SUMMARY
Keloids are benign dermal tumors occurring ~20 times more often in African- as compared to European-descent individuals. While most keloids occur sporadically, a genetic predisposition is supported by both familial aggregation of some keloids and large differences in risk among populations. Despite Africans and African Americans being at increased risk over lighter-skinned individuals, little genetic research exists into this phenotype. We reported, using a combination of admixture mapping and exome analysis, multiple common variants within chr15q21.2-22.3 associated with risk of keloid formation in African Americans. Here we describe a gene-based association analysis using 478 African American samples with exome genotyping data to identify genes containing low-frequency variants associated with keloids, with evaluation of genetically predicted gene expression in skin tissues using association summary statistics. The strongest signal from gene-based association was located in C15orf63 (p-value = 6.6×10−6) located at 15q15.3. The top result from gene expression was increased predicted DCAF4 expression (p-value = 5.5×10−4) in non-sun-exposed skin, followed by increased predicted OR10A3 expression in sun-exposed skin (p-value = 6.9×10−4). Our findings identify variation with putative role in keloid formation, enhanced by the use of predicted gene expression to support the biological roles of variation identified only though genetic association studies.
Keywords: African Americans, gene expression, genetic predisposition, keloid, skin
INTRODUCTION
Keloids are benign dermal tumors that form during prolonged wound healing as the result of a fibroproliferative process(Marneros & Krieg, 2004, Niessen et al., 1999). Although some cases of keloid formation may be due to somatic mutation(Saed et al., 1998, Ladin et al., 1998), multiple keloids in the same individual and evidence for a multicellular origin of keloids(Chevray & Manson, 2004, Moulton-Levy et al., 1984) argue against somatic mutation as the cause in most cases. Several lines of evidence support a genetic basis for keloids, including the occurrence of familial forms and racial and ethnic differences in prevalence(He et al., 2017). Keloid scarring is more prevalent in darker-skinned individuals(Ud-Din & Bayat, 2013) with approximately 20-fold higher prevalence in African Americans (AAs) compared to European Americans (EAs) in the US(Barrett, 1973).
Several studies have attempted to identify the genetic bases of keloids, but few associating genes have been identified (Brown et al., 2008, Halim et al., 2012, Shih & Bayat, 2010, Nakashima et al., 2010, Emami et al., 2012, Yan et al., 2007, Chen et al., 2007, Marneros et al., 2004). However, in the only published genome-wide association study (GWAS) of keloids, in a Japanese population(Nakashima et al., 2010), four single nucleotide polymorphisms (SNPs) at three loci (1q41, 3q22.3-23 and 15q21.3) showed significant association with keloid formation; two of these three (1q41 and 15q21.3) replicated in a Chinese population(Zhu et al., 2013). Using admixture mapping our group identified a genomic region on chr15q21.2-22.3 with increased local African ancestry that associated with keloid risk and that included multiple common genetic variants impacting keloid formation in AAs(Velez Edwards et al., 2014). The region identified through admixture mapping included NEDD4, a gene implicated in the prior Japanese keloid GWAS. However, the strongest associations were in a nearby gene, MYO1E.
Given the success of gene-based analyses for identifying genes harboring multiple rare variants associating with disease(Wessel et al., 2015, Huyghe et al., 2013, Do et al., 2015, Cirulli, 2016), we sought to extend our previous association analysis into less common genetic variants through the use of gene-based regression and genetically predicted gene expression (GPGE) in (non-keloid) skin tissues from GTEx(2015). This latter analysis can provide in silico supporting evidence by which genetic variation may impact the phenotype in a tissue-specific manner, particularly when functional studies in the samples of interest are difficult or impossible to perform.
METHODS
Sample Collection and Phenotyping
Samples used in this study have been previously described(Velez Edwards et al., 2014). These studies were approved by the Vanderbilt University Institutional Review Board. Briefly, the study included DNA from 71 AA keloid cases and 399 controls from the BioVU DNA Repository and 36 cases from the keloid fibroblast repository described in detail below. BioVU keloid cases were defined as AAs 18 years or older who were diagnosed with keloids in the electronic health record (EHR) of patients at Vanderbilt University Medical Center (VUMC) and who have at least two mentions of a keloid diagnosis in their record (either two diagnostic codes [International Classification of Diseases (ICD) version 9 = 701.4] or a code and mention of a keloid within their record).
Keloid cases (n=36) were also obtained from a repository (15-53 year olds) that includes cultured fibroblasts from normal and keloid scar tissue(Russell et al., 2010, Smith et al., 2008). The diagnosis was made both by the surgeon or dermatologist removing the tissue and by the pathologist who examined the tissue. The principal criterion used to differentiate keloid from other hypertrophic scars was the extent to which the scar exceeded the boundary of the initiatial wound. DNA was also obtained from another repository of blood samples of unrelated individuals (n = 21) who were part of multiplex families.
Controls were AA subjects 18 years and older who have had surgical procedures performed at VUMC that involved an open wound, such as breast surgery, cesarean section and open heart surgery, and have two years of follow-up in the EHR with no evidence of keloid formation. Controls were excluded if they had ICD-9 codes for other fibroproliferative diseases (asthma, nephrosclerosis, or fibroids) in their EHR, or if the words “excessive scarring” were present.
Genotyping
Genotyping and quality control procedures have also been described elsewhere(Velez Edwards et al., 2014). Briefly, DNA samples were isolated from whole blood using the Autopure LS system (QIAGEN Inc., Valencia, CA). We genotyped DNA from the 492 participants using the custom Affymetrix Axiom Exome Genotyping Array (Affymetrix Inc., Santa Clara, CA). The genomic DNA samples were processed according to standard Affymetrix procedures for processing of the assay and genotype calling was performed using the Affymetrix Power Tools software (APT, Affymetrix Inc., Santa Clara, CA).
Genotyping Quality Control
Quality control procedures included evaluation of all SNPs for deviation from Hardy–Weinberg equilibrium (HWE) using PLINK software (Purcell et al. 2007). SNPs with HWE p ≤ 1.0 × 10−6, low genotyping efficiency (<95 %), duplicates, non-autosomal locations, and those which were monomorphic were removed. After removal of subjects and SNPs for quality control, 478 subjects (122 cases and 356 controls) and 163,613 SNPs remained for analyses.
Statistical Analysis
The program EPACTS (Efficient and Parallelizable Association Container Toolbox; http://csg.sph.umich.edu/kang/epacts/index.html) was used for variant annotation (ANNOVAR). This annotation grouped the variants by gene for use in gene-based burden tests. The Optimal Sequence Kernel Association Test (SKAT-O) was also implemented in EPACTS for gene-based association among nonsynonymous (missense, nonsense and splice-site changes) variants. All nonsynonymous variants were used, with no allele frequency threshold for inclusion, as SKAT-O weights those variants with a minor allele frequency of less than 0.05 more heavily than common variation. Sex and 10 principal components to account for population stratification among the samples were included as covariates. The Bonferroni significance threshold for this analysis (p-value < 3.9×10−6) accounted for the number of tests, represented by the number of genes with more than one variant (N = 12,714)
In order to evaluate the genetic association results in the context of gene expression relevant to keloid scars, we employed the method S-PrediXcan(Barbeira et al., 2016), an extension of the PrediXcan method(Gamazon et al., 2015). This approach utilizes genetic association results in conjunction with GPGE levels to infer gene expression association with keloids. Briefly, all genetic variants within 1 MB of each gene are assessed to identify SNPs with an impact on that gene’s expression (cis-eQTL SNPs). Accounting for linkage disequilibrium, SNPs are weighted for their relative effect on gene expression and collapsed using LASSO into a single predictive model for each gene’s GPGE in each tissue. S-PrediXcan builds on this approach through the use of summary statistics, rather than individual level data. Given that each SNP also has statistics for their association with the phenotype of interest, the association between the GPGE levels and the phenotype (keloids) can be inferred. S-PrediXcan identifies regulatory mechanisms through which genetic variants affect phenotype (and the direction in which they do so) on the gene-level rather than the SNP, while largely avoiding reverse causality since predicted expression levels are based on germline variation, which are not affected by onset of disease. For the purposes of this study, we utilized predicted expression weights built for two tissues from GTEx: sun exposed skin from the lower leg and skin which was not sun-exposed (from the suprapubic region), as well as transformed fibroblast cells. The prediction models and covariance matrices used are available on PredictDB (http://predictdb.hakyimlab.org/). Gene prediction models were filtered to exclude those not passing a false discovery rate threshold of 5%. P-values < 0.05 from more than one analysis were considered to be suggestive.
RESULTS
The majority of subjects were female (70% cases and 62% controls). The mean age of study participants was 43±17 for cases and 52±18 controls.
Gene-based association analyses
We identified 12,714 genes in the exome chip data with at least two variants meeting the annotation criteria. The SKAT-O association results are summarized in Figure 1. The top result, though not reaching conservative statistical significance for the number of tests, was the open reading frame C15orf63 (p-value = 6.61×10−6), which contained two low-frequency nonsynonymous variants, one of which was seen only once (singleton). A total of 12 variants reached suggestive levels of significance (p-value < 1×10−4), these are presented in Table 2.
Figure 1. Summary of SKAT-O association results for keloids using exome chip variants.
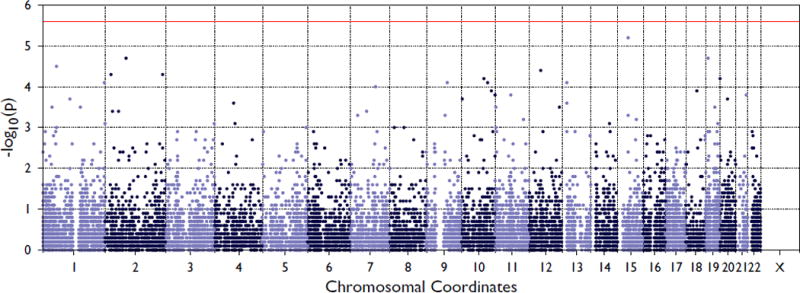
Red line represents Bonferroni significance threshold for multiple testing correction.
Table 2.
SKAT Results from exome-wide analyses.
| Chr. | Position (hg19) | Band | Gene | Fraction Rare† | N SNPs | N Singletons | p-value |
|---|---|---|---|---|---|---|---|
| 15 | 44092839–44093927 | 15q15.3 | C15orf63 | 0.08 | 2 | 1 | 6.41×10−6 |
| 19 | 11350326–11350935 | 19p13.2 | C19orf80 | 0.06 | 3 | 0 | 2.17×10−5 |
| 2 | 85570442–85578987 | 2p11.2 | RETSAT | 0.07 | 8 | 5 | 2.22×10−5 |
| 1 | 54417796–54433454 | 1p32.3 | LRRC42 | 0.02 | 2 | 0 | 3.41×10−5 |
| 2 | 232457693–232458648 | 2q37.1 | C2orf57 | 0.13 | 10 | 4 | 4.92×10−5 |
| 2 | 23977086–24090746 | 2p24.1–23.3 | ATAD2B | 0.08 | 8 | 1 | 5.57×10−5 |
| 10 | 90582744–90591710 | 10q23.31 | ANKRD22 | 0.006 | 2 | 1 | 6.94×10−5 |
| 1 | 246727700–246729395 | 1q44 | TFB2M | 0.008 | 3 | 2 | 7.62×10−5 |
| 13 | 20567666–20637020 | 13q12.11 | ZMYM2 | 0.07 | 4 | 2 | 8.01×10−5 |
| 10 | 106074366–106075500 | 10q25.1 | ITPRIP | 0.03 | 7 | 4 | 8.26×10−5 |
| 9 | 85863069–86153103 | 9q21.32 | FRMD3 | 0.03 | 5 | 0 | 8.31×10−5 |
| 7 | 102937991–102952123 | 7q22.1 | PMPCB | 0.07 | 6 | 2 | 9.79×10−5 |
Fraction of Samples w/a Rare Variant
Our previous admixture analysis and single-variant associations revealed a region of interest on chromosome 15, using common variants from the same exome chip. This extension of that work also had a suggestive hit on chromosome 15. However, when examining these SKAT-O results (which emphasize the uncommon variation) in the context of the admixture mapping, the top signals on the chromosome are proximal to the admixture mapping peak (Figure 2; Table 3). Further, the genes previously implicated (MYO1E in our previous work, and NEDD1 in a GWAS in Japanese subjects) are not associated in this gene-based analysis (p-value = 0.46 and 0.48, respectively), suggesting that the previously observed association at these genes are probably not due to known rare variants in those genes, but may be due to previously uncharacterized novel variants. Other genes reaching nominal significance in the admixture analyses from chromosome 15 are presented in Table 3. Nominally significant p-values for genes in this admixture mapping region ranged from <0.01 to 0.04.
Figure 2. Focused summary of SKAT-O gene-based association results on chromosome 15 overlaid on admixture mapping results.
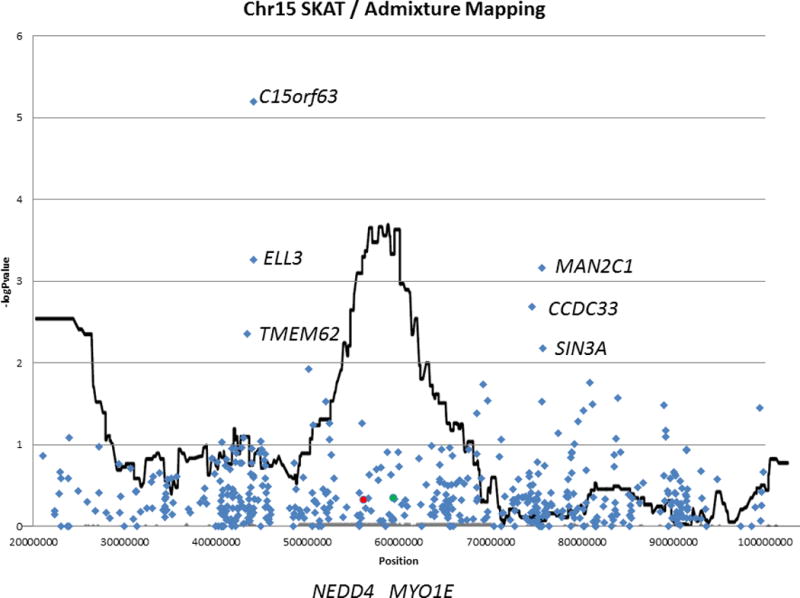
SKAT-O results for formerly detected genes of interest MYO1E and NEDD4 are indicated by green and red circles, respectively. Suggestive associations in this region flank the admixture mapping signal rather than occurring within the implicated region.
Table 3.
Suggestive (p-value < 0.005) S-PrediXcan results for skin predicted gene expression levels.
| Gene | Chr | Position | Band | Skin Tissue | z-score† | p-value | R2‡ | Q§ | N Variants¶ | Model N†† | SKAT p-value | SKAT N‡‡ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DCAF4 | 14 | 73404680–73425478 | 14q24.2 | Non sun-exposed | 3.45 | 5.52×10−4 | 0.045 | 0.002 | 2 | 33 | 0.242 | 10 |
| OR10A3 | 11 | 7960139–7961010 | 11p15.4 | Sun exposed | 3.40 | 6.86×10−4 | 0.012 | 0.018 | 1 | 45 | 0.150 | 8 |
| ANKRD7 | 7 | 117864712–117882784 | 7q31.31 | Sun exposed | 3.22 | 0.001 | 0.008 | 0.042 | 1 | 43 | NA | |
| OBFC1 | 10 | 105642542–105651971 | 10q24.33 | Sun exposed | −3.18 | 0.002 | 0.043 | 1.73×10−4 | 1 | 25 | 0.424 | 3 |
| ZNF580 | 19 | 56153418–56154836 | 19q13.42 | Non sun-exposed | 3.09 | 0.002 | 0.249 | 6.00×10−13 | 2 | 61 | 0.341 | 1 |
| GSTM1 | 1 | 110231746–110232965 | 1p13.3 | Non sun-exposed | −3.08 | 0.002 | 0.020 | 0.032 | 1 | 44 | 0.769 | 2 |
| TMEM253 | 14 | 21567096–21571883 | 14q11.2 | Sun exposed | −3.08 | 0.002 | 0.009 | 0.034 | 2 | 36 | NA | |
| TMEM229B | 14 | 67940444–67940444 | 14q24.1 | Sun exposed | 3.06 | 0.002 | 0.047 | 1.00×10−4 | 1 | 37 | 0.253 | 1 |
| HSPE1 | 2 | 198364721–198368187 | 2q33.1 | Non sun-exposed | −3.03 | 0.002 | 0.051 | 0.002 | 1 | 18 | NA | |
| PIGC | 1 | 172410967–172411374 | 1q24.3 | Sun exposed | −3.03 | 0.003 | 0.079 | 6.89×10−7 | 1 | 61 | 0.528 | 2 |
| NYAP1 | 7 | 100084515–100091399 | 7q22.1 | Non sun-exposed | 3.03 | 0.002 | 0.086 | 5.31×10−5 | 1 | 29 | 0.103 | 3 |
| QRICH2 | 17 | 74273284–74300497 | 17q25.1 | Sun exposed | −3.01 | 0.003 | 0.101 | 2.01×10−8 | 2 | 41 | 0.279 | 11 |
| NANP | 20 | 25596850–25597061 | 20p11.21 | Non sun-exposed | 2.99 | 0.002 | 0.052 | 0.001 | 2 | 62 | 1 | 2 |
| PNKP | 19 | 50364549–50370425 | 19q13.33 | Sun exposed | 2.98 | 0.003 | 0.034 | 7.32×10−4 | 1 | 5 | 0.052 | 6 |
| UBQLN4 | 1 | 156011427–156018299 | 1q22 | Non sun-exposed | 2.97 | 0.003 | 0.035 | 0.008 | 1 | 11 | 0.135 | 2 |
| SLPI | 20 | 43881680–43881682 | 20q13.12 | Sun exposed | 2.96 | 0.003 | 0.204 | 3.78×10−16 | 1 | 47 | 0.004 | 1 |
| ZNF432 | 19 | 52537202–52538294 | 19q13.41 | Non sun-exposed | 2.95 | 0.003 | 0.096 | 1.82×10−5 | 1 | 82 | 0.733 | 4 |
| ZNF471 | 19 | 57027720–57037309 | 19q13.43 | Sun exposed | 2.94 | 0.003 | 0.140 | 3.32×10−11 | 3 | 44 | 1 | 3 |
| CTC1 | 17 | 8131548–8151342 | 17p13.1 | Sun exposed | −2.91 | 0.004 | 0.016 | 0.011 | 1 | 14 | 0.336 | 10 |
| RNF222 | 17 | 8294023–8301144 | 17p13.1 | −2.91 | 0.003 | 0.028 | 0.014 | 1 | 43 | NA | ||
| DNAJB1 | 19 | 14625576–14640134 | 19p13.12 | Sun exposed | −2.89 | 0.004 | 0.121 | 4.68×10−9 | 1 | 31 | NA | |
| SLC6A11 | 3 | 10916706–10976854 | 3p25.3 | Non sun-exposed | −2.86 | 0.004 | 0.102 | 9.65×10−6 | 3 | 105 | 0.603 | 4 |
Z-score for association of predicted gene expression with increasing risk of keloids. Direction of effect of the expression is indicated by sign of the z-score+
Performance prediction R2
Prediction performance q-value.
Number of SNPs included in the prediction model for that gene available in the summary statistics.
Number of SNPs used to construct the prediction model for the gene in the tissue of interest using the GTEx data.
Number of SNPs within the gene included in the SKAT analysis. NA = gene was not available in this analysis
Evaluation of gene expression using GTEx
In order to evaluate the genetic association results in the context of gene expression relevant to keloid scars, we applied S-PrediXcan method to the single variant summary statistics. S-PrediXcan can identify regulatory mechanisms through which genetic variants effect the phenotype (and the direction in which they do so) on the gene-level rather than the SNP, while largely avoiding reverse causality since predicted expression levels are based on germline variation, which are not affected by onset of disease. GPGE in two skin tissues from GTEx was evaluated using the summary statistics from the single variant association analysis of keloids (Figure 3). The most significant result was with increased predicted DCAF4 expression in non-sun exposed skin tissue (from the suprapubic region; Table 3; p-value = 5.5×10−4). This gene is located on chromosome 14. The maximal GPGE result in sun-exposed skin (from the lower leg) was with increased OR10A3 on chromosome 11 (Table 3; p-value = 6.86×10−4).
Figure 3. S-PrediXcan results in three relevant tissues opposed from exome chip association results.
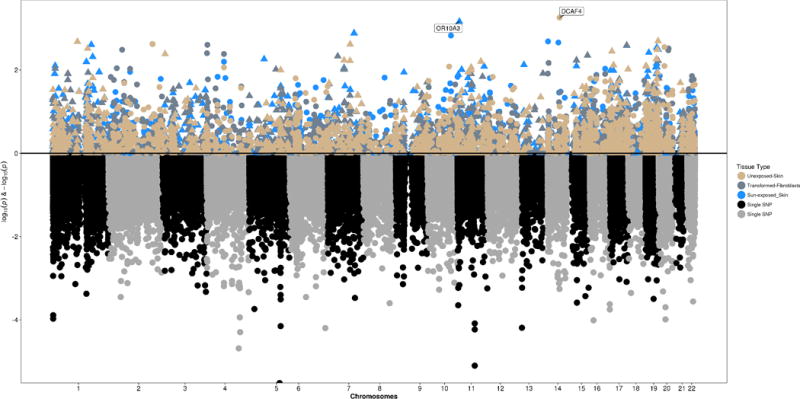
Upper panel displays genetically predicted gene expression results for sun-exposed and non-sun exposed skin, and transformed fibroblasts, while the bottom panel shows the genetic association results for keloid risk.
Overall, 23 genes had nominally significant (p<0.05) GPGE in both sun exposed and non-sun exposed skin, 10 from non-sun exposed tissue and 13 from sun exposed. There was no overlap of genes from the two tissues. Three of the 23 genes were also nominally associated through SKAT-O analysis of low frequency nonsynonymous variants (Table 4): SLPI and ZNF337 on chromosome 20, and ARFIP1 on chromosome 4. Most striking of these is SLPI (sun exposed p-value = 0.003, non-sun exposed p-value = 0.0392, SKAT-O p-value = 0.004), located on chromosome 20. Interestingly, increased predicted expression was associated with keloid risk in sun exposed skin, while decreased predicted expression associated with risk in non-sun exposed skin.
Table 4.
Selected genes with suggestive p-values (<0.05) from multiple analyses
| Gene | Chr | Position | Band | Sun-exposed Skin | Non-Sun-exposed Skin | SKAT | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| z-score | p-value | N Variants/Model N† | z-score | p-value | N Variants/Model N† | p-value | N‡ | ||||
| SLPI | 20 | 43881680–43881682 | 20q13.12 | 2.96 | 0.003 | 1/47 | −2.06 | 0.039 | 3/77 | 0.004 | 1 |
| ZNF337 | 20 | 25655765–25666737 | 20p11.1 | −2.65 | 0.008 | 1/82 | −2.65 | 0.008 | 1/28 | 0.032 | 4 |
| ARFIP1 | 4 | 153791955–153802203 | 4q31.3 | −2.21 | 0.027 | 1/15 | −2.21 | 0.027 | 1/14 | 0.027 | 1 |
| TMEM8B | 9 | 35829222–35854844 | 9p13.3 | −2.53 | 0.011 | 1/15 | NA | NA | NA | 0.004 | 5 |
| TOR3A | 1 | 179051112–179065129 | 1q25.2 | −2.25 | 0.025 | 1/20 | NA | NA | NA | 0.043 | 2 |
| MLXIPL | 7 | 73010191–73020440 | 7q11.23 | −2.62 | 0.009 | 3/16 | −2.76 | 0.006 | 5/48 | 0.679 | 6 |
| NAAA | 4 | 76836110–76862005 | 4q21.1 | 2.73 | 0.006 | 4/46 | 2.63 | 0.009 | 3/31 | 0.263 | 8 |
| ALDH16A1 | 19 | 49956613–49973578 | 19q13.33 | −2.70 | 0.007 | 3/21 | −2.30 | 0.022 | 2/58 | 0.423 | 14 |
| CDRT4 | 17 | 15341161–15341332 | 17p12 | 2.56 | 0.010 | 2/23 | 2.67 | 0.008 | 1/31 | 0.234 | 13 |
Boldface represents p-values < 0.05.
Number of SNPs included in the prediction model for that gene available in the summary statistics/Number of SNPs used to construct the prediction model for the gene in the tissue of interest using the GTEx data.
Number of SNPs with frequency <0.05 in the gene of interest and included in the analysis. NA = gene was not available in this analysis
A small number of the nominally significant GPGE results in either of the two skin tissues were located on chromosome 15, where the top SKAT-O signal and previously identified admixture mapping signal are located. FAM154B and RPAP1 were nominally (p-value < 0.05) associated in both sun-exposed skin and fibroblasts (data not shown), but not in non-sun exposed skin. Additional evaluation of the nominally significant results from SKAT-O the gene-based association testing revealed additional nominally significant GPGE results (Table 4).
DISCUSSION
This study sought to evaluate gene-based associations with keloids in AA through two mechanisms: the low-frequency coding variations assessed collectively for association with keloid risk, and prediction of keloid-associated genetic regulation of gene expression levels. The top result from the gene-based association tests was located in an open reading frame on chromosome 15. C15orf63, also known as HYPK (huntingtin interacting protein K), is a chaperone protein which interacts with huntingtin, the protein causing Huntington’s disease(Choudhury & Bhattacharyya, 2015, Raychaudhuri et al., 2014, Sakurai et al., 2014).
We also employed S-PrediXcan to evaluate the association of the genetic variants implicated in keloid risk with expression in skin tissues from GTEx (Figure 3). This technique summarizes all eQTL variants (within one megabase [MB] of the gene) impacting a gene’s expression into a single unit to infer association between GPGE in a given tissue and the outcome of interest. The most strongly associated GPGE with keloids was with increased DCAF4 in non-sun exposed skin tissue (Table 3; p-value = 5.5×10−4). This gene, DDB1 and CUL4 associated factor 4, is located on chromosome 14 and has been previously associated with leukocyte telomere length(Mangino et al., 2015) and lung cancer risk(Liu et al., 2017, Yan et al., 2017). Notably, the variants associated with telomere length were also eQTLs in sun exposed skin, while there was no association between keloid risk and predicted expression of DCAF4 in sun exposed skin in this study (p-value = 0.57).
Among the genes identified from gene-based association analyses, three had evidence from both skin tissues’ gene expression as well as gene-based burden tests (Table 4). Most notable of these is SLPI (secretory leukocyte peptidase inhibitor), for which evidence supports a role in several skin disorders(Schafer et al., 2014, Lancto et al., 2013, Ashcroft et al., 2012, Skrzeczynska-Moncznik et al., 2012, Meyer-Hoffert, 2009, Bando et al., 2007), as well as a wide variety of other fibrotic diseases (Habgood et al., 2016, Hentschel et al., 2015, Nair et al., 2013, Aozasa et al., 2012, Thijs et al., 2015) and cancer(Zheng et al., 2016, Noorlag et al., 2015, Zuo et al., 2015, Rosso et al., 2014, Timms et al., 2014). SLPI has also been found to reduce contractility of fibroblast-mediated collagen gel models of scarring, suggesting that it may have uses in promotion of scarless wound healing(Sumi et al., 2000). The discordant directions of effect for SLPI predicted expression is interesting (Table 4), however this may reflect differences in skin where keloids are more likely to occur. However, as keloids are more likely to be found on the head/neck and upper extremities, neither the sun-exposed skin from the lower leg nor non-sun exposed skin from the suprapubic region can provide a clear picture of the most appropriate direction for keloid risk. ZNF337 (zinc finger protein 337) and ARFIP1 (ADP ribosylation factor interacting protein 1) were also nominally associated in sun-exposed and non-sun exposed skin and SKAT-O. Both of these genes are expressed in a wide variety of tissues from GTEx, but neither has yet been implicated in disease pathophysiology.
Although these gene-based analyses did not support the previous findings with NEDD4 and MYO1E (Supplemental Table 1), it is important to note that those genes were implicated through mechanisms that emphasized the role of common variation (MAF>0.05), while the current study weighted only those uncommon variants that also were predicted to be functionally important. Furthermore, the GPGE analysis was unable to construct models for either NEDD4 or MYO1E GPGE. Therefore, we suggest that although common variation in NEDD4 and MYO1E may be important for the development of keloid scars, rare variation in these genes does not appear to have an impact, and the relationship between the genetic association with keloids and gene expression levels of these genes in lower leg or supra-pubic skin or fibroblast tissues remains undetermined.
Neither the gene-based association nor GPGE analyses reached multiple testing-corrected significance; however, considering convergent results which were suggestive in both analyses revealed genes with potential biological relevance to keloid risk. The lack of striking evidence for association in any one analysis may be due to small sample size and lack of comprehensive genomic evaluation. The association analyses presented here were limited to those known coding variants contained on the exome chip, which is known to be sub-optimal for non-EA populations(Nievergelt et al., 2014). The relatively small number of available SNPs also impacts the S-PrediXcan analysis, with only small proportions of the SNPs used in construction of the models available. Despite these challenges, studies of this type add new evidence to the body of literature evaluating the genetic component involved in the formation of keloid scarring in African American populations. Studies of genome-wide association in larger numbers of samples are necessary to refine the impact of genetic variation underlying keloid development across many population groups. This may be enabled by the growth of large biorepositories with genome-wide genotyping resources and linked to EHRs for ease of identification of cases and controls. Further knowledge may also be generated by obtaining eQTL databases for skin tissues more directly relevant to keloid formation, i.e. those located in chest/abdominal areas or face(Yedomon et al., 2012).
In summary, we evaluated whole-exome genotyping data for evidence of gene-based association with risk of keloids and observed modest evidence of association in several genes, based on both genetic and expression data. The finding that predicted expression of SLPI in skin tissue was associated with keloids is the first evidence of this type and is supported by the association with low frequency coding variants in that gene, as well as the known biology of this gene in skin disorders and potential role in scar formation. The results of this study indicate that the association of previously implicated genes NEDD4 and MYO1E is not due to underlying associations with low-frequency or rare coding variants, at least in this sample of AAs, though further evaluation of these and other genes in diverse populations will continue to inform. Finally, the identification of multiple genes associated with previously known pathways suggests a plausible role for genetic variants impacting keloid risk in AAs.
Supplementary Material
Supplemental Figure 1. Quantile-quantile plot for SKAT-O test.
Table 1.
Summary of demographic characteristics and ancestry estimates
| Keloid Cases (N = 122) |
Controls (N = 356) |
p-value | |
|---|---|---|---|
| Agea, years (Mean(SD)) | 43 (17) | 52 (18) | >0.0001 |
| Sex (% Female) | 70 | 62 | 0.13 |
| European Ancestry (%) | 19 | 21 | 0.011 |
Age was only available for samples obtained from BioVU
Acknowledgments
The authors gratefully acknowledge Ayush Giri for the provision of the R code used to generate Figure 3. Funding for this work was provided by NIH grant R21AR067938 to DRVE. The datasets used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding, the 1S10RR025141−01 instrumentation award, and by the Vanderbilt CTSA grant UL1TR000445 from NCATS/NIH. This project was also supported in part by the Vanderbilt Molecular and Genetic Epidemiology of Cancer (MAGEC) training program, which was funded by the US National Cancer Institute grant R25 CA160056 (PI: X.-O. Shu).
Footnotes
Author contributions: Study design: SBR, TLE, DRVE. Data Collection: SBR, SMW, TLE, DRVE. Data Analysis: JNH. Manuscript preparation: JNH, SBR, SMW, TLE, DRVE.
Conflict of Interest Statement: The authors declare no conflicts of interest
References
- Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (New York, NY) 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aozasa N, Asano Y, Akamata K, Noda S, Masui Y, Tamaki Z, Tada Y, Sugaya M, Kadono T, Sato S. Clinical significance of serum levels of secretory leukocyte protease inhibitor in patients with systemic sclerosis. Modern rheumatology/the Japan Rheumatism Association. 2012;22:576–83. doi: 10.1007/s10165-011-0553-1. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Jeong MJ, Ashworth JJ, Hardman M, Jin W, Moutsopoulos N, Wild T, Mccartney-Francis N, Sim D, Mcgrady G, Song XY, Wahl SM. Tumor necrosis factor-alpha (TNF-alpha) is a therapeutic target for impaired cutaneous wound healing. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2012;20:38–49. doi: 10.1111/j.1524-475X.2011.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Hiroshima Y, Kataoka M, Shinohara Y, Herzberg MC, Ross KF, Nagata T, Kido J. Interleukin-1alpha regulates antimicrobial peptide expression in human keratinocytes. Immunology and cell biology. 2007;85:532–7. doi: 10.1038/sj.icb.7100078. [DOI] [PubMed] [Google Scholar]
- Barbeira A, Dickinson SP, Torres JM, Torstenson ES, Zheng J, Wheeler HE, Shah KP, Edwards T, Consortium, G. Nicolae D, Cox NJ, Im HK. Integrating tissue specific mechanisms into GWAS summary results. bioRxiv 2016 [Google Scholar]
- Barrett J. Keloid. In: Bergsma D, editor. Birth Defect Compendium. Baltimore: Williams and Wilkins Company; 1973. Birth Defect Compendium. [Google Scholar]
- Brown JJ, Ollier W, Arscott G, Ke X, Lamb J, Day P, Bayat A. Genetic susceptibility to keloid scarring: SMAD gene SNP frequencies in Afro-Caribbeans. Experimental Dermatology. 2008;17:610–613. doi: 10.1111/j.1600-0625.2007.00654.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Gao JH, Yan X, Song M, Liu XJ. Location of predisposing gene for one Han Chinese keloid pedigree. Zhonghua Zheng Xing Wai Ke Za Zhi = Zhonghua Zhengxing Waike Zazhi = Chinese Journal of Plastic Surgery. 2007;23:137–140. [PubMed] [Google Scholar]
- Chevray PM, Manson PN. Keloid scars are formed by polyclonal fibroblasts. Annals of plastic surgery. 2004;52:605–608. doi: 10.1097/01.sap.0000099280.29831.6e. [DOI] [PubMed] [Google Scholar]
- Choudhury KR, Bhattacharyya NP. Chaperone protein HYPK interacts with the first 17 amino acid region of Huntingtin and modulates mutant HTT-mediated aggregation and cytotoxicity. Biochemical and biophysical research communications. 2015;456:66–73. doi: 10.1016/j.bbrc.2014.11.035. [DOI] [PubMed] [Google Scholar]
- Cirulli ET. The Increasing Importance of Gene-Based Analyses. PLoS genetics. 2016;12:e1005852. doi: 10.1371/journal.pgen.1005852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Stitziel NO, Won HH, Jørgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, Guella I, Asselta R, Lange LA, Peloso GM, Auer PL, Project NES, Girelli D, Martinelli N, Farlow DN, Depristo MA, Roberts R, Stewart AFR, Saleheen D, Danesh J, Epstein SE, Sivapalaratnam S, Hovingh GK, Kastelein JJ, Samani NJ, Schunkert H, Erdmann J, Shah SH, Kraus WE, Davies R, Nikpay M, Johansen CT, Wang J, Hegele RA, Hechter E, Marz W, Kleber ME, Huang J, Johnson AD, Li M, Burke GL, Gross M, Liu Y, Assimes TL, Heiss G, Lange EM, Folsom AR, Taylor HA, Olivieri O, Hamsten A, Clarke R, Reilly DF, Yin W, Rivas MA, Donnelly P, Rossouw JE, Psaty BM, Herrington DM, Wilson JG, Rich SS, Bamshad MJ, Tracy RP, Cupples LA, Rader DJ, Reilly MP, Spertus JA, Cresci S, Hartiala J, Tang WHW, Hazen SL, Allayee H, Reiner AP, Carlson CS, Kooperberg C, Jackson RD, Boerwinkle E, Lander ES, Schwartz SM, Siscovick DS, Mcpherson R, Tybjaerg-Hansen A, Abecasis GR, Watkins H, Nickerson DA, Ardissino D, Sunyaev SR, O'donnell CJ, Altshuler D, Gabriel S, Kathiresan S. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami A, Halim AS, Salahshourifar I, Yussof SJM, Khoo TL, Kannan TP. Association of TGFβ1 and SMAD4 variants in the etiology of keloid scar in the Malay population. Archives of dermatological research. 2012;304:541–547. doi: 10.1007/s00403-012-1262-0. [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habgood AN, Tatler AL, Porte J, Wahl SM, Laurent GJ, John AE, Johnson SR, Jenkins G. Secretory leukocyte protease inhibitor gene deletion alters bleomycin-induced lung injury, but not development of pulmonary fibrosis. Laboratory investigation; a journal of technical methods and pathology. 2016 doi: 10.1038/labinvest.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim AS, Emami A, Salahshourifar I, Kannan TP. Keloid scarring: understanding the genetic basis, advances, and prospects. Archives of plastic surgery. 2012;39:184–9. doi: 10.5999/aps.2012.39.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Deng Z, Alghamdi M, Lu L, Fear MW, He L. From genetics to epigenetics: new insights into keloid scarring. Cell proliferation. 2017 doi: 10.1111/cpr.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel J, Fischer N, Janhsen WK, Markert UR, Lehmann T, Sonnemann J, Boer K, Pfister W, Hipler UC, Mainz JG. Protease-antiprotease imbalances differ between Cystic Fibrosis patients' upper and lower airway secretions. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2015;14:324–33. doi: 10.1016/j.jcf.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stančáková A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, Chines PS, Teslovich TM, Romm JM, Ling H, Mcmullen I, Ingersoll R, Pugh EW, Doheny KF, Neale BM, Daly MJ, Kuusisto J, Scott LJ, Kang HM, Collins FS, Abecasis GR, Watanabe RM, Boehnke M, Laakso M, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladin DA, Hou Z, Patel D, Mcphail M, Olson JC, Saed GM, Fivenson DP. p53 and apoptosis alterations in keloids and keloid fibroblasts. Wound Repair and Regeneration: Official Publication of the Wound Healing Society [and] the European Tissue Repair Society. 1998;6:28–37. doi: 10.1046/j.1524-475x.1998.60106.x. [DOI] [PubMed] [Google Scholar]
- Lancto CA, Torres SM, Hendrickson JA, Martins KV, Rutherford MS. Altered expression of antimicrobial peptide genes in the skin of dogs with atopic dermatitis and other inflammatory skin conditions. Veterinary dermatology. 2013;24:414–21 e90. doi: 10.1111/vde.12034. [DOI] [PubMed] [Google Scholar]
- Liu H, Liu Z, Wang Y, Stinchcombe TE, Owzar K, Han Y, Hung RJ, Brhane Y, Mclaughlin J, Brennan P, Bickeboller H, Rosenberger A, Houlston RS, Caporaso N, Landi MT, Bruske I, Risch A, Wu X, Ye Y, Christiani DC, Amos CI, Wei Q. Functional variants in DCAF4 associated with lung cancer risk in European populations. Carcinogenesis. 2017 doi: 10.1093/carcin/bgx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangino M, Christiansen L, Stone R, Hunt SC, Horvath K, Eisenberg DT, Kimura M, Petersen I, Kark JD, Herbig U, Reiner AP, Benetos A, Codd V, Nyholt DR, Sinnreich R, Christensen K, Nassar H, Hwang SJ, Levy D, Bataille V, Fitzpatrick AL, Chen W, Berenson GS, Samani NJ, Martin NG, Tishkoff S, Schork NJ, Kyvik KO, Dalgard C, Spector TD, Aviv A. DCAF4, a novel gene associated with leucocyte telomere length. J Med Genet. 2015;52:157–62. doi: 10.1136/jmedgenet-2014-102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marneros AG, Krieg T. Keloids–clinical diagnosis, pathogenesis, and treatment options. Journal der Deutschen Dermatologischen Gesellschaft = Journal of the German Society of Dermatology: JDDG. 2004;2:905–913. doi: 10.1046/j.1439-0353.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- Marneros AG, Norris JE, Watanabe S, Reichenberger E, Olsen BR. Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. The Journal of investigative dermatology. 2004;122:1126–32. doi: 10.1111/j.0022-202X.2004.22327.x. [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U. Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Archivum immunologiae et therapiae experimentalis. 2009;57:345–54. doi: 10.1007/s00005-009-0045-6. [DOI] [PubMed] [Google Scholar]
- Moulton-Levy P, Jackson CE, Levy HG, Fialkow PJ. Multiple cell origin of traumatically induced keloids. Journal of the American Academy of Dermatology. 1984;10:986–988. doi: 10.1016/s0190-9622(84)80319-9. [DOI] [PubMed] [Google Scholar]
- Nair S, Saed GM, Atta HM, Rajaratnam V, Diamond MP, Curiel DT, Al-Hendy A. Towards gene therapy of postoperative adhesions: fiber and transcriptional modifications enhance adenovirus targeting towards human adhesion cells. Gynecologic and obstetric investigation. 2013;76:119–24. doi: 10.1159/000353426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, Hosono N, Kubo M, Nakamura Y, Zembutsu H. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nature Genetics. 2010;42:768–771. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- Niessen FB, Spauwen PH, Schalkwijk J, Kon M. On the nature of hypertrophic scars and keloids: a review. Plastic and reconstructive surgery. 1999;104:1435–1458. doi: 10.1097/00006534-199910000-00031. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Wineinger NE, Libiger O, Pham P, Zhang G, Baker DG, Schork NJ. Chip-based direct genotyping of coding variants in genome wide association studies: Utility, issues and prospects. Gene. 2014 doi: 10.1016/j.gene.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorlag R, Van Der Groep P, Leusink FK, Van Hooff SR, Frank MH, Willems SM, Van Es RJ. Nodal metastasis and survival in oral cancer: Association with protein expression of SLPI, not with LCN2, TACSTD2, or THBS2. Head & neck. 2015;37:1130–6. doi: 10.1002/hed.23716. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Banerjee R, Mukhopadhyay S, Bhattacharyya NP. Conserved C-terminal nascent peptide binding domain of HYPK facilitates its chaperone-like activity. Journal of biosciences. 2014;39:659–72. doi: 10.1007/s12038-014-9442-z. [DOI] [PubMed] [Google Scholar]
- Rosso M, Lapyckyj L, Amiano N, Besso MJ, Sanchez M, Chuluyan E, Vazquez-Levin MH. Secretory Leukocyte Protease Inhibitor (SLPI) expression downregulates E-cadherin, induces beta-catenin re-localisation and triggers apoptosis-related events in breast cancer cells. Biology of the cell/under the auspices of the European Cell Biology Organization. 2014;106:308–22. doi: 10.1111/boc.201300075. [DOI] [PubMed] [Google Scholar]
- Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, Broquist AH, Raju L, Williams SM. Epigenetically altered wound healing in keloid fibroblasts. The Journal of investigative dermatology. 2010;130:2489–96. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saed GM, Ladin D, Olson J, Han X, Hou Z, Fivenson D. Analysis of p53 gene mutations in keloids using polymerase chain reaction-based single-strand conformational polymorphism and DNA sequencing. Archives of dermatology. 1998;134:963–967. doi: 10.1001/archderm.134.8.963. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Sawai M, Ishikawa Y, Ota A, Kawahara E. Heat shock transcription factor HSF1 regulates the expression of the Huntingtin-interacting protein HYPK. Biochim Biophys Acta. 2014;1840:1181–7. doi: 10.1016/j.bbagen.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Schafer M, Willrodt AH, Kurinna S, Link AS, Farwanah H, Geusau A, Gruber F, Sorg O, Huebner AJ, Roop DR, Sandhoff K, Saurat JH, Tschachler E, Schneider MR, Langbein L, Bloch W, Beer HD, Werner S. Activation of Nrf2 in keratinocytes causes chloracne (MADISH)-like skin disease in mice. EMBO molecular medicine. 2014;6:442–57. doi: 10.1002/emmm.201303281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih B, Bayat A. Genetics of keloid scarring. Archives of dermatological research. 2010;302:319–339. doi: 10.1007/s00403-009-1014-y. [DOI] [PubMed] [Google Scholar]
- Skrzeczynska-Moncznik J, Wlodarczyk A, Zabieglo K, Kapinska-Mrowiecka M, Marewicz E, Dubin A, Potempa J, Cichy J. Secretory leukocyte proteinase inhibitor-competent DNA deposits are potent stimulators of plasmacytoid dendritic cells: implication for psoriasis. Journal of immunology (Baltimore, Md: 1950) 2012;189:1611–7. doi: 10.4049/jimmunol.1103293. [DOI] [PubMed] [Google Scholar]
- Smith JC, Boone BE, Opalenik SR, Williams SM, Russell SB. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. The Journal of investigative dermatology. 2008;128:1298–310. doi: 10.1038/sj.jid.5701149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi Y, Muramatsu H, Hata K, Ueda M, Muramatsu T. Secretory leukocyte protease inhibitor is a novel inhibitor of fibroblast-mediated collagen gel contraction. Exp Cell Res. 2000;256:203–12. doi: 10.1006/excr.2000.4815. [DOI] [PubMed] [Google Scholar]
- Thijs W, Janssen K, Van Schadewijk AM, Papapoulos SE, Le Cessie S, Middeldorp S, Melissant CF, Rabe KF, Hiemstra PS. Nasal Levels of Antimicrobial Peptides in Allergic Asthma Patients and Healthy Controls: Differences and Effect of a Short 1,25(OH)2 Vitamin D3 Treatment. PLoS One. 2015;10:e0140986. doi: 10.1371/journal.pone.0140986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms JF, Arslan-Low E, Kabir M, Worthington J, Camuzeaux S, Sinclair J, Szaub J, Afrough B, Podust VN, Fourkala EO, Cubizolles M, Kronenberg F, Fung ET, Gentry-Maharaj A, Menon U, Jacobs I. Discovery of serum biomarkers of ovarian cancer using complementary proteomic profiling strategies. Proteomics Clinical applications. 2014;8:982–93. doi: 10.1002/prca.201400063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Din S, Bayat A. Strategic management of keloid disease in ethnic skin: a structured approach supported by the emerging literature. The British journal of dermatology. 2013;169(Suppl 3):71–81. doi: 10.1111/bjd.12588. [DOI] [PubMed] [Google Scholar]
- Velez Edwards DR, Tsosie KS, Williams SM, Edwards TL, Russell SB. Admixture mapping identifies a locus at 15q21.2-22.3 associated with keloid formation in African Americans. Hum Genet. 2014;133:1513–23. doi: 10.1007/s00439-014-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, Brody JA, Dauriz M, Hivert MF, Raghavan S, Lipovich L, Hidalgo B, Fox K, Huffman JE, An P, Lu Y, Rasmussen-Torvik LJ, Grarup N, Ehm MG, Li L, Baldridge AS, Stancakova A, Abrol R, Besse C, Boland A, Bork-Jensen J, Fornage M, Freitag DF, Garcia ME, Guo X, Hara K, Isaacs A, Jakobsdottir J, Lange LA, Layton JC, Li M, Hua Zhao J, Meidtner K, Morrison AC, Nalls MA, Peters MJ, Sabater-Lleal M, Schurmann C, Silveira A, Smith AV, Southam L, Stoiber MH, Strawbridge RJ, Taylor KD, Varga TV, Allin KH, Amin N, Aponte JL, Aung T, Barbieri C, Bihlmeyer NA, Boehnke M, Bombieri C, Bowden DW, Burns SM, Chen Y, Chen YD, Cheng CY, Correa A, Czajkowski J, Dehghan A, Ehret GB, Eiriksdottir G, Escher SA, Farmaki AE, Franberg M, Gambaro G, Giulianini F, Goddard WA, 3rd, Goel A, Gottesman O, Grove ML, Gustafsson S, Hai Y, Hallmans G, Heo J, Hoffmann P, Ikram MK, Jensen RA, Jorgensen ME, Jorgensen T, Karaleftheri M, Khor CC, Kirkpatrick A, Kraja AT, Kuusisto J, Lange EM, Lee IT, Lee WJ, Leong A, Liao J, Liu C, Liu Y, Lindgren CM, Linneberg A, Malerba G, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nature communications. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bi L, Wang Y, Zhang X, Hou Z, Wang Q, Snijders AM, Mao JH. Integrative analysis of multi-omics data reveals distinct impacts of DDB1-CUL4 associated factors in human lung adenocarcinomas. Scientific reports. 2017;7:333. doi: 10.1038/s41598-017-00512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Gao JH, Chen Y, Song M, Liu XJ. Preliminary linkage analysis and mapping of keloid susceptibility locus in a Chinese pedigree. Zhonghua Zheng Xing Wai Ke Za Zhi = Zhonghua Zhengxing Waike Zazhi = Chinese Journal of Plastic Surgery. 2007;23:32–35. [PubMed] [Google Scholar]
- Yedomon GH, Adegbidi H, Atadokpede F, Akpadjan F, Mouto EJ, Do Ango-Padonou F. Keloids on dark skin: a consecutive series of 456 cases. Medecine et sante tropicales. 2012;22:287–91. doi: 10.1684/mst.2012.0052. [DOI] [PubMed] [Google Scholar]
- Zheng D, Gui B, Gray KP, Tinay I, Rafiei S, Huang Q, Sweeney CJ, Kibel AS, Jia L. Secretory leukocyte protease inhibitor is a survival and proliferation factor for castration-resistant prostate cancer. Oncogene. 2016 doi: 10.1038/onc.2016.13. [DOI] [PubMed] [Google Scholar]
- Zhu F, Wu B, Li P, Wang J, Tang H, Liu Y, Zuo X, Cheng H, Ding Y, Wang W, Zhai Y, Qian F, Wang W, Yuan X, Wang J, Ha W, Hou J, Zhou F, Wang Y, Gao J, Sheng Y, Sun L, Liu J, Yang S, Zhang X. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PloS One. 2013;8:e62377. doi: 10.1371/journal.pone.0062377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Zhang C, Ren C, Pang D, Li Y, Xie X, Tang Z, Jiang X. Secretory leukocyte protease inhibitor is a proliferation and survival factor for pancreatic cancer cells. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2015;17:314–21. doi: 10.1007/s12094-014-1232-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Quantile-quantile plot for SKAT-O test.


