Abstract
Intratumoral heterogeneity is among the greatest challenges in precision cancer therapy. However, developments in high-throughput single-cell RNA-sequencing may now provide the statistical power to dissect the diverse cellular populations of tumors. In the future, these technologies might inform selection of targeted combination therapies and enrollment criteria for clinical trials.
Precision cancer therapies exploit patient- and tumor-specific molecular features to attack transformed cells or manipulate the tumor microenvironment. However, the diversity of malignant and microenvironmental populations within a single tumor can complicate this approach. While collaborative molecular profiling efforts like The Cancer Genome Atlasi have enhanced our understanding of intertumoral heterogeneity across patients, efforts to dissect intratumoral heterogeneity are at a much earlier stage. Nonetheless, this information is likely crucial to the rational application of targeted therapies. The target of a given drug may only be present in a subset of cells, and even if the target is widely expressed, its role in driving a malignant phenotype may be limited to a specific subpopulation. For a given tumor type, there could be numerous potential therapies or combinations which could target, either by design or through a side-effect, both the malignant cells and the tumor microenvironment. While these complexities are widely appreciated, the tools to assess them comprehensively are just now becoming available.
The development of deep sequencing led to rapid advances in gene expression profiling by RNA sequencing (RNA-Seq). In its simplest form, RNA-Seq involves conversion of mRNA into a library of amplified cDNA fragments, which are then sequenced and quantified to obtain an expression profile. Soon after the introduction of RNA-Seq, high-sensitivity protocols for profiling individual cells were reported(1), and until recently, the scalability of these methods was limited to 10s–100s of cells per sample (Panel 1A). However, tumors are composed of numerous cellular populations. In addition to malignantly transformed cells, tumor microenvironments may contain diverse infiltrating immune populations (lymphocytes, myeloid cells, dendritic cells, etc.), cell types involved in the tumor’s blood supply, and other stromal populations. Furthermore, the malignant population may contain multiple genomic subclones superimposed on an aberrant developmental process, resulting in both immature, stem-like subpopulations and multiple lineages of more differentiated cells. Finally, malignant cells can occupy diverse physiological states resulting from stress (hypoxia, DNA damage, starvation), quiescence, or cell cycle stage. Acquiring the statistical power to identify and describe these complex populations requires profiling multiple cells of the same type in the same state. Thus, substantial throughput is needed to comprehensively dissect the phenotypic structure of a tumor. While the true throughput required to profile a given tumor is indeterminate, technologies that can sequence tens or even hundreds of individual cells per sample are likely insufficient.
While the majority of single-cell RNA-Seq (scRNA-Seq) studies in cancer have employed low-throughput methods (Panel 1A), these efforts already reveal the enormous potential of the approach. Pioneering work in glioblastoma demonstrated the ability to distinguish malignantly transformed tumor cells from untransformed cells in the tumor microenvironment by analyzing genetic alterations inferred from scRNA-Seq(2). This study revealed novel insights into the developmental and immunological programs of gene expression underlying glioblastoma. Building on this work, an impressive study in melanoma used scRNA-Seq to discover varying proportions of cells harboring drug-susceptible and drug-resistant phenotypes across patients(3). The authors used single-cell profiling to infer interactions between malignant cells and T cells, and identified expression patterns that correlate with T cell infiltration. More recently, a study in colorectal cancer classified individual tumor cells by their lineage resemblance to a large database of cell- and tissue-specific profiles, leading to new expression signatures that are predictive of prognosis(4). Despite this exciting progress, the experimental methods employed in these studies are relatively expensive, limiting the number of cells that can be profiled. Given the complex phenotypic landscapes of individual tumors and the potential clinical importance of rare populations, we believe that deploying new tools capable of inexpensive profiling from thousands of individual cells will be crucial to future clinical applications.
Recently, researchers have combined microfluidic tools with DNA barcoding to develop highly scalable platforms for genome-wide scRNA-Seq (Panel 1B)(5–7). These advances have made profiling thousands of individual cells from a single specimen routine and cost-effective. For example, we regularly use a system from our laboratory(8) to profile surgical tumor specimens with library preparation costs of ~5 cents per cell, overall cost of ~$1,400 per tumor including sequencing, and throughput of ~5,000 cells. High-throughput scRNA-Seq technologies use scalable microfluidics to co-encapsulate individual cells with single beads in microscale chambers, which may be microfabricated wells(7–9) or aqueous-oil emulsion droplets(5,6,10)(Panel 1B–C). The beads harbor oligonucleotides that are 3′-terminated with oligo(dT) for hybrid capture of polyadenylated mRNAs, and 5′-terminated with a universal adapter for PCR. Importantly, oligonucleotides contain a barcode sequence on the 5′-end of the oligo(dT) terminus that is unique to each bead. Therefore, after lysis and hybridization of the liberated mRNA from each cell to the oligonucleotides on the co-encapsulated bead, reverse transcription results in incorporation of a bead- (and therefore cell-) specific barcode sequence into the 5′-end of each cDNA molecule. In some cases, beads are intentionally chemically disrupted during lysis, releasing barcoded primers into solution(6,10). Parallel incorporation of cell-specific barcodes into cDNA from thousands of individual cells allows pooling of many cDNA libraries for downstream processing in a single vessel, dramatically reducing costs. Deep sequencing is then used to quantify gene expression and read out the barcodes that deconvolve the expression profiles of individual cells. These methods have reduced per-cell library preparation costs to a few cents, and because relatively few reads are required to saturate the resulting libraries (~50,000–100,000 reads per cell), per-cell sequencing costs are typically <$0.50. As a result, the number of cells that can be cost-effectively profiled across a cohort and within a single tumor sample is increased dramatically. Higher throughput not only increases statistical power to detect rare subpopulations, but also might allow researchers to analyze cellular population structure and interactions within a single tumor.
scRNA-Seq in tissues requires cellular dissociation, which is perturbative and disrupts valuable spatial information. Solid tumors generally contain multiple microenvironments with highly variable composition, and transformed cells in these complex niches could face disparate selection pressures that influence therapeutic response. Direct quantification of individual RNA molecules in cells can be achieved in intact tissue by fluorescence in situ hybridization (FISH). Until recently, RNA-FISH was limited to parallel quantification of a handful of genes (Figure 1D), but new approaches involving sequential hybridization and combinatorial barcoding have allowed profiling of >100 genes in complex tissues (Panel 1E)(11,12). We anticipate that these exciting new techniques will be complementary to scRNA-Seq in cancer, providing direct molecular characterization of tumor microenvironments and invaluable tools for targeted validation.
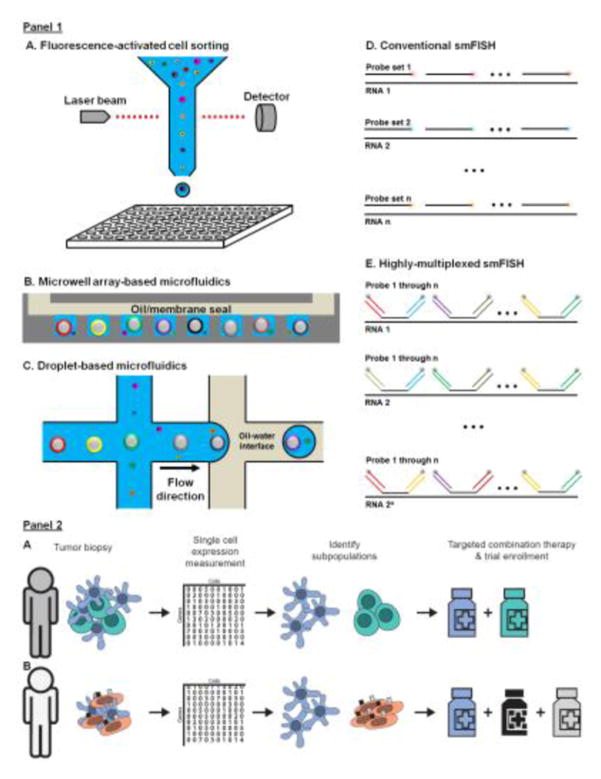
Figure 1 Key Figure. Single-cell transcriptomics technologies and applications in precision medicine.
Panel 1: technologies: A) In conventional scRNA-Seq, individual cells are flow sorted into multi-well plates. RNA-Seq libraries of individual cells are prepared, barcoded in separate wells (per cell reaction volume: ~ 10 μL) and pooled together for next generation sequencing. B) In high-throughput scRNA-Seq with microfabricated microwell arrays, individual cells are co-encapsulated with individual uniquely barcoded mRNA capture beads in physically-isolated microwells (per cell reaction volume: ~ 100 pL). Reverse transcription of bead-captured mRNA molecules results in the incorporation of a bead-specific barcode to each cDNA molecule. The barcoded cDNA molecules from all cells are then pooled together and converted into a single RNA-Seq library. C) High-throughput scRNA-Seq with droplet-based microfluidics is similar to B) except that the co-encapsulation occurs in droplets (per cell reaction volume: ~ 1 nL). D) In conventional RNA-FISH, multiple singly-labeled fluorescent probes are hybridized to transcripts encoding a specific target gene in situ. The fluorescent probes for each transcript share the same color. The number of transcript species that can be measured scales linearly with the number of imaging cycles/channels. E) In highly multiplexed RNA-FISH, probes against multiple genes are distinguished by unique combinations of secondary probes and read out by sequential hybridization. The number of transcript species that can be measured scales exponentially with the number of imaging cycles/channels.
Panel 2: applications Simplified illustrations of the potential application of single-cell transcriptomics in precision medicine: A) identification of distinct malignant subpopulations that could be effectively targeted by different drugs in combination and B) identification of distinct malignant subpopulations or cell types co-expressing targets in multiple redundant pathways that could be targeted by different drugs in combination.
How might single-cell transcriptomics fit into precision oncology? These new technologies could allow clinicians to determine the co-expression pattern of potential drug targets within both cells and tumors to inform selection of targeted therapies. Single-cell expression profiling can reveal whether a target is pervasively expressed or restricted to a rare subpopulation, and if potential targets for combination therapy are expressed in redundant pathways or separate subpopulations. While this may seem simple, these intratumoral features are rarely rigorously evaluated when enrolling patients in clinical trials. Co-expression analysis could be particularly useful for assigning combination therapies that target multiple cell types or pathways as highlighted in Panel 2. Finally, this type of analysis could prove valuable for assessing drug response, as one can directly determine whether specific subpopulation has been ablated or altered by treatment in comparison to an untreated specimen. As studies of this type mature and identify key markers of drug response, more targeted approaches to single-cell expression profiling such as the in situ methods described above could become sufficient. In sum, we believe that large-scale, single-cell transcriptomics will significantly impact the development and implementation of molecular enrollment criteria for clinical trials in cancer and evaluation of therapeutic response at the molecular level.
Footnotes
Resources
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 2.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49(5):708–18. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 5.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bose S, Wan Z, Carr A, Rizvi AH, Vieira G, Pe’er D, et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol. 2015;16(1):120. doi: 10.1186/s13059-015-0684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Sims PA. An Automated Microwell Platform for Large-Scale Single Cell RNA-Seq. Sci Rep. 2016;6:33883. doi: 10.1038/srep33883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gierahn TM, Wadsworth MH, 2nd, Hughes TK, Bryson BD, Butler A, Satija R, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14(4):395–8. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11(4):360–1. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348(6233):aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]