Abstract
Gametophytic development in Arabidopsis depends on nutrients and cell wall materials from sporophytic cells. However, it is not clear whether hormones and signaling molecules from sporophytic tissues are also required for gametophytic development. Herein, we show that auxin produced by the flavin monooxygenases YUC2 and YUC6 in the sporophytic microsporocytes is essential for early stages of pollen development. The first asymmetric mitotic division (PMI) of haploid microspores is the earliest event in male gametophyte development. Microspore development in yuc2yuc6 double mutants arrests before PMI and consequently yuc2yuc6 fail to produce viable pollens. Our genetic analyses reveal that YUC2 and YUC6 act as sporophytic genes for pollen formation. We further show that ectopic production of auxin in tapetum, which provides nutrients for pollen development, fails to rescue the sterile phenotypes of yuc2yuc6. In contrast, production of auxin in either microsporocytes or microspores rescued the defects of pollen development in yuc2yuc6 double mutants. Our results demonstrate that local auxin biosynthesis in sporophytic microsporocytic cells and microspore controls male gametophyte development during the generation transition from sporophyte to male gametophyte.
Author summary
Plant life cycle alternates between the diploid sporophyte generation and the haploid gametophyte generation. Understanding the molecular mechanisms governing the generation alternation impacts fundamental plant biology and plant breeding. It is known that the development of haploid generation in vascular plants requires the diploid tapetum cells to supply nutrients. Here we show that the male gametophyte (haploid) development in Arabidopsis requires auxin produced in the diploid microsporocytic cells. Moreover, we show that auxin produced in microsporocytic cells and microspore is also sufficient to support normal development of the haploid microspores. This work demonstrates that Arabidopsis uses two different diploid cell types to supply growth hormone and nutrients for the growth of the haploid generation.
Introduction
Life cycle of eukaryotes alternates between haploid and diploid generations. The alternation of generations is initiated by meiosis (2n to 1n) and gamete fusion (1n to 2n) [1]. In land plants, the multicellular diploid generation is called sporophyte, whereas the multicellular haploid organism is named gametophyte. In bryophytes (mosses and liverworts), haploid gametophyte is the dominant generation and represents the main plant. In vascular plants, including ferns, gymnosperms, and angiosperms, the diploid sporophyte generation is dominant, whereas the gametophyte generation is much reduced [1]. For example, in seed plants, both the female and male gametophytes develop within the sporophyte. Understanding the molecular mechanisms governing the generation alternation will impact fundamental plant biology and plant breeding.
Pollen grains, which are the male gametophyte in seed plants, are developed in locules encircled by four sporophytic cell layers: tapetum, middle layer, endothecium, and epidermis. Inside a locule, a diploid male meiocyte divides into a tetrad of four haploid microspores after meiosis [2, 3]. Each microspore then undergoes an asymmetric cell division (pollen mitosis I (PMI)), resulting in two structurally and functionally different daughter cells: the small generative cell and the large vegetative cell. The generative cell divides one more time (PMII) to produce two sperm cells whereas the vegetative cell no longer divides. The mature pollen grain contains two haploid sperm cells and one haploid vegetative cell [3, 4].
Genetic analyses have identified a number of genes that play important roles in pollen development [5]. These genes can be classified into two categories: gametophytic or sporophytic genes. Pollen development depends on coordinated expression of both sporophytic genes and gametophytic genes [3]. Most of the identified gametophytic genes are related to cell division and differentiation. For example, MOR1, a member of the microtubule-associated protein 215 (MAP215), is involved in the PMI asymmetric cell division [6, 7]. In the mor1/gem1 mutants, defects in microspore nucleus migration lead to altered division plane, and the formation of two equal- or similar sized cells [6, 7]. Genes including Two-in-one (TIO), GAMMA-TUBULIN 1 (TUBG1) and 2 essential for phragmoplast formation, localization and/or expansion, also affect male gametophyte development. Mutations in these genes cause incomplete cytokinesis in PMI and result in failure to produce the generative cell [8–13]. Several cell cycle factors have also been reported to be important for pollen mitosis. ICK4/KRP6 (Interactors of Cdc2 kinase 4/kip-related protein 6) is a cyclin-dependent kinase inhibitor. Timely degradation of ICK6/KRP6 by RHF1a/2a and SCFFBL17 E3 ligase is essential for the progression of pollen mitosis [14–16]. Cyclin-dependent kinase D (CDKD) was recently found to be essential for pollen mitosis. In the cdkd;1–1 cdkd;3–1 double mutants, microspore is defective in both PMI and PMII [17]. A main feature of the male gametophytic genes is that no viable mutant pollens can be generated and that no homozygous diploid mutants are available.
Sporophytic genes for pollen formation represent the contribution of sporophytic tissues including tapetum and microsporocyte for male gametophyte development. Tapetum directly provides pollen wall materials and nutrients including Magnesium for pollen development [18–22]. A series of transcription factors including DYT1, TDF1, AMS, MYB33/65, MS188/MYB80/MYB103, and MS1 have been shown to play essential roles in normal development of tapetum [23–32]. The defective tapetum caused by mutations in these genes results in pollen wall defects and leads to complete pollen abortion [23–32]. Enzymes involved in outer pollen wall formation are also expressed in tapetum and are essential for pollen development [33–41]. The pollen wall pattern is determined inside a tetrad that depends on the sporophytic genes expressed in microsporocytes, such as RPG1, NPU, NEF and DEX1 [42–45]. The sporophytic cells including tapetum and microsporocyte supply material and nutrients for pollen development and determine the pollen wall pattern. Unlike gametophytic genes, viable mutant pollen can be produced from heterozygous mutant plants, and homozygous mutant plants can be obtained. However, homozygous diploid mutants cannot produce viable pollens.
Plant hormones are essential for normal plant development. However, it is not clear whether plant hormones or other signaling molecules produced in sporophytic tissues are required for the development of male gametophyte. The plant hormone auxin plays critical roles in nearly all aspects of plant development including embryogenesis, organogenesis, gametophyte development, and various tropisms [46]. It is known that disruption of either auxin biosynthesis, or polar transport, or signaling causes defects in male gametophyte development and pollen formation. Indole-3-acetic acid (IAA), the primary natural auxin in plants, is mainly synthesized through a TAA/YUC two-step tryptophan-dependent pathway [47–51]. Simultaneously disruption of both YUC2 and YUC6 completely eliminates the production of viable pollen grains [49]. It was reported that the two atypical members of the PIN auxin efflux carriers, PIN5 and PIN8, which are believed to regulate intracellular auxin homeostasis and metabolism in pollen, also participate in the development of normal pollen morphology [52, 53]. Other auxin transporters including ATP-BINDING CASSETTE B19 (ABCB19)/MULTIDRUG RESISTANCE PROTEIN 1 (MDR1)/ P-GLYCOPROTEIN 19 (PGP19) and ABCB1/ PGP1 also play important roles in pollen development [54, 55]. The abcb1 abcb19 double mutants show precocious pollen maturation [54, 55]. Similar precocious pollen maturation takes place in tir1 afb2 afb3 triple and tir1 afb1 afb2 afb3 quadruple mutants, which are defective in auxin perception [54]. It is known that auxin biosynthetic, transport, and signaling genes are expressed during pollen development [49, 54].
Previous studies have clearly demonstrated that auxin is required for anther development and pollen formation [49, 52–55], but it was not understood the role of auxin for pollen development and in the transition from sporophytic generation to gametophyte generation. It was previously proposed that auxin produced in tapetum is required for pollen development [56]. Here we report that pollen development in the auxin biosynthetic mutants yuc2yuc6 failed to progress past the PMI, which is an early step in male gametophyte growth. Our genetic analyses demonstrated that both YUC2 and YUC6 function as sporophytic genes. We further show that auxin produced in sporophytic microsporocytes rather than tapetum is required for the early stages of pollen development, demonstrating that auxin produced in the diploid sporophytic cells plays a critical role in the haploid male gametophyte development.
Results
YUC2 and YUC6 are the main auxin biosynthetic flavin monooxygenases in anther
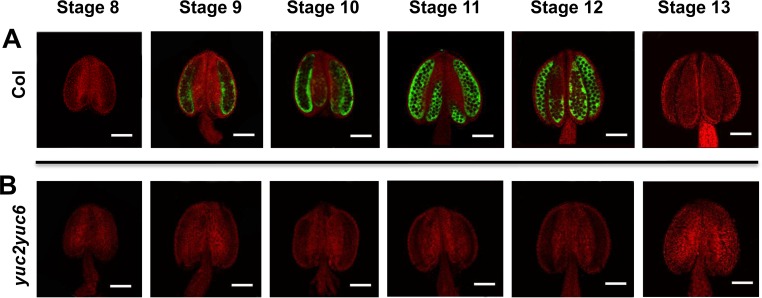
We previously showed that yuc2yuc6 double mutants were male sterile, and the expression of the bacterial auxin biosynthetic gene iaaM under the control of the YUC6 promoter fully restored the fertility of yuc2yuc6, indicating that the fertility defects of yuc2yuc6 were caused by partial auxin deficiency during anther development [49]. The auxin reporter DR5-GFP/GUS has been previously detected in anther from flower stages 10 to 12 [54, 57, 58]. It has also been reported that the DR5 activity is decreased in the yuc6 single mutant, although yuc6 did not display obvious reproductive defect [58]. To better understand the distribution patterns of DR5 in anthers of yuc2yuc6, we introduced the auxin reporter DR5-GFP into yuc2yuc6 plants and compared the GFP signals in yuc2yuc6 with those in WT plants. Consistent with previous findings [54, 57, 58], GFP signals in WT plants were detected in anthers from anther stages 9 to 12 (all the stages refer to anther stages in our results) (Fig 1A). The expression pattern of DR5-GFP in yuc2 and yuc6 anthers was similar to that in WT (S1 Fig). However, no substantial signals were detected in yuc2yuc6 anthers at the same developmental stages (Fig 1B). We also introduced the DR5-GUS into the yuc2yuc6 background. Similar to the DR5-GFP patterns, GUS signals were not detected in the yuc2yuc6 anthers (S1 Fig). Therefore, it appears that YUC2 and YUC6 are the main auxin biosynthetic genes responsible for auxin production during anther development.
Fig 1. The expression patterns of DR5:GFP in the anther of yuc2yuc6.
(A-B) Comparison of the auxin reporter DR5:GFP signal in anthers of wild type (A) and yuc2yuc6 (B) at different developmental stages. Reporter signal in anthers observed in wild type from stages 9 to 12 is completely absent in yuc2yuc6. Bars = 100 μm.
Expression of auxin reporters in unicellular microspores
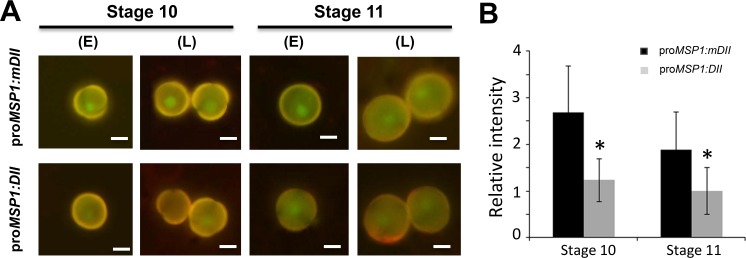
Because we hardly observed any DR5-GFP signal in pollen, we used DII and mDII auxin reporter lines as a proxy to determine the pattern of auxin-induced degradation of Aux/IAA repressors during pollen development [59]. We replaced the 35S promoter with the microspore-specific promoter proMSP1 to drive the DII-VENUS and mDII-VENUS expression units [60]. Our results showed that fluorescence signals in microspores/pollens of proMSP1:DII-VENUS transgenic plants were weaker than that in proMSP1:mDII-VENUS transgenic plants at stage 10 (proMSP1:DII-VENUS/proMSP1:mDII-VENUS = 0.45) and at stage 11 (proMSP1:DII-VENUS/proMSP1:mDII-VENUS = 0.54)) (Fig 2A and 2B). The results of the auxin response reporters are indicative that auxin accumulated significantly in unicellular microspores and bicellular pollens.
Fig 2. DII-VENUS for visualizing auxin-induced Aux/IAA degradation during pollen development.
(A) Fluorescence of proMSP1:mDII control and proMSP1:DII plants. MSP1 promoter (proMSP1) is a microspore-specific promoter. E: early, L: late. (B) Quantitative representation of the relative VENUS fluorescence intensity from nucleus of stage 10 and stage 11 pollen. The fluorescence intensity was measured using ImageJ software. Data shown in B represent mean with SD based on more than 20 pollen grains for each group (set as 1 for stage 11 pollen from proMSP1:DII). The symbol * indicates where the difference between proMSP1:DII and proMSP1:mDII at corresponding stage is significant in statistic test (p-value<0.05). Note that the fluorescence signals of DII in stage 10 and stage 11 were much weaker than that in mDII microspores. Bars = 7 μm.
To investigate whether signals of the auxin response reporters are correlated with the expression patterns of YUC2 and YUC6, we generated proYUC2-GFP and proYUC6-GFP transgenic plants. We found that both YUC2 and YUC6 were weakly expressed in microsporocytes, tetrad and microspores at stage 9 (S2A, S2B, S2C, S2I, S2J and S2O Fig) and strongly expressed in microspores from stages 10 to 13 (S2D–S2G and S2K–S2N Fig). We also found that YUC2 and YUC6 were strongly expressed in somatic cell layers including tapetum cells in anther (S2H, S2I and S2O Fig).
Early stages including pollen mitosis I (PMI) are defective in yuc2yuc6
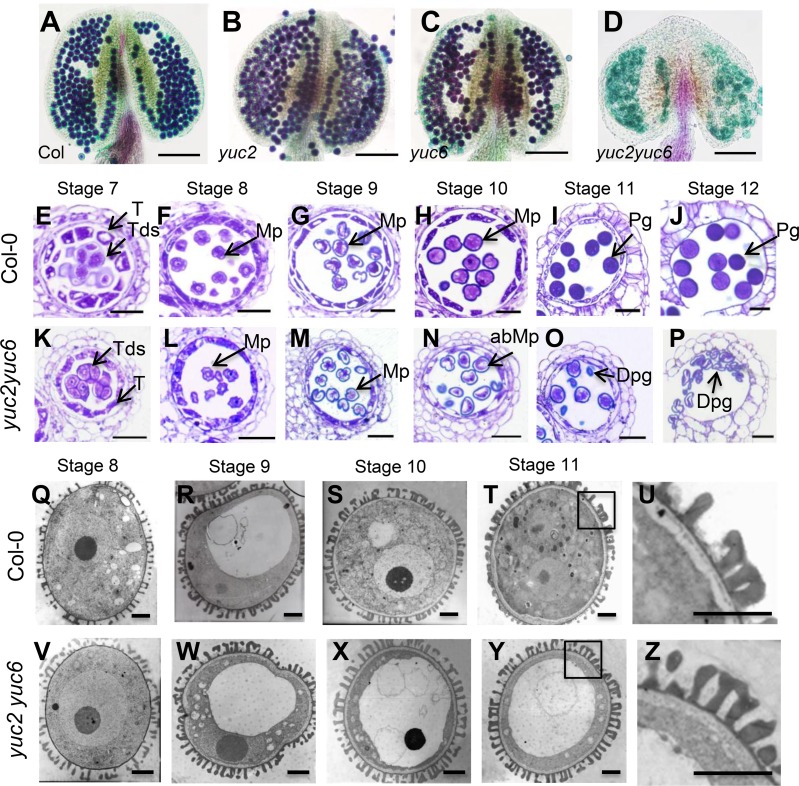
The yuc2yuc6 double mutants showed markedly reduced fertility with few viable pollen inside the locule [49, 54]. Pollination of the yuc2yuc6 pistil with WT pollen resulted in F1 plants with normal fertility (~50 seeds produced in each silique, n = 4), indicating that the female fertility of yuc2yuc6 was unaffected. Alexander staining was performed to understand the defects of pollen development in yuc2yuc6. Similar to those in wild type (WT), the anthers of yuc2 and yuc6 single mutants contained purple-stained viable pollen grains (Fig 3A–3C). Consistent with previous reports [49], Fig 3D showed that the yuc2yuc6 anthers did not produce viable pollens (Fig 3D). We then generated anther cross sections and used transmission electron microscopy to determine in which stage(s) the anther and pollen developmental defects took place in yuc2yuc6. We noticed that a normal tetrad was produced in yuc2yuc6 (Fig 3E and 3K), suggesting that the meiotic division progressed normally. After release from the tetrad, microspore development in yuc2yuc6 appeared similar to that of WT from stages 8 to 9 (Fig 3F, 3G, 3L, 3M, 3Q, 3R, 3V and 3W). However, at stage 10, WT microspores became vacuolated and contained a polarized nucleus (Fig 3H and 3S), whereas yuc2yuc6 microspores had an irregular shape and started to degenerate (Fig 3N and 3X). From stages 11 to 12, WT microspores underwent the first mitotic division and gradually developed into mature pollen (Fig 3I, 3J and 3T). In yuc2yuc6, microspores were severely degenerated and failed to form normal pollen (Fig 3O, 3P and 3Y). The pollen wall of yuc2yuc6 appeared normal as compared with that of WT (Fig 3U and 3Z).
Fig 3. The yuc2yuc6 double mutants were defective in pollen development.
(A-D) Alexander staining of anthers of wild type (A), yuc2 (B), yuc6 (C) and yuc2yuc6 (D). Note that yuc2yuc6 lacked viable pollen. (E-P) Semi-thin cross sections of anthers from wild type (E-J) and yuc2yuc6 (K-P) were stained with toluidine blue. Anthers at (E and K) stage 7; (F and L) stage 8; (G and M) stage 9; (H and N) stage 10; (I and O) stage 11; (J and P) stage 12. A microspore or pollen showing reduced or completely invisible of cytoplasm we defined as degeneration state. Note that the degenerated microspores were evident in yuc2yuc6 from stage 10 to 12. (Q-Z) Ultra-structures of pollen grains from wild type (Q-U) and yuc2yuc6 (V-Z) plants. Microspores from anthers at stage 8 (Q and V); stage 9 (R and W); stage 10 (S and X); stage 11 (T and Y) were shown. Enlarged images from T and Y were shown in (U and Z). It is clear that the pollen wall pattern in yuc2yuc6 appears similar to that in WT. Most of the cytoplasm is degenerated in the microspore from stage 10 and 11 anthers of yuc2yuc6 (X and Y). T, Tapetum; Tds, tetrads; Mp, microspore; abMp, abnormal microspore; Pg, pollen grain; Dpg, degenerated pollen grain. Bars = 100μm (A-D), 20 μm (E-P), 1 μm (Q-Z).
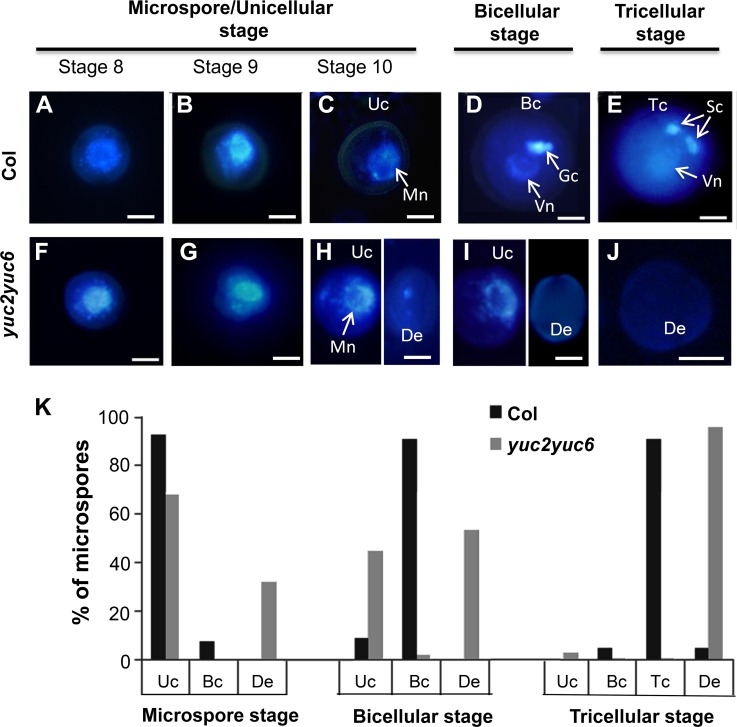
To obtain further insight into the microgametogenesis defects of yuc2yuc6, we stained nuclei with 4’, 6-diamidino-2-phenylindole (DAPI) in developing pollen. The microspores of yuc2yuc6 were similar to WT microspores at stages 8 to 9 (Fig 4A, 4B, 4F and 4G). However, at stage 10, some of the yuc2yuc6 microspores were degenerated, with little DAPI staining signal (Fig 4H right). At this stage, some normal yuc2yuc6 microspores with a nucleus located at one side of the microspore were still visible (Fig 4C and 4H left). Overall during the unicellular stage (from stages 8 to 10), most of the single haploid cells in both the WT (92.7%) and yuc2yuc6 (67.9%) showed a bright nucleus (Fig 4A–4C, 4F–4H and 4K), and about 30% of the microspores in yuc2yuc6 were degenerated (Fig 4H right, 4K). After PMI, 91.1% of the WT microspores divided asymmetrically, producing a large vegetative cell and a small generative cell engulfed in the cytoplasm of the vegetative cell (Fig 4D and 4K), which is called the bicellular stage. In contrast, only about 2.2% of the yuc2yuc6 microspores progressed past PMI to reach the bicellular stage (Fig 4K). Approximately 44.8% of the microspores in yuc2yuc6 remain arrested at the unicellular stage (Fig 4I left, 4K), and the rest (53%) became degenerated (Fig 4I right, 4K). At the tricellular stage, WT microspores fully developed into tricellular pollen, whereas yuc2yuc6 microspores (96%) became completely degenerated or still contained a very loosely packed DNA mass (Fig 4E, 4J and 4K). Therefore, we conclude that mutations in YUC2 and YUC6 lead to the defects in early stages including PMI of pollen development.
Fig 4. The early stages of male gametogenesis were defective in the yuc2yuc6 double mutants.
(A-J) DAPI staining of pollens from wild type (A-E) and yuc2yuc6 (F-J) at different developmental stages. Microspores from anthers at (A and F) stage 8; (B and G) stage 9; (C and H) stage 10 were shown. Some of the yuc2yuc6 microspores were degenerated and had little DAPI staining (H right). (D and I) pollens at the bicellular stage. Note that the nuclei of yuc2yuc6 were arrested at the unicellular state or degenerated and could not form two nuclei as in the wild type (I). (E and J) Pollen at the tricellular stage. The yuc2yuc6 pollen grains lacked genomic DNA (J) while WT pollen showed strong DAPI staining (E). (K) Quantitative analysis and comparison of pollen defects between wild type and yuc2yuc6 (n>600 for each stage). Uc, Unicellular pollen; Mn, microspore nucleus; Gc, generative cell; Vn, vegetative nucleus; Sc, sperm cell. Bc, Bicellular pollen; Tc, Tricellular pollen; De, Degenerated pollen. Bars = 5 μm.
We performed genetic complementation to determine whether mutations in the YUC genes are responsible for the sterile phenotype of yuc2yuc6. The DNA fragment including about 2 kb upstream sequence of YUC2 and the YUC2 open reading frame (ORF) was fused with GFP (named proYUC2:YUC2-GFP) and the construct was introduced into yuc2-/- yuc6+/- plants (S3 Fig). Two independent lines of yuc2yuc6 that contained the proYUC2:YUC2-GFP transgene were identified in T1 generation. Both lines of proYUC2:YUC2-GFP in the yuc2yuc6 background had normal fertility (S3B Fig). Alexander staining and DAPI staining results indicated that YUC2 could rescue the pollen development defects of yuc2yuc6 (S3C and S3D Fig and S5 Fig). The results also demonstrated that the YUC2-GFP fusion is functional.
Auxin produced in sporophytic cells controls gametophyte development
From our phenotypic analysis, it was clear that the yuc2yuc6 double mutants were defective in gametophyte development. Gametophyte development depends on the expression of both sporophytic and gametophytic genes. To determine whether YUC2 and YUC6 function as sporophytic or gametophytic genes, we analyzed the male transmission efficiency of yuc2yuc6. We used yuc2-/-yuc6+/- and yuc2+/-yuc6-/- plants as pollen donors to cross with WT plants. PCR genotyping revealed that approximately 50% of F1 progeny contained the yuc2yuc6 mutations (Table 1), suggesting that yuc2yuc6 microspores were transmitted normally. We next analyzed the segregation ratio of self-fertilized yuc2-/-yuc6+/-and yuc2+/-yuc6-/- plants. Consistent with the normal transmission efficiency, the segregation displayed a typical Mendelian ratio (3:1) in both cases (n>290 for each case). These results showed that YUC2 and YUC6 are sporophytic genes for pollen development, indicating that auxin produced in the sporophytic tissues by the YUCs is required for normal male gametophyte development.
Table 1. Effects of mutations in YUC2 and YUC6 on male transmission frequency.
| Parental Genotype (Male × Female) | Progeny | total | TE | χ2 | |||
|---|---|---|---|---|---|---|---|
| yuc2 +/- | yuc2 +/+ | yuc6 +/- | yuc6 +/+ | ||||
| yuc2 -/- yuc6 +/- × WT | 172 | 84 | 88 | 172 | 95.50% | 0.093 (p>0.05) | |
| yuc2 +/- yuc6 -/- × WT | 91 | 86 | 177 | 177 | 105.80% | 0.141 (p>0.05) | |
Transmission efficiencies (TE) = number of progenies with T-DNA insertion/number of progenies without T-DNA insertion. Expected values were based on the prediction that if the double mutant alleles were transmitted normally, about 50% of the progeny should receive the yuc2yuc6 knockout allele from the yuc2-/- yuc6+/- or yuc2-/+ yuc6-/- parents.
Ectopic expression of YUC2 in tapetum did not rescue the defects of pollen development in yuc2yuc6
It is known that YUC2/6 mRNA is expressed in meiocytes, microspores, tapetum, middle layer, and endothecium in anthers [54]. To investigate the source of auxin for pollen development, we used various promoters to drive the expression of YUC2-GFP fusion, which we have shown functional (S3 Fig). The DR5 auxin reporter line showed an extremely active auxin response in the tapetum cells during late developmental stages in Arabidopsis [54, 56, 58]. It was proposed that auxin is transported from tapetum cells into developing pollens [56]. To investigate whether the YUC genes expressed in tapetum are responsible for regulating microspore development and pollen formation, we generated transgenic lines that express YUC2-GFP in tapetum cells using specific tapetum promoters (proA9 and proATA7 (ARABIDOPSIS THALIANA ANTHER7)) in the yuc2yuc6 background [32, 61, 62] (Fig 5 and S4 Fig). We found that both proA9: YUC2-GFP (yuc2yuc6) (n = 6) and proATA7:YUC2-GFP (yuc2yuc6) (n = 7) T1 transgenic plants were still sterile (Fig 5B and S4 Fig). The pollen defects in yuc2yuc6 were not rescued by the YUC2-GFP transgene (Fig 5C, S4 and S5 Figs). We investigated whether the tapetum-specific promoters behaved as designed. RNA in situ hybridization data showed that YUC2-GFP was significantly transcribed in tapetum cells from microsporocytes stage to early microspore stage in proA9:YUC2-GFP (yuc2yuc6) plants (Fig 5D). The GFP signal in proA9:YUC2-GFP (yuc2yuc6) plants appeared in the tapetum layer at stages 8 and 9 (Fig 5E showed stage 9). Although the GFP fluorescence of proATA7:YUC2-GFP (yuc2yuc6) plants could not be detected (S4 Fig), the YUC2-GFP transcripts were detected in tapetum layer at stage 8 (S4 Fig). To rule out the possibility that the ATA7 promoter was too weak to drive adequate expression of YUC2-GFP in anther, we used real-time quantitative RT-PCR to analyze the transcript levels of YUC2 in WT and YUC2-GFP in the transgenic plants. The expression levels of YUC2-GFP in all of the analyzed transgenic plants were similar to or higher than YUC2 expression in WT (S5 Fig). These results suggest that the ATA7 and A9 promoters were able to drive YUC2-GFP expression in tapetum, but auxin production in tapetum is not sufficient to overcome the auxin deficiency in yuc2yuc6 microspores.
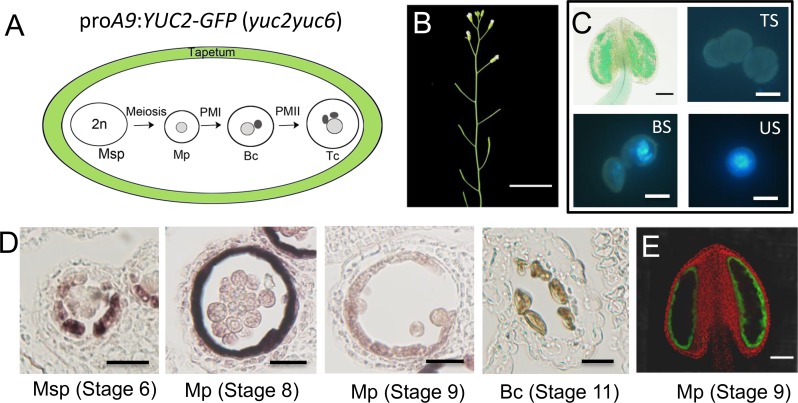
Fig 5. Localized auxin production in tapetum did not rescue the pollen defects in yuc2yuc6 mutants.
(A) The green color indicates the expression pattern of the A9 promoter. Msp, Microsporocytes; Mp, microspores; Bc, Bicellular pollen; Tc, Tricellular pollen. The A9 promoter cloned from Arabidopsis is used to drive YUC2-GFP expression specifically in tapetum cells from stages 6 to 9 [61, 62]. (B) Morphology of adult shoots (Bars = 2cm). (C) Alexander staining (Bars = 100 μm) and DAPI staining (Bars = 10 μm) of ProA9:YUC2-GFP (yuc2yuc6) anthers and pollens. Note that the sterility phenotype and pollen defects were not rescued in ProA9:YUC2-GFP (yuc2yuc6) transgenic plants. TS, Tricellular Stage; BS, Bicellular Stage; US; Unicellular Stage. (D) In situ hybridization of GFP in ProA9:YUC2-GFP (yuc2yuc6) transgenic plants (Bars = 20 μm). (E) Fluorescence images (Bars = 100 μm) of the YUC2-GFP fusion protein in anthers from yuc2yuc6 transformed with ProA9:YUC2-GFP. In ProA9: YUC2-GFP anther, GFP is significantly expressed in tapetum cell of microsporocyte stage and early microspore stage.
Expression of YUC2-GFP in microsporocyte and microspores rescued the defects of pollen development in yuc2yuc6
We next used promoters specific for microsporocytes and microspores (proARF17) [63] to drive the expression of YUC2-GFP in yuc2yuc6 plants (Fig 6). All of the 5 independent T1 proARF17:YUC2-GFP (yuc2yuc6) lines showed almost complete rescue of the sterile phenotypes of yuc2yuc6 (Fig 6A). In addition, the pollen defects were fully rescued in the transgenic plants (Fig 6A and S5 Fig). In the transgenic plants, YUC2-GFP expression was observed in microsporocytes and was significant in early stages of microspores (Fig 6A). Fig 6A also showed that the YUC2-GFP protein accumulated in early stages of microspores. Because ARF17 promoter drives gene expression in both microsporocytes and microspores, we further tested whether localized auxin biosynthesis in microspores is sufficient to rescue yuc2yuc6. We used the promoter proLAT52, which is specifically activated in male gametophyte [14, 64], to drive the expression of YUC2-GFP in yuc2yuc6 plants (Fig 6B). All of the 6 independent T1 proLat52:YUC2-GFP (yuc2yuc6) lines showed partial rescue of the fertility defects of yuc2yuc6 (Fig 6B). The proLat52:YUC2-GFP (yuc2yuc6) plants were fertile at a late reproductive development stage (Fig 6B). During late reproduction development, 30% to 70% of the pollen in the transgenic lines appeared normal in the anthers (Fig 6B and S5 Fig). At bicellular stage, 70.7% of the unicellular pollen can develop into bicellular pollen, and 18% of the microspores are arrested at unicellular stage. At tricellular stage, proLat52:YUC2-GFP (yuc2yuc6) produced 56.9% tricellular pollens, and 31.8% pollens were aborted (Fig 6B and S5 Fig). These results indicated that proLat52:YUC2-GFP could partially support the microspores development past PMI and PMII. In the proLat52:YUC2-GFP (yuc2yuc6) transgenic plants, YUC2-GFP was transcribed from late stages of microspores (Fig 6B). Meanwhile, YUC2-GFP protein was detected in pollen at stage 13 (Fig 6B). The observation that auxin produced in microspores partially rescued the sterile phenotype of yuc2yuc6 suggested that early stages of pollen development including PMI of unicellular microspores require auxin. The sterility rescue efficiencies of proARF17:YUC2-GFP (yuc2yuc6) and proYUC2:YUC2-GFP (yuc2yuc6) (S3 Fig) were significantly higher than that in proLat52:YUC2-GFP (yuc2yuc6) transgenic plants. We noticed that the expression of YUC2-GFP or GFP was at earlier stage of microspores in proARF17:YUC2-GFP (yuc2yuc6) and in proYUC2:GFP (S2 Fig) than that in proLat52:YUC2-GFP (yuc2yuc6) transgenic plants (Fig 6). Therefore, it is likely that auxin may be required at early phase of microspore development. Combined with the genetic data that YUC2 and YUC6 act as sporophytic genes, we conclude that the auxin synthesized in the sporophytic microsporocytes is essential for early stages of pollen development in plants. Therefore, auxin produced in sporophyte contributes to male gametophyte during the generation alternation in plant.
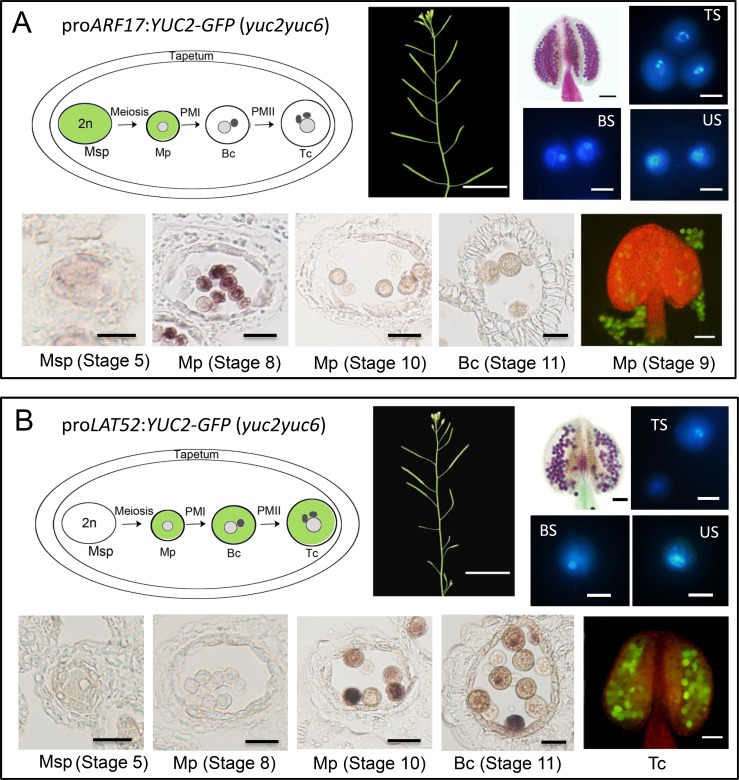
Fig 6. Localized auxin production in microsporocytes and microspores rescued the pollen defects in yuc2yuc6 mutants.
The green color indicates the expression pattern of the ARF17 (A) and LAT52 (B) promoter. The ARF17 promoter is known to be active in microsporocytes and microspores [63]. LAT52 is expressed in microspore and gametophyte specifically. Msp, Microsporocytes; Mp, microspores; Bc, Bicellular pollen; Tc, Tricellular pollen. Morphology of adult shoots (Bars = 2 cm), Alexander staining (Bars = 100 μm) of anthers and DAPI staining (Bars = 10 μm) of pollens from ProARF17:YUC2-GFP (yuc2yuc6) (A) and ProLAT52:YUC2-GFP (yuc2yuc6) (B) transgenic plants. Note that YUC2-GFP expression driven by ARF17 and LAT52 promoters are sufficient to rescue the yuc2yuc6 defects. In situ hybridization of GFP (Bars = 20 μm) and fluorescence images (Bars = 100 μm) of the YUC2-GFP fusion protein in anthers from yuc2yuc6 transformed with ProARF17:YUC2-GFP (A) and ProLAT52:YUC2-GFP (B). In ProARF17:YUC2-GFP (yuc2yuc6) anther, GFP is expressed in microsporocytes and in microspores at early stage (A). In ProLAT52:YUC2-GFP (yuc2yuc6) anther, GFP is significantly expressed in microspores at late stage and in bicellular pollens (B). TS, Tricellular Stage; BS, Bicellular Stage; US; Unicellular Stage.
Discussion
Both YUC2 and YUC6 are known required for pollen development and the yuc2yuc6 double mutants are male sterile. Here we further defined that the male sterility of yuc2yuc6 is caused by defects in early stages of pollen development including the first mitotic cell division (PMI) of microspores. Moreover, we show that early stages of microspore development and PMI require auxin produced in the diploid sporophytic microsporocytes, indicating that sporophytic cells not only provide nutrients and cell wall materials for pollen development, but also supply hormone and signaling molecules to haploid cells. Our results also demonstrate that different sporophytic cells play different roles in male gametophytic development. Tapetum cells provide nutrients, but auxin produced in tapetum cells is not sufficient to support early stages of pollen development. In contrast, auxin synthesized in sporophytic microsporocytes is necessary and sufficient for male gametophytic development.
Because yuc2yuc6 double mutants could undergo meiosis successfully and our results showed that supply of auxin after meiosis could partially rescue the pollen defects of yuc2yuc6 (Fig 6, ProLat52:YUC2-GFP (yuc2yuc6)), we conclude that auxin produced by YUC2 and YUC6 may not be required for male meiosis. Following meiosis, the microspore undergoes two rounds of mitosis to form mature pollen during anther development. PMI is the first round of mitosis for pollen formation. Several lines of evidence show that auxin produced by YUCs is essential for PMI. We showed that YUC2 and YUC6 are the main enzymes responsible for auxin synthesis in anther based on the expression of the auxin reporter DR5:GFP/GUS (Fig 1 and S1 Fig). Our results are consistent with previous studies showing DR5 activity in anthers possibly resulting from auxin synthesis [54]. Secondly, we found that the microspores of yuc2yuc6 were aborted and degenerated before PMI and failed to form mature pollen (Fig 3 and Fig 4). Consistent with this phenotype, we found significant AUX/IAA degradation, which is presumably induced by auxin, in unicellular microspores judging from the expression of the modified DII-Venus reporter (Fig 2). Lastly, we revealed that YUC2-GFP expressed in microspores at early gametophyte stages could partially rescue the yuc2yuc6 pollen defect (Fig 6, ProLat52:YUC2-GFP (yuc2yuc6)). These results demonstrated that auxin is required for early stages of pollen development. Because most of the microspores in yuc2yuc6 were arrested prior to PMI, we could not exclude the possibility that auxin is also required in subsequent steps such as PMII. Auxin regulates plant development mainly through auxin response factors (ARFs). Among the ARF genes, ARF6, ARF8, ARF10, ARF16, and ARF17 are expressed in unicellular microspores [65]. The arf17 mutant shows defective pollen formation [63], suggesting that auxin synthesized by YUCs may control male gametophyte development through ARF17. However, we noticed that pollen development defects in arf17 and yuc2yuc6 were not the same. The arf17 mutants displayed defective pollen coat formation whereas it was not the case for yuc2yuc6. Therefore, we believe that ARF17 may have unique functions in the earlier stages of pollen development that do not overlap with those of YUC2 and YUC6, such as controlling of the morphogenesis of pollen wall.
It was reported that pollen development in tir1 afb1 afb2 afb3 quadruple mutants was not significantly compromised, an observation that was quite different from the severe pollen defects found in yuc2yuc6. It is puzzling that disruption of auxin signaling and auxin biosynthesis resulted in different phenotypes. It was known that some of the tir1 afb1 afb2 afb3 plants can survive and produce seeds, but auxin transport mutants such as pin1 and auxin biosynthesis mutants including yuc1yuc4 were completely sterile [49, 66]. It is likely that other AFB proteins such as AFB4 and AFB5 may compensate the loss of tir1 afb1 afb2 afb3. In addition, we noticed that afb1 and afb3 in the quadruple mutants still produced truncated transcripts and might have produced residual protein activities [67]. The fact that tir1 afb1 afb2 afb3 were not a complete null in auxin receptor may account for the observed paradoxical results. AFB5 is expressed in bicellular pollens (S1 Table). Analysis of higher order tir1 afb mutants may clarify why auxin signaling mutants behaved differently from auxin biosynthesis and transport mutants.
Most of the known functions of auxin in flowering plants are related to sporophyte generation, which is the dominant generation. The auxin pathways are evolutionary conserved in the plant kingdom. The homologs of auxin biosynthetic genes such as TAAs and YUCs are also used for auxin biosynthesis in moss Physcomitrella patens and Liverwort Marchantia polymorpha [68, 69], whose haploid gametophyte is the dominant generation. In liverwort, auxin synthesized by YUCs is essential for normal gametophyte development and dormancy [69], implying that auxin may also play essential roles in gametophyte generation in flowering plants. However, it is difficult to study the roles of auxin in male gametophyte development because male gametophyte has been reduced to several cells within the diploid sporophyte. Our detailed analysis of the auxin biosynthetic mutants yuc2yuc6 revealed that auxin is required for male gametophyte development.
More importantly, we show that auxin produced in microsporocytes, which are sporophytic cells, is necessary for normal progression of the haploid microspores. Our genetic data suggest that YUC2 and YUC6 affect early stages of pollen development mainly via a sporophytic effect, implying that auxin required for pollen development may be synthesized by YUC2 and YUC6 in sporophytic anther tissues. These data are consistent with the transcription patterns of YUC2 and YUC6 in microsporocytes, microspores and anther somatic cell layers at premeiotic and meiotic stages [54]. In anther, tapetum cells and microsporocytes are closely related sporophyte cells involved in microspore/male gametophyte development. It was proposed that auxin could be transported to developing pollen from tapetum cells [56]. However, the expression of YUC2 in the tapetum cell of yuc2yuc6 could not lead to viable pollen (Fig 5 and S4 Fig), suggesting that the auxin from tapetum is not sufficient to support pollen development. On the other hand, the expression of YUC2-GFP in microsporocytes completely rescued the sterility phenotypes of yuc2yuc6 (Fig 6), demonstrating that auxin synthesized in microsporocytes is sufficient for pollen development.
We propose that auxin production in diploid microsporocytes is necessary and sufficient for the early stages of the development of the haploid cells during pollen development. Our conclusion is mainly based on two observations: 1) YUC2 and YUC6 are sporophytic genes and the yuc2yuc6 fail to produce viable pollen; 2) expression of YUC2 driven by specific promoters rescued yuc2yuc6 (Fig 6). The ARF17 promoter is active in both microsporocytes (sporophytic) and microspores (gametophytic) (Fig 6). Therefore, it is conceivable that the rescued yuc2yuc6 phenotypes by YUC2-GFP driven by ARF17 promoter were caused by ectopic auxin production in the haploid microspores. However, from our genetic analysis, it is clear that viable and fully functional pollen that lacks YUC2 and YUC6 can be produced from yuc2-/-yuc6+/- or yuc2+/-yuc6-/-, demonstrating that auxin does not have to be produced in microspores for the development of viable pollen. Rather auxin produced by YUCs in sporophytic cells (microsporocytes to be exact) is sufficient to guide the progressive development of microspores.
Microsporocytes developing into mature pollen through PMI and PMII is a continuing process. Production of auxin in microsporocytes is an efficient way to regulate newly formed microspores to undergo PMI and other processes of early male gametophyte development. Using two different diploid cell types (tapetum and microsporocytes) to provide nutrients and hormonal signals for male gametophyte development may also be advantageous in terms of efficiency and specificity.
Material and methods
Plant materials and growth conditions
All plants used in this study were in the Columbia-0 genetic background. The yuc2yuc6 mutants have been described previously [49]. All relevant primer sequences were listed in S2 Table. The DII-VENUS (N799173) and mDII-VENUS (N799174) seeds were ordered from the European Arabidopsis Stock Centre (NASC). Plants were grown under long-day conditions (16 hr light/8 hr dark) in a ~22°C growth room. The DR5:GFP and DR5:GUS reporter lines were introduced into yuc2yuc6 by genetic cross.
Microscopy
Plants were photographed using a Nikon digital camera (D-7000). Alexander solution was prepared as previously described [70]. Anthers were dissected and immersed in Alexander solution for 0.5 hr, and images were obtained under a microscope with an Olympus BX51 digital camera (Olympus, Japan). Plant materials for the semi-thin sections were prepared and embedded in Spurr resin as described in [25] and cut into 1-μm thick sections, stained with toluidine blue, then photographed with an Olympus BX51 digital camera. For transmission electron microscopic analysis, the same-stage anthers of wild type (WT) and yuc2yuc6 were fixed and embedded as previously described [25]. Green fluorescent protein (GFP) fluorescence in ProARF17:YUC2-GFP (yuc2yuc6), ProLAT52:YUC2-GFP (yuc2yuc6), ProMSP1:DII, and ProMSP1:mDII transgenic plants was detected under a fluorescence microscope (Olympus BX51). GFP fluorescence in ProA9:YUC2-GFP (yuc2yuc6) transgenic plants and WT or yuc2yuc6 expressing DR5:GFP was detected by confocal laser scanning microscopy (Carl Zeiss, LSM 5 PASCAL).
Plasmid construction and plant transformation
To generate YUC2-GFP, we amplified YUC2 cDNA without the stop codon from inflorescence mRNA by standard RT-PCR (see S2 Table for primers) for cloning into the modified pCAMBIA1300 binary vector (CAMBIA, Australia), pCAMBIA1300-GFP. The resulting construct was named pCAMBIA1300-YUC2-GFP. The promoter sequences of YUC2, YUC6, ARF17, ATA7 and A9 were PCR-amplified from genomic DNA of WT Col-0. The promoter sequence of LAT52 was PCR-amplified from genomic DNA of tomato. The primers for amplification were listed in S2 Table. The amplified sequences were first inserted into pMD19-T (Takara) for sequence verification, then subcloned into the vector pCAMBIA1300-YUC2-GFP for plant transformation. The promoter sequences for YUC2 and YUC6 were subcloned into pCAMBIA1300-GFP for plant transformation. The promoter sequence for MSP1 was PCR-amplified from genomic DNA of WT Col-0, then subcloned into the vector pCAMBIA1300 to obtain pCAMBIA1300-proMSP1. The DII-VENUS and mDII-VENUS sequences were amplified from genomic DNA from DII-VENUS and mDII-VENUS plants. The amplified sequences were inserted into pMD19-T (Takara) for verification, then subcloned into the vector pCAMBIA1300-proMSP1 for plant transformation. For plant transformation, all plasmids were introduced into the Agrobacterium strain GV3101 and transformed into plants by the floral dip method[71]. For tissue specific rescue experiments, all of the constructs were transformed into the offspring of yuc2-/-yuc6+/-. After collected the T1 seeds, we first selected transgenic plants by growing on MS media containing hygromycin. We then genotyped the transgenic plants for yuc2yuc6 double mutants[49].
In situ hybridization
The probe fragments were amplified from plasmid containing GFP (pCAMBIA1300-GFP) with primers GFP-F and GFP-R. The PCR products were cloned into the pBluescriptSK vector and confirmed by sequencing. Plasmid DNA was digested with HindIII or BamHI. The digestion products were used as templates for transcription into sense and antisense probes by T3 and T7 RNA polymerases, respectively (Roche). Oligonucleotide sequences of GFP-F and GFP-R are provided in S2 Table. Images were taken using the Olympus BX-51 microscope.
RNA extraction and real-time RT-PCR
Total RNA was extracted from the inflorescences of T1 transgenic plants by the Trizol method (Invitrogen, USA) following the manufacturer’s instructions. Quantitative real-time PCR involved an ABI PRISM 7300 detection system (Applied Biosystems, USA) with SYBR Green I master mix (Toyobo, Japan). Relevant primer sequences are in S2 Table. β-Tubulin was as a constitutive expression control. Three biological repeats were used for gene expression analysis.
Supporting information
(A) Expression of the DR5:GFP auxin-responsive reporter in yuc2 and yuc6 (Bars = 100 μm). (B) Expression of the DR5:GUS auxin-responsive reporter in wild type, yuc2, yuc6 and yuc2yuc6 flowers. Note that the GUS staining signal of DR5:GUS disappeared in the yuc2yuc6 mutant.
(TIF)
Fluorescence images of the proYUC2:GFP (A-G) and proYUC6:GFP (H-N). In situ hybridization of GFP in proYUC2:GFP transgenic plants (O). Bars = 50 μm for the anther in I. Bars = 10 μm for all the other images.
(TIF)
(A) Construct of the ProYUC2:YUC2-GFP plasmid. N, nopaline synthase terminator. (B) Morphology of adult shoots (Bars = 2cm) from yuc2yuc6 and yuc2yuc6 complemented with a YUC2-GFP under the control of the YUC2 promoter (ProYUC2:YUC2-GFP). The ProYUC2:YUC2-GFP can completely rescue the sterility phenotype of yuc2yuc6. (C and D) Alexander staining (Bars = 100 μm) (C) and DAPI staining (Bars = 10 μm) (D) of ProYUC2:YUC2-GFP (yuc2yuc6) anthers and pollens. The pollen defects are rescued in ProYUC2:YUC2-GFP (yuc2yuc6) plants.
(TIF)
The green color indicates the expression pattern of the ATA7 promoter. Msp, Microsporocytes; Mp, microspores; Bc, Bicellular pollen; Tc, Tricellular pollen. Morphology of adult shoots (Bars = 2cm). Alexander staining (Bars = 100 μm) and DAPI staining (Bars = 10 μm) of ProATA7:YUC2-GFP (yuc2yuc6) anthers and pollens. Note that the sterility phenotype and pollen defects were not rescued in ProATA7:YUC2-GFP (yuc2yuc6) transgenic plants. TS, Tricellular Stage; BS, Bicellular Stage; US; Unicellular Stage. Fluorescence images (Bars = 100 μm) showed that YUC2-GFP fusion protein was not observed in transgenic plants anthers. In situ hybridization of GFP (Bars = 20 μm) showed that GFP is weakly expressed in tapetum cell at stage 8 in ProATA7:YUC2-GFP (yuc2yuc6) anther.
(TIF)
(A)Quantitative analysis of pollen defects of transgenic plants (n>500 for each stage for each type of plants). Uc, Unicellular pollen; Bc, Bicellular pollen; Tc, Tricellular pollen; De, Degenerated pollen. (B) cDNA sample from the inflorescences of wild type Col, yuc2yuc6 and yuc2yuc6 transformed with ProARF17:YUC2-GFP, ProLAT52:YUC2-GFP, ProA9-:YUC2-GFP or ProATA7:YUC2-GFP. Data are mean±SD normalized to TUBULIN and compared with Col from three biological replicates. The transcript levels of YUC2 in all the transgenic plants were equal to or higher than that in Col.
(TIF)
These data are extracted from a published paper [65]. MS, microspores; BCP, bicellular pollen; TCP, tricellular pollen; MPG, mature pollen.
(XLSX)
(XLSX)
Acknowledgments
We thank Professor Xin Zhou for critical reading and revising of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the grants from the National Science Foundation of China (31401030) to XY and (31670314) to Z-NY and NIH grant R01GM114660 to YZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bowman JL, Sakakibara K, Furumizu C, Dierschke T. Evolution in the Cycles of Life. Annual review of genetics. 2016;50:133–54. doi: 10.1146/annurev-genet-120215-035227 [DOI] [PubMed] [Google Scholar]
- 2.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annual review of genetics. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
- 3.Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annual review of plant biology. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717 [DOI] [PubMed] [Google Scholar]
- 4.McCormick S. Control of male gametophyte development. The Plant cell. 2004;16 Suppl:S142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger F, Twell D. Germline specification and function in plants. Annual review of plant biology. 2011;62:461–84. doi: 10.1146/annurev-arplant-042110-103824 [DOI] [PubMed] [Google Scholar]
- 6.Park SK, Howden R, Twell D. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development. 1998;125(19):3789–99. [DOI] [PubMed] [Google Scholar]
- 7.Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, et al. MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nature cell biology. 2002;4(9):711–4. doi: 10.1038/ncb844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh SA, Johnson A, Smertenko A, Rahman D, Park SK, Hussey PJ, et al. A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Current biology: CB. 2005;15(23):2107–11. doi: 10.1016/j.cub.2005.10.044 [DOI] [PubMed] [Google Scholar]
- 9.Pastuglia M, Azimzadeh J, Goussot M, Camilleri C, Belcram K, Evrard JL, et al. Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. The Plant cell. 2006;18(6):1412–25. doi: 10.1105/tpc.105.039644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YR, Li Y, Liu B. Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. The Plant cell. 2007;19(8):2595–605. doi: 10.1105/tpc.107.050716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh SA, Bourdon V, Das 'Pal M, Dickinson H, Twell D. Arabidopsis kinesins HINKEL and TETRASPORE act redundantly to control cell plate expansion during cytokinesis in the male gametophyte. Molecular plant. 2008;1(5):794–9. doi: 10.1093/mp/ssn042 [DOI] [PubMed] [Google Scholar]
- 12.Zeng CJ, Lee YR, Liu B. The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. The Plant cell. 2009;21(4):1129–40. doi: 10.1105/tpc.109.065953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh SA, Allen T, Kim GJ, Sidorova A, Borg M, Park SK, et al. Arabidopsis Fused kinase and the Kinesin-12 subfamily constitute a signalling module required for phragmoplast expansion. The Plant journal: for cell and molecular biology. 2012;72(2):308–19. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Zhang Y, Qin G, Tsuge T, Sakaguchi N, Luo G, et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. The Plant cell. 2008;20(6):1538–54. doi: 10.1105/tpc.108.059741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, et al. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455(7216):1134–7. doi: 10.1038/nature07289 [DOI] [PubMed] [Google Scholar]
- 16.Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, et al. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PloS one. 2009;4(3):e4780 doi: 10.1371/journal.pone.0004780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takatsuka H, Umeda-Hara C, Umeda M. Cyclin-dependent kinase-activating kinases CDKD;1 and CDKD;3 are essential for preserving mitotic activity in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2015;82(6):1004–17. [DOI] [PubMed] [Google Scholar]
- 18.Blackmore S, Wortley AH, Skvarla JJ, Rowley JR. Pollen wall development in flowering plants. The New phytologist. 2007;174(3):483–98. doi: 10.1111/j.1469-8137.2007.02060.x [DOI] [PubMed] [Google Scholar]
- 19.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annual review of plant biology. 2011;62:437–60. doi: 10.1146/annurev-arplant-042809-112312 [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Fan XD. Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant molecular biology. 2013;83(3):165–75. doi: 10.1007/s11103-013-0085-5 [DOI] [PubMed] [Google Scholar]
- 21.Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. The Plant cell. 1993;5:1217–29. doi: 10.1105/tpc.5.10.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu XF, Wang B, Lou Y, Han WJ, Lu JY, Li DD, et al. Magnesium Transporter 5 plays an important role in Mg transport for male gametophyte development in Arabidopsis. The Plant journal: for cell and molecular biology. 2015;84(5):925–36. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen AM, Krober S, Unte US, Huijser P, Dekker K, Saedler H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. The Plant journal: for cell and molecular biology. 2003;33(2):413–23. [DOI] [PubMed] [Google Scholar]
- 24.Higginson T, Li SF, Parish RW. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2003;35(2):177–92. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. The Plant journal: for cell and molecular biology. 2007;52(3):528–38. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J, Zhang G, Chang Y, Li X, Yang J, Huang X, et al. AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Science China Life sciences. 2010;53(9):1112–22. doi: 10.1007/s11427-010-4060-y [DOI] [PubMed] [Google Scholar]
- 27.Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. The Plant journal: for cell and molecular biology. 2001;28(1):27–39. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, Shinozaki K. The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant & cell physiology. 2002;43(11):1285–92. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. The Plant cell. 2007;19(11):3549–62. doi: 10.1105/tpc.107.054536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. The Plant cell. 2007;19(11):3530–48. doi: 10.1105/tpc.107.054981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133(16):3085–95. doi: 10.1242/dev.02463 [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, et al. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. The Plant journal: for cell and molecular biology. 2008;55(2):266–77. [DOI] [PubMed] [Google Scholar]
- 33.Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, et al. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant journal: for cell and molecular biology. 1997;12(3):615–23. [DOI] [PubMed] [Google Scholar]
- 34.Morant M, Jorgensen K, Schaller H, Pinot F, Moller BL, Werck-Reichhart D, et al. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant cell. 2007;19(5):1473–87. doi: 10.1105/tpc.106.045948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, et al. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant cell. 2009;21(2):507–25. doi: 10.1105/tpc.108.062513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, et al. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant physiology. 2009;151(2):574–89. doi: 10.1104/pp.109.144469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, et al. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant physiology. 2010;153(3):937–55. doi: 10.1104/pp.110.157446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza Cde A, et al. Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. The Plant cell. 2010;22(12):4067–83. doi: 10.1105/tpc.110.080036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza Cde A, et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. The Plant cell. 2010;22(12):4045–66. doi: 10.1105/tpc.110.080028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lallemand B, Erhardt M, Heitz T, Legrand M. Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant physiology. 2013;162(2):616–25. doi: 10.1104/pp.112.213124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quilichini TD, Grienenberger E, Douglas CJ. The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry. 2015;113:170–82. doi: 10.1016/j.phytochem.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 42.Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant physiology. 2001;127(4):1739–49. [PMC free article] [PubMed] [Google Scholar]
- 43.Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, et al. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 2004;39(2):170–81. [DOI] [PubMed] [Google Scholar]
- 44.Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant physiology. 2008;147(2):852–63. doi: 10.1104/pp.108.118026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HS, Zhang C, Chang YH, Zhu J, Xu XF, Shi ZH, et al. No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant physiology. 2012;158(1):264–72. doi: 10.1104/pp.111.184853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136(6):1005–16. doi: 10.1016/j.cell.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y. Auxin biosynthesis and its role in plant development. Annual review of plant biology. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291(5502):306–9. doi: 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & development. 2006;20(13):1790–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(45):18512–7. doi: 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(45):18518–23. doi: 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dal Bosco C, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. The Plant journal: for cell and molecular biology. 2012;71(5):860–70. [DOI] [PubMed] [Google Scholar]
- 53.Ding Z, Wang B, Moreno I, Duplakova N, Simon S, Carraro N, et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nature communications. 2012;3:941 doi: 10.1038/ncomms1941 [DOI] [PubMed] [Google Scholar]
- 54.Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. The Plant cell. 2008;20(7):1760–74. doi: 10.1105/tpc.107.057570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cecchetti V, Brunetti P, Napoli N, Fattorini L, Altamura MM, Costantino P, et al. ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. Journal of integrative plant biology. 2015;57(12):1089–98. doi: 10.1111/jipb.12332 [DOI] [PubMed] [Google Scholar]
- 56.Aloni R, Aloni E, Langhans M, Ullrich CI. Role of auxin in regulating Arabidopsis flower development. Planta. 2006;223(2):315–28. doi: 10.1007/s00425-005-0088-9 [DOI] [PubMed] [Google Scholar]
- 57.Feng XL, Ni WM, Elge S, Mueller-Roeber B, Xu ZH, Xue HW. Auxin flow in anther filaments is critical for pollen grain development through regulating pollen mitosis. Plant molecular biology. 2006;61(1–2):215–26. doi: 10.1007/s11103-006-0005-z [DOI] [PubMed] [Google Scholar]
- 58.Cecchetti V, Celebrin D, Napoli N, Ghelli R, Brunetti P, Costantino P, et al. An auxin maximum in the middle layer controls stamen development and pollen maturation in Arabidopsis. The New phytologist. 2016. [DOI] [PubMed] [Google Scholar]
- 59.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482(7383):103–6. doi: 10.1038/nature10791 [DOI] [PubMed] [Google Scholar]
- 60.Honys D, Oh SA, Renak D, Donders M, Solcova B, Johnson JA, et al. Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC plant biology. 2006;6:31 doi: 10.1186/1471-2229-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paul W, Hodge R, Smartt S, Draper J, Scott R. The isolation and characterisation of the tapetum-specific Arabidopsis thaliana A9 gene. Plant molecular biology. 1992;19(4):611–22. [DOI] [PubMed] [Google Scholar]
- 62.Feng X, Dickinson HG. Tapetal cell fate, lineage and proliferation in the Arabidopsis anther. Development. 2010;137(14):2409–16. doi: 10.1242/dev.049320 [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Tian L, Sun MX, Huang XY, Zhu J, Guan YF, et al. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant physiology. 2013;162(2):720–31. doi: 10.1104/pp.113.214940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Twell D, Yamaguchi J, McCormick S. Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development. 1990;109:705–13. [DOI] [PubMed] [Google Scholar]
- 65.Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome biology. 2004;5(11):R85 doi: 10.1186/gb-2004-5-11-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. The Plant cell. 1991;3(7):677–84. doi: 10.1105/tpc.3.7.677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, et al. Plant development is regulated by a family of auxin receptor F box proteins. Developmental cell. 2005;9(1):109–19. doi: 10.1016/j.devcel.2005.05.014 [DOI] [PubMed] [Google Scholar]
- 68.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319(5859):64–9. doi: 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- 69.Eklund DM, Ishizaki K, Flores-Sandoval E, Kikuchi S, Takebayashi Y, Tsukamoto S, et al. Auxin Produced by the Indole-3-Pyruvic Acid Pathway Regulates Development and Gemmae Dormancy in the Liverwort Marchantia polymorpha. The Plant cell. 2015;27(6):1650–69. doi: 10.1105/tpc.15.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexander MP. Differential staining of aborted and nonaborted pollen. Stain technology. 1969;44(3):117–22. [DOI] [PubMed] [Google Scholar]
- 71.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant journal: for cell and molecular biology. 1998;16(6):735–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Expression of the DR5:GFP auxin-responsive reporter in yuc2 and yuc6 (Bars = 100 μm). (B) Expression of the DR5:GUS auxin-responsive reporter in wild type, yuc2, yuc6 and yuc2yuc6 flowers. Note that the GUS staining signal of DR5:GUS disappeared in the yuc2yuc6 mutant.
(TIF)
Fluorescence images of the proYUC2:GFP (A-G) and proYUC6:GFP (H-N). In situ hybridization of GFP in proYUC2:GFP transgenic plants (O). Bars = 50 μm for the anther in I. Bars = 10 μm for all the other images.
(TIF)
(A) Construct of the ProYUC2:YUC2-GFP plasmid. N, nopaline synthase terminator. (B) Morphology of adult shoots (Bars = 2cm) from yuc2yuc6 and yuc2yuc6 complemented with a YUC2-GFP under the control of the YUC2 promoter (ProYUC2:YUC2-GFP). The ProYUC2:YUC2-GFP can completely rescue the sterility phenotype of yuc2yuc6. (C and D) Alexander staining (Bars = 100 μm) (C) and DAPI staining (Bars = 10 μm) (D) of ProYUC2:YUC2-GFP (yuc2yuc6) anthers and pollens. The pollen defects are rescued in ProYUC2:YUC2-GFP (yuc2yuc6) plants.
(TIF)
The green color indicates the expression pattern of the ATA7 promoter. Msp, Microsporocytes; Mp, microspores; Bc, Bicellular pollen; Tc, Tricellular pollen. Morphology of adult shoots (Bars = 2cm). Alexander staining (Bars = 100 μm) and DAPI staining (Bars = 10 μm) of ProATA7:YUC2-GFP (yuc2yuc6) anthers and pollens. Note that the sterility phenotype and pollen defects were not rescued in ProATA7:YUC2-GFP (yuc2yuc6) transgenic plants. TS, Tricellular Stage; BS, Bicellular Stage; US; Unicellular Stage. Fluorescence images (Bars = 100 μm) showed that YUC2-GFP fusion protein was not observed in transgenic plants anthers. In situ hybridization of GFP (Bars = 20 μm) showed that GFP is weakly expressed in tapetum cell at stage 8 in ProATA7:YUC2-GFP (yuc2yuc6) anther.
(TIF)
(A)Quantitative analysis of pollen defects of transgenic plants (n>500 for each stage for each type of plants). Uc, Unicellular pollen; Bc, Bicellular pollen; Tc, Tricellular pollen; De, Degenerated pollen. (B) cDNA sample from the inflorescences of wild type Col, yuc2yuc6 and yuc2yuc6 transformed with ProARF17:YUC2-GFP, ProLAT52:YUC2-GFP, ProA9-:YUC2-GFP or ProATA7:YUC2-GFP. Data are mean±SD normalized to TUBULIN and compared with Col from three biological replicates. The transcript levels of YUC2 in all the transgenic plants were equal to or higher than that in Col.
(TIF)
These data are extracted from a published paper [65]. MS, microspores; BCP, bicellular pollen; TCP, tricellular pollen; MPG, mature pollen.
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.