Abstract
Background
Pancreatic duct adenocarcinoma (PDAC) remains a major health problem because conventional cancer treatments are relatively ineffective against it. Microarray studies have linked many genes to pancreatic cancer, but the available data have not been extensively mined for potential insights into PDAC. This study attempted to identify PDAC-associated genes and signaling pathways based on six microarray-based profiles of gene expression in pancreatic cancer deposited in the gene expression omnibus database.
Methods
Pathway network methods were used to analyze core pathways in six publicly available pancreatic cancer gene (GSE71989, GSE15471, GSE16515, GSE32676, GSE41368 and GSE28735) expression profiles. Genes potentially linked to PDAC were assessed for potential impact on survival time based on data in The Cancer Genome Atlas and International Cancer Genome Consortium databases, and the expression of one candidate gene (CKS2) and its association with survival was examined in 102 patients with PDAC from our hospital. Effects of CKS2 knockdown were explored in the PDAC cell lines BxPC-3 and CFPAC-1.
Results
The KEGG signaling pathway called “pathway in cancer” may play an important role in pancreatic cancer development and progression. Five genes (BIRC5, CKS2, ITGA3, ITGA6 and RALA) in this pathway were significantly associated with survival time in patients with PDAC. CKS2 was overexpressed in PDAC samples from our hospital, and higher CKS2 expression in these patients was associated with shorter survival time. CKS2 knockdown substantially inhibited PDAC cell proliferation in vitro.
Conclusions
Analysis integrating existing microarray datasets allowed identification of the “pathway in cancer” as an important signaling pathway in PDAC. This integrative approach may be powerful for identifying genes and pathways involved in cancer.
Electronic supplementary material
The online version of this article (10.1186/s40880-018-0289-9) contains supplementary material, which is available to authorized users.
Keywords: Pancreatic cancer, GEO, Pathway network, CKS2
Background
Pancreatic ductal adenocarcinoma (PDAC), which accounts for approximately 90% of all pancreatic tumors [1], is the ninth leading cause of cancer-related deaths in China [2]. Due to the lack of effective methods for early diagnosis and therapy, pancreatic cancer has the second highest ratio of mortality to incidence (approximately 88%) among all types of cancer [3]. Microarray platforms have produced numerous datasets of gene expression profiles in pancreatic cancer, which are publicly available through the gene expression omnibus (GEO) of the National Center for Biotechnology Information [4]. Studies of such data have identified several genes associated with pancreatic cancer, but these datasets have yet to be mined extensively to identify genes and pathways linked to PDAC.
To identify additional genes that may be associated with PDAC, we mined six frequently cited profiles of genes expressed in pancreatic cancer that have been deposited in the GEO database. We used pathway network analysis to identify core pathways that may affect pancreatic cancer development and progression. Our analysis of genes differentially expressed in pancreatic cancer took into account expression quantitative trait loci [5], copy number variation [6, 7], and DNA methylation [8], all of which can affect gene expression levels.
Methods
Affymetrix microarray data
The following pancreatic cancer gene expression profiles were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/): GSE71989, GSE15471, GSE16515, and GSE32676, which were obtained using an Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array; as well as GSE41368 and GSE28735, which were obtained using an Affymetrix Human Gene 1.0 ST Array.
Data processing
We downloaded the probe-level data from the GEO database as CEL files, then we used the GCBI on-line analysis tool (http://www.gcbi.com.cn) to normalize and log2-transform the data. Genes differentially expressed between tumor samples and non-tumor samples were filtered by requiring a fold difference of at least two and a maximum false discovery rate of 0.05. Pathway analysis was performed using DAVID software (https://david.ncifcrf.gov) as described [9].
Core pathway analysis
Pathways enriched for genes differentially expressed between tumor and non-tumor tissues were identified and connected in a pathway network to reveal relationships among the pathways. A pathway network was constructed using the GCBI on-line analysis tool, and for each pathway in the network, the number of upstream pathways (in-degree) and the number of downstream pathways (out-degree) were determined in order to assess the degree centrality of that pathway in the network. Greater degree centrality means that a pathway regulates or is regulated by more pathways, implying a more important role in the network.
CKS2 analysis based on database mining
Copy number variation of CKS2 in various types of cancer was analyzed using the cBioPortal database (http://www.cbioportal.org), and CKS2 overexpression in cancer was analyzed using the Oncomine database (http://www.omcomine.org). CKS2 expression levels in various cancer cell lines were analyzed using the CCLE database (https://portals.broadinstitute.org/ccle/home).
PDAC cell culture
Human PDAC cell lines BxPC-3 and CFPAC-1 were purchased from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology). The human immortalized pancreatic duct epithelial cell line HPDE6-C7 was purchased from Biotechnology Company. All cell lines were characterized by DNA fingerprinting and tested for mycoplasma and found to be free of infection.
BxPC-3 and HPDE6-C7 cells were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone, USA), while CFPAC-1 cells were cultured in IMDM (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS. All cell lines were grown without antibiotics in an atmosphere of 5% CO2 and 99% relative humidity at 37 °C.
Survival analysis of key genes
SurvExpress [10] was used to analyze survival associated with key genes found in this study.
Patient samples
Surgical biopsies of tumor tissues and paired non-tumor tissues from patients definitively diagnosed with PDAC based on histology were obtained from the Department of Hepatobiliary Surgery at Sun Yat-sen Memorial Hospital (Guangzhou, China). Tissues were preserved in liquid nitrogen immediately following surgical removal, paraffin-embedded and stored properly before use in the present study. None of the patients had received anticancer treatment before biopsy collection. Clinicopathology and follow-up data for patients are shown in Additional file 1: Table S1.
Western blotting for CKS2
Total protein (20 μg) was extracted from cells or patient tissue samples, resolved by SDS-PAGE and transferred to PVDF membranes (Millipore, Germany). Membranes were incubated with antibodies against CKS2 (ab155078; Abcam, Cambridge, UK) or β-actin (ab8227; Abcam), then with horseradish peroxidase-conjugated secondary antibody from the Phototope-HRP Western blot detection kit (Cell Signaling Technology, USA). Protein bands were visualized using the components in this kit.
CKS2 knockdown in PDAC cells
Short interfering RNA (siRNA) targeting CKS2 and non-targeting control siRNA (Additional file 2: Table S2) were purchased from GenePharma (Suzhou, China). PDAC cells were transfected with siRNA using Lipofectamine 2000 (Life Technologies, USA) according to the manufacturer’s instructions.
Proliferation of PDAC cell lines
Cells were seeded into 96-well plates (2 × 103 cells in 100 µL per well) and cultured for certain periods. Then cell viability was measured using a CCK-8 assay (Dojindo, Japan). Samples were analyzed as six replicates per experiment, and three independent experiments were performed.
RNA extraction and qRT-PCR analysis
Total RNA from pancreatic cancer cell lines was extracted using TRIzol regent (Invitrogen, USA). First-strand cDNA was synthesized using the PrimeScript™ RT reagent Kit (TaKaRa, Japan) and amplified by qRT-PCR on an ABI Prism 7900 sequence detection system (Applied Biosystems) using SYBR Green. Sequences of gene-specific primers are listed in Additional file 3: Table S3.
Statistical analyses
All statistical analyses were performed using Graphpad Prism 5.0 and P < 0.05 was considered significant. Differences between two groups were assessed for significance using Student’s t test. Kaplan–Meier survival estimates were plotted, and inter-group differences were assessed using the log-rank test.
Results
Genes differentially expressed in pancreatic cancer based on integrative analysis of GEO datasets
We examined six gene expression profiles from the GEO database (Additional file 4: Table S4) in order to identify genes differentially expressed between PDAC and normal tissues. One sample each from GSE15471 to GSE28735 were excluded from further analysis because they did not meet quality controls. Analysis of the remaining samples indicated a larger number of genes up-regulated in PDAC than down-regulated (Fig. 1).
Fig. 1.
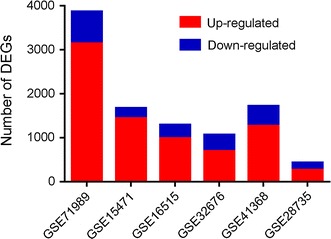
Genes differentially expressed between pancreatic cancer tissues and normal tissues. We analyzed differentially expressed genes in six GEO microarray datasets. Up-regulated: the number of genes whose expression levels were higher in pancreatic cancer tissues than in normal tissues. Down-regulated: the number of genes whose expression levels were lower in pancreatic cancer tissues than in normal tissues. GEO Gene Expression Omnibus, DEG differentially expressed gene
The “pathway in cancer” is the top core pathway in pancreatic cancer
Genes differentially expressed between PDAC and non-tumor tissues in each of the six profiles were assembled into functional pathways. As expected, the pathways that were enriched in differentially expressed genes varied among the six profiles (data not shown). We identified 32 significant pathways common to the six profiles (Fig. 2a). We also used network analysis to identify pathways common to the six profiles that were likely to be most important in PDAC, which led to 13 common core pathways (Fig. 2b): pathway in cancer, p53 signaling pathway, focal adhesion, cytokine–cytokine receptor interaction, regulation of actin cytoskeleton, glycolysis/gluconeogenesis, systemic lupus erythematosus, bladder cancer, small cell lung cancer, complement and coagulation cascades, ECM-receptor interaction, axon guidance, and renal cell carcinoma. Interestingly, all 13 common core pathways were among the 32 common significant pathways (Fig. 2c). Details of the 13 common core pathways in each profile are shown in Table 1. The pathway in cancer (KEGG ID: 05200) was the top significant pathway, with a total degree centrality of 116. This suggests that the pathway in cancer may play an important role in PDAC development and progression.
Fig. 2.
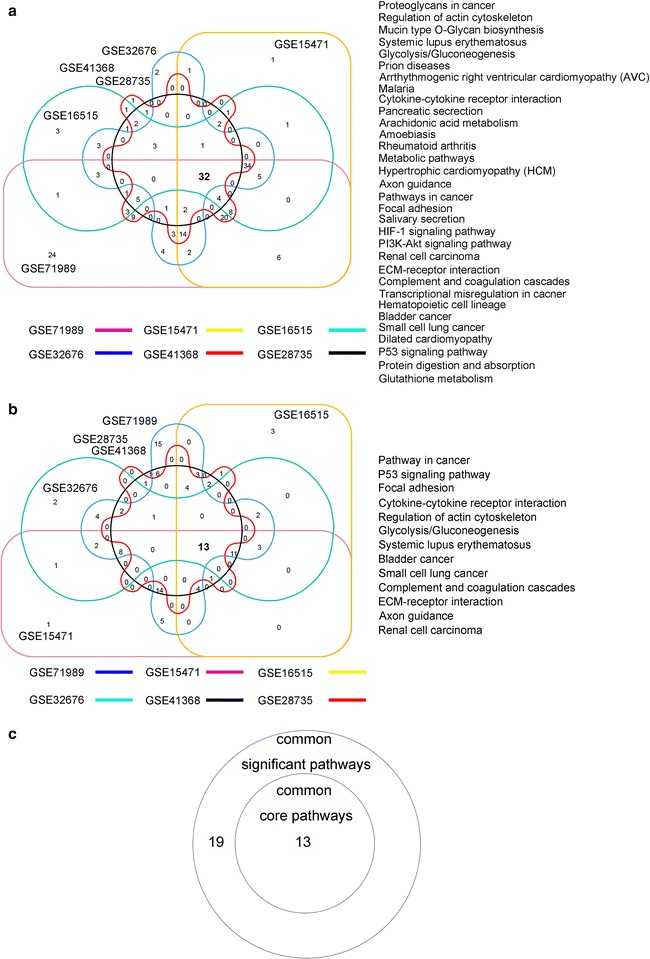
Core pathway analysis of six GEO datasets. a Overlap of significant pathways in each GEO dataset used in this study revealed 32 common significant pathways. Left, Venn diagram of the result. Right, names of common significant pathways. b Overlap of the core pathways of each GEO dataset used in this study revealed 13 common core pathways. Left, Venn diagram of the result. Right, names of common core pathways. c Relationship between the common significant pathways and common core pathways
Table 1.
Summary of 13 common core pathways associated with pancreatic cancer in six GEO microarray datasets
| Pathway ID | Pathway name | GSE71989 | GSE15471 | GSE16515 | GSE32676 | GSE41368 | GSE28735 | Total degree | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outd | Ind | Outd | Ind | Outd | Ind | Outd | Ind | Outd | Ind | Outd | Ind | |||
| 05200 | Pathway in cancer | 28 | 0 | 21 | 0 | 15 | 0 | 23 | 0 | 21 | 0 | 8 | 0 | 116 |
| 04115 | P53 signaling pathway | 2 | 16 | 2 | 12 | 2 | 6 | 1 | 13 | 1 | 11 | 0 | 3 | 69 |
| 04510 | Focal adhesion | 8 | 7 | 7 | 5 | 6 | 4 | 6 | 5 | 7 | 6 | 3 | 4 | 68 |
| 04060 | Cytokine-cytokine receptor interaction | 0 | 16 | 0 | 14 | 0 | 8 | 0 | 9 | 0 | 13 | 0 | 4 | 64 |
| 04810 | Regulation of actin cytoskeleton | 3 | 9 | 3 | 8 | 3 | 6 | 3 | 7 | 3 | 9 | 1 | 2 | 57 |
| 00010 | Glycolysis/Gluconeogenesis | 2 | 14 | 0 | 6 | 3 | 11 | 0 | 4 | 2 | 10 | 1 | 3 | 56 |
| 05322 | Systemic lupus erythematosus | 8 | 0 | 7 | 0 | 6 | 0 | 6 | 0 | 8 | 0 | 2 | 0 | 37 |
| 05219 | Bladder cancer | 6 | 1 | 5 | 1 | 5 | 1 | 5 | 1 | 4 | 1 | 1 | 1 | 32 |
| 05222 | Small cell lung cancer | 5 | 1 | 5 | 1 | 5 | 1 | 4 | 1 | 4 | 1 | 3 | 1 | 32 |
| 04610 | Complement and coagulation cascades | 1 | 7 | 0 | 6 | 0 | 2 | 0 | 2 | 1 | 6 | 0 | 1 | 26 |
| 04512 | ECM-receptor interaction | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 3 | 24 |
| 04360 | Axon guidance | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 2 | 0 | 17 |
| 05211 | Renal cell carcinoma | 4 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 0 | 1 | 17 |
Outd outdegree, the number of downstream signaling pathways connected to the node signaling pathway; Ind indegree, the number of upstream signaling pathways connected to node signaling pathway; Total degree: outdegree + indegree
Association of BIRC5, CKS2, ITGA3, ITGA6, and RALA with survival time in PDAC
To characterize the function of the pathway in cancer in PDAC, we analyzed the differentially expressed genes of this pathway in each of the six expression profiles. A total of 137 genes in this pathway were found to be differentially expressed in pancreatic cancer across all six datasets. Mining of The Cancer Genome Atlas (Internal ID: 811) and the International Cancer Genome Consortium database (Internal ID: 812) provided survival data connected with these 137 genes (Additional file 5: Table S5). We identified five genes that have been associated with patient survival time: baculoviral IAP repeat containing 5 (BIRC5), CDC28 protein kinase regulatory subunit 2 (CKS2), integrin subunit alpha 3 (ITGA3), integrin subunit alpha 6 (ITGA6), and RAS-like proto-oncogene A (RALA) (Fig. 3a). Patients with high expression of these five genes showed shorter survival. Consistent with these database results, we examined CKS2 expression in 102 patients with PDAC at our hospital and found that higher expression was associated with shorter survival (Fig. 3b).
Fig. 3.
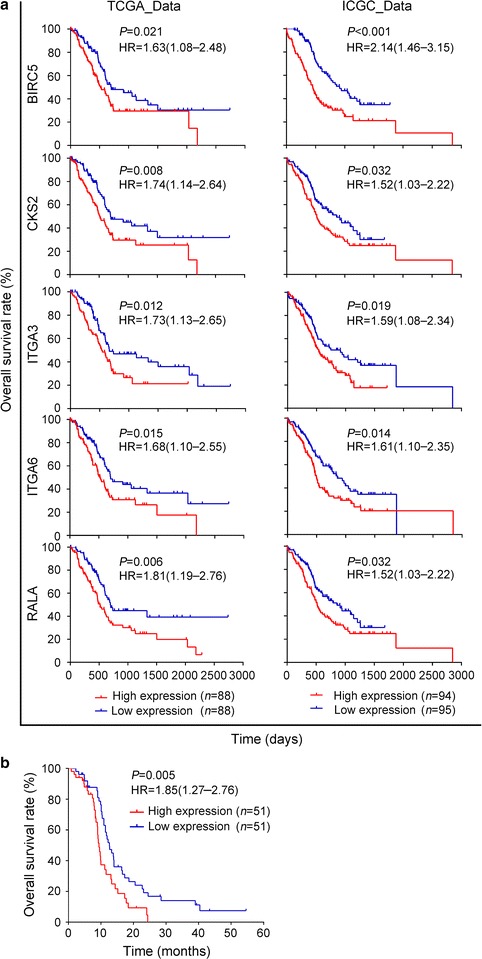
Identification of an association of BIRC5, CKS2, ITGA3, ITGA6 and RALA with survival time of patients with PDAC. a Kaplan–Meier survival curves of BIRC5, CKS2, ITGA3, ITGA6 and RALA in The Cancer Genome Atlas (Internal ID: 811) or International Cancer Genome Consortium database (Internal ID: 812). Survival data were obtained from the SurvExpress database. b Association of CKS2 expression with survival time in a cohort of 102 patients with PDAC at our hospital
Overexpression of CKS2 in PDAC
To determine which of these survival-related genes may be most important, we analyzed the fold-difference in the expression of each gene between PDAC and non-tumor tissues in the six expression profiles (Table 2). CKS2 showed the largest difference in expression, being up-regulated 4.82-fold in PDAC. Overexpression of CKS2 in PDAC compared with normal tissues has also been reported in the Oncomine database (Fig. 4), and data from the cbioportal database showed that CKS2 is amplified in many types of cancer (Fig. 5a). CKS2 expression levels positively correlate with copy number in 15 tumor cell lines (Fig. 5b).
Table 2.
Fold differences in expression of CKS2, ITGA3, BIRC5, RALA and ITGA6 between pancreatic cancer tissue and normal tissue in six GEO microarray datasets
| Gene | Fold differencea | Average fold difference | |||||
|---|---|---|---|---|---|---|---|
| GSE71989 | GSE15471 | GSE16515 | GSE32676 | GSE41368 | GSE28735 | ||
| CKS2 | 6.61 | 3.18 | 4.32 | 5.17 | 4.82 | ||
| ITGA3 | 2.32 | 2.51 | 3.01 | 3.49 | 4.74 | 7.95 | 4.00 |
| BIRC5 | 2.42 | 2.37 | 2.11 | 2.30 | |||
| RALA | 2.12 | 2.02 | 2.44 | 2.03 | 2.15 | ||
| ITGA6 | 2.01 | 2.20 | 2.11 | ||||
aPositive differences indicate up-regulation in pancreatic tumor tissue relative to non-tumor tissue. Fold differences of at least 2 were regarded as significant, so differences below 2 were left blank in this table
Fig. 4.
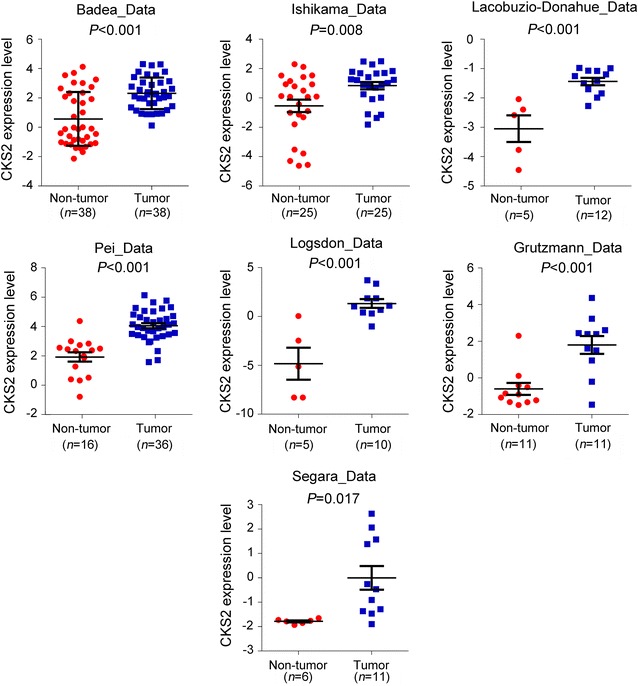
Overexpression of CKS2 in PDAC. CKS2 mRNA levels were significantly higher in PDAC tissues than in normal tissues according to the Oncomine database. Red color indicates CKS2 expression levels in normal pancreatic tissues, while blue color indicates CKS2 expression levels in pancreatic cancer
Fig. 5.

Genomic alterations of CKS2 in human cancers. a Summary of copy number variations of CKS2 across 49 cancer genomic studies (http://www.cbioportal.org). The results are displayed as a histogram of the frequencies of CKS2 copy numbers across cancer studies. b Correlation between CKS2 copy number and CKS2 expression level in 87 cancer cell lines derived from 15 types of human cancer (http://portals.broadinstitute.org/ccle/home). Pearson’s correlation coefficient (r) is shown. c Correlation of CKS2 expression and DNA methylation in 149 pancreatic cancer tissues in The Cancer Genome Atlas database. Pearson’s correlation coefficient (r) is shown
CKS2 expression negatively correlates with DNA methylation
Tumor cells show dysregulation of DNA methylation levels and patterns, leading to significant changes in the overall level of DNA methylation in tumors [11]. Data in The Cancer Genome Atlas for 149 PDAC tissue samples show that methylation levels at the CKS2 locus negatively correlate with CKS2 expression (r = − 0.368, P < 0.001; Fig. 5c).
CKS2 promotes PDAC cell proliferation
To characterize the potential functional role of CKS2, we analyzed genes whose expression parallels that of CKS2 based on RNA sequencing data from PDAC tissues in The Cancer Genome Atlas. A total of 325 genes co-expressed with CKS2 were identified (absolute r > 0.5; Additional file 6: Table S6), and these genes were enriched in the cell cycle pathway (Fig. 6a). Analysis of tumor and non-tumor tissues from our cohort of 102 patients with PDAC showed that CKS2 mRNA levels were higher in tumor tissues (Fig. 6b). CKS2 protein levels were higher in PDAC cell lines BxPC-3 and CFPAC-1 than in immortalized non-cancer pancreatic duct epithelial cell line HPDE6-C7 (Fig. 6c). Consistent with these results, CKS2 protein levels were higher in 20 samples of pancreatic cancer tissue than in the corresponding samples of adjacent normal tissue (Fig. 6d). Knockdown of CKS2 expression in the PDAC cell lines BxPC-3 and CFPAC-1 substantially suppressed proliferation (Fig. 6e, f).
Fig. 6.
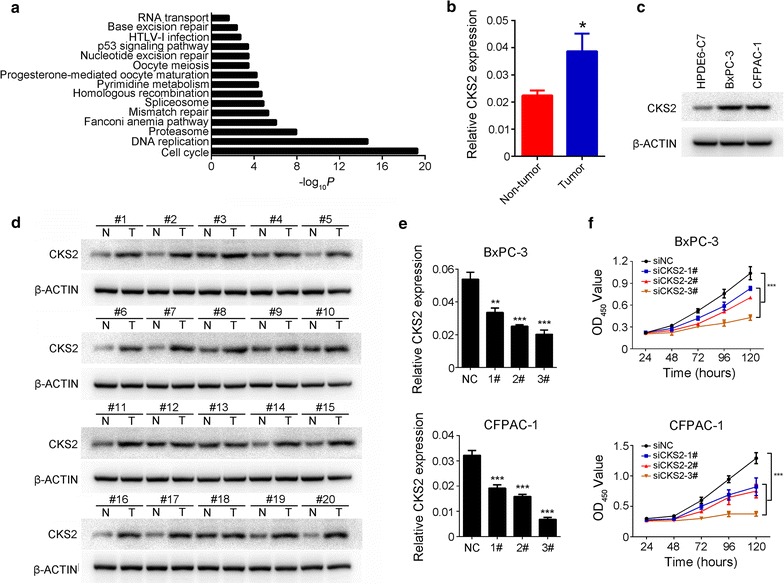
CKS2 promotes pancreatic cancer cell proliferation. a Significant pathways enriched in genes whose expression profile matches that of CKS2, based on DAVID software. b CKS2 mRNA levels in pancreatic cancer tissues and paired normal tissues from 102 patients with PDAC at our hospital, based on real-time PCR analysis. Results are mean ± SEM. *P < 0.05. c Western blotting of CKS2 in PDAC cell lines BxPC-3 and CFPAC-1, as well as in non-cancer pancreatic duct epithelial cell line HPDE6-C7. d Western blotting of CKS2 in PDAC (T) and paired normal tissues (N). Representative results of 20 pairs of clinical specimens are shown. e CKS2 knockdown in BxPC-3 and CFPAC-1 cells. Levels of mRNAs were determined by qRT-PCR. Results are mean ± SEM from three independent experiments. f CKS2 knockdown substantially reduced proliferation of BxPC-3 and CFPAC-1 cells, as determined using the CCK-8 assay. Results are mean ± SEM from three experiments, each conducted with six replicates of each sample. ***P < 0.001 vs. control
Discussion
In this study, we analyzed PDAC-associated genes and pathways in six frequently cited, published pancreatic cancer microarray datasets in the GEO database: GSE71989 [12, 13], GSE15471 [14], GSE16515 [15, 16], GSE32676 [17, 18], GSE41368 [19], and GSE28735 [18, 20]. We investigated differences in gene expression between pancreatic cancer tissues and normal tissues. Significant pathways enriched for differentially expressed genes were assembled into a pathway network, and core pathways showing high centrality degree in this network were chosen for further study. The pathway in cancer (KEGGID: 05200) was the top core pathway, and one gene in this pathway, CKS2, has been shown to be overexpressed in PDAC relative to normal tissue. CKS2 expression correlates positively with copy number and negatively with DNA methylation. CKS2 overexpression may be associated with shorter survival of patients with PDAC, probably due to CKS2 function in promoting proliferation of pancreatic cancer cells.
We used essentially all samples from these six GEO datasets to identify genes differentially expressed between PDAC tissue and normal tissue, although we excluded some samples to avoid the batch effect in previous studies [21]. This also allowed us to pool the results from each dataset to perform integrative analysis. As part of this analysis, we created pathway networks [22], which were introduced to mine GEO data. Molecules in different signaling pathways interact with one another, so the pathways in a cell can be connected into a system. This is, to the best of our knowledge, the first analysis of PDAC-associated genes at the level of pathway networks. We obtained different pathway networks from the six GEO datasets, reflecting differences in samples, testing platform and other reasons (Additional file 7: Figure S1). If a pathway is important for pancreatic cancer development or progression, it should appear consistently across microarray experiments. We found 32 significant pathways common to all six GEO datasets, and within those, 13 core pathways, which are particularly likely to be important in the development and progression of pancreatic cancer. Of these core pathways, the pathway in cancer (KEGG ID: 05200) showed the highest degree centrality, suggesting that it is the most important pathway in pancreatic cancer. The pathway in cancer is an integrated signaling pathway that includes RAS-mediated pathways [23]. KRAS is a well-known driver gene for pancreatic cancer, and mutation of this gene occurs in about 90% of patients [24]. This suggests that our pathway network analysis can give reliable results.
We then analyzed whether all differentially expressed genes in the pathway in cancer across six GEO datasets are associated with PDAC survival, based on data in The Cancer Genome Atlas and International Cancer Genome Consortium databases. In both databases, expression of BIRC5, CKS2, ITGA3, ITGA6 and RALA were significantly associated with survival time. We did not identify any expression quantitative trait loci associated with differential expression of these five genes in pancreatic tissues, based on data in the GTEx database [25]. This result may reflect the limited sample size. Of the five differentially expressed genes, CKS2 showed the greatest dysregulation in PDAC, which may be due to changes in copy number or/and DNA methylation (Fig. 5b, c).
CKS2, a gene encoding CDC28 protein kinase regulatory subunit, is located in chromosomal region 9q22.2. Previous studies have reported aberrant expression of this gene in cancers of the bladder [26], stomach [27], prostate [28], esophagus [29] and breast [30]. Our database mining showed that many types of tumor overexpress CKS2 (Additional file 8: Figure S2 and Additional file 9: S3). Although CKS2 has been shown to play an important role in many types of cancer, its function in PDAC has not been reported. Our database mining showed that the genes associated with CKS2 are involved mainly in cell proliferation processes, such as the cell cycle and DNA replication. This implies that CKS2 knockdown may inhibit PDAC cell proliferation, which we confirmed directly in cultures of PDAC cell lines. These results are consistent with those obtained with other types of cancer.
BIRC5 is a member of the inhibitor of apoptosis gene family, which is overexpressed in many types of tumors [31, 32]. BIRC5 expression can be regulated by STAT3 [33], and it can be suppressed by curcumin, which induces apoptosis and ultimately inhibits PDAC cell proliferation [34]. ITGA3 and ITGA6 are two members of the integrin alpha chain family of proteins. ITGA3 may promote proliferation or invasion in various types of cancer [35–37]. ITGA6 expression has been correlated with expression of migration-related genes [38], and it can promote the epithelial-mesenchymal transition [39] and tumor invasion [40]. ITGA6 also plays a role in tumorigenesis [41]. ITGA3 and ITGA6, both members of the integrin signaling pathway, are overexpressed in pancreatic cancer [42]. RALA may participate in pancreatic cancer initiation [43]. The details of how these genes participate in pancreatic cancer development and progression require further investigation.
Conclusions
Integrative analysis of six GEO microarray datasets of pancreatic cancer suggests that the pathway in cancer (KEGG ID: 05200) is an essential pathway in the development and progression of PDAC. High CKS2 expression is significantly associated with shorter survival time in patients with PDAC, which may reflect a function of CKS2 in promoting PDAC cell proliferation. These results demonstrate the potential of integrative data-mining from the GEO database in order to gain further insights into PDAC and other diseases.
Additional files
Additional file 1: Table S1. Characteristics of patients with pancreatic cancer in this study.
Additional file 2: Table S2. Short interfering RNA (siRNA) sequences used in this study.
Additional file 3: Table S3. Primers used for real-time PCR in this study.
Additional file 4: Table S4. Pancreatic cancer microarray datasets included in the study.
Additional file 5: Table S5. Hazard ratio (HR) of death for genes in the pathway in cancer that are differentially expressed in pancreatic cancer, based on data in two databases.
Additional file 6: Table S6. Genes associated with CKS2 expression level in pancreatic cancer tissues, based on data in The Cancer Genome Atlas.
Additional file 7: Figure S1. Pathway networks of all GEO datasets. Pathway networks for the datasets GSE15471 (A), GSE16515 (B), GSE32676 (C), GSE71989 (D), GSE28735 (E) and GSE42368 (F) were drawn using the GCBI online tool as described in Methods. Yellow dots indicate involvement of up- and down-regulated signaling pathway genes; red dots, up-regulated signaling pathway genes; and blue dots, down-regulated signaling pathway genes. The arrow points from the upstream toward the downstream signaling pathway.
Additional file 8: Figure S2. CKS2 expression in different types of cancer cell lines, based on data from the CCLE database (https://portals.broadinstitute.org/ccle/home).
Additional file 9: Figure S3. CKS2 expression across multiple types of cancer and corresponding normal tissues. The numbers of studies reporting up- or down-regulation is indicated, respectively, in red or blue boxes. Color intensity reflects the best gene rank percentile for analyses within the box. The following settings were used for the analysis: P=0.05, fold change=all, gene rank=all, and data type=mRNA (http://www.oncomine.org).
Authors’ contributions
DL conceptualized and supervised this study. JL contributed to the study design; JL, WenleT and LP conducted data analysis. JL and QC performed the proliferation assay. JianZ, JialiangZ and XH performed sample preparation and association analysis. JL, CW and WenT were involved in manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
We thank Prof. Rufu Chen from Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital to provide PDAC and adjacent normal samples for this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Consent for publication
We all consent for publication.
Ethics approval and consent to participate
Informed consent was obtained from all participants, and this study was approved by the Department of Pancreaticobiliary, Sun Yat-sen Memorial Hospital and Sun Yat-sen University Cancer Center. All experiments on the participants in this study were performed in accordance with the relevant guidelines and regulations.
Funding
This research was supported by the National Natural Science Foundation of China (No. 81572793, 81772586 and 81602461), a Grant (No. 2016A030313283) from the Natural Science Foundation of Guangdong Province and the Peking Union Medical College Doctor Innovation Fund (No. 2015-0710-17).
Abbreviations
- CCLE
cancer cell line encyclopedia
- FBS
fetal bovine serum
- GEO
gene expression omnibus
- ICGC
International Cancer Genome Consortium
- PDAC
pancreatic duct adenocarcinoma
- siRNA
short interfering RNA
- TCGA
The Cancer Genome Atlas
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40880-018-0289-9) contains supplementary material, which is available to authorized users.
Contributor Information
Jun Li, Email: junli198901@hotmail.com.
Wenle Tan, Email: tanwenle1991@163.com.
Linna Peng, Email: penglinna7@163.com.
Jialiang Zhang, Email: zhangjial@sysucc.org.cn.
Xudong Huang, Email: huangxudong1989@126.com.
Qionghua Cui, Email: cuiqionghua@hotmail.com.
Jian Zheng, Email: zhengjian@sysucc.org.cn.
Wen Tan, Email: tanwen68@hotmail.com.
Chen Wu, Email: chenwu@cicams.ac.cn.
Dongxin Lin, Phone: 10-87788715, Email: lindx@cicams.ac.cn.
References
- 1.Hackeng WM, Hruban RH, Offerhaus GJ, Brosens LA. Surgical and molecular pathology of pancreatic neoplasms. Diagn Pathol. 2016;11:47. doi: 10.1186/s13000-016-0497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, et al. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S, Liu Y, Jiang N, Chen J, Leach L, Luo Z, et al. Genome-wide eQTLs and heritability for gene expression traits in unrelated individuals. BMC Genomics. 2014;15:13. doi: 10.1186/1471-2164-15-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamazon ER, Stranger BE. The impact of human copy number variation on gene expression. Brief Funct Genomics. 2015;14:352–357. doi: 10.1093/bfgp/elv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CJ, Evans J, Kim K, Chae H, Kim S. Determining the effect of DNA methylation on gene expression in cancer cells. Methods Mol Biol. 2014;1101:161–178. doi: 10.1007/978-1-62703-721-1_9. [DOI] [PubMed] [Google Scholar]
- 9.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg. 2001;234:10–20. doi: 10.1097/00000658-200107000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giulietti M, Occhipinti G, Principato G, Piva F. Weighted gene co-expression network analysis reveals key genes involved in pancreatic ductal adenocarcinoma development. Cell Oncol. 2016;39:379–388. doi: 10.1007/s13402-016-0283-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhu T, Gao YF, Chen YX, Wang ZB, Yin JY, Mao XY, et al. Genome-scale analysis identifies GJB2 and ERO1LB as prognosis markers in patients with pancreatic cancer. Oncotarget. 2017;8:21281–21289. doi: 10.18632/oncotarget.15068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 15.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Zhu Z, Wang W, Shen P, Wei Z, Wang C, et al. Expression profiles analysis of pancreatic cancer. Eur Rev Med Pharmacol Sci. 2013;17:311–317. [PubMed] [Google Scholar]
- 17.Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JH, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L, et al. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS ONE. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frampton AE, Castellano L, Colombo T, Giovannetti E, Krell J, Jacob J, et al. MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology. 2014;146(268–77):e18. doi: 10.1053/j.gastro.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS ONE. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS ONE. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta B, Wallqvist A, Reifman J. PathNet: a tool for pathway analysis using topological information. Source Code Biol Med. 2012;7:10. doi: 10.1186/1751-0473-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 25.Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R, Feng C, Xu Y. Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J Int Med Res. 2011;39:533–540. doi: 10.1177/147323001103900222. [DOI] [PubMed] [Google Scholar]
- 27.Kang MA, Kim JT, Kim JH, Kim SY, Kim YH, Yeom YI, et al. Upregulation of the cyclin kinase subunit CKS2 increases cell proliferation rate in gastric cancer. J Cancer Res Clin Oncol. 2009;135:761–769. doi: 10.1007/s00432-008-0510-3. [DOI] [PubMed] [Google Scholar]
- 28.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–2825. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 29.Kita Y, Nishizono Y, Okumura H, Uchikado Y, Sasaki K, Matsumoto M, et al. Clinical and biological impact of cyclin-dependent kinase subunit 2 in esophageal squamous cell carcinoma. Oncol Rep. 2014;31:1986–1992. doi: 10.3892/or.2014.3062. [DOI] [PubMed] [Google Scholar]
- 30.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 31.Adida C, Berrebi D, Peuchmaur M, Reyes-Mugica M, Altieri DC. Anti-apoptosis gene, survivin, and prognosis of neuroblastoma. Lancet. 1998;351:882–883. doi: 10.1016/S0140-6736(05)70294-4. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127–134. [PubMed] [Google Scholar]
- 33.Glienke W, Hausmann E, Bergmann L. Targeting STAT3 signaling in pancreatic cancer promotes antiapoptotic gene expression. Pancreas. 2011;40:323–324. doi: 10.1097/MPA.0b013e318204ea7b. [DOI] [PubMed] [Google Scholar]
- 34.Glienke W, Maute L, Wicht J, Bergmann L. Curcumin inhibits constitutive STAT3 phosphorylation in human pancreatic cancer cell lines and downregulation of survivin/BIRC5 gene expression. Cancer Invest. 2010;28:166–171. doi: 10.3109/07357900903287006. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama SK, Olden K, Yamada KM. Fibronectin and integrins in invasion and metastasis. Cancer Metastasis Rev. 1995;14:173–189. doi: 10.1007/BF00690290. [DOI] [PubMed] [Google Scholar]
- 36.Koshizuka K, Hanazawa T, Kikkawa N, Arai T, Okato A, Kurozumi A, et al. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017;108:1681–1692. doi: 10.1111/cas.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakaguchi T, Yoshino H, Yonemori M, Miyamoto K, Sugita S, Matsushita R, et al. Regulation of ITGA3 by the dual-stranded microRNA-199 family as a potential prognostic marker in bladder cancer. Br J Cancer. 2017;116:1077–1087. doi: 10.1038/bjc.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golbert DC, Correa-de-Santana E, Ribeiro-Alves M, de Vasconcelos AT, Savino W. ITGA6 gene silencing by RNA interference modulates the expression of a large number of cell migration-related genes in human thymic epithelial cells. BMC Genomics. 2013;14(Suppl 6):S3. doi: 10.1186/1471-2164-14-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K, Myllymaki SM, Gao P, Devarajan R, Kytola V, Nykter M, et al. Oncogenic K-Ras upregulates ITGA6 expression via FOSL1 to induce anoikis resistance and synergizes with alphaV-Class integrins to promote EMT. Oncogene. 2017;36:5681–5694. doi: 10.1038/onc.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26. doi: 10.1186/s12943-016-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon J, Lee TS, Lee HW, Kang MC, Yoon HJ, Kim JH, et al. Integrin alpha 6: a novel therapeutic target in esophageal squamous cell carcinoma. Int J Oncol. 2013;43:1523–1530. doi: 10.3892/ijo.2013.2097. [DOI] [PubMed] [Google Scholar]
- 42.Van den Broeck A, Vankelecom H, Van Eijsden R, Govaere O, Topal B. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J Exp Clin Cancer Res. 2012;31:68. doi: 10.1186/1756-9966-31-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Characteristics of patients with pancreatic cancer in this study.
Additional file 2: Table S2. Short interfering RNA (siRNA) sequences used in this study.
Additional file 3: Table S3. Primers used for real-time PCR in this study.
Additional file 4: Table S4. Pancreatic cancer microarray datasets included in the study.
Additional file 5: Table S5. Hazard ratio (HR) of death for genes in the pathway in cancer that are differentially expressed in pancreatic cancer, based on data in two databases.
Additional file 6: Table S6. Genes associated with CKS2 expression level in pancreatic cancer tissues, based on data in The Cancer Genome Atlas.
Additional file 7: Figure S1. Pathway networks of all GEO datasets. Pathway networks for the datasets GSE15471 (A), GSE16515 (B), GSE32676 (C), GSE71989 (D), GSE28735 (E) and GSE42368 (F) were drawn using the GCBI online tool as described in Methods. Yellow dots indicate involvement of up- and down-regulated signaling pathway genes; red dots, up-regulated signaling pathway genes; and blue dots, down-regulated signaling pathway genes. The arrow points from the upstream toward the downstream signaling pathway.
Additional file 8: Figure S2. CKS2 expression in different types of cancer cell lines, based on data from the CCLE database (https://portals.broadinstitute.org/ccle/home).
Additional file 9: Figure S3. CKS2 expression across multiple types of cancer and corresponding normal tissues. The numbers of studies reporting up- or down-regulation is indicated, respectively, in red or blue boxes. Color intensity reflects the best gene rank percentile for analyses within the box. The following settings were used for the analysis: P=0.05, fold change=all, gene rank=all, and data type=mRNA (http://www.oncomine.org).
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


