Abstract
The Gram-positive lactic acid bacterium Lactobacillus buchneri CD034 is covered by a two-dimensional crystalline, glycoproteinaceous cell surface (S-) layer lattice. While lactobacilli are extensively exploited as cell surface display systems for applied purposes, questions about how they stick their cell wall together are remaining open. This also includes the identification of the S-layer cell wall ligand. In this study, lipoteichoic acid was isolated from the L. buchneri CD034 cell wall as a significant fraction of the bacterium’s cell wall glycopolymers, structurally characterized and analyzed for its potential to mediate binding of the S-layer to the cell wall. Combined component analyses and 1D- and 2D-nuclear magnetic resonance spectroscopy (NMR) revealed the lipoteichoic acid to be composed of on average 31 glycerol-phosphate repeating units partially substituted with α-d-glucose, and with an α-d-Galp(1→2)-α-d-Glcp(1→3)−1,2-diacyl-sn-Gro glycolipid anchor. The specificity of binding between the L. buchneri CD034 S-layer protein and purified lipoteichoic acid as well as their interaction force of about 45 pN were obtained by single-molecule force spectroscopy; this value is in the range of typical ligand–receptor interactions. This study sheds light on a functional implication of Lactobacillus cell wall architecture by showing direct binding between lipoteichoic acid and the S-layer of L. buchneri CD034.
Keywords: lactic acid bacteria, lipoteichoic acid, NMR, single-molecule force spectroscopy, S-layer binding interaction
Introduction
Lactic acid bacteria (LAB), particularly their cell surface, have been subject of major interest in various fields of applied research including medicine, food science and biotechnology (Leenhouts et al. 1999; Mao et al. 2016; Michon et al. 2016). This is primarily because of the combined benefits of organisms offering pro-/pre-biotic effects and generally regarded as safe (GRAS) status (Klaenhammer et al. 2005; Wedajo 2015). Protein cell surface display in LAB is achieved by attachment to the cytoplasmic membrane—using a transmembrane anchor or a lipoprotein anchor—or to the cell wall—by covalent linkage using sortase-mediated anchoring via the LPXTG motif, or by non-covalent liaisons employing binding domains such as LysM and WxL (Schneewind and Missiakas 2012; reviewed in Michon et al. 2016) or the cell wall binding region of cell surface (S-) layer proteins.
Bacterial S-layers are potent cell surface display systems; because of their intrinsic self-assembly ability they enable high-density display of proteins/epitopes with nanometer-scale periodicity (Messner et al. 2010; Sleytr et al. 2010; Fagan and Fairweather 2014). In Gram-positive bacteria, S-layer proteins are comprised of two distinct functional regions, one mediating protein self-assembly and the other enabling cell wall anchoring (Engelhardt and Peters 1998; Sára and Sleytr 2000). For this purpose, S-layers of so far investigated Bacillaceae strains associate with strain-specific peptidoglycan (PG)-bound cell wall glycopolymers (CWGs), which are structurally different from teichoic acids/teichuronic acids (Archibald et al. 1993; Mesnage et al. 2000; Sára 2001; Schäffer and Messner 2005; Messner et al. 2009; Schade and Weidenmaier 2016; Rajagopal and Walker 2017). The molecular interaction between S-layer proteins and the strain-specific CWG is frequently, but not necessarily, mediated by a repetitive, terminal structural motif within the S-layer proteins, termed “surface layer homology” (SLH) domain (Mesnage et al. 2000; Sára 2001).
In lactobacilli, S-layers are present on many species (e.g., Lactobacillus acidophilus ATCC 4356, Lactobacillus brevis ATCC 8287, Lactobacillus crispatus ZJ001 and JCM 5810, and Lactobacillus buchneri CD034) on top of a thick peptidoglycan sacculus (Sleytr and Messner 1983; Masuda and Kawata 1985; Åvall-Jääskeläinen et al. 2008; Heinl et al. 2012). Lactobacillus S-layer proteins differ from those of other bacteria in their smaller size (25–71 kDa) and higher pI values (9.4–10.4) (Malamud et al. 2017); further, they do not contain SLH-domains (Engelhardt and Peters 1998; Brechtel and Bahl 1999). In many of these S-layer proteins, positively charged amino acids are centered in the cell wall binding region, which might be located either N- or C-terminally, depending on the species. So far, mainly the C-terminal region of the L. acidophilus S-layer protein has been utilized to display heterologous proteins/epitopes by heterologous reattachment (Antikainen et al. 2002; Smit and Pouwels 2002; Hu et al. 2011), however without detailed knowledge of how S-layer anchoring to the cell wall is elaborated.
Based on reports in the literature it can be assumed that in Lactobacillus species, teichoic acids (TAs) are involved in S-layer protein anchoring. Lactobacillus cell walls usually harbor TAs, teichuronic acids, and other polysaccharides (Archibald et al. 1993; Messner et al. 2013; Rajagopal and Walker 2017). Teichoic acids are among the best characterized CWGs; usually they are present in two formats—wall teichoic acids (WTAs), which are covalently attached to the PG via a phosphodiester bond (Archibald et al. 1993; Brown et al. 2013) and lipoteichoic acids (LTAs), which are embedded in the cytoplasmic membrane via a glycolipid anchor (Delcour et al. 1999; Schirner et al. 2009; Reichmann and Gründling 2011; Schneewind and Missiakas 2014; Schade and Weidenmaier 2016). The repeating units of the WTA backbone consist of alditol phosphates linked by phosphodiester bonds, frequently glycerol phosphate (Gro-P) units, which may be further substituted by amino acids (d-alanine) and/or carbohydrate residues (Weidenmaier and Peschel 2008; Kleerebezem et al. 2010). Analysis of LTA structures of LAB showed that the length of the Gro-P chains and the degree and composition of substitution as well as the nature of the lipid anchor is highly diverse and varies between strains (Fischer et al. 1993; Schade and Weidenmaier 2016). In vitro binding studies of the S-layer proteins from L. acidophilus ATCC 4356 and L. crispatus indicated binding to native TAs as well as non-native LTAs (Antikainen et al. 2002; Smit and Pouwels 2002; Åvall-Jääskeläinen et al. 2008). On the other hand, CWGs other than TAs were described to interact with the S-layer of L. brevis ATCC 8287 and Lactobacillus hilgardii B706 via hydrogen bonding (Åvall-Jääskeläinen et al. 2008; Dohm et al. 2011), a fact, which also early studies on the L. buchneri S-layer may pinpoint (Masuda and Kawata 1981, 1985).
This study is focusing on S-layer anchoring of the industrially relevant strain L. buchneri CD034 (Heinl et al. 2011, 2012). Its S-layer is formed by self-assembly of the glycosylated S-layer protein SlpB (58.3 kDa; pI = 10.2) and exhibits oblique lattice symmetry (Anzengruber et al. 2014b). We isolated and structurally characterized LTA of L. buchneri CD034 as a significant fraction of the bacterium’s CWGs and investigated its possible biological function as a ligand of the bacterium’s S-layer protein SlpB. To obtain, for the first time for a Lactobacillus S-layer protein, distinct information about the specificity of binding to and the interaction force with its cell wall ligand, we applied the single-molecule force spectroscopy technique based on atomic force microscopy. This study contributes to our understanding of how L. buchneri CD034 maintains its cell wall integrity and provides general insight into the interaction of CWGs with cell surface proteins.
Results
Extraction of CWGs from L. buchneri cell walls using n-butanol
To define, if LTA serves as a cell wall ligand for the L. buchneri CD034 S-layer, CWGs were extracted with n-butanol from disrupted L. buchneri CD034 cells whose S-layer had been stripped-off before, and further separated by hydrophobic interaction chromatography (HIC) (Morath et al. 2001; Duda et al. 2016), yielding two pools. Fractions eluting from the HIC column isocratically with 15% n-propanol in 0.1 M ammonium acetate (pH 4.7) represented pool I and those eluting in the HIC gradient between 45% and 65% n-propanol in 0.1 M ammonium acetate (pH 4.7) were combined to pool II. Based on its elution interval from the HIC column, this pool was expected to contain LTA.
Component analyses of n-butanol extracted CWGs
Analyses of pool II revealed the presence of Glc and Gal (1117 and 92 nmol/mg, respectively), glycerol (Gro), glycerol-phosphate (Gro-P) and small amounts of glycerol-hexose (Gro-Hex). The absolute configuration of Glc and Gal was determined as d. Fatty acids were mainly 16:0 and 18:1 (169 and 89 nmol/mg) besides small amounts of 14:0, 18:0, 18:2 and cyclo-19:0. These data strongly indicated that pool II contained LTA; this pool was subsequently investigated in detail.
Analyses of pool I (material not bound to the HIC column) identified as major components Glc and Gal, a pyruvylated hexose as well as organic phosphate, but no fatty acids (E. Bönisch, K. A. Duda, C. Schäffer, P. Kosma, unpublished data). As concluded from NMR analysis (not shown) this pool was heterogeneous; pool I was not further investigated in the course of this study.
Complete structural analyses of purified L. buchneri LTA
To obtain all structural details of intact L. buchneri CD034 LTA (i.e., structure of the hydrophilic backbone, the linker anchoring the LTA to the membrane and the linkage of the poly(Gro-P) chain to the glycolipid anchor), 1D and 2D NMR experiments were carried out on native LTA (pool II as described in the previous paragraph), O-deacylated LTA, and the linker (representing 7% of total LTA, based on dry mass). The assignment of each spin system was obtained by analysis of COSY and TOCSY spectra; the HSQC spectrum allowed the identification of corresponding carbon atoms. The anomeric configuration of the sugar residues was defined based on the 3J1,2 coupling constants (Tables I–III). The sequence of residues could be determined by the analysis of the HMBC spectra and corroborated by inter residual NOE contacts observed in the ROESY spectra.
Table II.
1H, 13C and 31P NMR chemical shifts (δ, ppm) of O-deacylated LTA of L. buchneri CD034
| Residue | 1a | 1b | 2 | 3a | 3b | |
|---|---|---|---|---|---|---|
| A | 1H | 3.68 | 3.61 | 4.07 | 3.83 | 3.52 |
| Gro (linker) | 13C | 62.75 | 66.64 | 69.50 | ||
| Residue | 1 (3J1,2 Hz) | 2 | 3 | 4 | 5 | 6a | 6b | |
| B | 1H | 5.18 (3.4) | 3.54 | 3.77 | 3.40 | 3.94 | 3.88 | 3.76 |
| P-Gro-[Glcp]-P | 13C | 98.28 | 72.08 | 73.59 | 70.26 | 72.41 | 61.12 | |
| C | 1H | 5.15 (3.9) | 3.70 | 3.81 | 3.48 | 3.69 | 3.88 | 3.76 |
| 2-α-d-Glcp | 13C | 96.56 | 76.20 | 72.06 | 70.12 | 72.33 | 61.12 | |
| D | 1H | 5.12 (4.1) | 3.83 | 3.96 | 3.91 | 4.07 | 3.97 | 3.88 |
| P-6-α-d-Galp | 13C | 96.82 | 68.67 | 71.19 | 71.29 | 69.22 | 66.80 | |
| 31P | - | - | - | - | - | 1.2 | ||
| Residue | 1a | 1b | 2 | 3a | 3b | |
| E | 1H | 3.96 | 3.89 | 4.05 | 3.96 | 3.89 |
| P-Gro-P | 13C | 66.75 | 70.13 | 66.75 | ||
| 31P | 1.5 | - | 1.5 | |||
| F | 1H | 4.05 | 3.99 | 4.13 | 4.05 | 3,99 |
| P-Gro-[Glcp]-P | 13C | 65.08 | 75.85 | 65.08 | ||
| 31P | 1.4 | - | 1.4 | |||
Gro, P, FA—stands for glycerol, phosphate and fatty acids, respectively.
Table III.
1H, 13C NMR chemical shifts (δ, ppm) of the linker of LTA of L. buchneri CD034
| Residue | 1a | 1b | 2 | 3a | 3b | |
|---|---|---|---|---|---|---|
| A | 1H | 4.51 | 4.23 | 5.27 | 3.90 | 3.69 |
| Gro-FA | 13C | 64.51 | 72.14 | 67.71 | ||
| Residue | 1 (3J1,2 Hz) | 2 | 3 (3J3,4 Hz) | 4 | 5 | 6a | 6b | |
| C | 1H | 5.07 (3.6) | 3.61 | 3.78 (9.7) | 3.38 | 3.61 | 3.83 | 3.71 |
| 2-α-d-Glcp | 13C | 98.29 | 78.29 | 73.95 | 72.05 | 74.42 | 63.21 | |
| D | 1H | 5.04 (4.0) | 3.80 | 3.83 (3.0) | 3.92 | 4.12 | 3.75 | 3.72 |
| α-d-Galp | 13C | 98.76 | 70.85 | 71.91 | 71.70 | 73.17 | 63.46 | |
Gro, FA—stands for glycerol and fatty acids, respectively.
Table I.
1H, 13C and 31P NMR chemical shifts (δ, ppm) of LTA of L. buchneri CD034
| Residue | 1a | 1b | 2 | 3a | 3b | |
|---|---|---|---|---|---|---|
| A | 1H | 4.51 | 4.28 | 5.31 | 3.89 | 3.77 |
| Gro-FA | 13C | 63.64 | 71.35 | 66.37 | ||
| Residue | 1 (3J1,2 Hz) | 2 | 3 | 4 | 5 | 6a | 6b | |
| B | 1H | 5.19 (3.5) | 3.53 | 3.76 | 3.39 | 3.93 | 3.88 | 3.76 |
| P-Gro-[Glcp]-P | 13C | 98.35 | 72.16 | 73.62 | 70.33 | 72.49 | 61.19 | |
| C | 1H | 5.15 (3.5) | 3.68 | 3.77 | 3.52 | 3.64 | 3.89 | 3.77 |
| 2-α-d-Glcp | 13C | 96.46 | 75.87 | 71.96 | 69.89 | 72.41 | 60.99 | |
| D | 1H | 5.09 (3.8) | 3.80 | 3.97 | 3.90 | 4.05 | 3.95 | 3.88 |
| P-6-α-d-Galp | 13C | 96.78 | 68.73 | 72.50 | 71.36 | 72.16 | 68.72 | |
| 31P | - | - | - | - | - | 1.2 | ||
| Residue | 1a | 1b | 2 | 3a | 3b | |
| E | 1H | 3.96 | 3.88 | 4.05 | 3.96 | 3.88 |
| P-Gro-P | 13C | 66.99 | 70.31 | 66.99 | ||
| 31P | 1.49–1.24 | - | 1.49–1.24 | |||
| F | 1H | 4.05 | 3.99 | 4.12 | 4.05 | 3.99 |
| P-Gro-[Glcp]-P | 13C | 65.19 | 75.99 | 65.19 | ||
| 31P | 1.49–1.24 | - | 1.49–1.24 | |||
Gro, P, FA—stands for glycerol, phosphate and fatty acids, respectively.
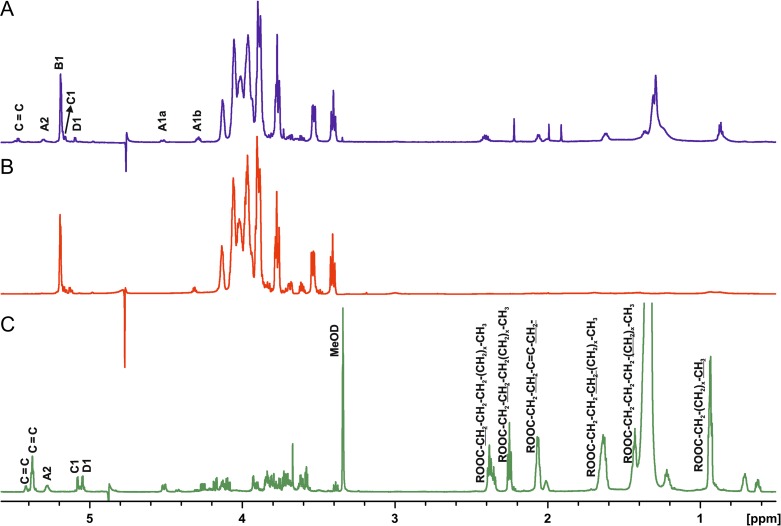
Figure 1 shows a comparison of 1H NMR spectra of the different preparations of L. buchneri CD034 LTA. The main difference between native LTA (Figure 1A) and O-deacylated LTA (Figure 1B) were missing signals at δ 2.44–2.36, δ 2.08–2.04, δ 1.32–1.27 and δ 0.89–0.85 corresponding to the –CH2–CH2–CO–, –CH2–CH2–CO–, –CH2– and terminal –CH3 in the O-deacylated sample. In the 1H spectrum of the linker, residues B, E and F (the two latter are not marked on the spectrum as they are present in the region of the ring carbons) were missing, and residues A, C and D became more prominent, which indicated residues B, E and F to be constituents of the hydrophilic chain of the LTA, and residues A, C and D parts of the linker.
Fig. 1.
1H NMR spectra of native LTA (A), O-deacylated LTA (B), recorded in D2O at 27°C and linker of LTA of L. buchneri CD034 (C) recorded in MeOD at 27°C on the 700 MHz Bruker spectrometer. Letters refer to the residues listed in Table I.
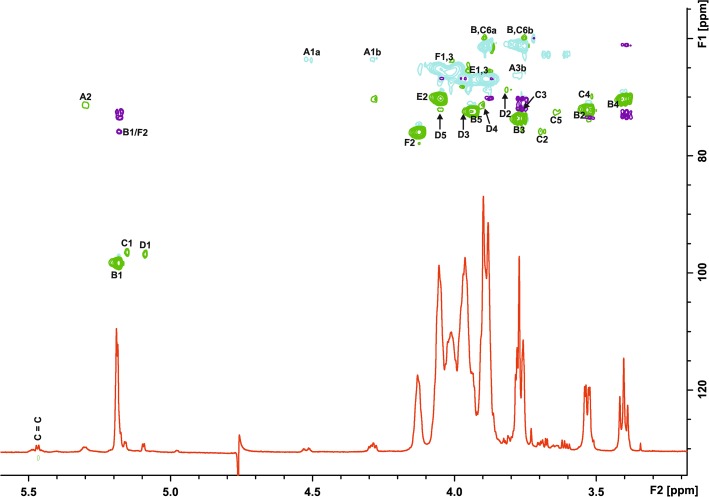
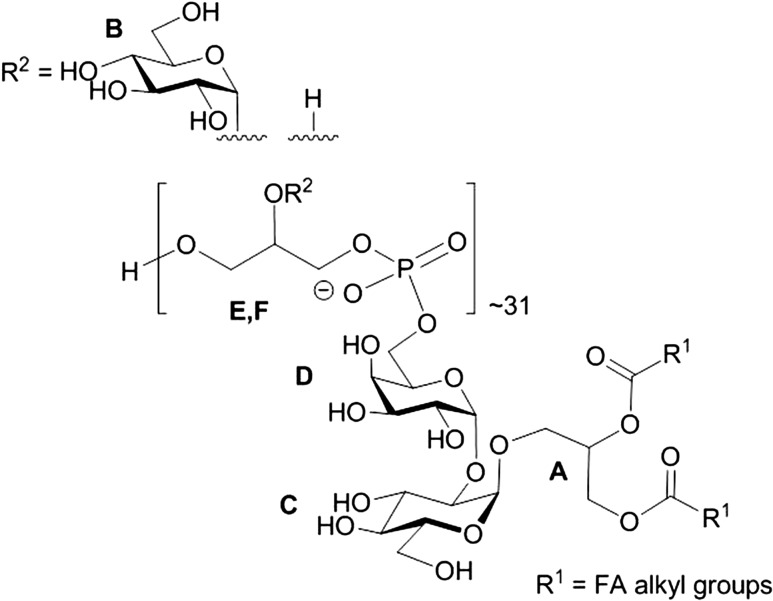
In general, all the information required for the complete LTA structure could be derived from spectra of native LTA, however, chemical shifts of the minor weak compounds were proven in the spectra of O-deacylated LTA and the linker. In detail, the 1H NMR and HSQC spectra of LTA (Figures 1A; 2) showed in the anomeric region a dominant signal at δH 5.19/δC 98.35 (residue B) and of much lower intensity signals at δH 5.15/δC 96.46 (C) and at δH 5.09/δC 96.78 (D). Additionally, the spectrum contained signals of the vicinal protons of the C = C group (δH 5.46) identified due to a characteristic carbon signal at 131.55 ppm. Cross-peaks observed in TOCSY of the signal at δH 5.31 to the signals of fatty acids at 1.32–1.27 ppm together with the δC value of 71.35 assigned the signal at δH 5.31 (A2) to H-2 of Gro-linked to fatty acid. In the region from δH 4.51 to δH 3.39/δC 61.19 to δC 75.99, signals of the ring carbons were observed. Residues B and C were identified as α-Glc due to the 3J1,2 values of 3.5 Hz and the presence of intra residual NOEs B H-2/H-4 (for residue C seen only in the ROESY spectrum of the linker). Residue D was assigned to α-Gal as proven by the measurement of the 3J1,2 value (3.8 Hz) and identification in the ROESY spectrum of the linker diagnostic for a galacto configuration D H-3/H-4 intra residual NOE signal. The dominant spin systems seen in the HSQC spectrum were attributed to unsubstituted P-Gro-P (E) and P-Gro-P substituted (F) based on the chemical shifts and correlation to P signals observed in the 31P TOCSY HMBC spectrum. The absence of downfield shifts of the ring carbon atom resonances of residue B indicated that α-Glc of the hydrophilic backbone was a side chain sugar unit. Downfield shifts of the C-2 carbon of residue C and carbon 6 of residue D suggested that α-Glc of the linker was substituted at position O-2 and α-Gal at position O-6. A cross peak B H-1/F C-2 in the HMBC spectrum and inter residual NOE F H-2/B H-1 confirmed the partial substitution of P-Gro-P with α-Glc (residue B). The inter residual NOEs D H-1/C H-2 and C H-1/A H-3a,b present in the ROESY spectrum of the linker determined the structure of the linker as α-d-Galp-(1→2)-α-d-Glcp-(1→3)-diacyl-sn-Gro. The correlation of the phosphate signal at δP 1.2 to H-6 of α-d-Gal, observed on the 1H,31P TOCSY HMBC spectrum of the native and O-deacylated LTA, identified the linkage of the hydrophilic backbone of LTA to O-6 of residue D of the linker. The ratio of the phosphate content in the LTA (2820 nmol/mg) to the α-Gal residue (92 nmol/mg), present only in the linker identified an average chain length of 31 Gro-P units. In summary, the data identified the complete structure of LTA of L. buchneri CD034 as shown in Figure 3.
Fig. 2.
Overlay of 1H NMR (red), HSQC (positive signals, green; negative signals, blue), and HMBC (violet) spectra of native LTA of L. buchneri CD034 recorded in D2O at 27°C (700 MHz). Letters refer to the residues listed in Table I.
Fig. 3.
Complete structure of L. buchneri CD034 LTA.
Single-molecule force spectroscopy of S-layer protein and deacylated LTA
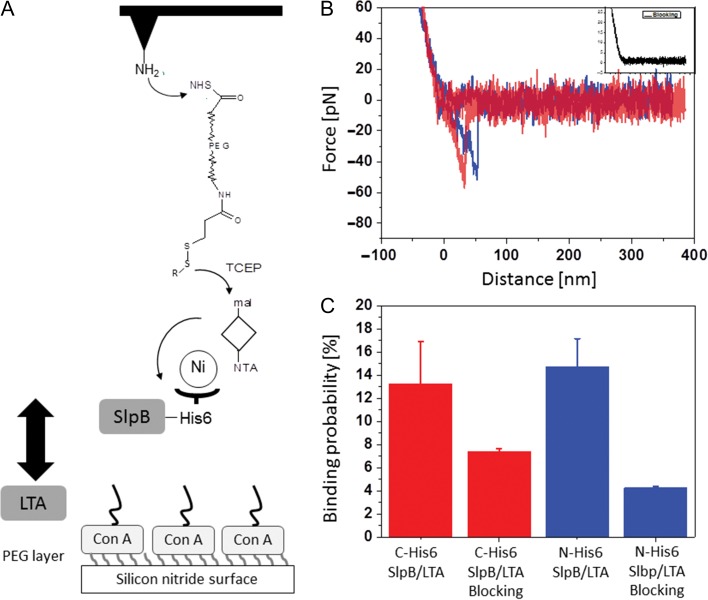
To gain detailed insight into the interaction between L. buchneri CD034 LTA and SlpB, single-molecule force spectroscopy was employed using recombinant, His-tagged S-layer protein in conjunction with O-deacylated LTA. Deacylated LTA was used to facilitate proper binding of the LTA’s carbohydrate portion to the wafer as required for the interaction measurements. Recombinant SlpB was immobilized on the AFM tip through a polyethylene glycol (PEG) linker with well-established surface chemistry (Figure 4) (Tang et al. 2009). In this set-up, N-His6-rSlpB and C-His6-rSlpB were compared in order to assess a possible influence of the orientation of the His6-tag on the S-layer protein on the data acquisition. The functionalized tip was used to detect binding events with deacylated LTA which had been surface-immobilized via Con A. The tip modified with recombinant S-layer protein approached the LTA-coated surface, allowing the interaction between them to occur. An increasing force was applied to the protein–LTA bond by pulling the complex until the dissociation occurred and the unbinding force was reached.
Fig. 4.
(A) Schematic design of single molecule force spectroscopy measurements. ConA, Concanavalin A; mal, maleimide; NTA, nitrilo acetic acid; TCEP, Tris(carboxyethyl)phosphine hydrochloride. (B) Typical force–distance curves (inset, blocking of the specific interaction). (C) Binding probability for C-His6-SlpB (red), N-His6-SlpB (blue) and addition of LTA to measurement solution for blocking SlpB on the AFM cantilever.
Figure 4A shows the schematic design of the tip chemistry used for covalent immobilization of His6-tagged rSlpB on the AFM tip via a flexible PEG linker. The PEG linker that connected the S-layer protein to the AFM tips ensured sufficient motional freedom for unconstrained interaction measurements. An LTA-layer was formed on Con A-bound silicon nitride surfaces, taking advantage of the strong interaction of the sugar residues of LTA and Con A.
Force–distance curves were measured by approaching the rSlpB-conjugated tip to the surface-bound LTA, followed by its retraction. Figure 4B shows the typical force–distance curves recorded using AFM cantilever tips functionalized with N-His6-rSlpB and C-His6-rSlpB, respectively. Single molecular unbinding events were evident from sharp spikes in the retraction curves (Figure 4B) at about 50 nm distance that corresponded to the dissociation of the tip-adorned S-layer protein from LTA on the wafer surface. Single unbinding events were measured with average unbinding force values of 44.7 ± 2.1 pN for C-His6-rSlpB, and 43.7 ± 3.2 pN for N-His6-rSlpB at a retraction velocity of 1000 nm/s. In most of the measurements, single unbinding events were observed. To prove that the measured forces were due to the specific interactions, blocking experiments were performed (Figure 4B, inset). Addition of O-deacylated LTA into the measurement solution resulted in a significant drop of the binding probability—from 13.26 ± 3.64% to 7.4 ± 0.25% for C-His6-rSlpB, and from 14.73 ± 2.41% to 4.25 ± 0.15% for N-His6-rSlpB (Figure 4C)—which represents the frequency of specific interaction events in force–distance curves, indicating the specificity of the interaction. The binding probability was determined as the percentage of 1000 force–distance curves displaying unbinding events with four different sample preparations.
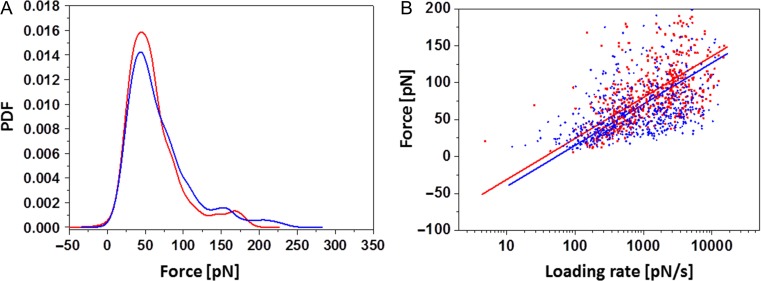
The distribution of unbinding forces between N-His6-rSlpB and C-His6-rSlpB is shown in Figure 5. The collected unbinding forces were gathered, summed up, and normalized to calculate the empirical probability density functions (PDF). The peak in this distribution represents the most probable measured unbinding force coming from a single receptor–ligand pair (Figure 5A), while the tail is indicative of occasionally occurring multiple binding events that are not taken into account for further analysis. Receptor–ligand interaction unbinding forces do not depend solely on the particular receptor–ligand pair but also on the loading rate of the experiment. With a single energy barrier the unbinding forces rise linearly with the logarithm of the loading rate (Bell 1978; Evans and Ritchie 1997). We characterized the molecular bonds further to attain the kinetic off-rate constant Koff and the length scale of the interaction energy potential Xβ. In our single-molecule force spectroscopy experiments, the pulling rate was varied and we plotted each measured unbinding force as a function of the logarithm of the loading rate (Figure 5B). The variance in determining the parameters Koff and Xβ comes from errors of the measurement system (i.e., spring constant and pulling speed), but mainly from the stochastic nature of the unbinding process. According to the kinetic models (Bell 1978; Evans and Ritchie 1997), a linear rise of the unbinding force with respect to a logarithmically increasing loading rate is expected when a single energy barrier is crossed in the thermally activated regime. Koff and Xβ were evaluated from data fits as shown in Figure 5B and yielded averages of Xβ 2.16 ± 0.6 Å (n = 2) and Koff 1.54 ± 0.48 1/s (n = 2) for C-His6-SlpB/LTA dissociation, and Xβ 1.59 ± 0.3 Å (n = 2) and Koff 1.93 ± 0.29 1/s (n = 2) for N-His6-SlpB/LTA dissociation, respectively. Both Xβ and Koff values were well comparable within the statistical error, indicating that the same structural epitope was probed.
Fig. 5.
(A) PDFs of unbinding forces at a retraction velocity of 1000 nm/s, and (B) a plot of unbinding force versus loading rate for the AFM tip containing S-layer protein dissociating from O-deacylated LTA to the surface. Blue, N-His6-SlpB; red, C-His6-SlpB.
Discussion
This study was designed to elucidate the structure and define, for the first time, the binding strength of LTA as a cell wall ligand for a Lactobacillus S-layer protein, exemplified with L. buchneri CD034, an organism known to possess a glycosylated S-layer (Anzengruber et al. 2014b).
On the basis of compositional data, NMR analyses, and reference information we conclude that the backbone of L. buchneri CD034 LTA is identical to that found in the most wide-spread type of LTAs in Gram-positive bacteria (Type I) (Fischer et al. 1990; Schneewind and Missiakas 2014). The hydrophobic backbone is composed of typical Gro-P residues, and about 30% of the Gro residues are decorated with α-d-Glc residues (Figure 1). Gro-P repeating units with substitution on the C-2 have already been reported in LTAs of other lactobacilli. Frequently, this position is substituted with d-alanyl esters (Neuhaus and Baddiley 2003; Perea Vélez et al. 2007; Räisänen et al. 2007; Shiraishi et al. 2013). Modification of LTAs by d-alanyl esters has been described to modulate the net negative charge of LTAs by providing positively charged amino groups as counteract to the negatively charged backbone phosphate groups, with effects on a number of cell morphology and surface properties. Interestingly, LABs with switched-off genes for d-alanylation showed increased levels of glucosylation, as evidenced with a dltD mutant of Lactobacillus rhamnosus GG (Perea Vélez et al. 2007) and a dltB mutant of Lactobacillus plantarum NCIMB8826 (Grangette et al. 2005). In the case of L. buchneri CD034, for which in the frame of the current study substitution of the LTA backbone with Glc but not with d-alanine was found, transcriptome analysis (Eikmeyer et al. 2015) identified the dlt genes which are principally required for d-alanylation, i.e., dltA (LBUCD034_0133; d-alanine-d-alanyl carrier protein ligase, subunit 1 (EC 6.1.1.13)), dltB (LBUCD034_0134; d-alanine transfer protein), dltC (LBUCD034_0135; d-alanine-poly(phosphoribitol)) ligase, subunit 2 (EC 6.1.1.13), dltD1 (LBUCD034_0136; d-alanyl transfer protein) and dltD3 (LBUCD034_0137; d-alanyl transfer protein), as oxygen-responsive transcripts (Eikmeyer et al. 2015). It remains to be investigated if all of these genes are translated into functional proteins and in case they are, which the targets of d-alanylation would be. It is very unlikely that under the chosen conditions of LTA preparation d-alanyl ester groups would have been unintentionally removed, since the pH value has been always kept below 6.2; dealanylation is known to occur at pH values above 8.0 (Morath et al. 2001; Räisänen et al. 2007). It is interesting to note that in early experiments with another L. buchneri strain—L. buchneri N.C.I.B. 8007—TA was investigated in the context of its proven function as a serogroup E antigen (Shaw and Baddiley 1964). This TA was found to be a polymer comprised of about 14 Gro-P residues, joined through phosphodiester linkages at positions 1 and 3 on each glycerol, with α-d-gluco-pyranosyl substituents probably randomly attached to the 2-position of four of these residues, and d-alanine ester residues attached to most of the remaining glycerol 2-hydroxyl groups.
Based on the analysis conducted in the course of this study, the glycolipid moiety of the L. buchneri CD034 LTA contains saturated and unsaturated fatty acids on the dihexosyl-diacylglycerol structure. The Hex2-diacylglycerol (DAG) glycolipid of L. buchneri LTA with the linking sugars d-Glc and d-Gal (Figure 1C) has been described for the first time by Fischer et al. (Fischer et al. 1978). Glc2-DAG appears to be a very common glycolipid anchor in Gram-positive bacteria. In lactobacilli, rather conserved Hex3 and unusual Hex4 structures with the linking sugars Glc and Gal have also been reported (Nakano and Fischer 1978; Fischer et al. 1990; Jang et al. 2011, Shiraishi et al. 2013).
For the assessment of the specific role of LTA in binding of the S-layer protein, we applied single-molecule force microscopy using an S-layer-functionalized AFM tip (Horejs et al. 2011) and the hydrophilic sugar portion of purified L. buchneri CD034 LTA as obtained upon deacylation, which was immobilized to the wafer via ConA (Figure 4). To account for the fact that His6-tagging, which was done for protein purification purposes, might influence the binding capacity of rSlpB to the ligand, we compared the behavior of N- and C-terminally His6-tagged S-layer protein (N-His6-SlpB versus C-His6-SlpB) in the experimental set-up. In the AFM experiment, C-His6-rSlpB and N-His6-rSlpB showed comparable unbinding forces and kinetic off rates. By blocking the AFM tip with deacylated LTA, the specificity of the interaction between the LTA and rSlpB was confirmed, revealing a decrease of the binding probability by 51% for rSlpB C-His6 and by 70% in the case of N-His6-rSlpB. The forces showed strictly mono-logarithmical dependence on the loading rate, suggesting that only one energy barrier was crossed (Figure 5). With the unbinding force being 45 pN and 44 pN for C-His6-rSlpB and N-His6-rSlpB, respectively, these values are comparable with reported binding forces between ligands and receptors at the single-molecule level. Previously, S-layer with attached Strep-tag or His6-tag showed interaction forces of 45 pN with streptavidin and of 160 pN with Tris-NTA, respectively, at comparable loading rates (Tang et al. 2008, 2009). The antibody–antigen interaction strength, for instance, ranges between 70 and 170 pN depending on the loading rate (Kienberger et al. 2005). The current hypothesis is that the binding between S-layer proteins and CWG is based on lectin-type interactions (Sára 2001). The unbinding forces of the CWG/S-layer complex are in the range of the antigen–antibody and S-layer Strep-tag experiments, indicating that a comparable force strength is needed for unbinding events (Horejs et al. 2011). In the case of L. buchneri CD034 (this study), the dissociation forces of S-layer protein and deacylated LTA are comparable regardless of the position of its His6-tag (Figure 5B). This indicates that the protein’s binding epitope is not affected by the tag.
Regarding the pool of CWGs that did not bind to the HIC column (pool I), it is an interesting future aspect to be investigated, if still another type of CWG, such as WTA or even neutral polysaccharides would be supportive to the LTA’s anchoring function of the S-layer.
In summary, we structurally characterized LTA of L. buchneri CD034 that resembles the typical Gro-P backbone with Glc as sole modification residue, and the lipid anchor with a linker disaccharide of Glc and Gal and fatty acids, yielding the overall structure Gro-(P-Gro*-P)∼31-6-α-d-Galp(1→2)-α-d-Glcp(1→3)-1,2-diacyl-sn-Gro (*H or ∼30% α-d-Glcp) (Figure 3). By single-molecule force spectroscopy we demonstrated a direct binding interaction between the hydrophilic portion of the LTA and the recombinant S-layer protein rSlpB from L. buchneri, supporting the hypothesis that LTA acts as cell wall ligand for the S-layer of L. buchneri CD034 in vivo exhibiting a binding strength that is common for ligand–receptor interactions.
Materials and Methods
Bacterial growth
Lactobacillus buchneri CD034 was cultivated at 37°C in MRS (De Man-Rogosa-Sharpe) broth (Carl Roth, Karlsruhe, Germany) under aerobic conditions for 24 h with gentle shaking (150 rpm). Bacteria were harvested by centrifugation (8600 × g, 4°C, 1 h) and washed twice with distilled water. The biomass was stored at –20°C until further use.
Isolation of L. buchneri CWGs by n-butanol extraction
Prior to extraction of CWGs, the S-layer of L. buchneri CD034 was removed from the bacterial cells by treatment with LiCl as described previously (Lortal et al. 1992). Briefly, 11 g of dry biomass was suspended in 100 mL of 5 M LiCl and kept at room temperature (RT = 22°C) for 15 min, followed by centrifugation (47,000 × g, 4°C, 30 min). The pellet of LiC1-extracted cells was washed with distilled water prior to resuspension in 80 mL of 20 mM phosphate buffer (pH 6.2) supplemented with 1 mM Mg2SO4 (Sigma-Aldrich, Vienna, Austria), and the cell walls were hydrolyzed with mutanolysin (50 U/mL; Sigma-Aldrich) and lysozyme (500 μg/mL; Sigma-Aldrich) at 37°C for 16 h. Nucleic acids were degraded for 3 h at 37°C, using DNase (25 μg/mL; Sigma-Aldrich) and RNase (25 μg/mL; Sigma-Aldrich), followed by treatment with proteinase K (1 mg/mL; Sigma-Aldrich) at 37°C for 16 h, and dialysis (MWCO 3500 Da; Carl Roth, Karlsruhe, Germany) against buffer. Cells were mechanically disrupted with a French Press (High pressure cell disruption TS 1.1 KW, Constant Systems Ltd., Low March, Daventry, Northants, UK). The pressure was adjusted to 40,000 psi and three cycles were applied in total.
For extraction of CWGs, disrupted L. buchneri cells were stirred with an equal volume of n-butanol (LiChrosolv, Merck, Darmstadt, Germany) at 22°C for 1 h (Morath et al. 2001). After centrifugation (7500 × g, 30 min), the aqueous phase containing CWGs was collected and dialyzed for 4 d against distilled water and lyophilized (Duda et al. 2016). Cell wall glycopolymers were purified from that phase by hydrophobic interaction chromatography (HIC) on a HiPrep column (16 mm × 100 mm, bed volume 20 mL) of octyl-Sepharose (GE Healthcare, Little Chalfont, UK) to separate LTA from co-extracted CWGs (Morath et al. 2001). Prior to applying the sample onto the column, it was dissolved in 15% (v/v) n-propanol in 0.1 M ammonium acetate (pH 4.7; buffer A) and filtered (0.45 μm, Pall Life Science, Vienna, Austria). Elution of material started isocratically over 20 min in buffer A, followed by a gradient from buffer A to 100% buffer B (60% [v/v] n-propanol in 0.1 M ammonium acetate [pH 4.7]). Fractions containing CWGs non-bound to the column (pool 1; eluting with buffer A) and those containing LTA (pool 2; eluting in the gradient) were combined according to their phosphate content (Lowry et al. 1954) and repeatedly lyophilized for removal of salt. To obtain a sufficient amount of LTA for analysis, the procedure was repeated several times.
Component analyses of L. buchneri cell wall glycopolymers
The components of L. buchneri CWGs were determined by methanolysis with 0.5 M HCl/methanol (MeOH) at 85°C for 45 min, followed by acetylation using acetic anhydride and pyridine (1:1 [v/v]) at 85°C for 10 min and detection by GLC and GLC-mass spectrometry (Agilent Technologies 7890 A gas chromatograph [Agilent Technologies, Wilmington, DE] equipped with a dimethylpolysiloxane column [Agilent, HP Ultra 1, 12 m × 0.2 mm × 0.33 μm film thickness] and a 5975 C series MSD detector with electron impact ionization [EI] mode) under autotune condition at 70 eV. The temperature program was 70°C for 1.5 min, then 60°C/min to 110°C and 5°C/min to 320°C (Duda et al. 2016).
Hexoses (Glc and Gal) were identified by GLC as alditol acetates after hydrolysis (2 M trifluoroacetic acid, 120°C, 2 h), reduction (NaBH4, 16 h in the dark), peracetylation (85°C, 10 min), and detection on the HP 5890 (series II) gas chromatograph with a flame-ionization detector and a column (30 m × 0.25 mm × 0.25 μm film thickness; Agilent) of 5% phenylmethylsiloxane (HP-5MS); helium was used as carrier gas (70 kPa). The temperature in GLC was 150°C for 3 min, and then increased with 3°C/min to 320°C. For quantification, xylose was used as internal standard and the sugars were identified by use of authentic standards (Duda et al. 2016). The absolute configuration of Glc and Gal was determined by GLC of the acetylated O-[(S)-2-butyl] glycoside after methanolysis (2 M HCl/MeOH, 85°C, 45 min), butanolysis (2 M HCl/(S)-2-BuOH, 65°C, 4 h) and peracetylation (85°C, 10 min), by comparison with authentic standards (Gerwig et al. 1979).
The content of organic phosphate in the CWGs was determined according to Lowry et al. using freshly prepared color reagent (1 mL 1 M sodium acetate, 1 mL 2.5% ammonium molybdate solution, 7 mL H2O, 1 mL freshly prepared ascorbic acid; 37°C for 90 min) (Lowry et al. 1954). The extinction of the ammonium molybdate-phosphate complex was measured at λ = 820 nm (HELIOS BETA 9423 UVB 1002E spectrophotometer, Thermo Electron Ltd., Altrincham, Cheshire, UK). For quantification, 5 mM Na2HPO4 solution was used as external standard (Lowry et al. 1954). For the calculation of the length of the hydrophilic chain of the LTA, LTA was hydrolyzed (62.7 mL H2O, 30.6 mL concentrated H2SO4, 6.7 mL 70% HClO4; 100°C for 1 h, followed by 165°C for 2 h) prior to determination of total phosphate.
Fatty acids of the isolated LTA glycolipid anchor (for isolation of the glycolipid linker see next paragraph) were detected in the chloroform phase (Wollenweber and Rietschel 1990) by GLC as methyl esters (diazomethane, 10 min, RT) after hydrolysis of LTA (4 M HCl, 100°C, 4 h), neutralization (5 M NaOH, 100°C, 30 min), and water-chloroform extraction, using an HP 6890 N gas chromatograph with FID and a column of phenylmethylsiloxane HP-5MS (Agilent Technologies, 30 m × 0.25 mm, 0.25 μm film thickness); the temperature program was 120°C for 3 min, and then with 5°C/min to 320°C.
Isolation of the LTA glycolipid anchor
To isolate the LTA glycolipid anchor, the Gro-P backbone of LTA was cleaved by treatment with 48% aqueous hydrofluoric acid (HF; 50 μL) at 4°C for 48 h. After repeated extraction with H2O/CHCl3/MeOH (3:3:1 [v/v/v]), the glycolipid was present in the organic phase. The combined organic phases were dried under N2, lyophilized and dissolved in MeOD prior to nuclear magnetic resonance spectroscopy (NMR) measurements.
Hydrazinolysis of LTA
To obtain O-deacylated LTA for NMR measurements and AFM analyses, 0.5 mL of absolute hydrazine were added to 5 mg of dried LTA (Holst 2000). The mixture was incubated at 37°C for 30 min. Subsequently, hydrazine was destroyed with 10 volumes of cold acetone, then the sample was centrifuged and the pellet was washed four times with acetone and then dissolved in D2O prior to NMR analysis.
NMR spectroscopy of LTA
NMR experiments of LTA and O-deacylated LTA were carried out in D2O at 27°C. NMR measurements of the glycolipid anchor were performed in MeOD at 27°C. All 1D and 2D NMR 1H, 1H COSY, TOCSY and ROESY as well as 1H, 13C HSQC, 1H, 13C HSQC–TOCSY, 1H, 13C HMBC and 1H,13P TOCSY HMBC experiments were recorded on a Bruker DRX Avance III 700 MHz spectrometer (operating frequencies of 700.75 MHz for 1H NMR, 176.2 MHz for 13C NMR and 283.7 MHz for 31P) and standard Bruker software (Bruker, Rheinstetten, Germany). COSY, TOCSY and ROESY experiments were recorded using data sets (t1 by t2) of 4096 by 515 points. The TOCSY experiments were carried out in the phase-sensitive mode with mixing times of 120 ms and ROESY of 300 ms. The 1H, 13C correlations measured in the 1H-detected mode via HMBC spectra were acquired using data sets of 4096 by 512 points and 128 (LTA) and 64 (O-deacylated LTA and linker) scans for each t1 value of 145 Hz and long range proton carbon coupling constant of 10 Hz. Chemical shifts of LTA and O-deacylated LTA are reported relative to external acetone (δH 2.225. δC 31.45) and those of linker relative to internal MeOD (δH 3.34, δC 49.86) (Fulmer et al. 2010).
Preparation of recombinant L. buchneri S-layer protein
For expression of recombinant S-layer protein rSlpB from L. buchneri CD034, expression plasmids were available in our laboratory from a previous study (Anzengruber et al. 2014a). Briefly, for purification and detection purposes, a hexa-hexahistidine tag was fused either to the N- or C-terminal sequence of the SlpB protein lacking the native signal peptide. The pET28a(+)-based constructs were transformed into Escherichia coli BL21 Star (DE3) (Thermo Fisher Scientific, Vienna, Austria) and the strain was grown in Erlenmeyer flasks containing 500 mL of LB medium supplemented with kanamycin (50 μg/mL) at 37°C and 200 rpm. Expression of recombinant protein was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at the mid exponential growth phase (OD600 ~0.6) and incubation was continued for additional 3 h. Cells were pelleted (10,000 x g, 20 min), suspended in lysis buffer (100 mM NaH2PO4, 10 mM Tris/HCl, 8 M urea [pH 8.0]) and stirred at RT for 60 min. The lysate was centrifuged at 48,000 × g for 30 min to pellet cellular debris. The clear lysate (~6 mL) was added to 1.5 mL of 50% Ni-NTA slurry (Qiagen, Hilden, Germany) and mixed gently by shaking (250 rpm on rotary shaker) at RT for 1 h. Lysate-resin was loaded into a column and washed twice with the buffer A (100 mM NaH2PO4, 10 mM Tris/HCl, 8 M urea, [pH 6.3]). Recombinant protein was eluted with buffer B (100 mM NaH2PO4, 10 mM Tris/HCl, 8 M urea [pH 5.9]), followed by buffer C (100 mM NaH2PO4, 10 mM Tris/HCl, 8 M urea [pH 4.5]), and combined. The recombinant His6-tagged proteins (named N-His6-rSlpB and C-His6-rSlpB) were dialyzed (MWCO 14–16 kDa) three times against 50 mM sodium citrate buffer (pH 5.5), 3 L, each.
Conjugation of S-layer protein to the AFM cantilever
Commercially available AFM cantilevers (MSCT, Bruker, Fremont, CA) with a nominal spring constant of 0.01–0.02 N/m were functionalized with amino groups by using a 3-aminopropyltriethoxysilane (APTES; Sigma-Aldrich) coating procedure (Ebner et al. 2007). The maleimide-poly(ethylene glycol; PEG) linker with a length of 6–9 nm was attached to the APTES-coated AFM cantilever by incubation for 2 h in 500 μL of CHCl3 containing 1 mg of maleimide-PEG-NHS (Polypure, Oslo, Norway) and 15 μL of trimethylamine. Then, the cantilever was washed three times with chloroform. After that, the cantilever was immersed for 2 h in a premixed solution that contained 50 μL of 4 mM thiol-TrisNTA in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-EDTA buffer, 2 μL of 100 mM EDTA, pH 7.5, 5 μL of 1 M HEPES buffer (pH 7.5), 2 μL of 100 mM Tris(carboxyethyl)phosphine hydrochloride, and 2 μL of 1 M HEPES buffer (pH 9.6), and then washed with HEPES buffer. Subsequently, the cantilevers were incubated for 3 h in a mixture of 4 μL of 5 mM NiCl2 and 100 μL of HEPES-buffered saline (HBS) containing 0.2 mg/mL of His6-tagged S-layer protein. After washing three times with HBS, the cantilevers were stored in HBS at 4°C (Oh et al. 2016).
Surface preparation for single-molecule force spectroscopy measurements
For the wafer surface, the silicon nitride surface was coated with APTES, and then a heterobifunctional aldehyde-PEG linker was attached via its NHS ester group and the aldehyde function on the free-tangling end of PEG was used for coupling of Concanavalin A (Con A; Sigma-Aldrich) via one of the lysine residues. The bond was fixed by reduction with NaCNBH3. The overall procedure was done as described before (Ebner et al. 2007). After three washing steps with phosphate buffer saline (PBS [pH 7.4]), deacylated LTA at 0.2 mg/mL (final concentration) was deposited on the Con A surface for 3 h, through interaction with L. buchneri CD034 LTA. Considering that in the in vivo context, the diacyl chains of intact LTA are integrated in the L. buchneri CD034 membrane, and, thus, not available for protein binding, the use of deacylated LTA in our studies is legitimate. Further, removal of the lipophilic part of the LTA is beneficial for its immobilization to the wafer.
Single-molecule force spectroscopy measurements
Force–distance curves were acquired by recording at least 1000 curves with vertical sweep rates between 0.5 and 10 Hz and at a z-range of typically 1000 nm, resulting in loading rates from 100 to 10,000 pN/s, using a commercial AFM (Keysight Technolgies, Santa Rosa, CA, USA). The relationship between experimentally measured unbinding forces and the interaction potential is described by kinetic models (Bell 1978; Evans and Ritchie 1997). Blocking of specific interaction between S-layer protein and LTA was done by injecting deacylated LTA into the bath solution which results in a non-activated S-layer protein on the tip. All force spectroscopy experiments were repeated on at least four different preparations of functionalized AFM cantilevers and sample surfaces.
Funding
This work was supported by the Austrian Science Fund FWF projects P24305-B20 (to P.M.), P21954-B22 and P27374-B22 (to C.S.) and the PhD Programme “Biomolecular Technology of Proteins” W1224.
Conflict of interest statement
None declared.
Abbreviations
AFM, atomic force microscopy; APTES, (3-aminopropyl)triethoxysilane; Con A, Concanavalin A; COSY, correlation spectroscopy; CWG, cell wall glycopolymer; DAG, diacylglycerol; EDTA, ethylenediaminetetraacetic acid; Gal, galactose; Glc, glucose; GLC, gas liquid chromatography; Gro-P, glycerol phosphate; HBS, HEPES-buffered saline; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HF, hydrofluoric acid; His6-tag, hexahistine tag; HMBC, heteronuclear multiple-bond correlation spectroscopy; HSQC, heteronuclear single-quantum correlation spectroscopy; IPTG, isopropyl-β-d-thiogalactopyranoside; Koff, kinetic off-rate constant; LAB, lactic acid bacteria; LB, lysogeny broth; LTA, lipoteichoic acid; MeOD, deuterated Methanol; NHS, N-hydroxysuccinimide; NOE, nuclear Overhauser effect spectroscopy; PDF, probability density function; PEG, polyethylene glycol; PG, peptidoglycan; PVDF, polyvinylidene fluoride; ROESY, rotating frame nuclear Overhauser effect spectroscopy; RT, room temperature; SD, standard deviation; SDS-PAGE, sodium dodecyl sulfatepolyacrylamide gel electrophoresis; S-layer, bacterial cell surface layer; SLH, S-layer homology; TA, teichoic acid; TOCSY, total correlation spectroscopy; WTA, wall teichoic acid; Xβ, interaction energy potential.
Acknowledgements
The authors would like to thank Katharina Jakob and Regina Engel (both Research Center Borstel) for excellent technical support and Heiko Kässner (Research Center Borstel) and Andreas Hofinger-Horvath (Universität für Bodenkultur Wien) for NMR recordings.
References
- Antikainen J, Anton L, Sillanpää J, Korhonen TK. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol. 46:381–394. [DOI] [PubMed] [Google Scholar]
- Anzengruber J, Courtin P, Claes IJ, Debreczeny M, Hofbauer S, Obinger C, Chapot-Chartier MP, Vanderleyden J, Messner P, Schäffer C. 2014. Biochemical characterization of the major N-acetylmuramidase from Lactobacillus buchneri. Microbiology. 160:1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzengruber J, Pabst M, Neumann L, Sekot G, Heinl S, Grabherr R, Altmann F, Messner P, Schäffer C. 2014. Protein O-glucosylation in Lactobacillus buchneri. Glycoconj J. 31:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald AR, Hancock IC, Harwood CR. 1993. Cell wall structure, synthesis and turnover In: Sonenshein A, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria. Washington, DC, USA: ASM Press; p. 381–410. [Google Scholar]
- Åvall-Jääskeläinen S, Hynönen U, Ilk N, Pum D, Sleytr UB, Palva A. 2008. Identification and characterization of domains responsible for self-assembly and cell wall binding of the surface layer protein of Lactobacillus brevis ATCC 8287. BMC Microbiol. 8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- Brechtel E, Bahl H. 1999. Thermoanaerobacterium thermosulfurigenes EM1 S-layer homology domains do not attach to peptidoglycan. J Bacteriol. 181:5017–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol. 67:313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour J, Ferain T, Deghorain M, Palumbo E, Hols P. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek. 76:159–184. [PubMed] [Google Scholar]
- Dohm N, Petri A, Schlander M, Schlott B, König H, Claus H. 2011. Molecular and biochemical properties of the S-layer protein from the wine bacterium Lactobacillus hilgardii B706. Arch Microbiol. 193:251–261. [DOI] [PubMed] [Google Scholar]
- Duda KA, Petersen S, Holst O. 2016. Structural characterization of the lipoteichoic acid isolated from Staphylococcus sciuri W620. Carbohydr Res. 430:44–47. [DOI] [PubMed] [Google Scholar]
- Ebner A, Wildling L, Kamruzzahan ASM, Rankl C, Wruss J, Hahn CD, Hölzl M, Zhu R, Kienberger F, Blaas D et al. . 2007. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjug Chem. 18:1176–1184. [DOI] [PubMed] [Google Scholar]
- Eikmeyer FG, Heinl S, Marx H, Pühler A, Grabherr R, Schlüter A. 2015. Identification of oxygen-responsive transcripts in the silage inoculant Lactobacillus buchneri CD034 by RNA sequencing. PLoS One. 10:e0134149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt H, Peters J. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J Struct Biol. 124:276–302. [DOI] [PubMed] [Google Scholar]
- Evans E, Ritchie K. 1997. Dynamic strength of molecular adhesion bonds. Biophys J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan RP, Fairweather NF. 2014. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 12:211–222. [DOI] [PubMed] [Google Scholar]
- Fischer W, Behr T, Hartmann R, Peter-Katalinić J, Egge H. 1993. Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid (C polysaccharide). Eur J Biochem. 215:851–857. [DOI] [PubMed] [Google Scholar]
- Fischer W, Laine RA, Nakano M. 1978. On the relationship between glycerophosphoglycolipids and lipoteichoic acids in Gram-positive bacteria. II. Structures of glycerophosphoglycolipids. Biochim Biophys Acta. 528:298–308. [DOI] [PubMed] [Google Scholar]
- Fischer W, Mannsfeld T, Hagen G. 1990. On the basic structure of poly(glycerophosphate) lipoteichoic acids. Biochem Cell Biol. 68:33–43. [DOI] [PubMed] [Google Scholar]
- Fulmer GR, Miller AJM, Sherden NH, Gottlieb HE, Nudelman A, Stoltz BM, Bercaw JE, Goldberg KI. 2010. NMR chemical shifts of trace impurities: Common laboratory solvents, organics, and gases in deuterated solvents relevant to the organometallic chemist. Organometallics. 29:2176–2179. [Google Scholar]
- Gerwig GJ, Kamerling JP, Vliegenthart JFG. 1979. Determination of the absolute configuration of mono-saccharides in complex carbohydrates by capillary g.l.c. Carbohydr Res. 77:10–17. [DOI] [PubMed] [Google Scholar]
- Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. 2005. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 102:10321–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinl S, Späth K, Egger E, Grabherr R. 2011. Sequence analysis and characterization of two cryptic plasmids derived from Lactobacillus buchneri CD034. Plasmid. 66:159–168. [DOI] [PubMed] [Google Scholar]
- Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, Linke B, Goesmann A, Grabherr R, Schwab H, Pühler A et al. . 2012. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol. 161:153–166. [DOI] [PubMed] [Google Scholar]
- Holst O. 2000. Deacylation of lipopolysaccharides and isolation of oligosaccharide phosphates In: Holst O, editor. Bacterial Toxins: Methods and Protocols. Totowa, New Jersey: Humana Press; p. 345–353. [DOI] [PubMed] [Google Scholar]
- Horejs C, Ristl R, Tscheliessnig R, Sleytr UB, Pum D. 2011. Single-molecule force spectroscopy reveals the individual mechanical unfolding pathways of a surface layer protein. J Biol Chem. 286:27416–27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Kong J, Sun Z, Han L, Kong W, Yang P. 2011. Heterologous protein display on the cell surface of lactic acid bacteria mediated by the S-layer protein. Microb Cell Fact. 10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K-S, Baik JE, Han SH, Chung DK, Kim B-G. 2011. Multi-spectrometric analyses of lipoteichoic acids isolated from Lactobacillus plantarum. Biochem Biophys Res Commun. 407:823–830. [DOI] [PubMed] [Google Scholar]
- Kienberger F, Kada G, Mueller H, Hinterdorfer P. 2005. Single molecule studies of antibody-antigen interaction strength versus intra-molecular antigen stability. J Mol Biol. 347:597–606. [DOI] [PubMed] [Google Scholar]
- Klaenhammer TR, Barrangou R, Buck BL, Azcarate-Peril MA, Altermann E. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol Rev. 29:393–409. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, Bron PA. 2010. The extracellular biology of the lactobacilli. FEMS Microbiol Rev. 34:199–230. [DOI] [PubMed] [Google Scholar]
- Leenhouts K, Buist G, Kok J. 1999. Anchoring of proteins to lactic acid bacteria. Antonie van Leeuwenhoek. 76:367–376. [PubMed] [Google Scholar]
- Lortal S, van Heijenoort J, Gruber K, Sleytr UB. 1992. S-Layer of Lactobacillus helveticus ATCC 12046 - Isolation, chemical characterization and re-formation after extraction with lithium-chloride. J Gen Microbiol. 138:611–618. [Google Scholar]
- Lowry OH, Roberts NR, Leiner KY, Wu M-L, Farr AL. 1954. The quantitative histochemistry of brain. I. Chemical methods. J Biol Chem. 207:1–17. [PubMed] [Google Scholar]
- Malamud M, Carasi P, Bronsoms S, Trejo SA, Serradell ML. 2017. Lactobacillus kefiri shows inter-strain variations in the amino acid sequence of the S-layer proteins. Antonie van Leeuwenhoek. 110:515–530. [DOI] [PubMed] [Google Scholar]
- Mao R, Wu D, Wang Y. 2016. Surface display on lactic acid bacteria without genetic modification: strategies and applications. Appl Microbiol Biotechnol. 100:9407–9421. [DOI] [PubMed] [Google Scholar]
- Masuda K, Kawata T. 1981. Characterization of a regular array in the wall of Lactobacillus buchneri and its reattachment to the other wall components. J Gen Microbiol. 124:81–90. [Google Scholar]
- Masuda K, Kawata T. 1985. Reassembly of a regularly arranged protein in the cell wall of Lactobacillus buchneri and its reattachment to cell walls: chemical modification studies. Microbiol Immunol. 29:927–938. [PubMed] [Google Scholar]
- Mesnage S, Fontaine T, Mignot T, Delepierre M, Mock M, Fouet A. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner P, Egelseer EM, Sleytr UB, Schäffer C. 2009. Bacterial surface layer glycoproteins and “non-classical” secondary cell wall polymers In: Moran AP, Brennan PJ, Holst O, von Itzstein M, editors. Microbial Glycobiology: Structures, Relevance and Applications. San Diego, USA: Academic Press; p. 109–128. [Google Scholar]
- Messner P, Schäffer C, Egelseer EM, Sleytr UB. 2010. Occurrence, structure, chemistry, genetics, morphogenesis, and functions of S-layers In: König H, Claus H, Varma A, editors. Prokaryotic Cell Wall Compounds – Structure and Biochemistry. Berlin: Springer-Verlag; p. 53–109. [Google Scholar]
- Messner P, Schäffer C, Kosma P. 2013. Bacterial cell-envelope glycoconjugates. Adv Carbohydr Chem Biochem. 69:209–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon C, Langella P, Eijsink VG, Mathiesen G, Chatel JM. 2016. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb Cell Fact. 15:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morath S, Geyer A, Hartung T. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J Exp Med. 193:393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Fischer W. 1978. Trihexosyldiacylglycerol and acyltrihexosyldiacylglycerol as lipid anchors of the lipoteichoic acid of Lactobacillus casei DSM 20021. Hoppe Seylers Z Physiol Chem. 359:1–11. [DOI] [PubMed] [Google Scholar]
- Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 67:686–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YJ, Hubauer-Brenner M, Gruber HJ, Cui Y, Traxler L, Siligan C, Park S, Hinterdorfer P. 2016. Curli mediate bacterial adhesion to fibronectin via tensile multiple bonds. Sci Rep. 6:33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea Vélez M, Verhoeven TLA, Draing C, von Aulock S, Pfitzenmaier M, Geyer A, Lambrichts I, Grangette C, Pot B, Vanderleyden J et al. . 2007. Functional analysis of d-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol. 73:3595–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal M, Walker S. 2017. Envelope structures of Gram-positive bacteria. Curr Top Microbiol Immunol. 404:1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann NT, Gründling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 319:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räisänen L, Draing C, Pfitzenmaier M, Schubert K, Jaakonsaari T, von Aulock S, Hartung T, Alatossava T. 2007. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on d-alanyl and α-glucose substitution of poly(glycerophosphate) backbones. J Bacteriol. 189:4135–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade J, Weidenmaier C. 2016. Cell wall glycopolymers of Firmicutes and their role as nonprotein adhesins. FEMS Microbiol Lett. 590:3758–3771. [DOI] [PubMed] [Google Scholar]
- Schirner K, Marles-Wright J, Lewis RJ, Errington J. 2009. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28:830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas DM. 2012. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 367:1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Missiakas D. 2014. Lipoteichoic acids, phosphate-containing polymers in the envelope of gram-positive bacteria. J Bacteriol. 196:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer C, Messner P. 2005. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology. 151:643–651. [DOI] [PubMed] [Google Scholar]
- Shaw N, Baddiley J. 1964. The teichoic acid from the walls of Lactobacillus buchneri N.C.I.B. 8007. Biochem J. 93:317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi T, Yokota S-i, Morita N, Fukiya S, Tomita S, Tanaka N, Okada S, Yokota A. 2013. Characterization of a Lactobacillus gasseri JCM 1131T lipoteichoic acid with a novel glycolipid anchor structure. Appl Environ Microbiol. 79:3315–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr UB, Egelseer EM, Ilk N, Messner P, Schäffer C, Pum D, Schuster B. 2010. Nanobiotechnological applications of S-layers In: König HC, Varma HA, editors. Prokaryotic Cell Wall Compounds - Structure and Biochemistry. Berlin: Springer; p. 459–481. [Google Scholar]
- Sleytr UB, Messner P. 1983. Crystalline surface layers on bacteria. Annu Rev Microbiol. 37:311–339. [DOI] [PubMed] [Google Scholar]
- Smit E, Pouwels PH. 2002. One repeat of the cell wall binding domain is sufficient for anchoring the Lactobacillus acidophilus surface layer protein. J Bacteriol. 184:4617–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sára M. 2001. Conserved anchoring mechanisms between crystalline cell surface S-layer proteins and secondary cell wall polymers in Gram-positive bacteria? Trends Microbiol. 9:47–49, discussion 49–50. [DOI] [PubMed] [Google Scholar]
- Sára M, Sleytr UB. 2000. S-Layer proteins. J Bacteriol. 182:859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Ebner A, Ilk N, Lichtblau H, Huber C, Zhu R, Pum D, Leitner M, Pastushenko V, Gruber HJ et al. . 2008. High-affinity tags fused to S-layer proteins probed by atomic force microscopy. Langmuir. 24:1324–1329. [DOI] [PubMed] [Google Scholar]
- Tang J, Ebner A, Kraxberger B, Leitner M, Hykollari A, Kepplinger C, Grunwald C, Gruber HJ, Tampé R, Sleytr UB et al. . 2009. Detection of metal binding sites on functional S-layer nanoarrays using single molecule force spectroscopy. J Struct Biol. 168:217–222. [DOI] [PubMed] [Google Scholar]
- Wedajo B. 2015. Lactic acid bacteria: benefits, selection criteria and probiotic potential in fermented food. J Prob Health. 3:2. [Google Scholar]
- Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 6:276–287. [DOI] [PubMed] [Google Scholar]
- Wollenweber HW, Rietschel ET. 1990. Analysis of lipopolysaccharide (lipid A) fatty acids. J Microbiol Meth. 11:195–211. [Google Scholar]