Abstract
Syndromes of hybrid dysgenesis (HD) have been critical for our understanding of the transgenerational maintenance of genome stability by piRNA. HD in D. virilis represents a special case of HD since it includes simultaneous mobilization of a set of TEs that belong to different classes. The standard explanation for HD is that eggs of the responder strains lack an abundant pool of piRNAs corresponding to the asymmetric TE families transmitted solely by sperm. However, there are several strains of D. virilis that lack asymmetric TEs, but exhibit a “neutral” cytotype that confers resistance to HD. To characterize the mechanism of resistance to HD, we performed a comparative analysis of the landscape of ovarian small RNAs in strains that vary in their resistance to HD mediated sterility. We demonstrate that resistance to HD cannot be solely explained by a maternal piRNA pool that matches the assemblage of TEs that likely cause HD. In support of this, we have witnessed a cytotype shift from neutral (N) to susceptible (M) in a strain devoid of all major TEs implicated in HD. This shift occurred in the absence of significant change in TE copy number and expression of piRNAs homologous to asymmetric TEs. Instead, this shift is associated with a change in the chromatin profile of repeat sequences unlikely to be causative of paternal induction. Overall, our data suggest that resistance to TE-mediated sterility during HD may be achieved by mechanisms that are distinct from the canonical syndromes of HD.
Author summary
Transposable elements (TE) can proliferate in genomes even if harmful. In response, mechanisms of small-RNA silencing have evolved to repress germline TE activity. Syndromes of hybrid dysgenesis in Drosophila—where unregulated TE activity in the germline causes sterility—have also revealed that maternal piRNAs play a critical role in maintaining TE control across generations. However, a syndrome of hybrid dysgenesis in D. virilis has identified additional complexity in the causes of hybrid dysgenesis. By surveying factors that modulate hybrid dysgenesis in D. virilis, we show that protection against sterility cannot be entirely explained by piRNAs that control known inducer TEs. Instead, spontaneous changes in the chromatin state of repeat sequences of the mother may also contribute to protection against sterility.
Introduction
Transposable elements are selfish elements that have the capacity to proliferate in genomes even if they are harmful [1]. In response to this threat, mechanisms of small-RNA based silencing have evolved to limit TE proliferation. In the germline of animals, Piwi-interacting RNAs (piRNAs) function to maintain TE repression through both transcriptional and post-transcriptional silencing [2]. Critically, the epigenetic and transgenerational nature of piRNA-mediated TE control has been revealed by syndromes of hybrid dysgenesis (HD) [3,4]. HD is a syndrome of TE-mediated sterility that occurs when males carrying active copies of TEs are crossed with females where such copies are rare or absent [5–7].
The hybrid dysgenesis syndrome (HD) is defined as a combination of various genetic disorders such as genic mutations and chromosomal aberrations that lead to sterility in the progeny of intraspecific crosses [5–7]. Sterility during HD is mediated by mobilization of certain TE families carried by the paternal genome and absent in the maternal genome [6,7]. To date, there are several independent HD systems in Drosophila melanogaster. The most well described are the I-R and P-M systems, controlled by the I-element (a non-LTR (long terminal repeat) retrotransposon) and the P-element (a DNA transposon), respectively [6–8]. Activation of paternally inherited TEs is explained by the fact that only the female maintains transgenerational TE repression via piRNAs transmitted through maternal deposition. When the female genome lacks certain TE families, female gametes also lack piRNAs that target these families. Thus, TE families solely transmitted through the male germline become de-repressed in the absence of repressive piRNAs inherited from the mother [2–4,9].
HD in D. virilis was initially observed when males of laboratory strain 160 and females of wild-type strain 9 were crossed. The F1 progeny exhibited up to 60% sterility, while sterility in the progeny of reciprocal crosses did not exceed 5–7% [10]. Similar to the D. melanogaster P-M system, the sterility of hybrids from dysgenic crosses is apparently the result of abnormal development (atrophy) of male and female gonads [10–12]. By analogy with the P-M system, strain 160 and strain 9 were called “P-like” (P) and “M-like” (M), respectively.
In contrast to I-R and P-M systems, the study of HD in D. virilis has demonstrated that multiple unrelated TEs belonging to different families are mobilized in dysgenic progeny [13–16]. The TEs presumably causal of dysgenesis and absent in M-like strain 9 include Penelope (a representative of the Penelope-like element (PLE) superfamily), Paris and Polyphemus (DNA transposons), as well as a non-LTR retrotransposon Helena [13–16]. A typical M-like strain 9 contains only diverged inactive remnants of these TEs. Additionally, piRNAs targeting Penelope, Paris, Polyphemus and Helena are highly abundant in the germline of strain 160 and are practically absent in strain 9 [17,18]. Thus, it has been suggested that the combined activity of these four asymmetric TEs, present only in strain 160, underlies gonadal atrophy and other manifestations of HD in D. virilis. This large asymmetry in TE abundance between strains suggests that HD in D. virilis may be considered a model for understanding the consequences of intermediate divergence in TE profiles within a species.
Nonetheless, recent studies have called into question whether the standard model of HD–described in D. melanogaster where sterility is caused by the absence of maternal piRNAs that target specific inducing TE families—applies in D. virilis [3,4,18,19]. This is because several “neutral” (N) strains exhibit “immunity” to HD in dysgenic crosses but lack maternal piRNA corresponding to Penelope elements, the presumptive primary driver of dysgenesis [19]. If Penelope is a key driver of dysgenesis, how do neutral strains exhibit "immunity" in the absence of maternally transmitted Penelope piRNA? Two fundamental issues arise. First, as observed in D. melanogaster, is there a single major element that serves as a key driver of HD in D. virilis? Second, do N-strains confer their resistance to HD solely through maternally provisioned piRNA or through alternate mechanisms? Despite significant progress in understanding the morphogenetic events occurring during gametogenesis and embryogenesis in the progeny of D. virilis dysgenic crosses, these questions still need to be answered [11,18].
To answer these questions, by using small RNA deep-sequencing and qPCR, we decided to perform a comparative survey of maternal piRNA profiles across several “neutral” strains of different origin that did not quite fit the HD paradigm developed in the previous studies of this phenomenon [3,4,9,19]. Additionally, we developed transgenic strains containing a presumptive causative TE and did not detect a cytotype change after its propagation in the genome. The accumulated data failed to pinpoint a single TE or specific set of TEs responsible for their “immunity” and support a model in which resistance to TE-mediated sterility during dysgenesis may be achieved by a mechanism that varies across strains. We thus propose an alternate model to explain resistance to TE mediated sterility in D. virilis. Instead of solely being explained by maternal piRNAs that target inducing TE families, the chromatin profile of repeats in the maternal genome may confer general immunity to the harmful effects of TE mobilization.
Results and discussion
A survey of neutral strain piRNA profiles reveals that there is no common pattern of maternal piRNAs derived from asymmetric TEs
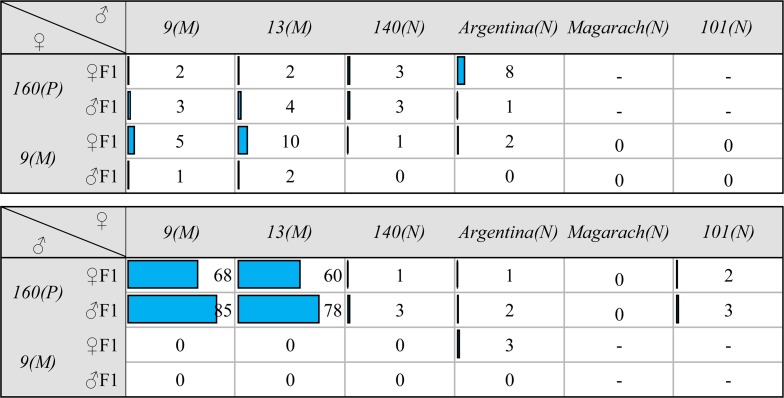
To characterize the piRNA profiles across diverse strains that vary in resistance to HD, we performed small RNA sequencing on six D. virilis strains obtained from various sources (see Materials and Methods) and maintained in our laboratory for more than 20 years. These strains exhibit different levels of gonadal atrophy when crossed with males of P-like strain 160. Two of them (9 and 13) represent strong M-strains (they exhibit up to 65% of gonadal atrophy in the F1 progeny of the dysgenic cross) and four (140, Argentina, Magarach and 101) behave as “neutral” or N-strains when crossed with strain 160 males and, hence, did not exhibit gonadal atrophy (less than 10% atrophied gonads) in such crosses (Fig 1).
Fig 1. The frequency (in %) of gonadal atrophy in the progeny of dysgenic and reciprocal crosses among the studied M-like and neutral strains.
The number of examined individuals was ~100–130 separately for females and males.
Previous studies suggest Penelope element as a key driver of HD in D. virilis [15,20,21]. However, while N-strains 140 and Argentina both carry Penelope elements, two other N-strains–Magarach and 101 contain neither functional Penelope copies nor Penelope-derived small RNAs [19]. This observation questions the key role of Penelope as a factor determining HD in D. virilis and suggests that piRNAs targeting other asymmetric TEs, e.g. Polyphemus, Helena and possibly Paris, may provide immunity to HD [14,15,17,21,22]. To explore this possibility we performed a comparative analysis of both classes of small RNAs (piRNAs and siRNAs) in the ovaries of all selected M- and N-strains using the extended list of TEs and other repeats recently defined in D. virilis genome [18].
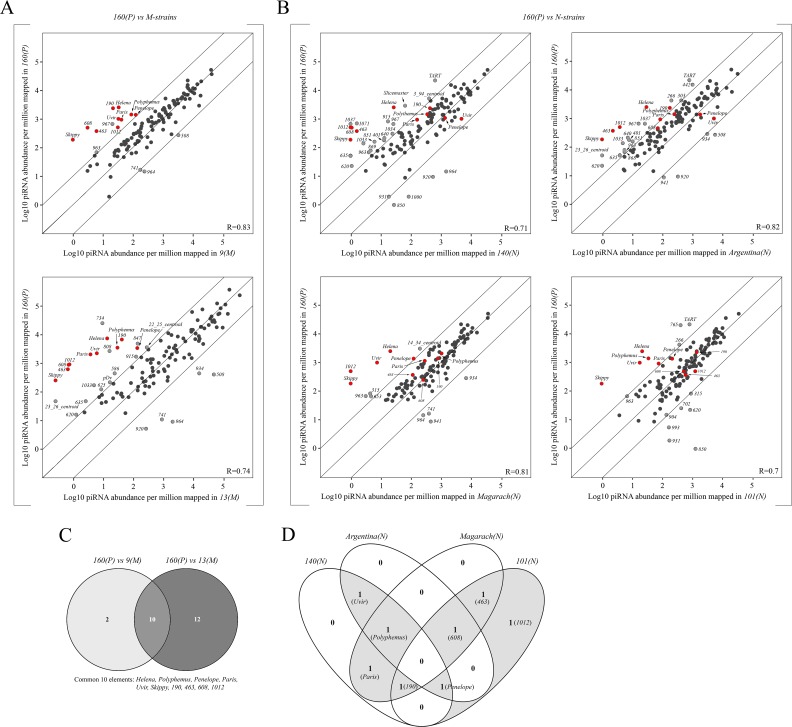
This analysis indicates that the total repertoire of targets for small RNA silencing in strain 160(P) is significantly higher than in all other studied strains (Figs 2A, 2B, S1A and S1B). Surprisingly, the global piRNA profile for known D. virilis TEs and other repeats is more similar between strain 160(P) and M-strains (R(160:9) = 0.83; R(160:13) = 0.74, Spearman’s correlation coefficient) than between strain 160(P) and several N-strains (R(160:140) = 0.71; R(160:101) = 0.7) (Fig 2A and 2B). This suggests the possibility that protection is not mediated by a general maternal piRNA profile, but rather to certain specific TEs yet to be identified. To identify such candidates, we compared sets of piRNA targets distinguishing strain 160(P) from both typical M-strains, 9 and 13, and obtained a list of ten TEs in common across comparisons (Fig 2C). These are TEs for which piRNAs are more abundant in strain 160(P) when compared to both M-strains: Polyphemus, Penelope, Paris, Helena, Uvir, Skippy, 190, 463, 608, and 1012. However, comparing 160(P) and N-strains, we find that piRNAs from Helena and Skippy are uniquely found at high levels in strain 160(P). Thus, if neutrality is conferred by piRNAs that uniformly target the same TE family or families, Helena and Skippy piRNAs are not likely to be required to prevent HD. However, among the eight remaining candidates, there is no shared family among the neutral strains (N-strains and 160(P)) that have a piRNA profile that is similar across strains. For example, in contrast to 160(P), Penelope-derived piRNAs are more lowly expressed in strain Magarach(N), Polyphemus-targeted piRNAs are more lowly expressed in strain 101(N) and, finally, Paris-related piRNAs are lowly expressed in strain Argentina(N) and in strain 101(N) (Fig 2D). Thus, we failed to detect one candidate causative TE or combinations of certain TEs present in all neutral strains whose piRNAs guarantee immunity to HD (Fig 2D). This suggests the possibility that maternal protection in crosses with strain 160(P) males may be conferred by different mechanisms across the strains.
Fig 2. Comparative analysis of the ovarian piRNA profiles between P-like strain 160 and both M- and neutral (N) strains studied.
A) and B) Scatter plots represent the result of pairwise comparison of normalized piRNAs (23–29 nt) in P-strain 160 versus M-like strains 9 and 13, and in P-strain 160 versus N-strains 140, Argentina, Magarach and 101, respectively. Diagonal lines indicate 10-fold levels of difference. All the TEs that exceed 10-fold line are marked as gray dots. The red dots indicate TEs that are shared between M-strains 9 and 13 in terms of their low expression levels in comparison with P-strain 160. Spearman’s correlation (R) is shown. C) Venn diagram depicting differences and similarities in a number of TEs exhibiting 10-fold lower piRNA expression level in M-strains 9 and 13 in comparison with P-strain 160. 10 families show the same pattern of deficit in strains 9 and 13, relative to strain 160. D) Venn diagram demonstrates distribution of piRNAs to eight essential elements distinguishing neutral strains from M-like in terms of piRNA-mediated silencing among studied N-strains.
A similar comparative analysis of siRNA expression between strain 160(P) and M-strains demonstrated that siRNAs complementary to only Penelope and Helena elements are absent in the ovaries of strain 9(M) and 13(M) (S1A and S1B Fig). However, we detected Penelope-homologous siRNAs only in half of the studied neutral trains i.e. strains Argentina and 140 (S1C Fig).
In the context of immunity to HD syndrome manifestations, probably the most important condition is to constantly maintain effective piRNA production in the germline. It is well known that ovarian piRNA pools consist of molecules generated by primary and secondary processing mechanisms. Due to germline expression of Ago3 and Aub proteins necessary for secondary processing (“ping-pong” amplification), the germline specific piRNA pool can be assessed quantitatively by counting of “ping-pong” pairs [2,23]. We analyzed the “ping-pong” signature of piRNAs to the selected TEs and showed that these piRNA species contain ping-pong pairs in varying degrees (S2 Fig). Importantly, all of them exhibit a signature of secondary piRNA processing indicating that production of these piRNAs takes place in the germline but each element lacks such a ping-pong signature in at least one or more of the neutral strains. In addition, Penelope expression was previously shown to be germline-specific by whole-mount RNA in situ hybridization [24]. In the present study, using the same technique with the ovaries of P-strain 160, we confirmed that Paris, Polyphemus and Helena elements exhibit germline-specific expression pattern as well (S3 Fig).
We further examined the pattern of divergence among piRNAs that map to the consensus TEs since piRNAs derived from divergent sequences are likely derived from degraded TE insertions. Among the selected HD-implicated TEs, the ovarian piRNA pool contains a very small amount of Paris-targeting piRNAs that were detected only in two studied N-strains—140 and Magarach. Interestingly, only 10% of both sense and antisense-oriented piRNAs apparently originate from modern active copies of Paris elements while the rest of the Paris-complementary piRNAs were produced from ancestral highly diverged ones (S4 Fig). The same applies to the Penelope-derived piRNAs in strain 101(N). All other piRNA species to HD-implicated TEs, especially in the antisense-orientation, in all studied neutral strains were practically identical to the consensus and, hence, apparently originated from active copies of these elements (S4 Fig). This analysis further indicates that there is no active candidate inducer family, represented by sequence similar piRNAs, shared across all six neutral strains.
Overall, these data indicate that, in terms of piRNA-mediated protection to HD in D. virilis neutral strains, there is no general rule in the context of ovarian piRNAs complementary to particular TEs implicated in HD. In other words, in neutral strains the maternally transmitted piRNA pool may include different amounts of piRNAs corresponding to various TEs and the repertoire of these TEs often radically differs between the strains with same cytotype.
Propagation and expression of Penelope does not change M-like cytotype
Syndromes of HD are explained by maternal protection against paternal induction and Penelope has long been considered the primary driver of paternal induction [18,20,22]. In the previous section we demonstrated that maternal piRNAs that target Penelope are not necessary to confer neutrality but, as neutrality may arise through different mechanisms, we sought to determine whether Penelope was either sufficient for induction or Penelope piRNA sufficient for protection. We thus characterized a simulation of natural invasion through the analysis of two transgenic strains of D. virilis containing full-size Penelope copies introduced into a typical D. virilis M-like strain 9 (the stock is assigned as w3) originally devoid of functional copies of this TE. Our previous experiments demonstrated that introduced Penelope underwent active amplification and occupied more than ten sites in the chromosomes of the transgenic strains [19]. However, at that time (in 2012) we did not detect any Penelope-derived small RNA species in these transgenic strains.
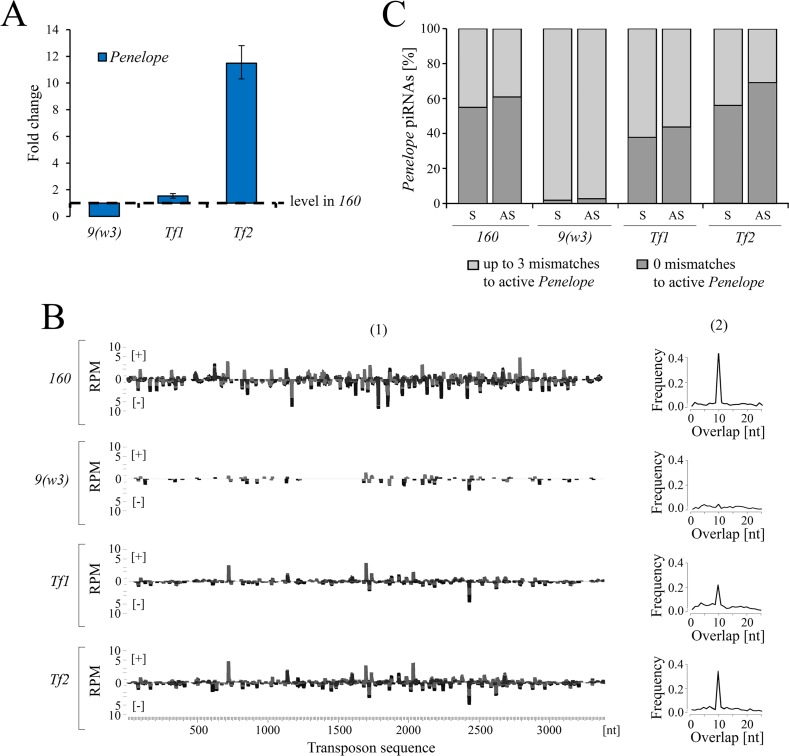
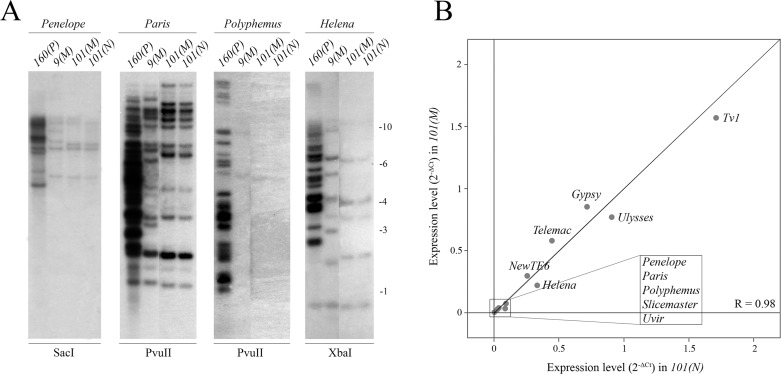
Subsequent to the early analysis performed in 2011–2012, we have now found that Penelope is actively transcribed in these two strains and exhibits steady-state RNA levels equal to or even higher than in strain 160 (Fig 3A). We further observed piRNAs in both transgenic strains, indicating that some of Penelope copies acquired the properties of piRNA-generating locus (Fig 3B). Thus, in strain Tf2 the level of piRNAs homologous to Penelope is only half as much as that observed in P-like strain 160. The analysis of Penelope-derived piRNAs indicates a distribution of piRNAs along the entire Penelope body and clear-cut ping-pong signature (Fig 3B).
Fig 3. Characterization of Penelope activity in the ovaries of Penelope-transformed strains.
A) Expression levels of Penelope in 9(w3), Tf1, Tf2 strains relative to P-strain 160. B) (1) The coverage of normalized Penelope-piRNA reads (23–29 nt) on the entire body of the element, across transformed strains. Sense reads are shown as [+], antisense as [–]. (2) The ping-pong signature of Penelope-derived piRNAs. C) Mapping proportions of Penelope-piRNAs to canonical sequence of the element with the perfect match and with the assumption of up to 3 mismatches.
Similar to strain 160, more than half of the Penelope-derived piRNAs in both strains originate from active and highly similar Penelope copies with few mismatches to the canonical sequence (Fig 3C). In contrast, Penelope piRNAs identified in the untransformed M-like strain 9(w3) are highly divergent and likely derive from inactivated Penelope copies (termed “Omegas”) located in heterochromatic regions of the genome [25,26]. Interestingly, the pool of Penelope derived small RNAs in transgenic strains are primarily piRNAs. This is in contrast to inducer strain 160 and D. melanogaster strains transformed with Penelope [19], where Penelope-derived siRNAs are the major class (S5 Fig).
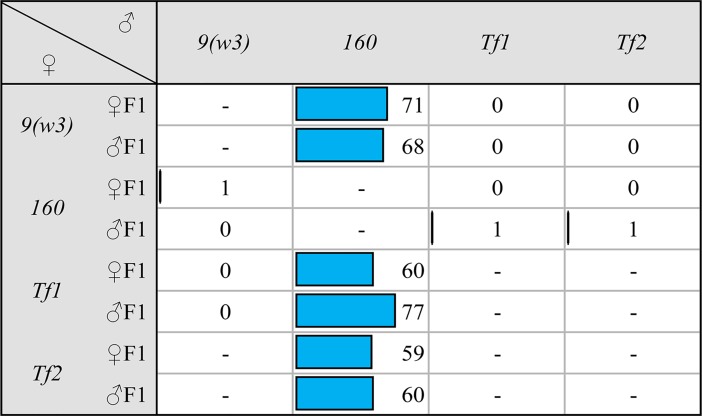
Surprisingly, both transgenic strains containing multiple Penelope copies and abundant piRNAs behave exactly as the original M-like strain 9 in dysgenic crosses (Fig 4). They neither have the capacity to induce HD paternally nor protect against HD maternally. Therefore, the introduction of full-size Penelope into an M-like strain accompanied by its propagation, active transcription and piRNAs production was not sufficient to modify the cytotype. These results also indicate that the presence of piRNA complementary to Penelope in the oocyte is not the only prerequisite to prevent gonadal sterility when crossed with males of P-like strain 160. Along these lines, it has been shown recently that the number of P-element and hobo copies per se has very little influence on gonadal sterility suggesting that HD is not determined solely by the dosage of HD-causative elements [27].
Fig 4. The frequency (in %) of gonadal atrophy in the progeny of dysgenic and reciprocal involving P-like strain 160 and Penelope-transformed strains.
The number of examined individuals was ~100 separately for females and males.
Strain cytotype can be altered without amplification of TEs implicated in HD
The above results demonstrate that the maternal piRNAs that target all, or even most, asymmetric TEs that likely cause dysgenesis are not necessary to confer neutral strain status (Fig 2). Furthermore, Penelope piRNAs are not sufficient for maternal protection and the presence of active Penelope copies is not sufficient for paternal induction (Figs 3 and 4). This begs the question: What are the necessary and sufficient factors of HD in D. virilis? Among the analyzed strains, neutral strain 101 represents a special case. This is due to the fact that the genome of this strain does not produce piRNAs to the most-described HD-implicated TEs, e.g. Paris, Helena, Polyphemus and a very small amount of divergent Penelope-homologous piRNAs (Figs 2 and S4).
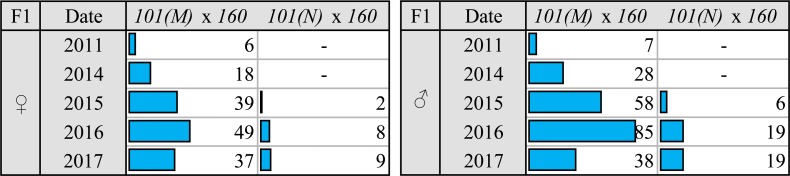
In the course of our long-term monitoring of the gonadal atrophy observed in the progeny of dysgenic crosses involving P-like strain and various laboratory and geographical strains of D. virilis, we often observed significant variation in the level of sterility in the progeny of the same crosses occurring with time. Strikingly, among these strains, we have identified a spontaneous change from neutral cytotype to M-like one. Thus, while an old laboratory strain 101 kept in the Stock Center of Koltzov Institute of Developmental Biology RAS maintained a neutral cytotype for the whole period of observation (2011–2017) the same strain kept in our laboratory gradually became M-like strain (Fig 5).
Fig 5. The alteration of the frequency (in %) of female and male gonadal atrophy in initially neutral strain 101.
The number of examined individuals was ~100 separately for females and males for each indicated time of monitoring.
We considered the possibility that this shift in cytotype could be explained by changes in the TE profile between the strains. Surprisingly, Southern blot and PCR analyses demonstrate that 101 N- and M- substrains have identical TE profiles for Penelope, Paris, Polyphemus and Helena (Figs 6A and S6). Additionally, qPCR analysis failed to detect any significant changes in the expression levels of the major asymmetric TEs as well as other described TEs in the compared variants (neutral vs M-like) of this strain (Fig 6B). These data rule out the possibility of strain contamination with a lab M-strain.
Fig 6. Genomic abundance and expression levels of putative HD-implicated TEs (Penelope, Paris, Polyphemus and Helena) in both cytotype variants of strain 101.
A) Southern blot analysis of genomic DNA of 160(P), 9(M), 101(M) and 101(N) strains. B) Expression levels of described D. virilis TEs in the ovaries of both variants of strain 101. Spearman’s rank test was used to calculate the correlation (R) between the strains studied.
Cytotype shift is accompanied by an altered piRNA and chromatin profile for a new set of repeats
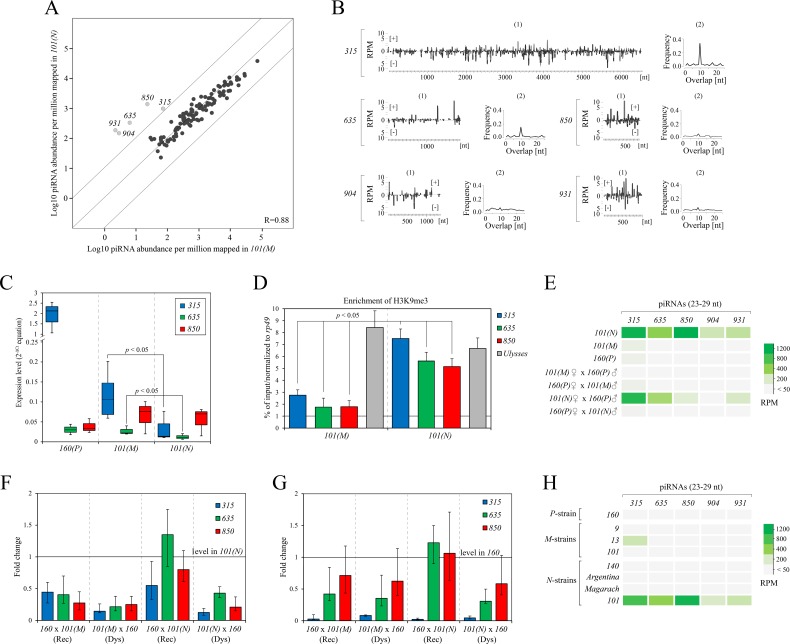
To understand the observed differences in the cytotype of strain 101 variants we performed additional small-RNA sequencing. Indeed, the piRNA profile of strain 101(N) has significantly higher piRNA levels (compared to 101(M)) for five previously undescribed repeats (315, 635, 850, 904 and 931) (Fig 7A), indicating that differences in cytotype could be attributed to these repeats. Among these piRNA species, only piRNAs targeting 315 and 635 elements comprise many ping-pong pairs and, hence, are generated predominantly by germline-specific secondary processing mechanism (Fig 7B). Based on sequence similarity to the TE consensus, at least 25% of antisense-oriented piRNA molecules apparently originated from modern active elements, with the exception of piRNAs targeting the 904-element (S7A Fig).
Fig 7. Characterization of small RNA mediated silencing in substrains of 101 exhibiting different cytotype.
A) Scatter plot represent the result of pairwise comparison of normalized piRNAs (23–29 nt) in M-like strain 101 versus its neutral variant. Diagonal lines indicate 10-fold levels of difference. Gray dots indicate the repeats exhibiting more than 10-fold greater piRNA level. Spearman’s rank test was used for correlation (R) calculation. B) (1) The coverage of normalized 315, 635, 850, 904 and 931-derived piRNA reads (23–29 nt) from the 101(N) strain on the entire body of correspondent elements. Sense reads are shown as [+], antisense as [–]. (2) The ping-pong signature of 315, 635, 850, 904 and 931-derived piRNAs. C) Expression levels of 315, 635, 850 elements in the ovaries of 160(P), 101(M) and 101(N) strains. P-values were calculated using t-test. D) Enrichment of H3K9me3 mark on the genomic loci carrying 315, 635 and 850 using ChIP-qPCR on ovaries of substrains 101. E) Heatmap of the ovarian expression of 315, 635, 850, 904, 931-derived piRNAs in dysgenic and reciprocal hybrids. F) Expression levels of 315, 635, 850 elements in the progeny from dysgenic and reciprocal crosses relative to parental 101(N) levels and G) to 160(P) levels. H) Heatmap represents expression pattern of 315, 635, 850, 904, 931-targeted piRNAs among the studied P-like, M-like and neutral strains.
Focusing on the three elements (315, 635, 850) with maximal piRNA expression levels, we compared both variants of strain 101 in more detail to determine if differences in repeat profile could explain differences in cytotype. Element 315 encodes three open reading frames (ORF). According to the protein-domain structure, two ORFs appear to encode gag and pol genes. The third ORF has no homology to the described TEs and possibly encodes an env gene. Thus, element 315 probably represents a retroelement. Since we failed to find any homology of the 315 element to the described families of TEs in Sophophora subgenus we propose that this element is an exclusive resident of Drosophila subgenus. Element 635 has some homology to the Invader element of D. melanogaster, which belongs to the Gypsy family of LTR-containing retrotransposons. However, it has no long terminal repeats (LTRs) in its sequence. Finally, short 850 element (749 nt) doesn’t encode any ORF and seems to be non-autonomous.
Importantly, based on Southern blot and PCR analysis, these particular repeats did not undergo amplification in the neutral variant of strain 101 and both compared substrains exhibit identical restriction patterns of these elements, similar to that of P-like strain 160 (S7B and S7C Fig). Hence, the observed cytotype shift as well as the differences in piRNA pool to these elements apparently do not stem from differences in copy number among 101 substrains. Interestingly, we observed a significant increase of expression levels of 315 and 635 elements (p < 0.05; t-test), but not 850, in the ovarian mRNA pool of M-like substrain 101 compared to the neutral substrain (Fig 7C). Overall, these results demonstrate that the capacity for these repeats to produce piRNAs is lower in the 101(M) strain, even in the absence of movement.
What could lead to differences in the piRNA profile for these repeats between the 101(N) and 101(M) strains in the absence of movement? Studies of piRNA-generating loci in Drosophila revealed that the H3K9me3 mark, which serves as a binding site to recruit HP1a and its germline homolog Rhino, is required for transcription of dual-strand piRNA-clusters and transposon silencing in ovaries [2,28,29]. We hypothesized that a shift of the chromatin state in strain 101 modified the ability of particular genomic loci, carrying 315, 635, 850 elements, to produce piRNA species. These changes in piRNA profile may be an indication of a chromatin-based modification that may confer resistance to HD sterility in the neutral 101 substrain. To test this hypothesis, we estimated the levels of H3K9me3 and HP1a marks by ChIP combined with qPCR analysis in the ovaries of two cytotype variants of strain 101. The analysis showed significant increase of H3K9me3 levels on genomic regions containing 315, 635 and 850 elements (enrichment > 2.5, p < 0.05) as well as slight increase of HP1a enrichment in the neutral variant of strain 101 compared to the M-like substrain (Figs 7D and S8). In turn, Ulysses carrying regions used as a control demonstrated equal levels of the H3K9me3 mark, consistent with Ulysses-targeting piRNA levels being almost equal in the strain 101 variants (Fig 7D). This indicates that certain repeats have experienced shift in their chromatin profile, but that this shift is not global. A similar phenomenon has been recently described in I-R HD system in D. melanogaster [30]. In that comparative analysis of two reactive strains (weak and strong), it was shown that despite having a similar number of copies of the I-element, these strains significantly differ by enrichment of Rhino at the 42AB piRNA-cluster containing I-elements remnants. Furthermore, a lower level of I-element targeted piRNA species was observed in the strong-reactive strain as a result [30].
Given these differences, it is possible that these elements are the primary drivers of dysgenesis in D. virilis. To further test the hypothesis that activation of these elements could contribute to HD, we compared first piRNA levels of all these elements in the ovaries of the F1 progeny from dysgenic-like and reciprocal crosses using variants of strain 101 and P-like strain 160. These experiments demonstrate that piRNAs targeting 315, 635, and 934 elements showed similar levels in the ovaries of F1 hybrids from dysgenic crosses (101(N) x 160) and parental neutral strain 101, but lower levels in progeny of reciprocal crosses where such piRNAs would not be maternal (160 x 101(N)) (Fig 7E). Thus, the maternally provisioned piRNAs complementary to 315, 635 and 931 elements are required to stimulate the generation of the corresponding piRNAs in the progeny, as shown in other systems of HD [3,4]. However, in the analysis of steady-state mRNA levels of these TEs in the ovaries of dysgenic and reciprocal progeny of crosses between 101 substrains and P-like strain 160, we failed to obtain any induction of 315, 635 and 850 elements exceeding their levels of parental strains (Fig 7F and 7G). On the contrary, the ovaries of F1 hybrids from the reciprocal (non-dysgenic) crosses involving strains 101(N) males and 160(P) females showed even significantly higher expression levels of these elements in comparison to dysgenic ones. Moreover, the dysgenic and reciprocal hybrids of M-like substrain 101 and strain 160(P) showed no differences in the mRNA levels of the studied elements (Fig 7F and 7G). These results indicate that activation of these elements per se is unlikely to be causative to HD because 101(N) and 101(M) have identical TE profiles. We therefore considered the possibility that what distinguishes strain 101(N) from 101(M) may have an epigenetic basis or, alternately, an unknown genetic change that alters repeat chromatin. If so, then lack of piRNAs to these elements in 101(M) could explain the M-cytotype. To test this, we compared piRNA levels and family level abundance with inducer strain 160(P). Critically, none of these elements show increased piRNA levels in strain 160(P) compared to strain 9(M) (Fig 7H). Thus, asymmetry in the piRNA pool for these particular elements is not a necessary condition for dysgenesis.
According to the recent studies differences in parental expression levels of genic piRNAs may contribute to the dysgenic manifestations in the progeny [18,30]. With this in mind, we compared the expression of genic piRNAs in the ovaries of both 101 substrains and did not observe significant differences in their levels (S9A Fig). Ping-pong of genic piRNA profiles are also exhibit high similarity between these strains (S9B Fig). Based on these data, we concluded that differences in genic piRNAs unlikely have impact on the observed cytotype shift.
Overall, we have shown that the enrichment of heterochromatic marks (H3K9me3 and HP1a) in the genomic regions containing 315, 635 and 850 elements is significantly lower in M-like variant of strain 101 compared to neutral one. Together, these data provide further evidence that the mechanism of maternal repression may significantly vary among strains. However, additional experiments involving Rhino ChIP and genome sequencing of strain 101 are needed to clearly prove this assumption and identify the loci responsible for the enhanced piRNA production in one of the two 101 substrains.
Ovarian levels of mRNA and complementary piRNAs for many asymmetric TEs are similar in dysgenic and reciprocal hybrids of D. virilis
One of the main consequences of activation of a particular asymmetric TE in the progeny of dysgenic crosses is their expression level excess compared to both paternal strains and reciprocal hybrids [3,15,18,31]. Studies of the I-R syndrome of HD in D. melanogaster demonstrate higher expression of the I-element in the F1 progeny from dysgenic crosses compared to reciprocal ones [3,30,31]. This is due to the maternal deposition of piRNAs targeting the I-element and its effective silencing in only one direction of the cross. Additionally, various studies of HD systems, including the D. virilis syndrome, demonstrated that transgenerational inheritance of piRNAs is able to trigger piRNA expression in the next generation by changing the chromatin of piRNA-clusters due to paramutation [3,4,32–34]. However, a pattern of higher TE expression in the absence of complementary maternal piRNA is less apparent in D. virilis. Despite strain asymmetry in genomic content and piRNA abundance of Penelope and several other TEs, germline piRNA pools do not differ drastically between reciprocal F1 progeny, with the exception of Helena element [18]. We therefore sought to determine whether this atypical pattern was also observed in crosses with other strains, focusing on asymmetric Penelope, Paris, Polyphemus and Helena as well as Ulysses present in all strains.
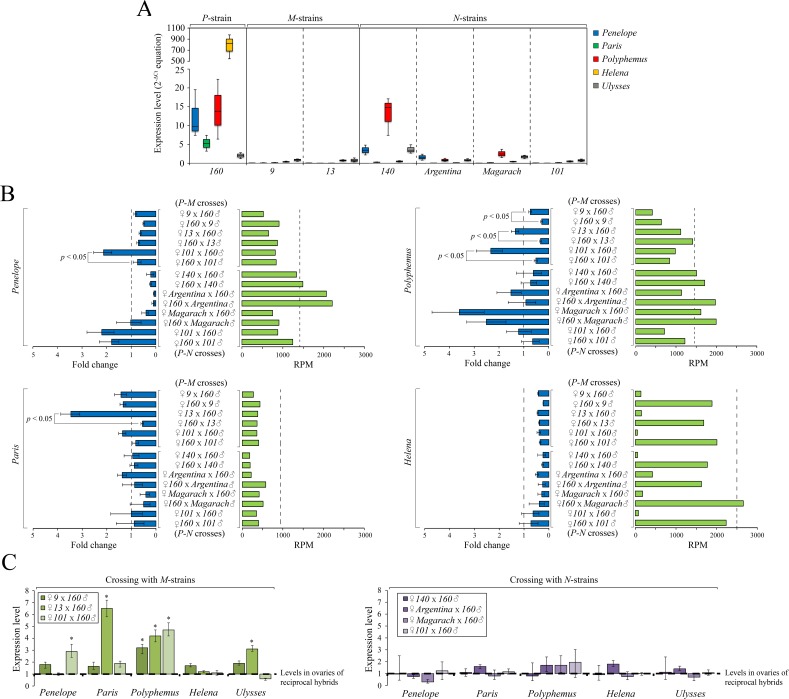
As expected, ovarian mRNA levels revealed a complete correspondence with the piRNA expression levels among strains (Figs 8A, 2A and 2B). For example, we detected both Penelope mRNA and piRNA expression in 140(N) and Argentina(N), but neither were evident in Magarach(N) and 101(N). However, in all cases when females from M-like strains are crossed with strain 160 males, ovarian levels of expression are uniformly significantly higher for only one asymmetric TE–Polyphemus (fold change 3, 5, 3.5, p < 0.05, t-test, in dysgenic hybrids with strains 9(M), 13(M) and 101(M), respectively) (Fig 8B and 8C). In most cases the observed differences in expression for Penelope and Paris elements in the ovaries of dysgenic and reciprocal hybrids were not dramatic and when exist rarely exceed 1.5–2 fold. Moreover, in the crosses involving neutral strains and strain 160, we failed to detect any characteristic differences in TEs expression between reciprocal hybrids (Fig 8B and 8C). Thus, independent of maternal piRNA profile, all reciprocal crosses with neutral strains show similar levels of expression. However, the two variants of strain 101 give different results when crossed with P-like strain 160. In spite of the fact that 101 substrains contain equal levels of piRNAs complementary to the HD-implicated TEs, in the case of the M-like variant we observed higher levels of expression in the dysgenic hybrids for Penelope (fold change 3; p < 0.05, t-test) and Polyphemus (fold change 3.5, p < 0.05, t-test). Moreover, increase of Ulysses element (found in all D. virilis strains) expression (fold change 3, p < 0.05, t-test) was demonstrated in the dysgenic ovaries of 13(M) and 160(P) hybrids (S10A Fig). These results demonstrate that factors other than maternal piRNA abundance lead to variation in resident TE expression in crosses between strain 160 and 101 substrains. For the neutral 101 strain, we failed to detect significant differences in the hybrids from both directions of crosses for any of TEs tested (Fig 8B).
Fig 8. Comparative analysis of a set of HD-implicated TEs in the ovaries of dysgenic and reciprocal hybrids.
A) Expression levels of Penelope, Paris, Polyphemus, Helena and Ulysses among the studied P-like, M-like and neutral strain. B) mRNA and piRNAs expression levels in the ovaries of the progeny from dysgenic and reciprocal hybrids. At the left—expression levels of indicated TEs relative to the level in P-strain 160. At the right–normalized piRNAs expression levels. The dotted line indicates level in P-strain 160. P-values were calculated using t-test. C) Expression level of Penelope, Paris, Polyphemus, Helena and Ulysses in the ovaries of dysgenic crosses compared to reciprocal ones. P-values were calculated using t-test.
With the exception of a few TEs and repeats, piRNA abundance in the ovaries from dysgenic and reciprocal progeny exhibited no drastic differences including piRNAs complementary to asymmetric TEs (Figs 8B and S11). Surprisingly, Helena, which maintains high level of asymmetry of the maternal pool of piRNAs in the progeny, exhibits very similar levels of correspondent mRNA expression in the hybrids obtained in both directions of crosses (Fig 8B). In spite of overall similarity, piRNA pools in the ovaries of F1 progeny are able to comprise significantly different number of ping-pong pairs to all of transposons studied (S10B Fig). For example, in the ovaries from dysgenic progeny (strain 160 males) with strains 9(M) and Argentina(N) females, the number of ping-pong pairs to Penelope, Paris and Polyphemus was 2-3-fold lower than in the ovaries from reciprocal hybrids (S10B Fig). We have also found that enrichment of the H3K9me3 mark on Penelope, Paris, Polyphemus and Helena sequences does not differ significantly in the F1 progeny of dysgenic and reciprocal crosses (S10C Fig). Thus, we propose that piRNA-mediated transcriptional gene silencing of these HD-implicated TEs is similar in both directions of crosses and maternally provisioned piRNAs to these TEs are not necessary to stimulate the production of correspondent piRNA species in the progeny. These results are in agreement with recently published data [18].
In summary, it should be emphasized that in contrast to the I-R system in D. melanogaster, where maternal deposition of I-element piRNAs results in dramatic increase of piRNA expression targeting I-element in the progeny and efficient suppression of I-element activity, in D. virilis maternally provisioned piRNAs do not always guarantee efficient generation of the correspondent piRNAs in the progeny to maintain silencing of complementary TEs and provide adaptive genome defense. We conclude that in D. virilis the determination of asymmetric TEs expression levels in the ovaries of the progeny from dysgenic and reciprocal crosses does not allow one to unambiguously assign causality for HD to specific TE families. This fact points to an alternate mode of HD in D. virilis.
Conclusions
The standard explanation for the phenomenon of hybrid dysgenesis is that TEs inherited paternally become germline activated in the absence of maternal piRNA. Here, however, we suggest that repression of paternal TEs by maternal piRNA may not be the sole mechanism of protection against this form of hybrid sterility. Using the D. virilis system of HD as a model, we have demonstrated that neutral strains exhibiting “immunity” to hybrid dysgenesis in D. virilis do not share a consistent pattern of piRNAs complementary to TEs likely causative of dysgenesis. Strikingly, the introduction and propagation of one presumably causal TE (Penelope) in the genome of M-like strain does not even change the cytotype of the transformed strains. Finally, we identified a shift of cytotype from N to M that occurred in a strain without changes in the expression or copy number of asymmetric TEs implicated in HD. The observed “immunity” of neutral strains to HD manifestations is apparently established by an increased repertoire of repeats that become targets for piRNA biogenesis as well as a modified chromatin state of several genomic regions compared to M-like strains. These studies suggest that hybrid dysgenesis in D. virilis cannot be solely explained by the well-established “hybrid dysgenesis paradigm”, developed in D. melanogaster. Rather, other properties of the genome contribute to maternal protection. The precise molecular mechanisms underlying “susceptibility” of D. virilis strains to TEs invasion requires further investigation.
Materials and methods
Fly strains and crosses
Seven D. virilis strains, namely 9 (Batumi, Georgia), 13 (Krasnodar, Russia), 101 (Japan), Argentina (Argentina), Magarach (Crimea, Russia), 140 (laboratory strain) and laboratory strain 160 were used in this study. Original fly stocks are available in the Stock Center of Koltzov Insitute of Developmental Biology RAS. In addition, two previously described 165 and 247 strains [19] were used, designated in this paper as Tf1 and Tf2, respectively. These strains were obtained as a result of transgenesis of the full copy of Penelope retroelement into 9 strain (19). All flies were reared on standard agar-yeast-sugar-raisins medium at the constant temperature regime (25°C). Since the start of monitoring for cytotype shift (2011) stocks of both substrains of 101 (from the Stock Center of Koltzov Institute of Developmental Biology RAS and ours) are reared on the same medium and an equivalent population size (not more than 20 flies per vial) has been maintained.
The dysgenic crosses involved males of P-like 160 strain and females of all aforementioned strains (9, 13, 101, Argentina, Magarach and 140). As a control, reciprocal crosses were performed. Monitoring and counting of gonadal atrophy were conducted as described [11].
Small RNA libraries sequencing and bioinformatic analysis
Total RNA was extracted from ovaries of 7–10 days old females using Extract RNA reagent (Evrogen, Russia). Total RNA was extracted from both 101 substrains after the second confirmation of cytotype shift in 2016. To prepare the small RNA fraction for cloning, total RNA from ovaries (~ 25 μg) was separated, using 15% polyacrylamide gel electrophoresis containing 8 M Urea. After incubation in an ethidium bromide solution (0.5 μg/ml), gel fragments corresponding to the small RNA fraction were excised, using chemically synthesized RNA corresponding to 21 and 29 nts as size markers. Cloning of small RNA libraries was performed by Illumina TruSeq Small RNA prep kit (Illumina, USA) according to the manufacturer’s protocol. Sequencing was conducted on an Illumina NextSeq 500 platform.
As a result of deep-sequencing we obtained 5–14 million reads of small RNAs for each library. Pre-processing procedure included: 3’-adapter trimming, filtration of reads by length (>18 nt) and quality (80% of nt have ≥ 20 Phred quality). Pre-processed reads were further subjected to subtraction of reads matching to all rRNA, tRNA, snRNA and miRNA sequences. The selected reads were mapped to the latest release of D. virilis genome by Bowtie [35], requiring a perfect match. In order to identify siRNAs and piRNAs, the sequenced reads were mapped to the canonical sequences of TEs obtained from combined libraries of annotated and computationally predicted D. virilis transposons and repeats sequences [18]. In addition, recently described DAIBAM MITE (GenBank: EU280326) and Tetris (GenBank: KF723713.1; KF723710.1) elements sequences were considered [36,37]. For genic piRNAs mapping the latest annotation of D. virilis transcripts was used (r1.06) and the alignment performed with requirement of perfect match. Less than 25 CPM of mapped siRNAs and piRNAs per transposons or transcripts were considered as lowly expressed and discarded. Length distribution and counting of siRNAs and piRNAs reads, nucleotide biases, ping-pong signatures, coverage of transposon sequence by piRNAs were analyzed with accordance to the well-described technique [23,38] using custom scripts written in Python. Venn diagrams were made using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/). Scatter plots were created using Plotly (https://plot.ly).
Quantitative PCR
For analysis, cDNA was prepared from 2 μg Turbo-DNAase (Ambion, USA) treated total RNA using oligo(dT) primer and MMLV reverse transcriptase (Evrogen, Russia). PCR was performed on ABI PRISM 7500 System (Applied Biosystems, USA). Detection of amplification products was carried out using SYBR Green 1 with the presence of ROX reference dye (Evrogen, Russia) in accordance with the manufacturer’s protocol. Quantification was normalized to ubiquitously expressed rp49 gene and calculation of relative expression levels was performed using the equation 2-ddCt. Specificity of amplified products was verified by sequencing as well as melting curve analysis. The resulting value of the expression level for each sample was determined basing on at least three biological replicates. Sequences of used primers are shown in S1 Table.
Whole mount in situ RNA hybridization
Dissection of ovaries, fixation, Proteinase K treatment, re-fixation and hybridization steps were performed as described [24].
Matrices for probe preparation were prepared by PCR (sequences of used primers are shown in S1 Table) of genomic DNA of 160 strain. Labeling of RNA probes with DIG-11-UTP (Roche, France) was made by MAXIscript T7 kit (Ambion, USA). Anti-DIG-AP antibodies (Roche, France) were used in 1:2000 dilution. Images obtained by binocular microscope Nikon Alphaphot-2 YS2 (Japan).
Southern blotting
To analyze genomic TE profiles, DNA samples (~ 10 μg) were digested by restriction enzymes (Penelope–SacI; Paris, Polyphemus and 850 –PvuII; Helena–XbaI; 315 –NcoI; 635 –PvuI), fractionated through 0.8% agarose gel and transferred onto Hybond-XL membrane (Amersham Biosciences, USA). Hybridization was carried out at 60°C overnight in solution containing 6xSSC, 10 mM EDTA, 0.5% SDS and 5x Denhardt’s solution. The PCR-prepared matrices were used for probe preparation. Primers can be found in S1 Table. Labeling of probe with (P32)-dATP was performed using Decalabel kit (Thermo Scientific, USA).
Semiquantitative PCR of genomic DNA
PCR was performed, in addition to Southern analysis, to compare the presence of full copies of TEs in the genomes of 160, 9, 101(N) and 101(M) strains. Amplification was performed using ScreenMix kit (Evrogen, Russia). PCR products were separated in 1.5% agarose gel including ethidium bromide (0.5 μg/ml) for detection. Reaction using primers to rp49 gene represents a loading control. Used primers were the same as for probe preparation in previously described hybridization procedure.
Chromatin immunoprecipitation and quantitative PCR analysis
Chromatin from Drosophila ovaries (~ 100 pairs for each IP experiment) was extracted and immunoprecipitated according to the published protocol [39]. ChIP experiments were carried out using commercially available antibodies anti-H3K9me3 (ab8898) (Abcam, UK) and anti-HP1a (C1A9) (DSHB). To bind antibodies Pierce protein A/G agarose (Thermo Fisher Scientific, USA) was used. Quantitative PCR was applied to evaluate the protein enrichment in the genomic loci. Percent of precipitated chromatin was calculated according to input values following normalization to actively transcribed rp49 gene. The resulting value represents the mean of two biological replicates. The error is indicated by the standard error of mean (SEM). Primer sequences are presented in S1 Table.
Statistical analysis
Student’s t-test was used to compare groups with each other (qPCR and ChIP-qPCR data). P-values ≤ 0.05 were considered statistically significant.
Supporting information
A) and B) Scatter plots represent the result of pairwise comparison of normalized siRNAs (21 nt) in P-strain 160 versus M-like strains 9 and 13, and in P-strain 160 versus N-strains 140, Argentina, Magarach and 101, respectively. Diagonal lines indicate 10-fold levels of difference. Gray dots indicate the TEs exhibiting more than 10-fold greater level of siRNAs between the pair of comparison. The results of Spearman’s correlation tests (R) are demonstrated. C) Venn diagram depicts differences and similarities in a number of TEs exhibiting 10-fold greater siRNA expression level in M-strains 9 and 13 in comparison with P-strain 160.
(TIF)
M-strains were not plotted due to a very small expression of piRNAs and inability to calculate ping-pong signal.
(TIF)
(TIF)
Number of mismatches (up to 3) to active element are indicated; piRNAs are split into sense (S) and antisense (AS) species.
(TIF)
(TIF)
Primers correspond to the middle region of indicated TEs. Rp49 gene serves as a loading control.
(TIF)
A) Mapping of 315, 635, 850, 904 and 931-piRNAs to canonical sequence of the element in strain 101(N). B) Southern-blot analysis of genomic DNA of 160(P), 101(M) and 101(N) strains. C) Semiquantitative PCR of 315, 635 and 850 elements of genomic DNA of 160(P), 101(M) and 101(N) strains. Primers correspond to the middle region of indicated TEs. Rp49 gene serves as a loading control.
(TIF)
(TIF)
A) Scatter plot represents the result of comparison of normalized piRNAs (23–29 nt) in 101M and 101N strains. Diagonal lines indicate 10-fold levels of difference. The results of Spearman’s correlation tests (R) are demonstrated. B) At the left–number of consitituted ping-pong pairs; at the right–frequency of overlapped piRNAs.
(TIF)
A) mRNA and piRNAs expression levels in the ovaries of the progeny from dysgenic and reciprocal hybrids. At the left—expression levels of indicated TEs relative to the level in P-strain 160. At the right–normalized piRNAs expression levels. The dotted line indicates level in P-strain 160. P-values were calculated using t-test. B) The ratio of constituted ping-pong pairs between the dysgenic and reciprocal hybrids. C) H3K9me3 ChIP-qPCR on ovaries of dysgenic and reciprocal hybrids involving P-strain 160 and M-like strain 9.
(TIF)
A) and B) Scatter plots represent the result of pairwise comparison of normalized piRNAs (23–29 nt) between dysgenic and reciprocal ovaries of strain 160 and M-strains and 160 and N-strains, respectively. Diagonal lines indicate 10-fold levels of difference. All the TEs that exceed 10-fold line are marked as gray dots. The results of Spearman’s correlation tests (R) are shown.
(TIF)
(XLSX)
Acknowledgments
We thank the members of Molecular genetics of cell laboratory in Institute of Molecular Genetics RAS and, personally, Vladimir Gvozdev, Sergei Ryazansky, Mikhail Klenov, Ludmila Olenina for helpful discussion on the study. We are grateful to Alla Kalmykova for ideas and critical comments on the manuscript. We commend Yury Funikov for helping with Bash scripts. We thank Olga Zatcepina and Anastasiya Snezhkina for technical assistance in small RNA sequencing and helpful advices. We are also grateful to Nikolay Rozhkov for his preliminary studies of Magarach strain. The main part of this work (sequencing of small RNA libraries) was performed using the equipment of Engelhardt Institute of Molecular Biology RAS Genome center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Data Availability
Sequences reported in this study can be accessed using GEO accession number: GSE108298; GSE22067; PRJNA288729.
Funding Statement
This work was supported by the Russian Science Foundation grant (No. 14-50-00060). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annual Review of Genetics 1989: 23:251–287. doi: 10.1146/annurev.ge.23.120189.001343 [DOI] [PubMed] [Google Scholar]
- 2.Toth KF, Pezic D, Stuwe E, Webster A (2016) The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv Exp Med Biol 886: 51–77. doi: 10.1007/978-94-017-7417-8_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A et al. (2008) An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science 322: 1387–1392. doi: 10.1126/science.1165171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC et al. (2011) Adaptation to P Element Transposon Invasion in Drosophila melanogaster. Cell 147: 1551–1563. doi: 10.1016/j.cell.2011.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidwell MG (1983) Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc Natl Acad Sci USA 80: 1655–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucheton A, Busseau I, Teninges D (2002) I elements in Drosophila melanogaster Mobile DNA II. (Craig NL, Craigie R, Gellert M, Lambowitz AM eds). Washington, DC: ASM Press; pp. 796–812. [Google Scholar]
- 7.Kidwell MG, Lisch DR (2002) Transposable elements as sources of genomic variation Mobile DNA II. (Craig NL, Craigie R, Gellert M, Lambowitz AM eds). Washington, DC: ASM Press; pp. 796–812. [Google Scholar]
- 8.Castro JP, Carareto CM (2004) Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica 121(2): 107–18. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher ES (2016) Reexamining the P-Element Invasion of Drosophila melanogaster Through the Lens of piRNA Silencing. Genetics 203(4): 1513–31. doi: 10.1534/genetics.115.184119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozovskaya ER, Scheinker VS, Evgen'ev MB (1990) A hybrid dysgenesis syndrome in Drosophila virilis. Genetics 126(3): 619–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sokolova MI, Zelentsova ES, Shostak NG, Rozhkov NV, Evgen'ev MB (2013) Ontogenetic consequences of dysgenic crosses in Drosophila virilis. Int J Dev Biol 57(9–10): 731–9. doi: 10.1387/ijdb.120189me [DOI] [PubMed] [Google Scholar]
- 12.Dorogova NV, Bolobolova EU, Zakharenko LP (2016) Cellular aspects of gonadal atrophy in Drosophila P-M hybrid dysgenesis. Dev Biol 424(2): 105–112. doi: 10.1016/j.ydbio.2017.02.020 [DOI] [PubMed] [Google Scholar]
- 13.Scheinker VS, Lozovskaya ER, Bishop JG, Corces VG, Evgenev MB (1990) A long terminal repeatcontaining retrotransposon in mobilized during hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci USA 87: 9615–9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER (1995) Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci USA 92: 8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evgen’ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, et al. (1997) Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci USA 94: 196–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenstiel JP (2014) Whole genome sequencing in Drosophila virilis identifies Polyphemus, a recently activated Tc1-like transposon with a possible role in hybrid dysgenesis. Mob DNA 5: 6 doi: 10.1186/1759-8753-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R et al. (2010) Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 16: 1634–1645. doi: 10.1261/rna.2217810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erwin AA, Galdos MA, Wickersheim ML, Harrison CC, Marr KD et al. (2015) piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS Genet 11: e1005332 doi: 10.1371/journal.pgen.1005332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozhkov NV, Schostak NG, Zelentsova ES, Yushenova IA, Zatsepina OG et al. (2013) Evolution and dynamics of small RNA response to a retroelement invasion in Drosophila. Mol Biol Evol 30: 397–408. doi: 10.1093/molbev/mss241 [DOI] [PubMed] [Google Scholar]
- 20.Evgen'ev M, Zelentsova H, Mnjoian L, Poluectova H, Kidwell MG (2000) Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma 109(5): 350–7. [DOI] [PubMed] [Google Scholar]
- 21.Vieira J, Vieira CP, Hartl DL, Lozovskaya ER (1998) Factors contributing to the hybrid dysgenesis syndrome in Drosophila virilis. Genet Res 71(2): 109–17. [DOI] [PubMed] [Google Scholar]
- 22.Blumenstiel JP, Hartl DL (2005) Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci USA 102: 15965–15970. doi: 10.1073/pnas.0508192102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. doi: 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- 24.Rozhkov NV, Zelentsova ES, Shostak NG, Evgen'ev MB (2011) Expression of Drosophila virilis retroelements and role of small RNAs in their intrastrain transposition. PLoS One 6(7): e21883 doi: 10.1371/journal.pone.0021883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zelentsova H, Poluectova H, Mnjoian L, Lyozin G, Veleikodvorskaja V, et al. (1999) Distribution and evolution of mobile elements in the virilis species group of Drosophila. Chromosoma 108(7): 443–56. [DOI] [PubMed] [Google Scholar]
- 26.Lyozin GT, Makarova KS, Velikodvorskaja VV, Zelentsova HS, Khechumian RR et al. (2001) The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. Journal of Molecular Evolution 52: 445–456. doi: 10.1007/s002390010174 [DOI] [PubMed] [Google Scholar]
- 27.Srivastav SP, Kelleher ES (2017) Paternal Induction of Hybrid Dysgenesis in Drosophila melanogaster Is Weakly Correlated with Both P-Element and hobo Element Dosage. G3 (Bethesda) 7(5): 1487–1497. doi: 10.1534/g3.117.040634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klattenhoff C, Xi H, Li C, Lee S, Xu J, et al. (2009) The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. doi: 10.1016/j.cell.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangan P, Malone C, Navarro C, Newbold S, Hayes P, et al. (2011) piRNA production requires heterochromatin formation in Drosophila. Curr Biol 21: 1373–1379. doi: 10.1016/j.cub.2011.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryazansky S, Radion E, Mironova A, Akulenko N, Abramov Y, et al. (2017) Natural variation of piRNA expression affects immunity to transposable elements. PLoS Genet 27,13(4): e1006731 doi: 10.1371/journal.pgen.1006731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelleher ES, Edelman NB, Barbash DA (2012) Drosophila interspecific hybrids phenocopy piRNA pathway mutants. PLoS Biol 10: e1001428 doi: 10.1371/journal.pbio.1001428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L et al. (2012) Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490: 112–115. doi: 10.1038/nature11416 [DOI] [PubMed] [Google Scholar]
- 33.Le Thomas A, Stuwe E, Li S, Du J, Marinov G et al. (2014) Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. doi: 10.1101/gad.245514.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Thomas A, Marinov GK, Aravin AA (2014) A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep 8: 1617–1623. doi: 10.1016/j.celrep.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3): R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fonseca NA, Vieira CP, Schlötterer C, Vieira J (2012) The DAIBAM MITE element is involved in the origin of one fixed and two polymorphic Drosophila virilis phylad inversions. Fly (Austin) 6(2): 71–4. doi: 10.4161/fly.19423 [DOI] [PubMed] [Google Scholar]
- 37.Dias GB, Svartman M, Delprat A, Ruiz A, Kuhn GC (2014) Tetris is a foldback transposon that provided the building blocks for an emerging satellite DNA of Drosophila virilis. Genome Biol Evol 6(6): 1302–13. doi: 10.1093/gbe/evu108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J, Liu J, Schnakenberg SL, Ha H, Xing J, Chen KC (2014) Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol Evol 29,6(10): 2786–98. doi: 10.1093/gbe/evu217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nègre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G (2006) Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol 4(6): e170 doi: 10.1371/journal.pbio.0040170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) and B) Scatter plots represent the result of pairwise comparison of normalized siRNAs (21 nt) in P-strain 160 versus M-like strains 9 and 13, and in P-strain 160 versus N-strains 140, Argentina, Magarach and 101, respectively. Diagonal lines indicate 10-fold levels of difference. Gray dots indicate the TEs exhibiting more than 10-fold greater level of siRNAs between the pair of comparison. The results of Spearman’s correlation tests (R) are demonstrated. C) Venn diagram depicts differences and similarities in a number of TEs exhibiting 10-fold greater siRNA expression level in M-strains 9 and 13 in comparison with P-strain 160.
(TIF)
M-strains were not plotted due to a very small expression of piRNAs and inability to calculate ping-pong signal.
(TIF)
(TIF)
Number of mismatches (up to 3) to active element are indicated; piRNAs are split into sense (S) and antisense (AS) species.
(TIF)
(TIF)
Primers correspond to the middle region of indicated TEs. Rp49 gene serves as a loading control.
(TIF)
A) Mapping of 315, 635, 850, 904 and 931-piRNAs to canonical sequence of the element in strain 101(N). B) Southern-blot analysis of genomic DNA of 160(P), 101(M) and 101(N) strains. C) Semiquantitative PCR of 315, 635 and 850 elements of genomic DNA of 160(P), 101(M) and 101(N) strains. Primers correspond to the middle region of indicated TEs. Rp49 gene serves as a loading control.
(TIF)
(TIF)
A) Scatter plot represents the result of comparison of normalized piRNAs (23–29 nt) in 101M and 101N strains. Diagonal lines indicate 10-fold levels of difference. The results of Spearman’s correlation tests (R) are demonstrated. B) At the left–number of consitituted ping-pong pairs; at the right–frequency of overlapped piRNAs.
(TIF)
A) mRNA and piRNAs expression levels in the ovaries of the progeny from dysgenic and reciprocal hybrids. At the left—expression levels of indicated TEs relative to the level in P-strain 160. At the right–normalized piRNAs expression levels. The dotted line indicates level in P-strain 160. P-values were calculated using t-test. B) The ratio of constituted ping-pong pairs between the dysgenic and reciprocal hybrids. C) H3K9me3 ChIP-qPCR on ovaries of dysgenic and reciprocal hybrids involving P-strain 160 and M-like strain 9.
(TIF)
A) and B) Scatter plots represent the result of pairwise comparison of normalized piRNAs (23–29 nt) between dysgenic and reciprocal ovaries of strain 160 and M-strains and 160 and N-strains, respectively. Diagonal lines indicate 10-fold levels of difference. All the TEs that exceed 10-fold line are marked as gray dots. The results of Spearman’s correlation tests (R) are shown.
(TIF)
(XLSX)
Data Availability Statement
Sequences reported in this study can be accessed using GEO accession number: GSE108298; GSE22067; PRJNA288729.