Abstract
The spatial organization of chromosomes in the nuclear space is an extensively studied field that relies on measurements of structural features and 3D positions of chromosomes with high precision and robustness. However, no tools are currently available to image and analyze chromosome territories in a high-throughput format. Here, we have developed High-throughput Chromosome Territory Mapping (HiCTMap), a method for the robust and rapid analysis of 2D and 3D chromosome territory positioning in mammalian cells. HiCTMap is a high-throughput imaging-based chromosome detection method which enables routine analysis of chromosome structure and nuclear position. Using an optimized FISH staining protocol in a 384-well plate format in conjunction with a bespoke automated image analysis workflow, HiCTMap faithfully detects chromosome territories and their position in 2D and 3D in a large population of cells per experimental condition. We apply this novel technique to visualize chromosomes 18, X, and Y in male and female primary human skin fibroblasts, and show accurate detection of the correct number of chromosomes in the respective genotypes. Given the ability to visualize and quantitatively analyze large numbers of nuclei, we use HiCTMap to measure chromosome territory area and volume with high precision and determine the radial position of chromosome territories using either centroid or equidistant-shell analysis. The HiCTMap protocol is also compatible with RNA FISH as demonstrated by simultaneous labeling of X chromosomes and Xist RNA in female cells. We suggest HiCTMap will be a useful tool for routine precision mapping of chromosome territories in a wide range of cell types and tissues.
Keywords: Chromosome territory, X-chromosome inactivation, High-throughput imaging, Chromosome paint, Random forest, Machine learning
1. Introduction
The organization of the eukaryotic genome extends from DNA that is coiled around histone octamers to form nucleosomes, which are further folded into a chromatin fiber, to ultimately form chromosomes [1–3]. Chromosomes are the largest unit of genome organization and are confined to distinct regions within the nucleus known as chromosome territories (CTs) [4]. The spatial organization of CTs is non-random, and CTs occupy preferred nuclear positions [5]. Changes in genome spatial organization are observed during stages of differentiation, transcription, and diseases [6–9]. Furthermore, chromosome position is cell type- and tissue-specific [10–13].
Fluorescence in situ hybridization (FISH) of entire chromosomes using specific DNA probes, also known as chromosome paints, provides vital insight into defining chromosome architecture and position within the 3D space of the nucleus [14]. By measuring the radial position of a chromosome relative to the center of the nucleus, or their relative position to each other, it has been recognized that CTs are non-randomly organized in the 3D space of the nucleus [10,15–17]. These experiments show that chromosomes have a characteristic distribution of positioning within the cell nucleus, however, the preferential positioning of chromosomes is not absolute and varies probabilistically between individual cells in a population [3,5].
Because of the stochastic distribution of each chromosome in the cell population, accurate mapping of chromosome positions requires analysis of a relatively large number of cells. Due to limitations in imaging throughput of conventional microscopy methods, the number of cells commonly analyzed for chromosome paint experiments are often limited, typically on the order of a hundred cells or less.
We have developed HiCTMap, an unbiased and systematic method for the quantitative detection of CTs using high-throughput imaging that enables routine analysis of hundreds of nuclei and CTs per sample in a single experiment. The HiCTMap protocol consists of fixation of interphase cells in 384-well plates, followed by DNA FISH staining using chromosome paint probes. Large image data sets are acquired using automated 3D confocal high-throughput microscopy and analyzed with a custom high-content image analysis workflow to determine the spatial positioning and features of chromosomes in multiple imaging channels. In these workflows, we coupled existing image processing algorithms for segmentation of nuclei [18–20] and CTs [21] with supervised classifiers [22,23] for filtering out mis-segmented nuclei and non-CT regions. The nucleus segmentation algorithm in our custom workflows follows a similar approach as other programs developed for studying nuclear architecture [24,25]. As proof-of-principle, we applied HiCTMap to measure the size and positioning of chromosomes 18, X, and Y in normal male and female human primary skin fibroblasts. We demonstrate that chromosome X has a more peripheral position than chromosomes 18 and Y, as previously shown by others [16,17,26,27]. Moreover, we combined RNA FISH and chromosome paint in a high throughput format to differentiate active and inactive X chromosomes and demonstrate accurate detection of the reduced size and a slightly more peripheral positioning of the inactive X chromosome.
2. Materials and methods
2.1. Cell culture
Primary human skin fibroblasts were isolated as follows: a sterile 3 mm skin punch biopsy was obtained from the inside forearm of each subject and placed immediately in complete media: DMEM (ThermoFisher Scientific, Cat # 10566-016) supplemented with 20% fetal bovine serum (ThermoFisher Scientific, Cat # 10082-147) and with 50 U/ml penicillin/streptomycin (ThermoFisher Scientific, Cat # 15070-030). The biopsy was then dissected into approximately 9–12 pieces; these pieces were secured to the bottoms of T25 flasks, covered with complete media, and grown at 37 °C and 5% CO2. Flasks were monitored frequently and media was changed at least once a week. Fibroblasts were expanded to passage 1 (p1) when their outgrowth covered approximately one third of the flask, and/or when the fibroblast outgrowth on any portion of the flask appeared to be approaching confluency, to avoid contact inhibition. Expansion was done by removing media, washing cells with PBS (Phosphate Buffer Saline, ThermoScientific, Cat 10010-023) and adding TryLE Express (ThermoFisher, Cat # 12604-013). Expanded fibroblasts were seeded into new T25 or T75 flasks, ensuring confluency remained above 10%. Fibroblast freezes were prepared from flasks that were 80–95% confluent. Freezing media contained complete media supplemented with 10% DMSO (Sigma, Cat # D2650).
Female (XX) and male (XY) primary human skin fibroblast were thawed in DMEM media supplemented with 20% FBS, 2 mM Gluta-MAX™, and 1% penicillin/streptomycin (ThermoFisher Scientific, Cat # 15140122) at 37 °C and 5% CO2. Cells were split 1:2 every 3–4 days and kept at a low passage (P3-5). We find that more than 40% of cells are Ki67 positive, demonstrating they are neither senescent nor quiescent. Cells were plated in PerkinElmer Cell Carrier Ultra 384-well plates (PerkinElmer, Cat # 6057300) at a density of 4000 cells/well and grown overnight, then fixed for 10 min in 4% PFA (Electron Microscopy Sciences, Cat # 15710), washed, and stored in DPBS (Dulbecco’s Phosphate Buffer Saline) (Lonza, Cat # 17-512F) at 4 °C.
2.2. Chromosome paint FISH in 384-well plates
Whole chromosome paint probes were generated in house to human chromosomes 18 (HSA-18), X (HSA-X), and Y (HSA-Y) using Spectrum Orange (Abbott Molecular, Abbot Park, IL, USA, Cat # 02N33-050), Dy505 (Dyomics, Jena, Germany, Cat # Dy505-dUTP), and Dy651 (Dyomics, Jena, Germany, Cat # Dy651-dUTP), for labeling of HSA18, HSA-X and HSA-Y, respectively. Detailed protocols are available online at (https://ccr.cancer.gov/Genetics-Branch/thomas-ried, Resources). Probe mix for chromosome paint probes consisting each of 300 ng DNA, 30 μg salmon sperm DNA (Ambion, Cat # AM9680), and 1 μg human COT1 Human DNA (Sigma-Aldrich Roche, Cat # 11581074001) per well was ethanol precipitated at −80 °C for 25 min and re-suspended in 12 μL of hybridization buffer per well (50% deionized formamide, pH7 (Ambion, Cat # AM9342), 20% dextran sulfate (Milipore, Cat # S4030), and 2X SSC (KSD Scientific, Cat # B3821-1000).
Fixed cells were permeabilized for 20 min in 0.5% Triton X-100 (Sigma-Aldrich, Cat # T9234-500ML) in DPBS, washed in 0.05% Triton X-100 in DPBS, incubated for 10 min in 0.1 N HCl, neutralized for 5 min in 2X SSC, and equilibrated for at least 30 min in 50% formamide in 2X SSC prior to probe addition. Probes and nuclear DNA were denatured at 85 °C for 7 min and plates were immediately moved to a 37 °C humidified chamber for overnight hybridization. The next day, plates were washed 3 times for 5 min each in 50% formamide (Sigma-Aldrich, Cat # 47670-1L-F) in 2X SSC at 42 °C, 1X SSC at 60 °C, and 0.1X SSC at 60 °C.
A DesignReady Xist RNA probe designed by Stellaris (25 μMconcentration; LGC Biosearch Technologies, Petaluma, CA, Cat # VSMF-2431-5) was diluted 1:10 in TE buffer, pH 8 and diluted again 1:3 in 12 μL of RNA hybridization buffer (10% formamide, 10% dextran sulfate in 2XSSC). Plates were hybridized at 37 °C in a humidified chamber for at least 4 h. Plates were then washed 3 times for 5 min with 2X SSC at 37 °C, nuclear DNA was stained with 1.25 μg DAPI per well for 1 min in DPBS, and imaged. For combined DNA-RNA FISH, DNA FISH was completed as described above at 37 °C overnight with corresponding DNA FISH washes, immediately followed by RNA FISH as described above for 4 h at 37 °C with corresponding RNA FISH washes.
2.3. High-throughput imaging
Imaging was performed using a 40X (0.95NA) air objective lens on a high-throughput spinning disk confocal microscope (Yokogawa CV7000). For excitation, four lasers lines (405, 488, 561, 640 nm) were used, in conjunction with a quad-band excitation dichroic mirror (405/488/561/640 nm), a fixed short pass emission dichroic mirror (568 nm), and emission band-pass filters (filters for DAPI: BP445/45, Green: BP525/50, Red: BP600/37, and FarRed: BP676/29) in an automated and sequential mode. Image acquisition was configured to capture three-dimensional Z-stacks for each channel with 8–9 Z-sections at 0.5 μm intervals without pixel binning. The X-Y pixel size was 162.5 nm and the size of each field of view was 416 × 351 μm (2560 × 2160 pixels). 12–13 fields of view were imaged per well in 15 wells per karyotype, producing approximately 15,000 16-bit images in approximately 1 h.
2.3.1. Image analysis tools
The image processing steps described in the subsequent section (s) were implemented into four (4) bespoke image analysis workflows in the Konstanz Information Miner (KNIME) [28] Analytics Platform (Version 3.2.1, 64-bit) using compatible KNIME Image Processing Nodes (KNIP) [29]. We chose KNIME Analytics Platform primarily because it is open-source and readily supports development of reproducible workflows by capturing data flow. In addition, KNIME-based workflows are operating system agnostic and can scale extremely well from desktops to high-performance batch clusters.
The first workflow executes the image preprocessing and nuclear segmentation component of the pipeline (refer to Section 2.3.2) and allows the end-user to annotate segmented objects into good (visually accurate segmentation) and bad (over/under segmentation) classes, calculates their morphological features, and trains a binary random forest (RF) classifier for rejecting missegmented nuclei (bad class). The second workflow segments nuclei in 2D, applies the trained RF classifier from previous workflow to reject mis-segmented nuclei in 2D, segments nuclei in 3D by extending the 2D nucleus mask in Z-direction of the DAPI channel (refer to Section 2.3.4), and crops CT images from corresponding channels using the binary nucleus mask in both 2D and 3D. The third workflow is responsible for CT detection training (refer to Section 2.3.5) and is designed to annotate segmented objects from the CT channels into good (visually accurate segmentation of CTs) and bad (background regions and/or mis-segmented CTs) classes, calculates their intensity and morphological features, and trains a binary RF classifier for each CT channel. The fourth KNIME workflow segments CTs (Xist), implements training from the third KNIME workflow to filter CTs (Xist) from multiple spectral channels in 2D and 3D, and extracts parameters required for studying spatial organization and architectural features of CTs (i.e. radial positioning, area, and volume).
All KNIME workflows were either executed on a workstation running Microsoft Windows 2012 Server R2 (64-bit) with 16-cores of AMD Opteron 6212 processor (2.7 GHz) and 256 GB RAM or on a dedicated high-performance batch cluster compute node running RedHat Enterprise Linux 6.9 (Santiago) with 28-cores (56-processors) of Intel X2680, 196 RAM and 300 GB of Solid State Storage.
2.3.2. 2D nucleus segmentation
The nuclei from the maximum intensity projected DAPI channel were segmented using a seeded watershed algorithm [18]. The preliminary segmentation boundaries from the seeded watershed are further refined using ultrametric contour maps (UCMs) to minimize over-segmentation [19]. Briefly, UCMs achieve this by combining several types of low-level image information (e.g., gradients and intensity) to construct hierarchical representation of the image boundaries. Under this representation boundary pixels along the nucleus periphery, typically, receive higher score than other pixels associated with internal structures of the nucleus. Applying global thresholding (e.g., Otsu [20]) on UCMs should eliminate weaker (internal) boundaries, thus, minimizing over-segmentation.
2.3.3. Random forest for filtering overlapping nuclei
We used a binary (two classes: good and bad) supervised classification algorithm based on RF [22,23] to filter out under/over segmented nuclei from subsequent analysis. A 14-dimensional numeric vector describing the morphology of segmented objects (e.g., circularity, solidity, area, perimeter, and major elongation) was used for discriminating good (accurately segmented) and bad segmented objects. The first KNIME workflow was used to manually annotate 100 (good-65; bad-35) segmented objects drawn randomly from images from different wells of a plate that was imaged during the plate optimization process. Subsequent nodes in the KNIME workflow then autonomously train a RF classifier on this annotated data. We used a RF classifier with 1000 decision trees, where the nodes in each decision tree were split based on the information gain ratio. This resulted in a RF classifier with out-of-bag accuracy of ~92%. The trained RF classifier was subsequently used for filtering out bad-class of segmented objects for all other plates referenced in this work without further (re)training.
2.3.4. 3D nucleus segmentation
For segmentation of nuclei in 3D, i.e., along the Z-stack of the DAPI channel, we first extended the 2D binary mask of nuclei along the Z-direction. This extended 3D binary mask was applied to the Z-stack of DAPI channel for extracting 3D voxels of the nucleus. The intensity voxels were then thresholded using Otsu [20] followed my morphological operations to generate the 3D binary segmentation mask of the nucleus.
2.3.5. 2D CT segmentation
Prior to segmenting CTs, pixel intensity values of CT channel corresponding to the 2D nucleus segmentation mask were rescaled to range from 0 to 1 using min-max normalization, meaning the lowest pixel intensity value was set to zero and the highest intensity value was set to one. The min-max normalization step was added to minimize the influence of intensity fluctuations in the CT channels between wells within a plate and across plates. Post intensity normalization, CTs in each spectral channel were segmented using the undecimated wavelet-based spot detection technique [21]. We used up to four (4) wavelet scales for segmenting CTs. The per-scale threshold factors for the algorithm were manually fixed so that CTs in all three channels (18, X, and Y/Xist) were reliably detected with the same set of values. These per-scale threshold factors were intentionally set low to enable detection of all CTs. Such low values, as expected, resulted in segmentation of background (non-CT) regions.
2.3.6. Random forest for filtering CTs
To filter out background regions segmented by the undecimated wavelet-based spot detection technique [21], we again used a binary, supervised RF classifier similar to the one described earlier for filtering over/under segmented nuclei. We used an appropriate interactive KNIME workflow to randomly sample 2%–4% of the detected nuclei for each karyotype in the plate. CTs in each spectral channel were detected as described above (Section 2.3.5). Our rational was to determine if the RF classifier, trained on annotated data from just one replicate, can robustly filter segmented CT objects from two biological replicates in the sample plate. If a plate contained wells with replicates, we ensured that stratified random sampling of nuclei was restricted to just one replicate. A user manually annotated correctly (class-good CT) and incorrectly (class-bad CT) segmented CT regions via the interactive nodes in this workflow – approximately 100 objects per class for each spectral channel (green, red and far-red). Next, we extracted morphometric and (normalized) intensity-based features for the objects and trained a RF classifier with 1000 decision trees and node splitting governed by information gain ratio. The out-of-bag accuracy of the RF classifiers was approximately 95.6%, 99.1% and 95.4% for CTs in green, red and far-red channels, respectively. Various efforts (e.g., standardization and normalization of features) to combine three separate RF classifiers into one, generally, resulted in lower out-of-bag accuracy (data not shown). Similarly, classifier(s) trained on one plate, typically, resulted in poor filtering of incorrectly segmented (class-bad CT) CT regions when applied to plates with different karyotype(s). Hence, we decided to generate a separate classifier for each channel for each plate.
2.3.7. 3D CT segmentation
The CTs were segmented in 3D using the following approach. First, we propagate the 2D binary mask of the CT along the Z-direction. This extended 3D mask is used to extract the 3D intensity voxels of the CT from the Z-stack images using a simple image masking operation. Next, Otsu thresholding was applied to the extracted 3D voxels followed by a morphological closing operation, to clean-up isolated voxels, for generating the 3D binary masks for CTs.
2.4. Statistical analysis
All statistical measurements and graphics was generated using R version 3.3.2, 64-bit with ggplot2 graphics package (version 2.2.1, [30]). P-values were calculated using either the Wilcoxon-Mann-Whitney test [31] or the two sample Kolmogorov-Smirnov (KS) test [32,33].
The number of CTs detected was calculated on a per chromosome basis in two biological replicates containing 3 technical replicate wells using R and plotted using Microsoft Excel. For nuclear volume and area as well as CT area and volume box plots, were graphed on a per CT per karyotype basis in technical triplicates. For Xist area box plots, Xist signal intensity was measured on a per X CT basis in triplicates. Normalized radial positioning of CT centroids was calculated using KNIME. Statistical analysis of Xist staining was done in technical triplicates.
2.5. Data and software availability
KNIME analysis workflows and R scripts are available on Github (https://github.com/CBIIT/Misteli-Lab-CCR-NCI/tree/master/Jowhar_Methods_2017).
3. Results
3.1. High-throughput detection of CTs
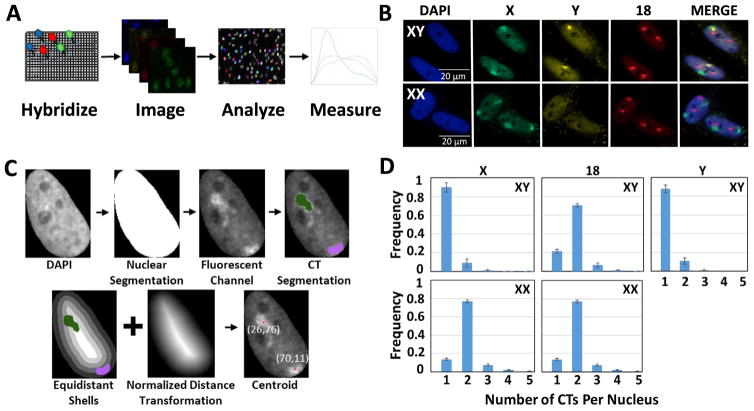
We sought to develop an assay for the rapid, robust, and accurate identification of CTs at high-throughput to overcome the significant labor required for the acquisition, detection, and analysis of sufficiently large numbers of CTs for statistical analysis of their organization and localization (Fig. 1A, B). Our experimental pipeline includes staining of CTs in a 384-well plate format using chromosome paint FISH probes that were generated as previously described ([34–36], see Materials and Methods). To adapt the standard CT FISH protocol to a high-throughput format, incubation volumes, incubation times and washing steps were empirically optimized (see Materials and Methods). We applied this assay to the detection of human chromosomes 18 (HSA-18), X (HSA-X), and Y (HSA-Y) in normal human primary skin fibroblasts from female (XX) and male (XY) individuals (Fig. 1B) (see Materials and Methods). Typically, 12–13 fields per well were imaged per karyotype acquiring ~4000/cells per well using a high-throughput spinning disk confocal microscope. For image analysis, we used a custom KNIME-based image analysis workflow (see Materials and Methods) to identify nuclei with high precision using a seeded watershed and ultrameric contour map for nucleus segmentation, followed by a random forest (RF) classifier for filtering out overlapping and mis-segmented nuclei. We typically analyzed a minimum of 1600 cells in two biological replicates containing three technical replicate wells per karyotype (Sup Fig. 1). A separate customized KNIME workflow was used to first detect CTs within the segmented nuclei using an undecimated wavelet-based spot/vesicle detection approach, and to then filter out background (non-CT) regions using a separate RF classifier for each CT channel. In addition, this workflow calculates various geometric and intensity-related features of the segmented CTs, including chromosome area and volume, as well as their position (only for 2D analysis) with respect to the nucleus border expressed either as a normalized Euclidean distance transform (nEDT) of the CT’s centroid or as the percent of the segmented CT region in each of five equidistant shells (Fig. 1C).
Fig. 1.
HiCTMap imaging, analysis and measurement. (A) HiCTMap pipeline. Cells are cultured in 384-well imaging plates and DNA FISH is carried out using chromosome paints, followed by automated image acquisition using high-throughput microscopy. Image analysis by KNIME segments the nuclei and detects CTs in three channels. CT features are measured and plotted using the R software. (B) CT signal detection. Representative maximal projections of images acquired in three channels. Maximal projections of 40X confocal images z-stacks in 4 channels of XX and XY skin fibroblast stained with DAPI (4′,6-diamidino-2-phenylindole) (Blue-408) and chromosome paint probes (X- Green (Alexa488), 18- Red (Dy505), and Y- FarRed (Dy651)). Scale bar: 20 μm. (C) KNIME automated image segmentation pipeline. Nuclei are segmented using the DAPI channel, thus generating a segmentation mask. The fluorescent channel of each chromosome paint probe is then used to segment CTs of HSA-18, -X, and -Y within the nucleus mask. Positioning of segmented chromosomes using normalized distance transformation and equidistant shells is calculated using the KNIME image analysis pipeline (see Materials and Methods for details). (D) Histogram showing the number of CTs detected per nucleus in XX and XY cells using 2D segmentation. Values represent averages from two biological replicates containing 3 technical replicates ± SD. Histograms represent data from approximately 3800 XY nuclei and XX nuclei each. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Using HiCTMap, we measured the expected number of HSA-X in male and female cells. We detected the correct number of X chromosomes in 90 ± 6.0% SD of XY cells and 77 ± 1.7% SD of XX cells in 2D (Fig. 1D). Similarly, HSA-Y was correctly detected in 88 ± 2% SD of XY fibroblasts (Fig. 1D). Two copies of HSA-18 were detected in 70 ± 1.7% SD XY cells and 68 ± 2% SD in XX cells. In many cases where the number of CTs detected was less than the expected (under-detection of CTs), it was not due to limitations in the segmentation algorithm since fewer copies of the corresponding chromosomes were also detected by visual inspection in those nuclei. In some cases, under-detection was due to clustering of the two homologous chromosomes and in others due to incomplete FISH hybridization (Sup Fig. 2). Less commonly, we observed that the number of CTs detected was greater than expected (over-detection of CTs), which was in part due to a combination of complex CT structure, the wrapping of CTs around nuclear structures (i.e. nucleoli), or the sub-optimal binding of chromosome paint probes to CTs, resulting in apparent fragmentation of the signals. For all further analyses, nuclei that contained an incorrect number of detected CTs for each channel were excluded. This population was typically under 15–20% of analyzed nuclei.
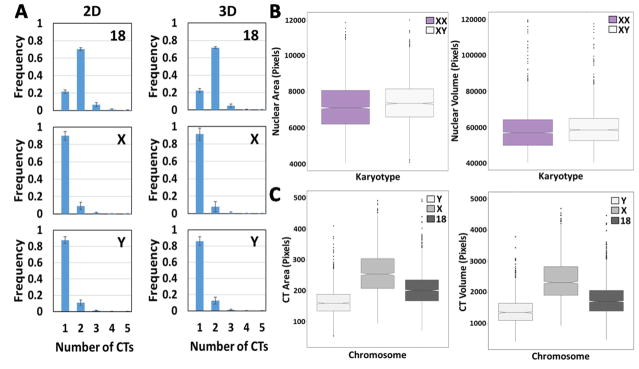
Fig. 2.
2D vs. 3D image analysis of segmented CTs and nuclei. (A) Histograms showing the number of chromosomes HSA-18, -X and -Y per nucleus in XY cells using 2D or 3D analysis. Values represent averages from three experiments ± SD. (B) Top left: Box plots of measured nuclear area of XX and XY cells using 2D nuclear segmentation. Top right: Box plots of measured nuclear volume of XX and XY cells using 3D nuclear segmentation. (C) Bottom left: Box plots of measured CT area of chromosomes HSA-18, -X and -Y in XY cells using 2D CT segmentation. Bottom right: Box plot of measured CT volume of chromosomes HSA-18, -X and -Y in XY cells using 3D CT segmentation method. Boxes show the 25th, 50th (median) and 75th percentile of the distributions and whiskers extend to 1.5 x inter-quantile range (IQR), outliers are represented as dots. Notches indicate the estimated 95% confidence interval of the median (CI). Distributions represent data from approximately 5000 nuclei.
3.2. 2D vs 3D analysis of CT features
To determine how similar the detection of CTs is in 2D compared to 3D, we systematically compared the percent of correctly segmented CTs using either 2D or 3D image analysis (Fig. 2A). For 2D analysis, 9–10 Z-planes were imaged, maximally projected in 2D and used to segment CTs using the undecimated wavelet-based spot detection method (see Materials and Methods). For 3D segmentation of CTs, we propagated the 2D binary mask along the Z-direction to extract 3D intensity voxels of the CT and then applied Otsu thresholding on these voxels to generate the 3D binary masks for the CTs (see Materials and Methods). We found remarkable correspondence in the number of detected CTs for HSA-18, X, and -Y using either 2D or 3D analysis (Fig. 2A). We used 2D and 3D analysis, respectively, to calculate nuclear area and volume of XX and XY cells (Fig. 2B). The 2D nuclear area of XY cells was slightly larger than that of XX cells (7107 vs 7369 average pixels per nucleus, 3.7% difference, Wilcoxon test p-value = 3.5e–21) (Fig. 2B, top left), and similarly we detect a 3.7% increase in nuclear volume of XX nuclei compared to XY nuclei when analyzed in 3D (Wilcoxon test p-value = 1.4e–11) (Fig. 2B, top right) in roughly 5000 nuclei. With regards to CT features in male skin fibroblasts, as expected, the CT area and volume of HSA-X was the largest (2D: 254 ± 73 pixels SD, 3D: 2320 ± 657 pixels SD), HSA-Y the smallest (2D: 160 ± 43 pixels SD, 3D: 1362 ± 417 pixels SD) and HSA-18 intermediate (2D: 201 ± 54 pixels SD, 3D: 1716 ± 490 pix els SD). The overall trends in our 2D measurements were replicated in our measurements in 3D of nuclear size and CT size (Fig. 2B, C), and show agreement between 2D and 3D analysis as shown previously [37]. Given the reduced computing time in 2D when compared to the similar analysis task in 3D, all subsequent image analysis of nuclei and CTs were done using 2D analysis.
3.3. Measurement of Xa and Xi chromosome size
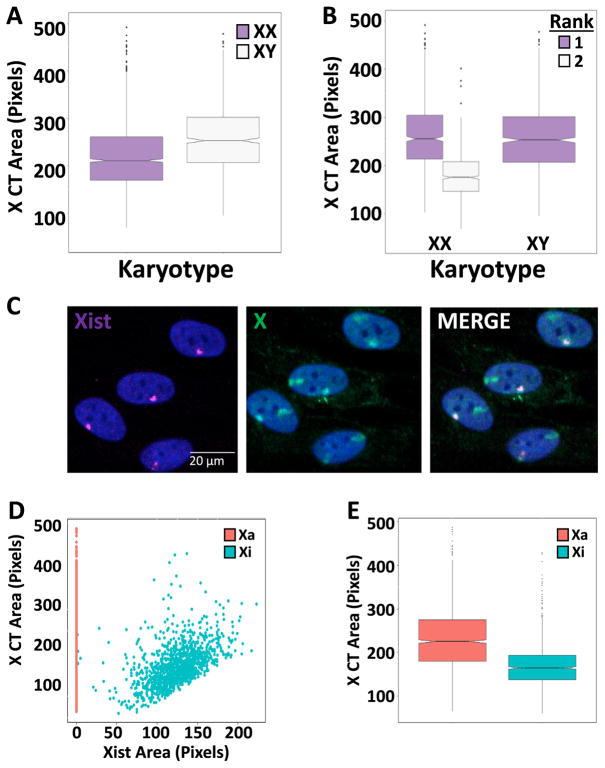
We then used HiCTMap to probe the size variations of HSA-X in populations of XY and XX fibroblasts. We observed that the HSA-X area in XY fibroblasts is significantly larger (median = 254 ± 73 pix els) compared to XX fibroblasts (median = 212 ± 72 pixels SD) (Wilcoxon test p-value: 1.74e–11) as expected, reflecting the well-established inactivation and compaction of one of the HSA-X chromosomes in female cells (Fig. 3A) [38–41]. Using HiCTMap we can reliably detect both the transcriptionally active chromosome (Xa), which is decondensed, and the transcriptionally inactive chromosome (Xi), which is more compacted. To distinguish between the two HSA-X in female cells, we ranked the CTs by size on a per nucleus basis (Fig. 3B). The area of the larger HSA-X CT in XX fibroblasts was comparable to the single X CT in XY cells (256 ± 68 SD vs. 254 ± 73 pixels SD) (Wilcoxon test p-value > 0.05), whereas the smaller HSA-X CT, which in most cases corresponds to the inactive HSA-X (see below), showed a 31% decrease in area compared to the larger HSA-X (176 ± 45 pixels SD) (Wilcoxon test p-value: 2e–89) (Fig. 3B).
Fig. 3.
Detection and measurement of Xa and Xi (A) Box plots of X CT area per nucleus in XX and XY cells. (B) Box plots of X CT area of the largest (1) and second (2) largest ranked X CT per nucleus in XX and XY cells. (C) Representative images of combined DNA-RNA FISH (Scale bar: 20 μm). (D) Scatter plot of X CT area in pixels vs. the segmented Xist area normalized to nuclear size (multiplied by 100) in XX cells (Xa = active X chromosome, Xi = inactive X chromosome). Note that all Xa are aligned at 0 due to the lack of Xist staining. (E) Box plots of CT area in Xi vs. Xa per nucleus in XX cells on a per CT basis. Boxes show the 25th, 50th (median) and 75th percentile of the distributions and whiskers extend up to 1.5* inter-quantile range, outliers are shown as dots. Notches indicate the estimated 95% confidence interval of the median (CI). Distributions represent data from approximately 2500 nuclei.
The well characterized mechanism of HSA-X chromosome inactivation is initiated and maintained by the Xist lncRNA [42,43]. Xist coats the inactive HSA-X chromosome (Xi) in cis and promotes repressive histone modifications, while the active HSA-X (Xa) is primarily used as the transcribed chromosome [44,45]. To visualize and unambiguously identify the inactive HSA-X chromosome, XX cells were processed for sequential DNA/RNA FISH using chromosome-paint probes to label the HSA-X CTs and fluorescently labelled oligonucleotides to target Xist RNA (see Materials and Methods). This approach enables high-throughput visualization of simultaneously labelled X chromosomes and Xist RNA in the same nucleus (Fig. 3C). Any nucleus containing more than one Xist signal was excluded from analysis. As expected, HSA-X CTs can be separated into two distinct populations according to Xist staining (Fig. 3D). While we find a robust single Xist RNA signal in nuclei from XX cells, no Xist RNA signals are detected in XY cells as expected (data not shown). In line with our size analysis, we find that HSA-X CTs that lack Xist are typically larger (225 pixels ± 71 SD) compared to Xist containing CTs (164 ± 45 pixels SD) (Wilcoxon test p-value: 8.23e–121, Fig. 3E). These observations demonstrate accurate detection and the ability to distinguish HSA-Xi and HSA-Xa based on Xist staining and they establish the feasibility of using HiCTMap to combine DNA and RNA FISH in high-throughput imaging approaches.
3.4. Chromosome positioning analysis
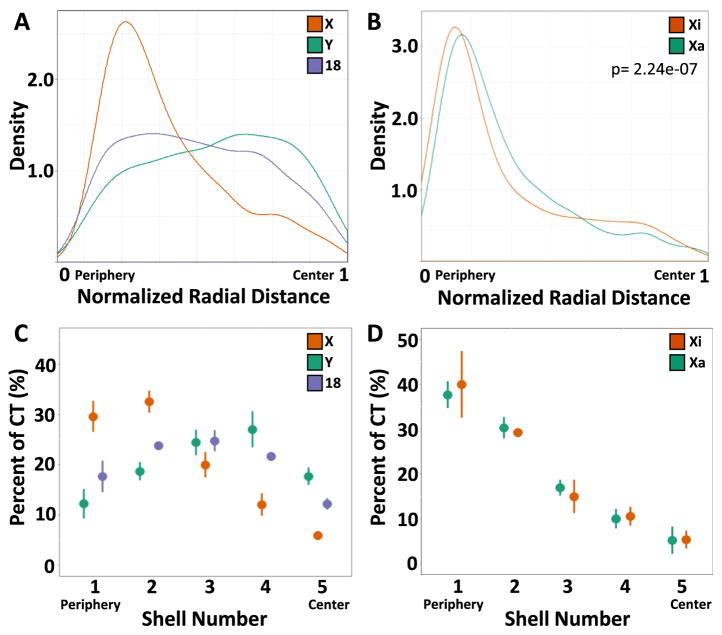
We finally applied HiCTMap to determine the nuclear positioning of HSA-18, -X, and -Y in XY fibroblasts. We used two approaches to determine chromosome positioning. First, we determined the centroid of the segmented CT defined as the geometric center of the segmented CT and measured its normalized radial distance from the nucleus border using Euclidean distance transformation as previously described [46]. The distribution of the centroid radial distance for each chromosome was distinct (Fig. 4A). HSA-X showed a strong preferential distribution to the periphery of the nucleus in XY cells (median normalized radial distance = 0. 34), whereas HSA-Y exhibited a slight preference for positioning to the center of the nucleus (median normalized radial distance = 0.58) and HSA-18 showed an even distribution of positions (0.48) (Fig. 4A), in line with previous observations showing a non-peripheral localization of HSA-18 in fibroblasts [26]. Comparison of the radial positioning of the centroid of Xa and Xi in XX fibroblasts, showed a small, but significant, more peripheral position of the Xist-marked Xi centroid (median normalized radial dis tance = 0.25) compared to the Xa (median normalized radial dis tance = 0.29) (KS test p-value: 2.24e–07) (Fig. 4B). Measurements of center of gravity, which accounts for intensity values in the CT, gave similar results (data not shown).
Fig. 4.
Radial positioning of CTs (A) Density curves for normalized radial distance distributions for the indicated chromosome’s centroid. (B) Density curves for normalized radial distance distributions of the centroids of Xa and Xi. (C) The percentage of CTs of HSA-18, -X, and -Y in 5 concentric equidistant nuclear shells. (D) The percentage of Xi and Xa CT in 5 concentric equidistant nuclear shells. Values represent averages from three technical replicates ± SD. Distributions represent data from at least 2500 CTs.
Since using centroids as a proxy for the spatial localization of an entire CT has the potential to generate misleading results, given the irregular and diffuse structure of chromosome territories, we also used an alternative method to more faithfully determine chromosome location. In this previously established approach, each nucleus is divided into five equidistant shells, and the percentage of the CT falling in each of the equidistant shells is measured [10,17,38,47]. When chromosome positioning was measured using equidistant shells, we observed similar results to the centroid analysis: a strong preferential positioning of the HSA-X to the periphery of the nucleus with the highest average percent in shells 1 and 2 (30% and 32%, respectively), a slight preference for positioning of HSA-Y to the center of the nucleus with the highest average percent in shells 3 and 4 (24% and 27%, respectively) and even positioning of HSA-18 with the highest average percent in shells 2 and 3 (24% and 25%, respectively) (Fig. 4C). Similar to the results obtained with centroids, in XX fibroblasts, both Xa and Xi were peripherally positioned, and Xa was slightly more internal (38% in shell 5) compared to Xi (42% in shell 5) (Fig. 4D). Our data are in line with previous observations demonstrating a more peripheral localization of the Xi chromosome compared to the Xa [17,41] and as such validate the ability of HiCTMap to accurately detect and measure chromosome territory positioning.
Taken together, these results establish a high-throughput imaging pipeline for the detection of CTs and the determination of structural features of chromosomes including area, size and radial position. In addition, we show that chromosome painting in a high throughput format can be used to detect multiple chromosome combinations in the same experiment and can be combined with RNA FISH. We suggest that HiCTMap is a versatile tool for the study of chromosomes at a high-throughput scale.
4. Discussion
Here, we describe a systematic and quantitative method for the detection of multiple chromosome territories using high-throughput imaging. Our approach, referred to as HiCTMap, enables collection of imaging data on large numbers of cells and quantitative measurements of chromosome features including size and positioning using automated image analysis tools. HiCTMap overcomes the constraints of standard CT analysis [10,16,26,38], particularly the reliance on a relatively small number of cells due to technical limitations in sample preparation and extensive imaging time using conventional microscopy approaches.
Due to its high-throughput nature, HiCTMap is well suited to acquire images for several thousands of cells per experimental condition in a short time, typically about seven minutes per well. In contrast to most FISH approaches that rely on visual inspection of relatively small numbers of samples and require significant user input, automated imaging of thousands of cells per sample generates a more faithful representation of the frequency of chromosomal architecture and positioning in the population and eliminates user biases in selection of nuclei for quantitative analysis. We demonstrate that chromosome painting at high-throughput can robustly detect the expected number of CTs simultaneously in three channels. We also establish the accuracy of HiCTMap in measuring the position of chromosomes using two methods of radial distance measurements, equidistant shells and centroids and we show its compatibility with RNA FISH. The fact that we can detect with equal accuracy the transcriptionally active and open Xa chromosome and the transcriptionally silent highly compacted Xi demonstrates the usability of HiCTMap to a wide range, likely all, chromosomes.
As previously reported [37] for traditional imaging methods, we find little differences when analyzing CTs in 2D or 3D. The number of chromosomes detected in male (XY) and female (XX) cell populations was similar in 2D and 3D analysis as were nuclear size and chromosome size. The relatively large size of chromosomes in the nuclear space provides a lower threshold for accurate detection, segmentation and analysis than for the precise and meticulous accuracy of single gene FISH in 2D versus 3D [37]. It is worth noting that use of high-magnification imaging provides better resolution of chromosome architecture, however, 60X image acquisition yielded similar results to our standard 40X magnification used and provided no further benefits to measurements in 2D or 3D.
HiCTMap is sufficiently sensitive to detect the expected differences in X chromosome size in XY and XX cells, due to the presence of a mixed population of smaller Xi and larger Xa chromosomes. While the population of X chromosomes in XY cells is relatively homogeneous, a more heterogeneous distribution of X chromosome size was found in XX cells, consistent with the well-established smaller size of the inactive X chromosome [38]. The identity of the consistently smaller X chromosome as the Xi was confirmed by sequential Xist RNA FISH and chromosome painting (Fig. 3E). Chromosome painting at high-throughput in combination with Xist RNA FISH offers advantages over conventional RNA FISH followed by imaging and subsequent DNA FISH, since it allows for immediate acquisition of both RNA and DNA FISH in one imaging step, eliminating the possibility of misaligning or incorrectly identifying nuclei when plates or cover slips are removed for DNA FISH and re-imaged. In addition, in HiCTMap, multiple chromosomes can be observed in combination with the Xist RNA probe, thus making the method suitable for use in basic research applications to probe aneuploidies of the genome.
Furthermore, we show robust detection by HiCTMap of chromosome positioning using both centroid and equidistant shells approaches. We detected differences in nuclear positioning of Xi and Xa using combined Xist RNA and CT FISH, further demonstrating the suitability of HiCTMap for chromosome position mapping. Similar trends were observed by measuring the centroid location or equidistant shells radial positioning. Our findings are in line with the reported observation of a more peripherally positioned Xi and its proposed anchoring to the nuclear lamina [48]. Unlike previous observations where a much greater difference in positioning is observed between the two X chromosomes [16,17], our results may be different due to the use of cells freshly derived from tissue or the large number of nuclei measured that represents the population more faithfully.
The limitations of HiCTMap are relatively minor. First, CT segmentation requires user input to determine the ground truth necessary to implement the random forest filtration. This step typically requires 30 min of training and harbors the possibility of biases, which, however, can be eliminated by careful supervision of the user and determination of the ground truth by multiple users. Second, our imaging conditions were not optimized for maximal resolution in 3D. However, we did not observe improved resolution in 3D after increasing the magnification from 40X to 60X, suggesting this is the maximal resolution for this microscope.
Taken together, we describe here HiCTMap, a method for chromosome painting in high-throughput. Given its versatility and its compatibility with RNA FISH, we anticipate that HiCTMap will be of considerable use in future analyses of chromosome organization and function. HiCTMap is primed for accurate detection and characterization of numerical and structural chromosomal aberrations in diseases where aneuploidy is frequent, such as in cancer.
Supplementary Material
Acknowledgments
The authors thank the CBIIT Server Team, NCI, National Institutes of Health (NIH) and the High Performance Computing Group, CIT, NIH for computational support. This research was supported by funding from the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and Center for Cancer Research.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ymeth.2018.01.013.
References
- 1.Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356(Pt 2):297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 3.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Exp Cell Res. 2004;296(1):64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Bartova E, Kozubek S, Jirsova P, Kozubek M, Gajova H, Lukasova E, Skalnikova M, Ganova A, Koutna I, Hausmann M. Nuclear structure and gene activity in human differentiated cells. J Struct Biol. 2002;139(2):76–89. doi: 10.1016/s1047-8477(02)00560-9. [DOI] [PubMed] [Google Scholar]
- 7.Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242(4886):1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- 8.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories–a functional nuclear landscape. Curr Opin Cell Biol. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1(2):137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 10.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145(6):1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5(7):R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornforth MN, Greulich-Bode KM, Loucas BD, Arsuaga J, Vazquez M, Sachs RK, Bruckner M, Molls M, Hahnfeldt P, Hlatky L, Brenner DJ. Chromosomes are predominantly located randomly with respect to each other in interphase human cells. J Cell Biol. 2002;159(2):237–244. doi: 10.1083/jcb.200206009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neusser M, Schubel V, Koch A, Cremer T, Muller S. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 2007;116(3):307–320. doi: 10.1007/s00412-007-0099-3. [DOI] [PubMed] [Google Scholar]
- 14.Cremer M, Grasser F, Lanctot C, Muller S, Neusser M, Zinner R, Solovei I, Cremer T. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- 15.Misteli T. Spatial positioning; a new dimension in genome function. Cell. 2004;119(2):153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 16.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10(3):211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 17.Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Ried T, Robert-Nicoud M, Cremer C, Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135(6 Pt 1):1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent L, Soille P. Watersheds in digital spaces – an efficient algorithm based on immersion simulations. IEEE Trans Pattern Anal. 1991;13(6):583–598. [Google Scholar]
- 19.Arbelaez P. Boundary extraction in natural images using ultrametric contour maps. Proceedings of the 2006 Conference on Computer Vision and Pattern Recognition Workshop, IEEE Computer Society; 2006; p. 182. [Google Scholar]
- 20.Sezgin M. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging. 2004;13(1):146–168. [Google Scholar]
- 21.Olivo-Marin JC. Extraction of spots in biological images using multiscale products. Pattern Recogn. 2002;35(9):1989–1996. [Google Scholar]
- 22.Ho TK. The random subspace method for constructing decision forests. IEEE Trans Pattern Anal. 1998;20(8):832–844. [Google Scholar]
- 23.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 24.Ollion J, Cochennec J, Loll F, Escude C, Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29(14):1840–1841. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulet A, Arganda-Carreras I, Legland D, Probst AV, Andrey P, Tatout C. NucleusJ: an ImageJ plugin for quantifying 3D images of interphase nuclei. Bioinformatics. 2015;31(7):1144–1146. doi: 10.1093/bioinformatics/btu774. [DOI] [PubMed] [Google Scholar]
- 26.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3(5):e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belmont AS, Bignone F, Ts’o PO. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp Cell Res. 1986;165(1):165–179. doi: 10.1016/0014-4827(86)90541-0. [DOI] [PubMed] [Google Scholar]
- 28.Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B. KNIME: the Konstanz information miner. In: Preisach C, Burkhardt H, Schmidt-Thieme L, Decker R, editors. Data Analysis, Machine Learning and Applications: Proceedings of the 31st Annual Conference of the Gesellschaft für Klassifikation e.V; Albert-Ludwigs-Universität Freiburg. March 7–9, 2007; Berlin, Heidelberg: Springer Berlin Heidelberg; 2008. pp. 319–326. [Google Scholar]
- 29.Dietz C, Berthold MR. KNIME for open-source bioimage analysis: a tutorial. In: De Vos WH, Munck S, Timmermans J-P, editors. Focus on Bio-Image Informatics. Springer International Publishing; Cham: 2016. pp. 179–197. [DOI] [PubMed] [Google Scholar]
- 30.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- 31.Neuhäuser M. Wilcoxon–Mann–Whitney test. In: Lovric M, editor. International Encyclopedia of Statistical Science. Springer; Berlin, Heidelberg: 2011. pp. 1656–1658. [Google Scholar]
- 32.Smellie A, Wilson CJ, Ng SC. Visualization and interpretation of high content screening data. J Chem Inf Model. 2006;46(1):201–207. doi: 10.1021/ci050404g. [DOI] [PubMed] [Google Scholar]
- 33.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306(5699):1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 34.Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T. Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J. 2014;28(8):3423–3434. doi: 10.1096/fj.14-250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85(23):9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum Genet. 1988;80(3):235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- 37.Finn EH, Pegoraro G, Shachar S, Misteli T. Comparative analysis of 2D and 3D distance measurements to study spatial genome organization. Methods. 2017;123:47–55. doi: 10.1016/j.ymeth.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bischoff A, Albers J, Kharboush I, Stelzer E, Cremer T, Cremer C. Differences of size and shape of active and inactive X-chromosome domains in human amniotic fluid cell nuclei. Microsc Res Tech. 1993;25(1):68–77. doi: 10.1002/jemt.1070250110. [DOI] [PubMed] [Google Scholar]
- 39.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Lee JT. X-chromosome inactivation: a multi-disciplinary approach. Semin Cell Dev Biol. 2003;14(6):311–312. doi: 10.1016/j.semcdb.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 41.Teller K, Illner D, Thamm S, Casas-Delucchi CS, Versteeg R, Indemans M, Cremer T, Cremer M. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at >/=20 Mb genomic length scales. Nucleus. 2011;2(5):465–477. doi: 10.4161/nucl.2.5.17862. [DOI] [PubMed] [Google Scholar]
- 42.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2(1):59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 43.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 44.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 45.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meaburn KJ, Gudla PR, Khan S, Lockett SJ, Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187(6):801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanabe H, Muller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci USA. 2002;99(7):4424–4429. doi: 10.1073/pnas.072618599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.