Summary
The chromatin remodeling factor SMARCAD1, an SWI/SNF ATPase family member, has a role in 5′ end resection at DNA double-strand breaks (DSBs) to produce single-strand DNA (ssDNA), a critical step for subsequent checkpoint and repair factor loading to remove DNA damage. However, the mechanistic details of SMARCAD1 coupling to the DNA damage response and repair pathways remains unknown. Here we report that SMARCAD1 is recruited to DNA DSBs through an ATM-dependent process. Depletion of SMARCAD1 reduces ionizing radiation (IR)-induced repairosome foci formation and DSB repair by homologous recombination (HR). IR induces SMARCAD1 phosphorylation at a conserved T906 by ATM kinase, a modification essential for SMARCAD1 recruitment to DSBs. Interestingly, T906 phosphorylation is also important for SMARCAD1 ubiquitination by RING1 at K905. Both these post-translational modifications are critical for regulating the role of SMARCAD1 in DNA end resection, HR-mediated repair, and cell survival after DNA damage.
Subject Areas: In Vitro Toxicology Including 3D Culture, Bioengineering, Tissue Engineering
Graphical Abstract

Highlights
-
•
DNA damage induces ATM-dependent phosphorylation of SMARCAD1 at T906
-
•
Ubiquitination by RING1 at K905 stabilizes SMARCAD1-pT906
-
•
SMARCAD1-pT906 and ubiquitination are necessary for DSB recruitment and DNA resection
-
•
Post-translational SMARCAD1 modifications are critical for DSB repair by HR
In Vitro Toxicology Including 3D Culture; Bioengineering; Tissue Engineering
Introduction
Repair of DNA double-strand breaks (DSBs) is essential for maintaining genomic integrity and cell survival. Chromatin remodeling factors are key components of the DNA DSB repair process since repair takes place within a dynamic chromatin context, and several ATP-dependent chromatin remodeling factors have been implicated in DNA repair pathways including the homologous recombination (HR), non-homologous end joining (NHEJ), and nuclear excision pathways (Deem et al., 2012, Gravel et al., 2008, Hunt et al., 2013, Lan et al., 2010, Lukas et al., 2011, Manning and Peterson, 2013, Mimitou and Symington, 2008, Morrison and Shen, 2005, Pandita and Richardson, 2009, Peterson and Almouzni, 2013, Polo and Jackson, 2011, Price and D'Andrea, 2013, Raschella et al., 2017, van Attikum and Gasser, 2005, Zhu et al., 2008).
The SMARCAD1 protein is a member of the SNF2 helicase subfamily and the SWI/SNF family of ATPase containing DEAD/H box helicase domain proteins (Adra et al., 2000). The ATP-dependent SWI/SNF family members are critical for DNA replication and HR (Collins et al., 2002, Morrison et al., 2004, van Attikum et al., 2004). SMARCAD1 also functions in the propagation and organization of epigenetic patterns following DNA replication (Rowbotham et al., 2011) as well as in heterochromatin maintenance and inheritance (Taneja et al., 2017). Functionally, SMARCAD1 is involved in the regulation of global histone acetylation with a suggested role in histone deacetylation by interacting with HDAC1 during replication (Horikoshi et al., 2013, Rowbotham et al., 2011). SMARCAD1 is also recruited to newly synthesized DNA and facilitates histone deacetylation, histone H3K9 trimethylation (H3K9me3), and efficient HP1 recruitment through a mechanism coupled to ATP hydrolysis (Rowbotham et al., 2011). Previously it has been shown by immunofluorescence co-staining of SMARCAD1 and γ−H2AX along laser micro-irradiation tracks that SMARCAD1 localizes to the vicinity of DSB sites (Costelloe et al., 2012). Moreover, SMARCAD1 was suggested to have a role in DNA end resection because RPA foci formation in depleted cells was decreased. The timing of SMARCAD1 function appears to be early during repair because it has been reported that SMARCAD1 binds to H2A-Ub at the DNA damage sites and is required for 53BP1 repositioning after BRCA1 recruitment (Densham et al., 2016). A proteomics survey of ionizing radiation (IR)-induced phosphopeptides identified SMARCAD1 as a possible ATM substrate with potential sites at S132 and S302 (Matsuoka et al., 2007). However, further confirmation of these sites or their biological function has not been reported but does suggest potential SMARCAD1 coupling to IR-induced ATM activity.
Further details about the mechanism and functional significance of SMARCAD1 regulation during DNA repair are limited. In this study, we identified two previously unknown post-translational SMARCAD1 modifications, ATM-mediated phosphorylation of Thr906 (T906) and ubiquitination of Lys905 (K905) by RING1, which are critical steps required for subsequent binding of DNA repair factors and healing of DNA lesions. Depletion of SMARCAD1 or loss of these post-translational modifications decreased DNA end resection, reduced DNA DSB repair by HR, and increased IR-induced chromosome damage and cell death.
Results
SMARCAD1 Localizes to DSBs Preferentially during S-Phase
SMARCAD1 has been implicated in DNA resection, a critical step in homologous-recombination-mediated DNA DSB repair, which primarily occurs during the cell cycle S-phase. Therefore we measured SMARCAD1 recruitment to a DSB site during the cell cycle, before and after induction of the DSB, by chromatin immunoprecipitation (ChIP)/PCR using specific primers at different distances from a defined l-Scel-induced DSB. Exponentially growing cells were enriched in G1-, S-, or G2/M-phase as described previously (Gupta et al., 2014, Rodrigue et al., 2006), induced for l-Scel site cleavage, and analyzed by ChIP for γ-H2AX, KU80, RAD51, and SMARCAD1. As expected, exponentially growing cells showed recruitment of SMARCAD1 as well as γ-H2AX, KU80, and RAD51 to the DSB produced by l-Scel cleavage (Figure 1A). Irrespective of the cell phase, the surrogate DSB marker γ-H2AX was largely recruited to the l-Scel DNA site only after break induction (Figures 1B–1D). In G1-phase cells, increased localization of KU80 was observed, whereas levels of RAD51 and SMARCAD1 at the break site were significantly less (Figure 1B). Conversely, significant recruitment of RAD51 and SMARCAD1 was observed in S-phase cells, whereas KU80 levels were relatively lower (Figure 1C). In G2/M-phase cells, the levels of SMARCAD1 were once again low when compared with the KU80 levels, which were significantly greater (Figure 1D). Therefore SMARCAD1 is recruited to DSBs predominantly during the S-phase, and the results are similar to those of the known HR repair component RAD51. Interestingly, there were no differences in cell cycle distribution between control small interfering RNA (siRNA)-transfected cells (G1 57.36%, S 23.56%, G2 19.08%) and SMARCAD1-depleted cells (G1 54.87%, S 26.45%, and G2 20.67%), suggesting that SMARCAD1 depletion does not interfere with the cell cycle status in the absence of damage.
Figure 1.
Recruitment of Repair Proteins at DSB
SMARCAD1 is recruited to DSB site: Detection of repair proteins by ChIP at I-SceI site before and after induction of DSB. Cell synchronization, cell cycle analysis, I-SceI-induced DSB, and ChIP analysis were done according to the described procedure (Gupta et al., 2014, Rodrigue et al., 2006). The closest PCR products to the l-Scel-induced DSB site are 94–378, 675–1044, and 901–1210 (away from the I-SceI-induced break site).
(A–D) (A) Exponential phase cells, (B) G1-phase cells, (C) S-phase cells, and (D) G2/M-phase cells.
SMARCAD1 Depletion Decreases IR-Induced Foci Formation by Proteins Involved in DSB Repair by Homologous Recombination
To determine the impact of SMARCAD1 on repairosome kinetics, we examined IR-induced γ-H2AX, 53BP1, BRCA1, RPA, and RAD51 foci formation by immunofluorescent staining of cells with and without SMARCAD1 depletion (Figure 2). Depletion of SMARCAD1 increased the frequency of irradiated cells with residual γ-H2AX (Figures 2A and 2B) and 53BP1 foci (Figures 2C and 2D), whereas the number of cells with BRCA1 (Figures 2E and S1C), RPA (Figures 2F and S1B), and RAD51 (Figures 2G and 2H) foci was significantly reduced, without any change in the overall cellular levels of BRCA1, RPA, or RAD51 protein (Figures S1D, S1E, S1F, and S1G).
Figure 2.
Immunofluorescence Measurement of Repairosome Foci Formation
SMARCAD1 depletion affects repairosome formation: (A) immunofluorescent visualization of γ-H2AX at different time points after 2 Gy irradiation.
(B) Quantification of cells for IR-induced foci formation with, and without, SMARCAD1 depletion.
(C) Immunofluorescent visualization of 53BP1 at different time points after 5 Gy irradiation.
(D) Quantification of cells for IR-induced foci formation with, and without, SMARCAD1 depletion.
(E) Quantification of cells for IR-induced BRCA1 foci formation with, and without, SMARCAD1 depletion.
(F) Quantification of cells for IR-induced RPA foci formation with, and without, SMARCAD1 depletion.
(G) Immunofluorescent visualization of RAD51 at different time points after 5 Gy irradiation.
(H) Quantification of cells for IR-induced RAD51 foci formation with, and without, SMARCAD1 depletion.
For cells with foci, at least 100 cells were examined, and the mean presented is from three to four experiments (*p < 0.05; **p < 0.01; ***p < 0.001 as determined by Student's t test).
The 53BP1 and BRCA1 balance at a DSB site determines the repair pathway choice, and SMARCAD1 depletion appeared to impede/delay 53BP1 removal from damage sites (Figures 2C and 2D) while correspondingly decreasing/delaying BRCA1 recruitment (Figure S1C). Immunofluorescence co-localization studies demonstrated that 53BP1/BRCA1 foci were present in control cells at 120 min post-irradiation, but by 240 min, there was little foci overlap and most of the 53BP1 signal had dispersed. Depletion of SMARCAD1 led to mainly 53BP1 foci being present, with occasional 53BP1/BRCA1 co-foci, at 120 min post-irradiation (Figure S1H). At 240 min, there were still 53BP1 foci present, but limited co-localization of BRCA1/53BP1 foci were detected (Figure S1H). These results are consistent with a model in which SMARCAD1 facilitates BRCA1 displacement of 53BP1 from DSB sites (Densham et al., 2016) to further HR-mediated repair.
SMARCAD1 Recruitment at DNA DSBs Is ATM Dependent
Eukaryotic cells respond to DNA damage with a rapid activation of the ATM/ATR protein kinase signaling cascade to induce cell cycle arrest and activate the checkpoint kinases required to maintain genome integrity (Lee and Paull, 2004, Shiloh, 2003, Shiloh and Ziv, 2013). To determine the impact of ATM or ATR inactivation on the recruitment of SMARCAD1 at DSBs, cells were treated with an inhibitor of ATM (KU55933) or ATR (VE-821) and then examined for SMARCAD1 DSB recruitment by ChIP, as described above. Exponential phase cells treated with ATM inhibitor had a significant decrease of SMARCAD1 association with the DSB, whereas no impact was observed in cells treated with ATR (Figure 3A). The decrease in SMARCAD1 association with DSB was maximum in S-phase cells after ATM inhibitor treatment, whereas ATR had no effect, suggesting that ATM-dependent modification(s) may be required for the association of SMARCAD1 with the DNA DSBs (Figures 3B, 3C, and 3D).
Figure 3.
Impact of SMARCAD1 on Resection
SMARCAD1 phosphorylation is critical for DNA end resection: (A–D) effect of ATM inhibitor KU55933 or ATR inhibitor on SMARCAD1 recruitment at DSB in cells growing in exponential phase (A), G1-phase (B), S-phase (C), and G2/M-phase (D). Note that cells in S-phase have reduced amount of SMARCAD1 associated with DSB.
(E) Western blot showing phosphorylation of SMARCAD1 after cells are exposed to 10 Gy ionizing radiation.
(F and G) Schematic diagram of ER-AsiSI DNA end resection assay based on ChIP/qPCR quantitation of ssDNA (F), and 4-Hydroxytamoxifen (OHT) inducible as visualization by immunofluorescence (G).
(H) Western blot showing MRE11 depletion by specific siRNA.
(I) SMARCAD1 depletion or expression of the T906A, T906E, and K528R mutations decreases ssDNA formation. MRE11 depletion is a positive control for the resection assay. DNA resection assay was performed in U2OS cells using qPCR.
The mean presented is from three to four experiments (*p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0005 as determined by Student's t test).
IR activates ATM (Bakkenist and Kastan, 2003, Pandita et al., 2000), which then phosphorylates specific SQ/TQ sites on downstream DNA damage signaling/repair effector proteins (Matsuoka et al., 2007). SMARCAD1 has multiple SQ motifs and one TQ motif, the core consensus sequence sites phosphorylated by ATM/ATR (Beli et al., 2012, Matsuoka et al., 2007). Phosphorylation was further confirmed by western blot analysis of SMARCAD1 immunoprecipitates from cell extracts with a phospho-SQ/TQ antibody, which detected IR-induced phosphorylation within 30 min post-irradiation (Figure 3E).
SMARCAD1 T906 Phosphorylation Is a Prerequisite for Efficient DNA End Resection
Based on our observations that SMARCAD1 had only one potential ATM TQ site and that it was conserved in higher vertebrates, including humans (Figures S2A and S2B), the single SMARCAD1 TQ (T906Q907) motif was mutated (T906A and T906E) and the effect of mutations on resection was determined in cells depleted of endogenous SMARCAD1 (Figures S2C, S2D, and S2E). Resection was measured as ssDNA production at 0.3, 1.6, and 3.5 kb from an AsiSI DNA cleavage site in SMARCAD1-depleted cells by using a previously described technique (Zhou et al., 2014) (Figures 3F and 3G). Consistent with a major role for SMARCAD1 in resection, we found that ssDNA levels at all distances from the DSB were decreased upon SMARCAD1 depletion, which are results similar to that produced by MRE11 nuclease depletion (Figures 3H and 3I). The reduction in DNA resection after endogenous SMARCAD1 depletion could not be rescued by SMARCAD1-T906A expression but was rescued by the phospho-mimetic T906E mutant (Figure 3I), confirming the importance of ATM-mediated SMARCAD1 phosphorylation in resection. Likewise, resection after SMARCAD1 depletion was not rescued by expression of the ATPase mutant K528R, suggesting that the ATPase activity of SMARCAD1 is also required during DNA end resection (Figure 3I).
Ionizing Radiation-Induced SMARCAD1 Ubiquitination by RING1
The interplay between post-translational phosphorylation and ubiquitination has emerged as an important means of regulation in eukaryotic cell signaling pathways (Hunter, 2007) where phosphorylation can serve as a marker to trigger ubiquitination (Fuchs et al., 1996, Magnani et al., 2000, Treier et al., 1994). Ubiquitination itself can provide a switching mechanism to turn kinases on or off (Witowsky and Johnson, 2003) as well as provide interaction sites for additional regulatory proteins that contain ubiquitin-binding domains (UBDs). SMARCAD1 contains two N-terminal CUE domains (CUE1, CUE2), which are equivalent to UBD. Such UBDs are often key regulators in mediating protein/protein interaction during repairosome assembly (Messick and Greenberg, 2009, Schwertman et al., 2016). We determined whether SMARCAD1 was subject to post-translational ubiquitination by immunoprecipitating SMARCAD1 from cell extracts after irradiation followed by western blot analysis with a pan-ubiquitin antibody (Figure 4A). Significant levels of ubiquitinated SMARCAD1 (ub-SMARCAD1) were not detected in control cells or in cells 0.5 hr post-irradiation. However, 2 hr post-irradiation, ub-SMARCAD1 was easily detectable and still present, but it declined at 3 hr post-irradiation before becoming undetectable at 4 hr. The ubiquitin linkage is mainly K63 (Figure 4B), although some K48 linkage can be detected particularly 2 hr after irradiation (Figure 4C) when K63-linked SMARCAD1 has declined (Figure 4B). This pattern and type of ubiquitination is consistent with a regulatory role early in the DDR and not a degradation function, particularly as there is no change in total cellular SMARCAD1 levels over the time period examined (0–240 min) (Figures 4A and 4B).
Figure 4.
Ubiquitination of SMARCAD1
IR-induced K63-linked ubiquitination of SMARCAD1 by RING1: immunoprecipitation (under denaturing conditions), followed by western blot analysis with the indicated antibodies
(A) IR-dependent ubiquitination of SMARCAD1 (pan Ub antibody).
(B) IR-induced K63-linked ubiquitination of SMARCAD1.
(C) IR-induced SMARCAD1 K48-linked ubiquitination.
(D) Mutation of the SMARCAD1 (T906A) ATM phosphorylation site blocks ubiquitination.
(E) Mutation of SMARCAD1 lysine 905 (K905R) blocks ubiquitination.
(F) Phosphorylation of the SMARCAD1 ubiquitination site (K905R) mutant.
(G) BARD1 depletion has no impact on IR-induced SMARCAD1 ubiquitination.
(H) IR-induced SMARCAD1 ubiquitination is lost in cells depleted of the E3 Ub ligase RING1.
The K63 ubiquitination of SMARCAD1 at 2 hr post-irradiation (Figure 4B) was dependent on IR-induced ATM phosphorylation, as the T906A SMARCAD1 mutant was not ubiquitinated (Figure 4D). SMARCAD1 has a potential ubiquitination site at lysine 905, and K905R mutation led to the loss of IR-induced ubiquitination (Figure 4E). Initially, K905R is phosphorylated after irradiation, but the phosphorylation does not persist over time (60 min onward) (Figure 4F). These data suggest that SMARCAD1 ubiquitination is required for the stabilization of SMARCAD1 phosphorylation, which is also supported by our K63 ubiquitination data (Figure 4B).
The BRCA1-BARD1 complex ubiquitinates histone H2A, and SMARCAD1 binds to H2A-ubiquitin through its two CUE domains, which facilitates localization to DNA damage sites (Densham et al., 2016). However, disruption of the BRCA1-BARD1 complex by BARD1 depletion had no effect on SMARCAD1 ubiquitination (Figure 4G), suggesting that it is not a BRCA1 substrate. A previous mass-spectrometric analysis of proteins co-immunoprecipitating with SMARCDA1 identified several known E3 ligases including RING1 (Rowbotham et al., 2011). Analysis of potential ligases by siRNA-specific depletion indicated that only RING1 loss correlated with decreased SMARCAD1 ubiquitination (Figure 4H).
SMARCAD1 Post-translational Modifications Affect Repair Factor Recruitment to DSBs
To test the functional consequences of SMARCAD1 ubiquitination status, we analyzed DNA DSB resection in cells depleted of RING1 and in cells expressing the SMARCAD1-K905R mutant that is not ubiquitinated. Resection was reduced both in cells expressing SMARCAD1-K905R and in those depleted of RING1 (Figures S3A and S3B). The failure of SMARCAD1 and RING1 co-depletion to further decrease the level of resection suggests that both act in the same pathway (Figure S3B). These results, together with the additional data described above, suggest that an obligatory, at least two-step series of SMARCD1 post-translational modifications is required for DSB end resection: phosphorylation (ATM) and ubiquitination (RING1).
We determined the importance of post-translational modifications for SMARCAD1 recruitment to l-Scel DSBs by expressing FLAG-tagged wild-type and mutant SMARCAD1 in cells depleted of endogenous SMARCAD1 (Figures S2D and S2E). The only mutation in a potential ATM phosphorylation site (S132, S302, and T906) that reduced SMARCAD1 accumulation at the DSB was T906A; recruitment to the DSB also was not altered by the SMARCAD1 ATPase mutation K528R (Figure 5A). Neither SMARCAD1 depletion nor SMARCAD1 mutant expression altered H2AX phosphorylation at DSB sites (Figure 5B). We also observed that the recruitment of BRCA1 at DSBs was affected, as cells expressing the ATPase mutant (SMARCAD1 K528R), like SMARCAD1 T906A-expressing or SMARCAD1-depleted cells, had significantly reduced BRCA1 levels at DSBs (Figure 5C). DR95 cells with and without SMARCAD1 depletion were examined for γ-H2AX accumulation at a site-specific DSB induced by l-SceI expression. There was SMARCAD1 co-localization with γ-H2AX foci in cells with SMARCAD1, whereas no co-localization of SMARCAD1 with γ-H2AX foci was observed in SMARCAD1-depleted cells (Figure 5D). These results are consistent with the ChIP data showing that SMARCAD1 recruits to DSB sites.
Figure 5.
Impact of SMARCAD1 Mutation on Recruitment
(A–D) Post-translational modification of SMARCAD1 recruitment to a DSB as well as recruitment of BRCA1: Analysis by ChIP/qPCR demonstrating that (A) SMARCAD1 recruitment to a DSB is blocked by the T906A mutation but not the S32A, S302A, or K528R mutation. (B) Amount of γ-H2AX at the DSB site is unaltered by SMARCAD1 mutant expression. (C) BRCA1 localization at break site is decreased by the T906A and K528R mutations but not the S302A mutation. (D) SMARCAD1 co-localizes with γ–H2X at a unique DSB site induced by ISce1 cleavage. Scale bars shown are 10 um. The mean presented is from three to four experiments (***p < 0.001, as determined by Student's t test).
Depletion of RING1 decreased the accumulation of SMARCAD1 at DSBs (Figure 6A) but had no effect on the accumulation of γ-H2AX at DSBs (Figure 6B). Consistent with these results, the accumulation of phosphomimetic SMARCAD1 mutant (Flag-SMARCAD1-T906E) was comparable with wild-type SMARCAD1; however, reduced accumulation at DSBs was observed for SMARCAD1 K905R (Figure 6C). These results suggest that phosphorylation of SMARCAD1 T906 and also ubiquitination are essential for the recruitment of SMARCAD1 to DNA DSBs.
Figure 6.
Impact of RING1 on SMARCAD1 Recruitment
(A–C) Impact of RINGI depletion on recruitment of SMARCAD1 at DSB: (A) RING1 depletion affects SMARCAD1 localization at the DSBs. (B) Localization of γ-H2AX at the DSBs in the presence or absence of RING1 siRNA is not affected. (C) Phosphomimetic mutant FLAG-SMARCAD1 T906E goes to the DSBs but not the Ub-specific mutant FLAG-SMARCAD1 K905R. The mean presented is from three to four experiments (***p < 0.001, as determined by Student's t test).
SMARCAD1 Depletion or Lack of Post-translational Modifications Increase Cell Killing after Genotoxic Stress
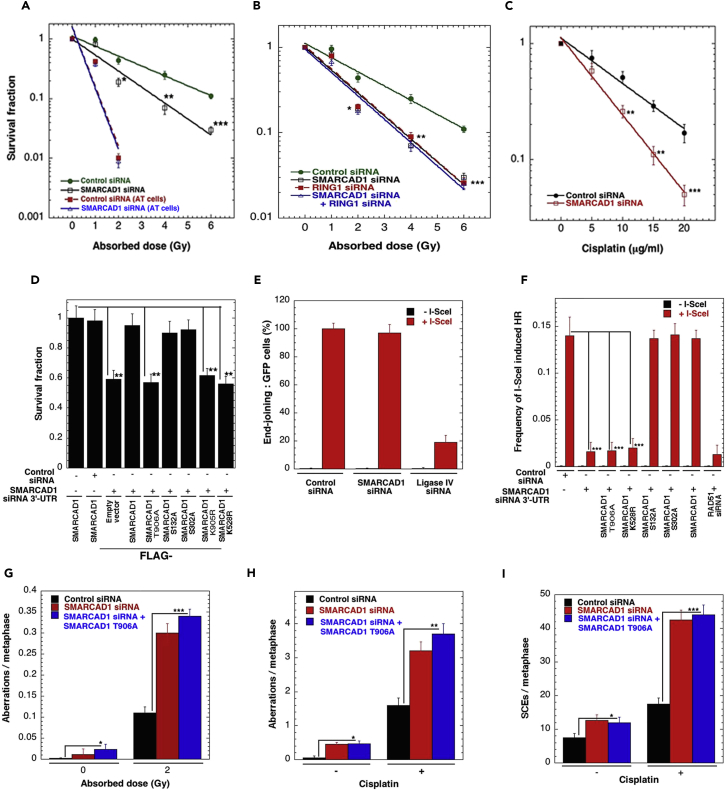
To determine the contribution of post-translational SMARCAD1 modifications to radiosensitivity, clonogenic cell survival assays were performed (Figure 7A). Depletion of SMARCAD1 decreased H1299 cell survival after irradiation but had no additional impact on survival of ATM-deficient (AT-5) cells, suggesting that SMARCAD1 and ATM act in the same repair pathway. Depletion of RING1 with siRNA also decreased the post-irradiation cell survival, but depletion of both RING1 and SMARCAD1 had no additional impact on cell survival (Figure 7B), also indicating a common pathway. Decreased survival was also observed in SMARCAD1-depleted cells after cisplatin treatment (Figure 7C). Cells expressing either SMARCAD1 T906A or SMARCAD1 K528R also had decreased survival following irradiation, at levels similar to those of SMARCAD1-depleted cells (Figure 7D).
Figure 7.
SMARCAD1 Phosphorylation and Ubiquitination Contributes to Cell Survival and DNA Damage Repair by HR
(A) Clonogenic survival in cells with and without SMARCAD1 depletion after IR treatment and when compared with ATM-deficient (AT-5) cells.
(B) Decreased survival of RING1-depleted cells after IR treatment.
(C) Decreased survival of SMARCAD1-depleted cells after cisplatin treatment, clonogenic survival.
(D) Expression of T906A, K528R, or K905R decreases cell survival after IR (1 Gy) but not S132A or S302A.
(E) SMARCAD1 depletion does not alter GFP-based NHEJ repair.
(F) Loss of SMARCAD1, or expression of T906A, K528R, or K905R, decreases DSB repair by HR, DR-GFP-based HR assay. DR, direct repeats.
(G) Cells depleted of SMARCAD1 or expressing Flag-T907A have increased S-phase-specific chromosome aberrations, as indicated by the presence of radials, breaks, and gaps, both constitutively and after irradiation.
(H) Cells depleted of SMARCAD1 or expressing Flag-T906A have increased metaphase chromosome aberrations both constitutively and after treatment with cisplatin.
(I) Sister chromatid exchanges (SCEs) increase in cells depleted of SMARCAD1 or expressing Flag-T906A in the presence or absence of cisplatin. The mean presented is from three to four experiments (*p < 0.05; **p < 0.01; ***p < 0.001 as determined by Student's t test).
Next we tested the effect of SMARCAD1 mutations on the efficiency of DSB repair by using standard GFP gene reporter l-Scel-inducible assays (Gupta et al., 2014). Cells depleted for SMARCAD1 did not show any significant defect in DSB repair by NHEJ (Figure 7E). In contrast, cells depleted of endogenous SMARCAD1 had significantly decreased HR repair, which was not restored by expression of either SMARCAD1 T906A or SMARCAD1 K528R but was restored by exogenous wild-type or SMARCAD1 T906E expression (Figure 7F). These data suggest that ATM-mediated phosphorylation of SMARCAD1 at T906, as well as SMARCAD1 ATPase activity, are required for efficient DNA DSB repair by the HR pathway.
Role of SMARCAD1 Modifications in S-Phase-Specific Chromosome Repair
We measured the level of IR-induced G1-, S-, and G2-phase-specific chromosomal aberrations in exponentially growing cells (Figures S4A, S4B, and S4C) and observed an increase in SMARCAD1-depleted cells (Figure S4D). The results described above indicated that SMARCAD1 depletion or mutation affected DNA DSB resection during HR-mediated repair, a largely S-phase cell-specific pathway. Therefore we analyzed chromosome aberrations in each specific cell phase. SMARCAD1-depleted cells had higher levels of IR-induced S-phase-specific chromosome aberrations (Figure 7G), whereas G1- and G2-phase aberrations were similar to those of control cells (Figures S4E and S4F), supporting the model that SMARCAD1 is not involved in DSB repair in G1-phase where NHEJ is the predominant repair. These results are consistent with DNA DSB end joining assay (Figure 7E). In addition, cells depleted for SMACAD1 but expressing SMARCAD1 T906A also had a high frequency of IR-induced S-phase aberrations (Figure 7G). Similarly, cisplatin treatment induced a higher frequency of chromosome aberrations in cells depleted of SMARCAD1 or expressing SMARCAD1 T906A than in control cells (Figure 7H). These results are consistent with an S-phase role for SMARCAD1, where HR repair is the predominant mode of DNA repair. Furthermore, we found that even in the absence of induced DNA damage, SMARCAD1-depleted cells had elevated levels of spontaneous sister chromatid exchanges (SCEs), which was further enhanced upon cisplatin treatment (Figures S4G, S4H, and 7I). This possibly indicates hyper-recombination or increased crossover as previously observed in yeast (Chen et al., 2012, Wallis et al., 1989) and in BLM helicase-deficient human cells (Yamagata et al., 1998).
Discussion
SMARCAD1 has been reported to play a role in the DSB end resection step required for subsequent loading of checkpoint and repair proteins during DNA damage repair (Chen et al., 2012, Chen et al., 2016, Costelloe et al., 2012). In this study, we demonstrate the role and complex regulatory pattern for SMARCAD1 in multiple steps of HR-mediated DNA repair. During the process of DSB repair, chromatin structure influences the docking of signaling and repair proteins. Our data provide insight into this mechanism in that DNA damage induces SMARCAD1 phosphorylation by ATM, promoting a cascade of HR repair functions where SMARCAD1 is directly involved.
DNA end resection in eukaryotes is a two-step process (Gravel et al., 2008, Mimitou and Symington, 2008). To initiate DNA end resection, the first step is to displace 53BP1 by BRCA1 (Isono et al., 2017). Our results suggest that SMARCAD1 works early in the resection pathway along with ATM and therefore its role differs from the yeast ortholog Fun30, which is needed primarily for the extensive resection mediated by Exo1 or Sgs1 (Chen et al., 2012, Costelloe et al., 2012). It has been reported that SMARCAD1 is a substrate of ATM (Densham et al., 2016, Matsuoka et al., 2007). We report here that the post-translational phosphorylation of the highly conserved T906Q motif is a key to SMARCAD1 function in DNA end resection and DSB repair by the HR pathway. Phosphorylation at this site was not detected in the previous studies (Chen et al., 2012, Costelloe et al., 2012, Densham et al., 2016), which detected only Ser132 and Ser302 phosphorylation (Matsuoka et al., 2007). The ER-AsiSI DNA resection system that we utilized has limitations in measuring long-range resection beyond 3.5 Kb, not reaching the 5–10 Kb resection range. Our results, however, suggest that SMARCAD1 is involved in short-range resection.
We have demonstrated that SMARCAD1 facilitates the recruitment of BRCA1 at the DNA break site and that SMARCAD1 phosphorylation at T906 and its ATPase activity are required for BRCA1 localization at break sites. Recent studies have shown that BRCA1-BARD1 ligase activity modifies chromatin, which affects the accumulation of SMARCAD1 and the repositioning of 53BP1 to allow for resection completion (Densham et al., 2016). Disruption of the BRCA1-BARD1 complex by depleting BARD1, which is an E3 ligase, did not affect SMARCAD1 ubiquitination, and we instead identified RING1 as the potential SMARCAD1 E3 ligase that modifies K905. Loss of RING1 abolishes SMARCAD1 ubiquitination and affects its function during DNA end resection and cell survival, suggesting that both act in the same pathway with SMARCAD1 as a substrate downstream of RING1. These findings are an addition to the previously known functions of RING1 during DNA damage signaling, ubiquitination of H2A at K118 and K119 (Cao et al., 2005, Stock et al., 2007). Our data suggest that coincident phosphorylation and ubiquitination stabilizes SMARCAD1 (2 hr post-irradiation), contributing to the process of BRCA1 localization at the break site by replacing 53BP1. We also find that the post-translational modification of SMARCAD1 is essential to carry out additional steps in HR repair since specific combinations of SMARCAD1 post-translational modifications allow binding of DNA repair factors and healing of the DNA lesion. Immunofluorescence data and ChIP data are consistent with a model in which SMARCAD1 interacts with and facilitates BRCA1 displacement of 53BP1 from DSB sites.
In conclusion, our findings reveal that the recruitment of SMARCAD1 to DSB sites is ATM dependent as shown in the graphic model (Figure 8), thus SMARCAD1 is an upstream DDR fidelity protein. SMARCAD1 is subject to IR-induced post-translational modifications carried out by ATM kinase and RING1 E3 ligase, both of which are important for proper DSB end resection. These SMARCAD1 post-translational modifications then affect downstream events in HR-mediated DNA repair, and their loss ultimately contributes to decreased HR and cell survival after genotoxic injury.
Figure 8.
SMARCAD1 Post-translational Modifications Impact DNA Resection
Model Showing SMARCAD1 Regulation during Repair: ATM-dependent phosphorylation and RING1-dependent ubiquitination of SMARCAD1 helicase regulates the DNA end resection step of repair by homologous recombination.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We are thankful to Grzegorz Ira and Ralph Scully for their helpful suggestions and discussion. We thank T.T. Paull for providing the reagents to perform the resection assay. Thanks to the members of the Pandita laboratory for their suggestions and comments on the paper. This work was supported by The Houston Methodist Research Institute and NIH RO1 CA129537 and RO1 GM109768 (T.K.P.).
Author Contributions
S.C., S.P.I., C.R.H., and T.K.P. designed the experiments. S.C., R.K.P., K.M.A., S.H., V.C., A.R.M., and C.R.H. performed the experiments. S.C., C.R.H., and T.K.P. wrote the manuscript.
Declaration of Interests
The authors declare that they have no conflict of interest.
Published: April 27, 2018
Footnotes
Supplemental Information includes Transparent Methods and four figures and can be found with this article online at https://doi.org/10.1016/j.isci.2018.03.016.
Contributor Information
Sharmistha Chakraborty, Email: schakraborty2@houstonmethodist.org.
Tej K. Pandita, Email: tpandita@houstonmethodist.org.
Supplemental Information
References
- Adra C.N., Donato J.L., Badovinac R., Syed F., Kheraj R., Cai H., Moran C., Kolker M.T., Turner H., Weremowicz S. SMARCAD1, a novel human helicase family-defining member associated with genetic instability: cloning, expression, and mapping to 4q22-q23, a band rich in breakpoints and deletion mutants involved in several human diseases. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Beli P., Lukashchuk N., Wagner S.A., Weinert B.T., Olsen J.V., Baskcomb L., Mann M., Jackson S.P., Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Tsukada Y., Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Chen X., Cui D., Papusha A., Zhang X., Chu C.D., Tang J., Chen K., Pan X., Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Niu H., Yu Y., Wang J., Zhu S., Zhou J., Papusha A., Cui D., Pan X., Kwon Y. Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 2016;44:2742–2753. doi: 10.1093/nar/gkv1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Poot R.A., Kukimoto I., Garcia-Jimenez C., Dellaire G., Varga-Weisz P.D. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- Costelloe T., Louge R., Tomimatsu N., Mukherjee B., Martini E., Khadaroo B., Dubois K., Wiegant W.W., Thierry A., Burma S. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A.K., Li X., Tyler J.K. Epigenetic regulation of genomic integrity. Chromosoma. 2012;121:131–151. doi: 10.1007/s00412-011-0358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham R.M., Garvin A.J., Stone H.R., Strachan J., Baldock R.A., Daza-Martin M., Fletcher A., Blair-Reid S., Beesley J., Johal B. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016;23:647–655. doi: 10.1038/nsmb.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S.Y., Dolan L., Davis R.J., Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Hunt C.R., Hegde M.L., Chakraborty S., Udayakumar D., Horikoshi N., Singh M., Ramnarain D.B., Hittelman W.N., Namjoshi S. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014;8:177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi N., Kumar P., Sharma G.G., Chen M., Hunt C.R., Westover K., Chowdhury S., Pandita T.K. Genome-wide distribution of histone H4 Lysine 16 acetylation sites and their relationship to gene expression. Genome Integr. 2013;4:3. doi: 10.1186/2041-9414-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C.R., Ramnarain D., Horikoshi N., Iyengar P., Pandita R.K., Shay J.W., Pandita T.K. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013;179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S.Y., Obuse C., Nishi R. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017;18:520–532. doi: 10.1016/j.celrep.2016.12.042. [DOI] [PubMed] [Google Scholar]
- Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S.M., Ogiwara H., Kohno T., Kanno S. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lukas J., Lukas C., Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Magnani M., Crinelli R., Bianchi M., Antonelli A. The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-kB (NF-kB) Curr. Drug Targets. 2000;1:387–399. doi: 10.2174/1389450003349056. [DOI] [PubMed] [Google Scholar]
- Manning B.J., Peterson C.L. Releasing the brakes on a chromatin-remodeling enzyme. Nat. Struct. Mol. Biol. 2013;20:5–7. doi: 10.1038/nsmb.2482. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Messick T.E., Greenberg R.A. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Morrison A.J., Shen X. DNA repair in the context of chromatin. Cell Cycle. 2005;4:568–571. [PubMed] [Google Scholar]
- Pandita T.K., Lieberman H.B., Lim D.S., Dhar S., Zheng W., Taya Y., Kastan M.B. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19:1386–1391. doi: 10.1038/sj.onc.1203444. [DOI] [PubMed] [Google Scholar]
- Pandita T.K., Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L., Almouzni G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb. Perspect. Biol. 2013;5:1–12. doi: 10.1101/cshperspect.a012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Jackson S.P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price B.D., D'Andrea A.D. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschella G., Melino G., Malewicz M. New factors in mammalian DNA repair-the chromatin connection. Oncogene. 2017;36:4673–4681. doi: 10.1038/onc.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A., Lafrance M., Gauthier M.C., McDonald D., Hendzel M., West S.C., Jasin M., Masson J.Y. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham S.P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W.R., Webster J., Oxley D. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Schwertman P., Bekker-Jensen S., Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Stock J.K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A.G., Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Taneja N., Zofall M., Balachandran V., Thillainadesan G., Sugiyama T., Wheeler D., Zhou M., Grewal S.I. SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell. 2017;66:50–62 e56. doi: 10.1016/j.molcel.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Staszewski L.M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- Wallis J.W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Witowsky J.A., Johnson G.L. Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J. Biol. Chem. 2003;278:1403–1406. doi: 10.1074/jbc.C200616200. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato J., Shimamoto A., Goto M., Furuichi Y., Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Caron P., Legube G., Paull T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.