWCO) (ThermoFisher Scientific) at room temperature overnight.
Evaporate the peptide solution in a 0.5 mL microcentrifuge tube to dryness by speedvac.
Subject to the click reaction with Man9-azide sugar as described above, and check glycosylated products by SDS-PAGE with visualization by autoradiography or fluorography.
-
Optional procedure: For some peptides, not all portions go to completion of click glycosylation. In that case, affinity purification for 2G12 removes most of the incompletely glycosylated products, as well as other impurities, improving the maximum fraction bound and the accuracy of KD measurement (figure 8A). Affinity purification for 2G12 can be done as follows. Dilute glycosylated peptide (< 4 pmol) with 100 nM 2G12 in selection buffer (40 μL) at room temperature. Tumble the solution for 30 minutes with 0.12 mg of equilibrated Dynabeads Protein G to capture 2G12–glycopeptide complex. Wash the beads three times with 40 μL of selection buffer and resuspend in 10 μL of selection buffer. Heat the bead suspension at 70 °C for 30 minutes to denature 2G12 and elute glycopeptides, chill on ice for 5 minutes, and tumble at room temperature for 10 minutes. Put the tube onto a magnetic rack for 1 minute and pick up the supernatant. Rinse the beads with 10 μL of selection buffer. Combine the supernatant and the rinsed solution as the purified glycopeptide fraction. Measure the yield by liquid scintillation counting.
Note: The recovery of radioactivity is typically in the range of 25–55 %.
Figure 8.
Analysis of individual glycopeptides identified in the selection targeting 2G12. (A) Comparison of peptides before and after 2G12-affinity purification by SDS-PAGE. One clone from each library (10F6 and 10V1) is shown as a representative. Peptides were labeled with 3H-histidine. Gels and standards are the same as Figure 6A. (B) Binding curves of 10F6 and 10V1 with 2G12. Markers show averages at each concentration and error bars are standard errors.
5.2. Measurement of binding constants by radiometric bead-binding assay
In this assay, radioactive glycopeptides in complex with various concentrations of 2G12 are captured on magnetic protein G beads. The bound and unbound fractions are separated and the radioactivity of each is measured to calculate the percent bound for each concentration of 2G12 for curve-fitting. This protocol is designed to measure apparent KDs in the range of approximately 1–10 nM. The concentration of the target and glycopeptides, as well as bead amounts, can be adjusted accordingly depending on expected KD; for an accurate KD value, the range of target concentrations tested should ideally include an order of magnitude above and below the KD. Use an 8-channel pipette whenever possible to minimize pipetting effort.
-
Using 8-strip PCR tubes, mix 0.12–0.2 nM radioactive glycopeptides and 0 or 0.25–64 nM 2G12 in 40 μL of selection buffer and incubate at room temperature for 1 hour. Prepare samples at least in triplicate. Typical volumes to work with are shown below. 2G12 is diluted to 1 μM in selection buffer and stored as a stock solution at 4 °C, and 8 and 128 nM 2G12 solutions are freshly diluted from the stock for each assay. Glycopeptides are also diluted immediately before the assay.
Final 2G12 concentration (nM) 0 0.25 0.5 1 2 4 selection buffer 20 18.75 17.5 15 10 - 8 nM 2G12 in selection buffer - 1.25 2.5 5 10 20 glycopeptide (≤0.4 nM) 20 20 20 20 20 20
Total 40 40 40 40 40 40 Final 2G12 concentration (nM) 8 16 32 64 selection buffer 17.5 15 10 - 128 nM 2G12 in selection buffer 2.5 5 10 20 glycopeptide (≤0.4 nM) 20 20 20 20
Total 40 40 40 40 While incubating the glycopeptide with 2G12, prepare equilibrated protein G Dynabeads (Invitrogen) as follows. Put 4 μL each of protein G Dynabeads into 8-strip PCR tubes. Add 80 μL of selection buffer, briefly spin down the suspension using a mini-centrifuge, place on a magnetic rack for 1 minute, and remove supernatant. Repeat this step twice. Final removal of the buffer is done immediately prior to the next step.
After 2G12-glycopeptide mixed solutions are incubated, transfer the solutions to the equilibrated protein G Dynabeads, and tumble at room temperature for 30 minutes.
Place on a magnetic rack for 1 minute and isolate supernatants. Resuspend the beads in 40 μL of selection buffer, briefly spin down the suspension using a mini-centrifuge, place on a magnetic rack for 1 minute, and isolate the rinse as a residual part of the unbound fraction. Repeat this step twice. Combine the supernatant and subsequent rinses as the unbound fraction and measure the radioactivity for each 2G12 concentration by liquid scintillation counting.
-
Resuspend the beads in 40 μL of selection buffer, heat at 70 °C for 30 minutes to elute the bound glycopeptides, chill on ice for 5 minutes, tumble at room temperature for 10 minutes and isolate the eluent. Rinse the beads with 40 μL of selection buffer and isolate the residual eluent. Combine the eluents and measure the radioactivity for each 2G12 concentration by liquid scintillation counting. Resuspend the beads in 40 μL of selection buffer, and measure the radioactivity of the bead suspension for each 2G12 concentration by liquid scintillation counting. Radioactivity of eluted fractions and bead suspensions are combined as the apparent bound fraction.
Note: Glycopeptides are eluted from the beads prior to radioactivity detection because liquid scintillation counting with glycopeptides on the beads results in partially suppressed detection, particularly in the case of 3H-label.
Calculate fraction bound to 2G12 (Fbound 2G12) as follows:
First, calculate fraction bound to beads in the presence of 2G12 (Fbound 2G12 and beads).
Then, subtract the fraction bound to beads in the absence of 2G12 (Fbound beads) as background, which typically ranges from 0 to 6 % of the total radioactivity, to determine the actual fraction bound to 2G12 (Fbound 2G12).
Finally, fit a curve to the formula below to determine KD and Fmax (maximum fraction bound).
Representative plots and fitted curves are shown in Figure 8B. Calculated KDs and Fmaxs of the interactions between individual glycopeptides and 2G12 are summarized in Table 1.
6. Discussion, summary and conclusions
Using glycopeptide mRNA display, we successfully identified glycopeptides that bind to a target monoclonal antibody, 2G12, with sub nM to low nM KDs, comparable to the strength of the interaction of 2G12 with its natural binding partner, gp120. The number of glycans on selected glycopeptides was 3–5, similar to the number of glycans on gp120 that bind to 2G12.
We also observed that some selected glycopeptides contained a consensus motif MxSIP(–/x)YTY(L/xW)(–/x)P, where M denotes a potential Man9 glycosylation site. This consensus motif was present in all Library #2 (“Variable”) sequences, but in only some Library #1 (“Fixed”) sequences. It is possible that the prevalence of the consensus sequence in Library #2 is due to the biased codons, which not only increase HPG frequency, but simultaneously increase frequency of three other amino acids within the consensus sequence (Ile, Thr and Ser). Although we did not predict a consensus sequence, these results suggest that use of multiple library designs is beneficial to avoid getting results that are dominated by one major motif, even when using long and diverse libraries. Moreover, in these libraries, the theoretical peptide sequence space (2030–32) is so much greater than the actual library size (~1013), that it may be preferable to sample that space in two different random fashions, rather than to prepare a single library of twice the size.
There are several reports of mRNA display that involve modification by posttranslational treatment including cyclization mediated by bis-NHS crosslinking, bis-alkylation, lanthionine formation, CuAAC, or disulfide bond(s) assisted by protein disulfide isomerase, or conjugation of a small molecules (Fiacco et al., 2016; Guillen Schlippe et al., 2012; Hacker, Hoinka, Iqbal, Przytycka, & Hartman, 2017; Hofmann et al., 2012; Li & Roberts, 2003; Millward, Fiacco, Austin, & Roberts, 2007; Yamaguchi et al., 2009). The present method is similar to those, with the striking differences that our library modifications (oligosaccharides) are much larger and are derived from precious azide-modified sugar reagents synthesized in-house. With the low-volume glycosylation procedures reported herein, this method requires less than < 1 mg of Man9-azide sugars per library for the entire selection. Therefore, this method is in principle applicable to conjugation of other valuable precursors to peptides for mRNA display selection, including various oligosaccharides. Thus, this directed evolution method should be broadly applicable to evolution of multivalent glycoligands for use in glycobiology, or multivalent ligands for other fields. Additionally, this protocol has successfully yielded libraries of ~1013 sequences of the libraries, which is at the larger end of the library sizes reported in mRNA display selections, even though relatively long peptide sequences with 30 or more random amino acids were used. Therefore, this protocol will also be useful for peptide selections of unmodified unnatural or natural peptides using mRNA display.
Acknowledgments
This work is supported by the NIH (R01-AI090745 and R01-AI113737). We thank Dr. Sebastian Temme for helpful discussions and the synthesis of Man9-azide, and Dr. Yollete Guillen and Prof. Jack Szostak for their generous support in the development stages of this methodology.
7. Appendix
In this section, we describe precautions for nuclease contamination and protocols used in our group to prepare components for the PURE system.
7.1. RNA precautions
RNA is handled throughout most parts of this method. Therefore, except in the procedures handling E. coli itself, we strongly recommend work be done in an RNase-free environment. All buffers are sterilized by filtration through 0.22 μm filters, except solutions that are purchased as “RNA” grade. Also, it is important to frequently check the integrity of RNA-peptide fusions by PAGE. When RNA degradation is suspected, RNase contamination in reagents should be tested. There are various ways to assay RNase activity, including commercially available kits such as the RNaseAlert kit (Ambion or IDT), which is a convenient fluorescence-based system.
7.2. Preparation of PURE system components
7.2.1. Preparation of His6-tagged proteins
There are several detailed protocols available for the purification of PURE system proteins. We typically purify MTF and 9 translation factors using either of the following methods: a) capture on Ni-NTA agarose followed by elution with a step-wise gradient of imidazole, which is described in this section, or b) an FPLC purification similar to that described by Ueda’s group, capturing proteins on a HiTrap Chelating column with Ni2+, followed by elution with a linear gradient of imidazole (Shimizu & Ueda, 2010). For purification of 20 AARSs, we usually utilize a simpler method similar to the one described by Hartman’s group, in which elution from Ni-NTA agarose is done once with a high concentration of imidazole (Z. Ma & Hartman, 2012). After all proteins are prepared, we make MTF and factor mix stock solutions and 20 AARS mix stock solutions as described below.
We have transformed E. coli with the plasmids that express His6-proteins for the PURE system (Josephson et al., 2005; Shimizu et al., 2001), and we maintain the transformants as frozen stocks, which can be stably stored for years at –80 °C. The frozen stocks are used directly to inoculate LB for protein expression and purification.
-
1
Preparation of frozen stocks is done as follows: transform the plasmids that express His6-tagged proteins into E. coli BL21 Star (DE3) (Invitrogen) (for pET vector) or M15[pREP4] (for pQE30 vector) and obtain single colonies on LB plates containing an appropriate antibiotic.
-
2
Pick single colonies and inoculate 4 mL LB containing an appropriate antibiotic. If clone screening is planned (as in steps 6–11), pick and inoculate multiple (usually 3–4) colonies independently per transformant.
-
3
Incubate at 37 °C with shaking at 250 rpm.
-
4
When OD600 reaches 0.6–1, add glycerol to a final concentration of 15 % (v/v) or the same volume of 20 % (v/v) glycerol.
-
5
Quickly freeze the cells on dry ice and store at −80 °C.
-
6
As an optional procedure, clone screening can be done to pick a colony with optimal expression, as follows. Inoculate 5 mL LB containing an appropriate antibiotic with each glycerol stock and incubate overnight at 37 °C with shaking at 250 rpm.
-
7
For the overnight culture from each glycerol stock, inoculate two portions of 5 mL fresh LB containing an appropriate antibiotic with 100 μL starter culture and grow at 37 °C with shaking at 250 rpm.
-
8
Once OD600 reaches 0.6–0.8, harvest cells equivalent to 1 mL of 0.8 OD600 from one portion as pre-induced culture by centrifugation at 16,000–21,000g for 2.5 minutes and store the pellet at −20 °C until lysis. Induce the other 5 mL of portion with 0.1 mM isopropyl-β-D-thiogalactoside (IPTG).
-
9
Incubate induced culture for an additional 4 hours, then harvest cells equivalent to 1 mL of 0.8 OD600 culture as described above.
-
10
Resuspend each pellet in 100 μL 1x Laemmli Sample Buffer with 2.5 % (v/v) β-mercaptoethanol and heat samples at 95 °C for 10 minutes to lyse cells.
-
11
After cooling to room temperature, analyze the samples (15 μL each) by SDS-PAGE to compare expression of the protein of interest before and after induction. Choose the glycerol stock(s) that show the greatest expression levels for protein(s) of interest, and use them for the next step.
-
12
Expression and purification of proteins are done as follows; Inoculate 50 mL LB containing an appropriate antibiotic with frozen stocks of interest and incubate at 37 °C with shaking at 250 rpm overnight.
-
13
Inoculate 1 L fresh LB containing an appropriate antibiotic with 20 mL of overnight culture.
-
14
Incubate cultures at 37 °C with shaking at 250 rpm.
-
15
When OD600 reaches 0.6–0.8 (usually in 2–3 hours), induce protein expression by adding IPTG to a final concentration of 0.1 mM.
-
16
Incubate at 37 °C for an additional 4 hours shaking at 250 rpm.
-
17
Harvest cells by centrifugation at 4,000g for 20 minutes at 4 °C.
-
18
Store cell pellets at −20 °C.
-
19
Resuspend frozen cells in 32 mL B-PER Bacterial Protein Extraction Reagent (Thermo Fisher) for cell lysis and transfer to a Nalgene Oakridge centrifuge tube. Incubate 15 minutes at room temperature with gentle swirling.
Note: B-PER is a solution containing a nonionic detergent in 20mM Tris-HCl (pH 7.5). Alternatively, salts can be added to B-PER. In that case, we normally use B-PER II (Thermo Fisher), which is twice as concentrated as B-PER, as follows: Freshly combine 16 mL B-PER II with an equal volume of 600 mM NaCl to make 32 mL of 300 mM NaCl in B-PER. Add 7 mM β-mercaptoethanol immediately prior to use. -
20
Centrifuge the lysate at 16,000g for 20 minutes at 4 °C.
-
21
In the following procedure, keep the lysate and/or protein solutions at 4 °C or on ice except during analysis or storage steps. Transfer supernatant to a 50 mL conical centrifuge tube.
-
22
Add 1 mL Ni-NTA Agarose (Qiagen) and elution buffer to a final concentration of 10 mM imidazole.
Elution buffer: 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 250 mM imidazole (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use. -
23
Tumble for 1 hour.
-
24
Load approximately half of the suspension onto a 20 mL Econo-Pac® Chromatography Column (Bio-Rad). Allow to drain by gravity, then add the rest of the solution and drain again.
-
25
Wash the resin with 10 mL wash buffer, rinsing the conical centrifuge tube previously containing the lysate. Wash the resin twice more with 10 mL wash buffer, without rinsing the tube.
Wash buffer: 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 20 mM imidazole (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use. -
26
Elute protein from the resin using a step gradient of 5 mL each of 50 mM imidazole, 100 mM imidazole and 250 mM imidazole in a buffer containing 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 7 mM β-mercaptoethanol, collecting 1 mL fractions in separate 1.5 mL microcentrifuge tubes.
Note: The imidazole solutions are made from elution buffer and wash buffer described in steps 22 and 25. The step gradient can be altered depending on the protein of interest. Proteins need to be sufficiently concentrated to make MTF and factor mix as described below. A steeper gradient may be considered for some proteins. Alternatively, concentration by ultrafiltration using centrifugal filters such as Amicon Ultra (Millipore) may be possible, but we usually obtain sufficiently concentrated proteins using either the step gradient shown here or one that is about twice as steep. -
24
Analyze the eluted fractions by SDS-PAGE and choose the fractions that exhibit an intense band for the protein of interest, with few undesired protein bands.
Note: Normally, we use a 12 % gel. In the case of IF1, the molecular weight is too small for the range of separation in a 12 % gel, so we use a 4–20 % Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad). -
25
Combine the fractions for dialysis in a Slide-A-Lyzer dialysis cassette (Thermo Fisher Scientific) overnight and dialyze against a > 100-fold larger volume of dialysis buffer.
Dialysis buffer: 50 mM HEPES-KOH (pH 7.6), 100 mM KCl, 10 mM MgCl2 (make as 10–20x and store at 4 °C), with dilution to 1x and 7 mM β-mercaptoethanol addition immediately prior to use.Slide-A-Lyzer dialysis cassette: Use cassettes with a 3–12 mL volume range and 20,000 molecular weight cut off membrane, except IF1, for which the 7,000 molecular weight cut off membrane is used. -
26
Move the dialysis cassette to > 100-fold excess volume of storage buffer and continue dialysis for an additional 6–8 hours.
Storage buffer (except for MetRS): Dialysis buffer, with 30 % (v/v) glycerol and 7 mM β-mercaptoethanol added immediately prior to use. In the case of MetRS, use 50 % (v/v) glycerol instead of 30 % (v/v). -
27
Transfer protein solution from the dialysis cassette to a 15 mL conical centrifuge tube.
-
28
Measure the OD280 to estimate concentration using the calculated protein extinction coefficient.
-
29
For proteins except MetRS, make small aliquots (usually 50 μL) and place on dry ice to freeze for storage at −80 °C. For MetRS, make ~500 μL aliquots and store at −20 °C.
-
30
After all proteins are prepared, make MTF and factor mix solution and 20 AARS mix solution as follows:
MTF and factor mix: Determine the concentration of each protein by Bradford Protein Assay (Bio-Rad) using Bovine Serum Albumin Standard (Pierce). Combine factors such that the mixture is as concentrated as allowable without adding any dilution buffer. The final (1X) concentration of the proteins in the translation reaction is 20 μg/mL MTF, 10 μg/mL IF1, 40 μg/mL IF2, 10 μg/mL IF3, 100 μg/mL EF-Tu, 50 μg/mL EF-G, 50 μg/mL EF-Ts, 10 μg/mL RF1, 10 μg/mL RF3, 10 μg/mL RRF. The ideal concentration of a stock solution is 8x or higher. Store as small aliquots at −80 °C. Typical aliquot size is an amount sufficient for 250 μL of translation reaction.
20 AARS mix: Mix equal volumes of each of the 19 AARS protein solutions (except MetRS) regardless of concentrations (in our hands, the concentration of each AARS prior to mixing has varied in the range of 4–260 μM based on extinction coefficient). Store as 38 μL aliquots at −80 °C. Add 2 μL MetRS, which is stored at −20 °C in a buffer with 50 % glycerol, to a 38 μL aliquot of 19 AARS mix just prior to translation reaction.
7.2.2. Preparation of ribosomes
Ribosomes are prepared from the RNase I-defective E. coli strain A19. There are several protocols available. Conventionally, ribosomes for the PURE system are purified using ultracentrifugation with a sucrose density gradient (Shimizu et al., 2001; Spedding, 1990) or sucrose cushion (Josephson et al., 2005; Subtelny, Hartman, & Szostak, 2008). Alternatively, a method using hydrophobic chromatography followed by sucrose cushion has been developed to reduce nuclease contamination (Ohashi, Shimizu, Ying, & Ueda, 2007). We combine two protocols: first, cells are lysed with a bead beater homogenizer (Z. Ma & Hartman, 2012; Subtelny et al., 2008), and then the ribosomes are purified by hydrophobic chromatography in FPLC (Ohashi et al., 2007; Shimizu & Ueda, 2010).
-
Make glycerol stocks of A19 as described above, but without antibiotics.
Note: The A19 strain can be obtained at the Coli Genetic Stock Center at Yale University. Inoculate 100 mL LB with A19 glycerol stock and incubate at 37 °C with shaking at 250 rpm overnight.
Inoculate 4 L fresh LB with a total of 60 mL starter culture.
Incubate at 37 °C with shaking at 250 rpm until OD600 reaches 0.6–0.8.
Harvest cells by centrifugation at 4,000g for 20 minutes at 4 °C.
-
Combine all cells into one centrifuge bottle using ~150 mL suspension buffer, and centrifuge at 4,000g for 20 minutes at 4 °C.
Suspension buffer: 10 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM Mg(OAc)2 (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use.Note: The bacterial pellet can be frozen at −80 °C overnight at this stage. In this case, leave the pellet at room temperature until thawed and then keep on ice before the next step. Lyse cells using a BeadBeater (Bio Spec) as follows. Transfer cells to the 15 mL blender chamber on ice using a total of 10 mL ice-cold suspension buffer. Fill the chamber with pre-chilled 0.1 mm glass beads to the top of the chamber (until a meniscus forms) and close the lid. Place the chamber into the larger cooling chamber (350 mL) filled with crushed ice and water and securely set on the motor base. Lyse by pulsing 6 times for 20 seconds, with 40 second intervals of rest in between each blending pulse.
Transfer the lysate together with glass beads to a Nalgene Oakridge tube on ice using a total of 10 mL ice-cold suspension buffer.
Centrifuge glass beads and lysate at 20,000g for 20 minutes at 4 °C.
-
Immediately after the centrifugation, decant the supernatant into another Nalgene Oakridge tube. Measure the volume using a serological pipet and add an equal volume of suspension buffer (high salt) so that the final concentration of ammonium sulfate is 1.5 M.
Note: Contamination with a small amount of the beads in the decanted supernatant is acceptable because the beads are more tightly pelleted and easily eliminated in the next centrifugation step.
Suspension buffer (high salt): 10 mM HEPES-KOH (pH 7.6), 50 mM KCl, 3 M (NH4)2SO4, 10 mM Mg(OAc)2 (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use. Centrifuge at 20,000g for 20 minutes at 4 °C, and immediately decant the supernatant to a 50 mL conical centrifuge tube. Filter through 0.45 μm membrane just prior to FPLC.
-
Purify ribosomes using a 25 mL butyl column, which consists of 5 serially connected 5 mL HiTrap Butyl FF columns (GE Healthcare), on FPLC at 4 °C using the following procedure. Monitor ABS260 or ABS280 during the FPLC procedure. Equilibrate the column with buffer C. Load solution onto the column. Wash with 50 mL of [100 % buffer C/0 % buffer D] followed by 62.5 mL of [80 % buffer C/20 % buffer D]. Elute ribosomes with 75 mL of [50 % buffer C/50 % buffer D] followed by 62.5 mL of [0 % buffer C/100 % buffer D] at a flow rate of 5 mL or less per minute. Save flow-through from injection and collect all washes and eluents in 4 mL fractions.
Buffer C: 20 mM HEPES-KOH (pH 7.6), 1.5 M (NH4)2SO4, 10 mM Mg(OAc)2 (make fresh and degas), with 7 mM β-mercaptoethanol added immediately prior to use.Buffer D: 20 mM HEPES-KOH (pH 7.6), 10 mM Mg(OAc)2 (make fresh and degas), with 7 mM β-mercaptoethanol added immediately prior to use. Combine fractions containing ribosomes, which are collected during the first peak of UV absorbance after switching to [50 % buffer C/50 % buffer D], on ice for ultra-centrifugation. The total volume is typically ~76 mL.
-
Place 30 mL cushion buffer in each of two ultracentrifuge tubes (70 mL nominal volume) on ice and carefully layer eluent over sucrose cushions, ~38 mL each.
Cushion buffer: 20 mM HEPES-KOH (pH 7.6), 30 mM NH4Cl, 10 mM Mg(OAc)2, 30 % (w/v) sucrose (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use. Using Beckman type 45 Ti rotor, centrifuge at 100,000g for 16 hours at 4 °C.
-
Remove supernatant and briefly rinse the surface of each clear ribosome pellet three times with 1 mL each of ice-cold ribosome buffer.
Ribosome buffer: 20 mM HEPES-KOH (pH 7.6), 30 mM KCl, 6 mM Mg(OAc)2 (store at 4 °C), with 7 mM β-mercaptoethanol added immediately prior to use. Resuspend each ribosome pellet with 50–100 μL ribosome buffer using a mini stir bar (3 mm diameter x 10 mm length) on ice at the slowest setting on a magnetic stir plate.
Combine resuspended ribosomes into one tube on ice, rinsing the empty centrifuge tube with 40–100 μL ribosome buffer.
Measure ABS260 using a 1:100 dilution with ribosome buffer and calculate the concentration. An absorbance of 10 for the diluted solution corresponds to a 23 μM concentration of undiluted ribosome solution.
Divide into aliquots (typically ~300 pmol each) and immediately freeze on dry ice. Store at −80 °C.
7.2.3. Other PURE System components
Translation buffer: Make 8x concentrated solution containing 400 mM HEPES-KOH (pH 7.6), 96 mM Mg(OAc)2, 800 mM potassium glutamate, 16 mM spermidine. Store in aliquots at −80 °C.
Dithiothreitol (DTT): Freshly dissolve to 500 mM in water immediately prior to setting up translation reaction.
-
Protease inhibitor cocktail: Suspend one cOmplete ULTRA Tablet, EDTA-Free (Roche, 5892791001) in 1 mL water to make 10x suspension and store in aliquots at −20 °C.
Note: Although the PURE system is essentially protease-free, we have observed fragmentation of translated peptides, which are likely to be products from cleavage by potential protease contamination. To prevent this, protease inhibitor cocktail is added. ATP and GTP mix: ATP and GTP are purchased as sodium salts (Sigma, A26209 and Sigma, G8877, respectively). Combine 200 μL of freshly dissolved 100 mM ATP or GTP with 20 μL 3.81 M KCl and 600 μL ethanol and mix by vortexing for 1 minute. Centrifuge at 16,000–21,000g for 10 minutes at room temperature. Discard the supernatant and rinse the pellets with 1 mL 70 % (v/v) ethanol. Centrifuge at 16,000–21,000g for 5 minutes. Remove the supernatant and dry the pellets. Resuspend the pellets in 50 μL water and neutralize pH with KOH. Use a 1:500 dilution in water to measure the ABS260 to determine the concentration. Absorbances of 30.8 for the diluted ATP solution and 27.4 for the diluted GTP solution correspond to 1M concentrations of the undiluted solutions. Make a solution containing 25 mM each ATP and GTP and store as aliquots at −80 °C.
Creatine phosphate: Dissolve creatine phosphate dipotassium salt (Calbiochem, 237911) in water to 0.5 M and store as aliquots at −80 °C.
19 amino acid mix: Either cysteine or histidine is not added to the mix but separately to the translation reaction because the amount varies for radiolabeling purposes. Natural amino acids are supplied as a kit of separately packaged solids (Sigma, LAA21). At first, make the 17 amino acid mixture (20 natural amino acids minus methionine, cysteine and histidine) as follows. Combine and dissolve in water L-alanine, L-arginine hydrochloride, L-asparagine, L-aspartic acid, L-glutamic acid, L-glutamine, glycine, L-isoleucine, L-leucine, L-lysine hydrochloride, L-phenylalanine, L-proline, L-serine, L-threonine, L-tryptophan, L-tyrosine, and L-valine at 3 mM each, adjusting pH to 7.6 with KOH. Store as aliquots at −20 °C. Individually dissolve L-homopropargylglycine (Chiralix, CX90138) and L-histidine hydrochloride in water to 300 mM and store at −80 °C. Freshly dissolve L-cysteine in water to 300 mM immediately prior to setting up the translation reaction. Then, combine 98 parts 3 mM 17 amino acid mixture, 1 part 300 mM HPG, and 1 part 300 mM histidine or cysteine (whichever of these two is not added at this stage will be added at the last moment in radiolabeled form).
Radiolabeled cysteine or histidine: To prepare aliquots of 35S-cysteine, pipet 10 μL L-[35S]-cysteine (Perkin Elmer, NEG022T) into a 0.5 mL microcentrifuge tube, gently blow argon into the tube for 5 seconds, immediately close the lid and place on dry ice to freeze (Note: This procedure is performed in a fume hood to avoid any conceivable exposure to volatile decomposition products such as radioactive H2S). Repeat these steps to aliquot all of the solution and store at −80 °C. Use only freshly thawed aliquot(s) in the translation reaction. [2,5-3H]-L-Histidine (Moravek, MT905) is used as radioactive histidine and stored at 4 °C without freezing or −20 °C after quickly freezing on dry ice.
-
E. coli total tRNA: Dissolve lyophilized powder of tRNA from E. coli MRE600 (Roche, 10109541001) in 500 μL water. Check ABS260 using 1:100 dilution. Dilute further until ABS260 reaches 20 for 1:100 dilution. Store as aliquots at −80 °C.
Note: In one case, commercial tRNA was found to contain RNase activity. As a precaution, this can be assayed with RNase Alert kit (Ambion or IDT).
-
5,10-methenyltetrahydrofolate (5,10-MeTHF): Suspend (6R, S)-5,10-methenyl-5,6,7,8-tetrahydrofolic acid chloride (Schircks Laboratories, 16.230) in water to ~5 mg/mL. Add 1 N HCl until dissolved. Add DTT to a final concentration of 20 mM, and water and additional HCl until 5,10-MeTHF is 2 mg/mL. The pH is typically below 2.5. Depending on the batch, this reagent may not dissolve completely. In that case, use as a suspension. Store as aliquots at −80 °C. 5,10-MeTHF is converted to 10-formyltetrahydrofolate at the higher pH of the translation conditions (Stover & Schirch, 1992).
Note: We have experienced a gradual increase of precipitate after repeated freeze/thaw cycles. However, this does not seem to affect its performance in translation.
Creatine kinase: Freshly prepare a 10 ABS280 solution of creatine kinase from rabbit muscle (Roche, 10127566001) on ice in a buffer containing 10 mM Tris-HCl, 10 mM Mg(OAc)2, 100 mM NH4Cl, pH 7.5 at 37 °C with freshly supplemented 1 mM DTT. Store for up to 1 week at 4 °C.
Nucleoside 5′-diphosphate kinase from bovine liver (Sigma, N2635). Store at 4 °C.
Myokinase from rabbit muscle (Sigma, M3003). Store at 4 °C.
Inorganic pyrophosphatase: Dissolve inorganic pyrophosphatase from E. coli lyophilized powder (Sigma, I5907) to a final concentration of 100 U/mL in buffer containing 20 mM Tris-HCl, 0.1 mM EDTA, 1 mM DTT, 100 mM KCl, 50 % (v/v) glycerol, pH 8.0. Store aliquots at −80 °C. Once an aliquot is thawed, store at −20 °C.
References
- Arai K, Tsutsumi H, Mihara H. A monosaccharide-modified peptide phage library for screening of ligands to carbohydrate-binding proteins. Bioorganic & Medicinal Chemistry Letters. 2013;23:4940–4943. doi: 10.1016/j.bmcl.2013.06.059. [DOI] [PubMed] [Google Scholar]
- Bailey JK, Nguyen DN, Horiya S, Krauss IJ. Synthesis of multivalent glycopeptide conjugates that mimic an HIV epitope. Tetrahedron. 2016;72:6091–6098. doi: 10.1016/j.tet.2016.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Takahashi TT, Balakin A, Roberts RW. Selection of RNA-binding peptides using mRNA-peptide fusions. Methods. 2001a;23:287–293. doi: 10.1006/meth.2000.1139. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Takahashi TT, Ren J, Xia T, Roberts RW. Large libraries reveal diverse solutions to an RNA recognition problem. Proceedings of the National Academy of Sciences of the United States of America. 2001b;98:12374–12378. doi: 10.1073/pnas.221467798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. Journal of Virology. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annual Review of Immunology. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Chaput JC, Szostak JW. Evolutionary optimization of a nonbiological ATP binding protein for improved folding stability. Chemistry & Biology. 2004;11:865–874. doi: 10.1016/j.chembiol.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Cho G, Keefe AD, Liu R, Wilson DS, Szostak JW. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. Journal of Molecular Biology. 2000;297:309–319. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- Çelik E, Fisher AC, Guarino C, Mansell TJ, DeLisa MP. A filamentous phage display system for N-linked glycoproteins. Protein Science. 2010;19:2006–2013. doi: 10.1002/pro.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, et al. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. International Journal of Molecular Sciences. 2016;17:144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiacco SV, Kelderhouse LE, Hardy A, Peleg Y, Hu B, Ornelas A, et al. Directed Evolution of Scanning Unnatural-Protease-Resistant (SUPR) Peptides for in Vivo Applications. Chembiochem. 2016;17:1643–1651. doi: 10.1002/cbic.201600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Weissbach H, Blacklow SC. A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Analytical Biochemistry. 2001;297:60–70. doi: 10.1006/abio.2001.5329. [DOI] [PubMed] [Google Scholar]
- Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Luo D, Xu J. Cell-free protein expression under macromolecular crowding conditions. PLoS ONE. 2011;6:e28707. doi: 10.1371/journal.pone.0028707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm SK, Battles MB, Ackerman ME. Directed evolution of a yeast-displayed HIV-1 SOSIP gp140 spike protein toward improved expression and affinity for conformational antibodies. PLoS ONE. 2015;10:e0117227. doi: 10.1371/journal.pone.0117227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen Schlippe YV, Hartman MCT, Josephson K, Szostak JW. In vitro selection of highly modified cyclic peptides that act as tight binding inhibitors. Journal of the American Chemical Society. 2012;134:10469–10477. doi: 10.1021/ja301017y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker DE, Hoinka J, Iqbal ES, Przytycka TM, Hartman MCT. Highly Constrained Bicyclic Scaffolds for the Discovery of Protease-Stable Peptides via mRNA Display. ACS Chemical Biology. 2017;12:795–804. doi: 10.1021/acschembio.6b01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann FT, Szostak JW, Seebeck FP. In vitro selection of functional lantipeptides. Journal of the American Chemical Society. 2012;134:8038–8041. doi: 10.1021/ja302082d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angewandte Chemie International Edition. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S, Bailey JK, Temme JS, Guillen Schlippe YV, Krauss IJ. Directed evolution of multivalent glycopeptides tightly recognized by HIV antibody 2G12. Journal of the American Chemical Society. 2014a;136:5407–5415. doi: 10.1021/ja500678v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiya S, MacPherson IS, Krauss IJ. recent strategies targeting hiV glycans in vaccinedesign. Nature Chemical Biology. 2014b;10:990–999. doi: 10.1038/nchembio.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K, Hartman MCT, Szostak JW. Ribosomal synthesis of unnatural peptides. Journal of the American Chemical Society. 2005;127:11727–11735. doi: 10.1021/ja0515809. [DOI] [PubMed] [Google Scholar]
- Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discovery Today. 2014;19:388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Ogawa K, Hatta T, Goshima N, Natsume T. Directed Evolution of a Cyclized Peptoid-Peptide Chimera against a Cell-Free Expressed Protein and Proteomic Profiling of the Interacting Proteins to Create a Protein-Protein Interaction Inhibitor. ACS Chemical Biology. 2016;11:1569–1577. doi: 10.1021/acschembio.5b01014. [DOI] [PubMed] [Google Scholar]
- Keefe AD. Protein selection using mRNA display. Current Protocols in Molecular Biology. 2001;24(5):24.5.1–24.5.34. doi: 10.1002/0471142727.mb2405s53. [DOI] [PubMed] [Google Scholar]
- Kiessling LL, Young T, Gruber TD, Mortell KH. Multivalency in Protein–Carbohydrate Recognition. In: Fraser-Reid BO, Tatsuta K, Thiem J, editors. Glycoscience Chemistry and Chemical Biology. 2. Vol. 1. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008. pp. 2483–2523. [Google Scholar]
- Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie International Edition. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kurz M, Gu K, Lohse PA. Psoralen photo-crosslinked mRNA-puromycin conjugates: a novel template for the rapid and facile preparation of mRNA-protein fusions. Nucleic Acids Research. 2000;28:E83. doi: 10.1093/nar/28.18.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane MD, Seelig B. Advances in the directed evolution of proteins. Current Opinion in Chemical Biology. 2014;22:129–136. doi: 10.1016/j.cbpa.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Roberts RW. A novel strategy for in vitro selection of peptide-drug conjugates. Chemistry & Biology. 2003;10:233–239. doi: 10.1016/s1074-5521(03)00047-4. [DOI] [PubMed] [Google Scholar]
- Liu R, Barrick JE, Szostak JW, Roberts RW. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods in Enzymology. 2000;318:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- Ma Z, Hartman MCT. In vitro selection of unnatural cyclic peptide libraries via mRNA display. Methods in Molecular Biology. 2012;805:367–390. doi: 10.1007/978-1-61779-379-0_21. [DOI] [PubMed] [Google Scholar]
- Millward SW, Fiacco S, Austin RJ, Roberts RW. Design of Cyclic Peptides That Bind Protein Surfaces with Antibody-Like Affinity. ACS Chemical Biology. 2007;2:625–634. doi: 10.1021/cb7001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Letters. 1997;414:405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- Ng S, Lin E, Kitov PI, Tjhung KF, Gerlits OO, Deng L, et al. Genetically encoded fragment-based discovery of glycopeptide ligands for carbohydrate-binding proteins. Journal of the American Chemical Society. 2015;137:5248–5251. doi: 10.1021/ja511237n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi H, Shimizu Y, Ying BW, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochemical and Biophysical Research Communications. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Packer MS, Liu DR. Methods for the directed evolution of proteins. Nature Reviews Genetics. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- Passioura T, Suga H. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chemical Communications. 2017;53:1931–1940. doi: 10.1039/c6cc06951g. [DOI] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angewandte Chemie International Edition. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sau SP, Larsen AC, Chaput JC. Automated solid-phase synthesis of high capacity oligo-dT cellulose for affinity purification of poly-A tagged biomolecules. Bioorganic & Medicinal Chemistry Letters. 2014;24:5692–5694. doi: 10.1016/j.bmcl.2014.10.065. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. Journal of Virology. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig B. mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nature Protocols. 2011;6:540–552. doi: 10.1038/nprot.2011.312. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. PURE technology. Methods in Molecular Biology. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nature Biotechnology. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and Protein Synthesis A Practical Approach. New York, NY: Ribosomes and Protein Synthesis; 1990. pp. 1–29. [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover P, Schirch V. Synthesis of (6S)-5-formyltetrahydropteroyl-polyglutamates and interconversion to other reduced pteroylpolyglutamate derivatives. Analytical Biochemistry. 1992;202:82–88. doi: 10.1016/0003-2697(92)90210-x. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Hartman MCT, Szostak JW. Ribosomal synthesis of N-methyl peptides. Journal of the American Chemical Society. 2008;130:6131–6136. doi: 10.1021/ja710016v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme JS, Krauss IJ. SELMA: Selection with Modified Aptamers. Current Protocols in Chemical Biology. 2015;7:73–92. doi: 10.1002/9780470559277.ch140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme JS, Drzyzga MG, MacPherson IS, Krauss IJ. Directed evolution of 2G12-targeted nonamannose glycoclusters by SELMA. Chemistry - a European Journal. 2013;19:17291–17295. doi: 10.1002/chem.201303848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme JS, MacPherson IS, DeCourcey JF, Krauss IJ. High temperature SELMA: evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnAb 2G12. Journal of the American Chemical Society. 2014;136:1726–1729. doi: 10.1021/ja411212q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. Journal of Virology. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hest JCM, Kiick KL, Tirrell DA. Efficient incorporation of unsaturated methionine analogues into proteins in vivo. Journal of the American Chemical Society. 2000;122:1282–1288. [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Shoji I, Miyagawa S, Kawakami T, Katoh T, Goto Y, Suga H. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chemistry & Biology. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Yamaguchi J, Naimuddin M, Biyani M, Sasaki T, Machida M, Kubo T, et al. cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucleic Acids Research. 2009;37:e108. doi: 10.1093/nar/gkp514. [DOI] [PMC free article] [PubMed] [Google Scholar]