Abstract
Circulating tumor cells (CTCs) are cancer cells shredded from either a primary tumor or a metastatic site and circulate in the blood as the potential cellular origin of metastasis. By detecting and analyzing CTCs, we will be able to noninvasively monitor disease progression in individual cancer patients and obtain insightful information for assessing disease status, thus realizing the concept of “tumor liquid biopsy”. However, it is technically challenging to identify CTCs in patient blood samples because of the extremely low abundance of CTCs among a large number of hematologic cells. In order to address this challenge, our research team at UCLA pioneered a unique concept of “NanoVelcro” cell-affinity substrates, in which CTC capture agent-coated nanostructured substrates were utilized to immobilize CTCs with remarkable efficiency. Four generations of NanoVelcro CTC assays have been developed over the past decade for a variety of clinical utilities. The 1st-gen NanoVelcro chips, composed of a silicon nanowire substrate (SiNS) and an overlaid microfluidic chaotic mixer, were created for CTC enumeration. The 2nd-gen NanoVelcro chips (i.e., NanoVelcro-LMD), based on polymer nanosubstrates, were developed for single-CTC isolation in conjunction with the use of the laser microdissection (LMD) technique. By grafting thermoresponsive polymer brushes onto SiNS, the 3rd-gen Thermoresponsive NanoVelcro chips have demonstrated the capture and release of CTCs at 37 and 4 °C respectively, thereby allowing for rapid CTC purification while maintaining cell viability and molecular integrity. Fabricated with boronic acid-grafted conducting polymer-based nanomaterial on chip surface, the 4th-gen NanoVelcro Chips (Sweet chip) were able to purify CTCs with well-preserved RNA transcripts, which could be used for downstream analysis of several cancer specific RNA biomarkers. In this review article, we will summarize the development of the four generations of NanoVelcro CTC Assays, and the clinical applications of each generation of devices.
Graphical abstract
1. Introduction
1.1. Circulating tumor cell (CTC)
The gold standard for cancer diagnosis is based on pathological analysis of tumor tissues, which relies upon tissue specimens acquired by invasive procedures, e.g., surgical excision or needle biopsy. Crucial information including histopathology and molecular profiling can be generated to achieve accurate diagnosis and classification of the disease. However, these invasive procedures impose risks to cancer patients. First, the invasive procedures can be quite costly. The risk of injury to the patient may limit the implementation of the invasive procedures (e.g., pneumothoraxes that can be caused by lung biopsies). Further, certain malignancies pose technical challenges due to the anatomical locations of metastasis. For instance, advanced prostate cancer metastases are commonly found in the bone and are sclerotic in nature. In such cases, typical small needle biopsies are avoided and larger, drill-based sampling is required. Moreover, the well-recognized tumor temporospatial heterogeneity[1–7] raises severe concerns over how accurately a given biopsied sample represents a disease whose biological and molecular nature varies from site to site and changes over time in the course of treatment interventions. Despite its difficulty, a re-biopsy procedure is often recommended to detect a possible new biology profile of cancer cells during the clinical treatment course in some solid tumors (e.g. lung cancer).
As a non-invasive alternative to tumor biopsy, researchers have been exploring the use of circulating tumor cells (CTCs) as “liquid biopsies” of solid tumors. CTCs are blood borne tumor cells shed from either primary or metastatic sites. Through a simple blood draw, CTCs can be detected and recovered throughout the course of disease development without needing invasive and painful biopsy procedures. In addition to conventional diagnostic imaging and serum marker detection, detecting and characterizing CTCs in patient blood provides an opportunity for early diagnosis of cancer metastasis. Further, serial CTC tests can be performed over the disease progression with relatively high frequency, creating an opportunity to perform real-time, dynamic monitoring of an evolving and adapting malignant process[8, 9]. To address this unmet need, there have been significant research endeavors[10], especially in the fields of chemistry, materials science, and bioengineering, devoted to developing CTC detection, isolation, and characterization technologies[11]. However, identifying CTCs in blood samples has been technically challenging due to the extremely low abundance (a few to hundreds per milliliter) of CTCs among a large number (109 mL−1) of hematologic cells in the blood. Initial CTC studies focused on enumeration and protein expression analysis [12–14]. More recent research efforts have demonstrated that CTCs and their matching tumor tissues share significant similarities at the genomic[15–17] and transcriptomic[18, 19] levels. Mounting evidence has consistently shown CTCs to be a powerful tool with which we can study the underlying biology of cancer, guide therapeutic interventions, and monitor the progression of disease.
1.2. Existing CTC assays
In order to effectively conduct detection and analysis of CTCs, a variety of methodologies have been developed. (i) Immunomagnetic separation: Positive selection/enrichment of CTCs[13, 20, 21] can be achieved using capture agent-labeled magnetic beads. For epithelial-origin solid tumors, anti-EpCAM [22] is the most widely used capture agent. Alternatively, negative depletion [23, 24] of white blood cells (WBCs) using magnetic beads tagged by anti-CD45 can result in purified CTCs. CellSearch™ [12–14] is the only FDA-approved CTC assay with prognostic utility in metastatic breast, colorectal, and prostate cancers. CellSearch™ Assay enriches CTCs with magnetic beads tagged with anti-EpCAM, followed by immunocytochemistry (ICC) treatment to distinguish CTCs (DAPI+/cytokeratin−CK+/CD45−) from WBCs (DAPI+/CK−/CD45+) and cell debris in the background. New immunomagnetic technologies including MagSweeper[25], magnetic sifters[26], AdnaGen/Qiagen[27], IsoFlux[28], VerIFAST[29] and Cynvenio[30], were developed to improve the speed, efficiency, and cost of CTC detection and characterization. (ii) Flow cytometry: Although one of the most commonly used cell-sorting technologies for the analysis of sub-populations of fluorescently labeled cells [31, 32], flow cytometry lacked the ability to reveal sufficient morphological information to satisfy the standards set by pathologists for CTCs. The development of ensemble decision aliquot ranking (eDAR) [33, 34] addressed this drawback. (iii) Microfluidics-enabled CTC assays: Massachusetts General Hospital (MGH) developed the microfluidic devices [35], propelling recent research efforts focusing on the development of microfluidic CTC assays. Their 1st-generation (gen) microchips[35] (i.e., CTC-Chip) covalently attached anti-EpCAM to silicon microposts, thus maximizing the contact between the flow-through cells and device surfaces. Biocept’s CTC assay [36] and GEDI approach [37] were created using similar device configurations. Moreover, utilizing microposts (self-assembled from magnetic beads) in a microchip, the “Ephesia” approach [38] demonstrated the advantages of both immunomagnetic and microchip-based methods. The MGH team’s 2nd-gen microchips [39] (i.e., herringbone-chip) featured herringbone structures micro-engineered into a polydimethylsiloxane (PDMS) block to enhance contact between CTCs and the antibody-tagged device surfaces. Most recently, the MGH team developed the 3rd-gen iChip [40], which integrates inertial focusing of blood cells with immunomagnetic WBC depletion in a microfluidic chip. Notably, “Unmanipulated” CTCs can be recovered from whole blood samples with well-preserved viability and molecular integrity, enabling CTC-based transcriptomic profiling [18], drug susceptibility testing, [41] and ex vivo culturing. However, it was shown that the sorting mechanism of iChips might hamper recovery of CTC clusters[42]. “Cluster-Chip”, was developed to overcome this problem[43]. Several other types of microfluidics-based CTC assays, including cell rolling[44], micro-nuclear magnetic resonance (μNMR) [45], Vortex technology[46, 47], and lipid bilayer-deposited microchips [48] have also been developed. Moreover, Velocity Valley Chip was developed to separate CTCs into subpopulations [49] according to their EpCAM expression levels. (iv) Microscopy imaging [50–52]: Using high-speed fluorescent microscopy, CTCs can be identified in smeared peripheral blood mononuclear cell (PBMC) samples, with high-quality morphological features. Epic Sciences’ CLIA-certified (Clinical Laboratory Improvement Amendments) services establish them as one of the current leaders in this technology. Similarly, RareCyte’s fluorescence microscopy-based platform has demonstrated CTC identification in clinical samples [53], as well as the feasibility of performing single-CTC genotyping.
While all the aforementioned technologies utilize capture agents to isolate CTCs (e.g., EpCAM), CTC assays are not inherently required to use such markers. Instead, these “label-free” technologies exploit the physical properties of CTCs. (v) CTC filters: Filter-based techniques [54–58] are established to sort CTCs based on their size difference. A variety of systems/kits including Clearbridge [56, 57], Rarecells [59], Creatv MicroTech, ScreenCell [60], Celsee [61] are now commercially available to support utilities in research laboratories. Since these filter devices only capture CTCs, which exhibit larger sizes than WBCs, small-sized CTCs might be overlooked in this kind of approach. (vi) Dielectrophoresis: A dielectrophoretic field can be used to sort CTCs apart from the background WBCs as a result of CTCs’ differential dielectric properties (which depend on their diameter, membrane area, density, conductivity and volume). ApoCell’s technology[62] introduces a dielectrophoretic field in a microchip to achieve CTC sorting. DEPArray™ (Silicon BioSystems) combines dielectrophoretic sorting and microscopy imaging [63] to isolate pre-sorted CTC mixtures, allowing for downstream CTC-based molecular characterizations with single-cell resolution. DEPArray™ is frequently used as a downstream single-CTC isolation technique using the recovered CTC samples from CellSearch™ Assays.[64] (vii) Several outstanding review articles[65–67] that highlight CTC detection and characterization technologies provide different coverage and scope from this review article.
2. NanoVelcro CTC Assays
2.1. Development and evolution
Over the last few decades, medical research in the field of nanotechnology has made significant progress in reducing the costs of CTC characterization [68–70]. The goal of personalized care is one-step closer with the deployment of emerging advances in oncology. Knowing that the nanoscale tissue microenvironment (particularly the cellular membrane and extracellular matrix) can help mediate cellular behavior, the Tseng Lab at UCLA launched the nanosubstrate microfluidic platform (i.e., silicon nanowire substrate, SiNS[71]) for capturing CTCs, colloquially deemed the “NanoVelcro” assay [71, 72].
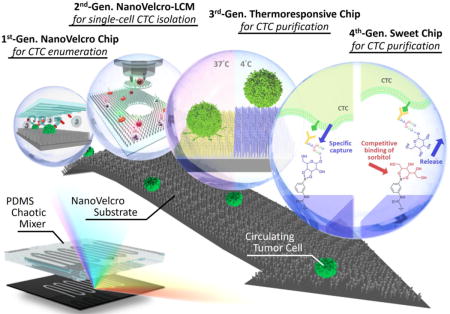
This novel approach utilized capture agent-coated nanosubstrates to immobilize CTCs with high efficiency, resembling the working mechanism of Velcro™. In much the same way that two fabric strips of a Velcro fastener attach tightly together, the NanoVelcro cell-affinity substrates interact with CTCs to form strong binding. The team developed a three-color ICC protocol [73] with DAPI, anti-CK, and anti-CD45 to identify CTCs immobilized on the nanosubstrate. Nonspecifically captured WBCs (DAPI+/CK−/CD45+, sizes<12 μm) are differentiated from CTCs (DAPI+/CK+/CD45−, sizes>6 μm) by cell imaging criteria with CK/CD45 expression, object size, and DAPI staining. In the time since the initial proof-of-concept of the NanoVelcro substrates and optimization of the ICC protocol, four generations of NanoVelcro CTC Assays have been developed since the launch of the first model for a variety of clinical utilities[74, 75].(Figure 1)
Figure 1.
Graphic illustration and table comparison of the NanoVelcro CTC Assays among four different generations.
2.2. Proof-of-Concept demonstration of NanoVelcro Cell-Affinity substrates - stationary NanoVelcro CTC Assay
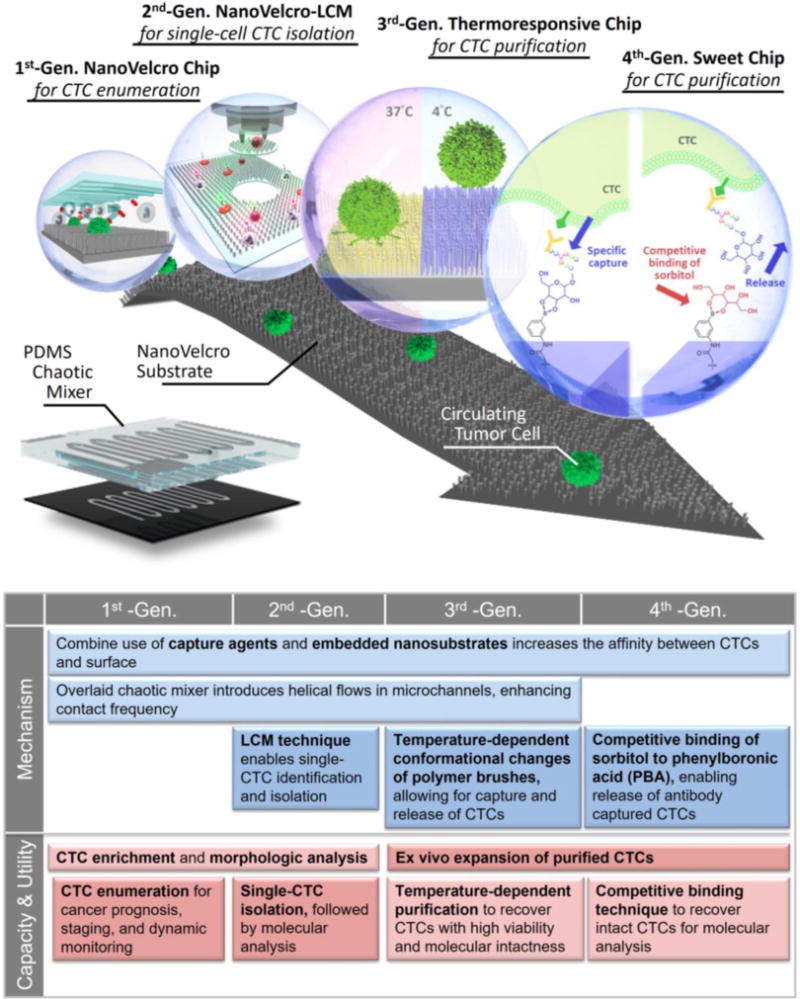
The working mechanism (Figure 2a) of NanoVelcro assay [71, 76–78] lies in the use of SiNS, which allows for Velcro-like interactions [79] between SiNS and nanoscale cell-surface components. Anti-EpCAM was selected as the capture agent for its specificity for capturing CTCs and drastically improved capture efficiency compared to flat SiNS. (Figure 2b). The anti-EpCAM capture agent is grafted onto SiNS via biotin-streptavidin conjugation (Figure 2c). CTC capture in whole blood samples was achieved in a stationary device setting via comprehensive optimization. SEM characterization of CTCs on SiNS (Figure 2d) and imprinted PLGA NanoVelcro substrates (Figure 2e) supported the effectiveness of the proposed NanoVelcro working mechanism. Through testing, NanoVelcro cell-affinity substrates demonstrate CTC capture efficiencies ranging from 40 to 70% in a stationary device setting. [69]
Figure 2. Velcro-like working mechanism of NanoVelcro cell-affinity substrates.
a) An anti-EpCAM-coated SiNS was employed to achieve significantly enhanced capture of CTCs in contrast to b) an anti-EpCAM-coated flat silicon substrate. c) Anti-EpCAM is grafted onto SiNS to confer specificity for recognizing CTCs. d) and e) SEM images of a SiNS and PLGA NanoVelcro substrate, on which MCF7 cells and prostate cancer CTCs were captured, respectively.
2.3. General applicability of the concept of NanoVelcro Cell-Affinity Assay
In addition to SiNS[71], extensive research endeavors have been devoted to exploring the use of different nanomaterials on the NanoVelcro cell-affinity assay. Such materials include TiO2 nanowires[77], polymer dots[76], nanopillars[78], nanoparticles[80], Fe3O4 nanoparticles[81], gold clusters on silicon nanowires[82], layer-by-layer-assembled nanostructures[83], graphene oxide nanosheets[84], and DNA networks[85]. These efforts have demonstrated the ability to efficiently capture CTCs and other types of rare cells. Other groups of researchers have utilized nanomaterials for rare cell assays. Dr. Wang’s group [86] at the Chinese Academy of Science focused on structural adjustments and introduced a hierarchically-assembled ITO nanowire array with vertical and horizontal branches fabricated as a three-dimensional fractal nano-biointerface for CTC capture with enhanced efficiency. Dr Cui’s group at Stanford employed a dual-function lipid coating on traditional nanostructured surfaces. Based on their investigation of interactions between cells and the nanopillar array, both prevention of nonspecific cell adhesion and CTC capture by a functionalized layer of antibodies were demonstrated. [87, 88] A capture-agent free approach was carried out by Dr. Fu’s group[89] at the University of Michigan. A nanoroughened glass substrate on slides exhibited a differential adhesion affinity for CTCs rather than normal blood cells. This approach demonstrated CTC capture ability independent of the surface biomarker[22].
2.4. 1st-Gen NanoVelcro Chip
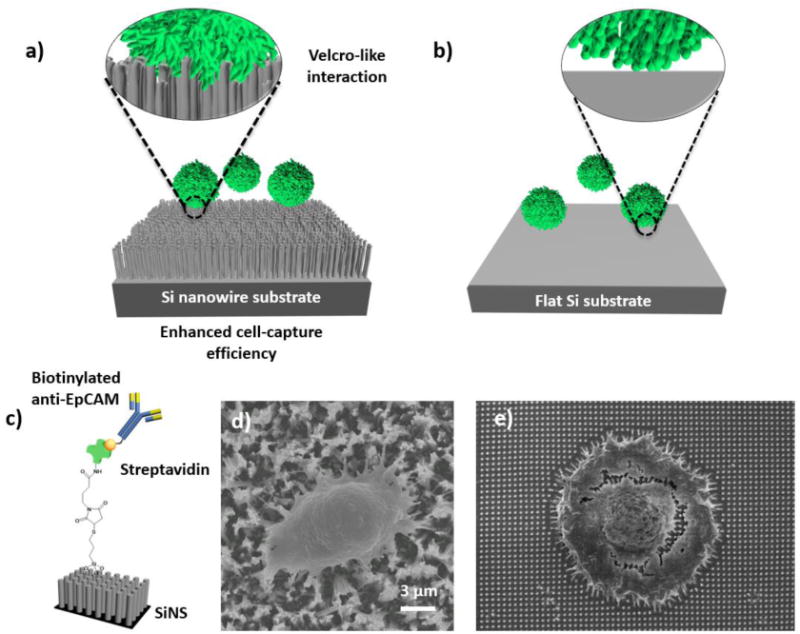
On the basis of the stationary NanoVelcro cell-affinity substrates [71, 76–78], we sought for further improvement of the capture performance. This effort led to the development of the 1st-gen NanoVelcro Chip[8, 90], which consists of a lithographically patterned SiNS and an overlaid microfluidic chaotic mixer (Figure 3a and b). The PDMS chaotic mixer has herringbone microstructures on the roof and induces[91] vertical flows in the microchannel, which increases the contact frequency between cells and nanosubstrates. CTCs captured by this device are stained by a 3-color ICC protocol with DAPI, anti-CK and anti-CD45. Single-cell image cytometry [72] (Figure 3c and d) analyzing CK/CD45 expressions and CTC footprint sizes is employed to identify CTCs from nonspecifically captured WBCs and cellular debris. Using artificial CTC samples, we achieved >85% capture efficiency with the 1st-gen NanoVelcro Chips. After performing side-by-side validation studies using patient blood samples, we found that the sensitivity of the 1st-gen NanoVelcro Chip [8] is superior to the FDA-cleared CellSearch™ Assay. The 1st-gen NanoVelcro Chips along with the ICC protocol have since been used for CTC enumeration in multiple cancers.
Figure 3. Workflow of NanoVelcro and Imaging CTC System.
a) and b) Configuration of 1st-gen NanoVelcro CTC chip. The device is composed of a patterned NanoVelcro substrate and an overlaid PDMS chaotic mixer. c) Using a 3-color ICC protocol, CTCs (DAPI+/CK+/CD45-, sizes>6 μm) can be clearly distinguished from nonspecifically captured WBCs (DAPI+/CK−/CD45+, sizes<12 μm [72] by a 3-color ICC protocol, d) Fluorescent images of 2 prostate cancer CTCs captured on the substrate along with non-specifically captured WBCs.
2.5. Alternative capture agents: Aptamers
Despite being the most widely utilized CTC capture agent, anti-EpCAM possesses several shortcomings, including its high cost and poor stability. These constrain the use of CTC-based diagnostics, especially in low-resource environments. Among other choices, aptamers stood out as a strong candidate for CTC-based diagnostics because of their many desirable properties. First, the ease of selection and synthesis make aptamers more affordable than antibodies like anti-EpCAM. Second, they possess comparable binding affinity and specificity to antibody-based capture agents. Third, low immunogenicity of aptamers erases concerns for compatibility issues[92]. Recently, we have generated two singled-stranded DNA aptamers via an in vitro cell-SELEX[93] (systematic evolution of ligands by exponential enrichment) that has shown success in capturing non-small cell lung cancer (NSCLC) CTCs. With the aptamer-grafted NanoVelcro Chip (Figure 4), we demonstrated high efficiency of capturing NSCLC CTCs from blood. We were also able to recover the nanosubstrate-bounded CTCs using enzymatic treatment. Our recent study suggests that rationally-designed aptamer cocktails may elicit a synergistic effect for capturing CTCs in a microfluidic setting, achieving enhanced and differential capture of CTCs for non-small cell lung cancer (NSCLC) patients[94].
Figure 4.
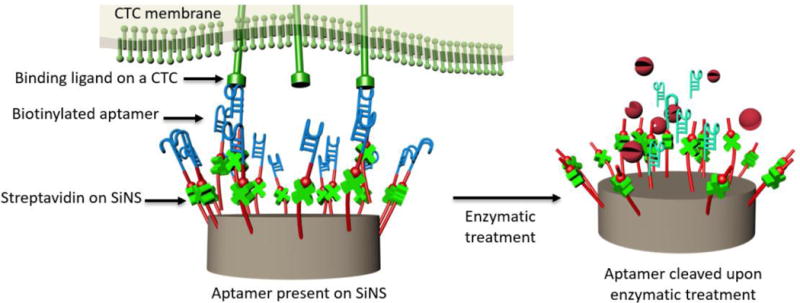
Molecular mechanism governing the capture and enzymatic release of NSCLC CTCs from the aptamer-coated SiNS.
2.6. 2nd-Gen NanoVelcro Chip
The 2nd-gen NanoVelcro Chip (Figure 5) was developed to address the need to isolate CTCs for subsequent molecular analysis[16, 95, 96]. Known as the NanoVelcro-LCM (laser capture microdissection) approach, we engineered a polymer-based CTC capture substrate, which can be dissected by an LCM microscope for molecular analysis, especially at the genomic level.
Figure 5. Illustrations of 2nd-Gen NanoVelcro-LCM technology and its application of single-CTC isolation.
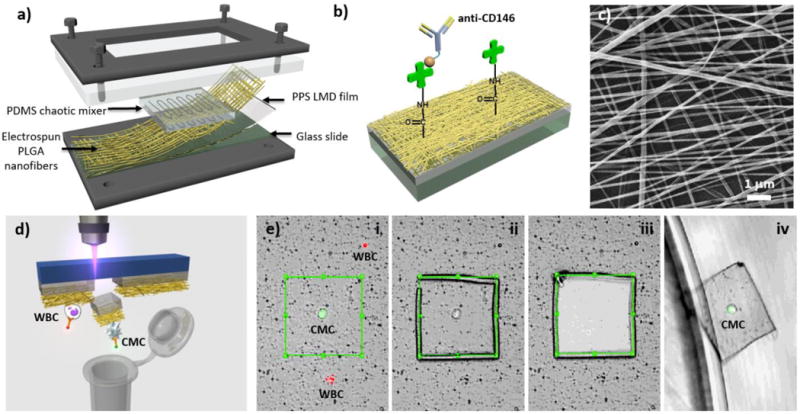
(a) The PDMS chaotic mixer is laid on top of PLGA nanofibers [96] to increase cell capture efficiency. (b) Streptavidin is covalently bound to PLGA nanofibers to interact and bind with biotinylated capture agents. (e.g., anti-CD146 for Circulating Melanoma Cells (CMCs) and anti-EpCAM for pancreatic cancer CTCs). (c) Image of PLGA nanofibers under scanning electron microscope. (d) The graphic illustration of single CMC/CTC isolation from NanoVelcro chips with the use of LMD. (e) Steps of single cell isolation: (i) CMC/CTC identification (ii) CMC/CTC isolation with laser cutting the PLGA around the cell and followed by (iii) moving the PLGA piece containing CMC/CTC from the chip (iv) into a 200 μL PCR tube.
The commercial LCM slides are polymer coated microscope slides with their thermoplastic membrane dissectible by laser dissection microscopes[97]. LCM technology was used in dissecting visually selected areas of tissues from the pathological samples, and the quality of the genomic materials was well preserved. By electrospinning nanofibers made of PLGA (poly(lactic-co-glycolic acid)), onto the LCM slides (Figure 5a-c), the PLGA NanoVelcro substrates were able to interact with nanoscale cellular structures in much the same way as the SiNS on the 1st-gen NanoVelcro Chips, while remaining transparent and laser dissectible. The negatively charged PLGA reduced non-specific adsorption of cell-free DNA by the substrate, thus decreasing the background contamination in subsequent analysis[98]. The PLGA nanofibers were then covalently attached to streptavidin, followed by conjugation of biotinylated antibodies to confer specific capture of CTCs from whole blood samples. (Figure 5b) As with the 1st-gen NanoVelcro Chip, CTC enrichment was achieved by introducing an overlaid PDMS microfluidic chaotic mixer.
Once CTCs were immobilized on the substrate, the CTC-bearing substrate was fixed with methanol and stained with anti-cytokeratin (CK) and anti-CD45. DAPI nuclear staining was excluded from the process to avoid interference in the genome amplification process. Under an LCM microscope, single-CTCs were visually identified by their CK+/CD45- characteristics and were specifically isolated. (Figure 5d and e) To analyze the genomes of single cells, whole genome amplification (WGA) was employed prior to sequencing. Initially, we used PCR-based WGA, and were successful in performing Sanger sequencing targeting a short fragment[96]. However, similar technology only yielded 25–80% coverage in whole exome sequencing (WES), and significantly limited analysis[32]. To improve our sequencing quality, we incorporated a multiple displacement amplification (MDA) reaction to amplify CTC DNA in long fragments. MDA was better than PCR-based WGA in reducing amplification errors[99]. An additional rigorous quality check was employed to select high-quality CTC samples using a multiplexed PCR based on eight housekeeping genes. The successful amplification of the housekeeping genes directly correlated with the final coverage of sequencing. By choosing only the best quality CTC samples, we avoided the unnecessary waste of the sequencing power, and demonstrated the successful whole genome sequencing (WGS) of 4 CTCs with more than 95% coverage[16].
In addition to CTC analysis, our team utilized the 2nd-gen NanoVelcro Assay for enrichment, isolation and characterization of circulating fetal nucleated cells (CFNCs) in maternal blood, showing the feasibility of non-invasive prenatal diagnostics (NIPD)[100]. Using the newly developed imprinted PLGA nanosubstrate with the optimized capture condition and immunocytochemistry (ICC) protocol, this 2nd-gen NanoVelcro Assay was able to effectively enrich a subcategory of CFNCs, i.e., circulating trophoblasts (cTBs) from maternal blood. After we captured and identified single cTBs (Hoechst+/CK7+/HLA−G+/CD45−, 20 μm >sizes> 12 μm), we utilized similar LCM techniques for cTB isolation, followed by downstream genetic testing by array Comparative Genomic Hybridization (aCGH) and Short Tandem Repeats (STR) analysis. Using maternal blood samples collected from expectant mothers with a single fetus, the cTB-derived aCGH data was able to detect fetal genders as well as chromosomal aberrations, which had been confirmed by standard clinical practice. This non-invasive prenatal testing (NIPT) method could potentially replace the current gold standard for diagnosing fetal genetic abnormalities, which involves invasive and higher-risk procedures such as amniocentesis and chorionic villus sampling.
2.7. 3rd-Gen NanoVelcro Chips
Since functional[41] and molecular[15] analyses of captured CTCs have gained more recognition recently, the need for isolating high quality CTCs with higher efficiency and simpler techniques has become essential. Although our 2nd-gen NanoVelcro-LCM approach[71, 101] demonstrated great precision in single-CTC isolation, problems stemming from the time and labor-intensive process and poor viabilities of isolated cells still need to be overcome. As we continued to advance our CTC capture technology with improved efficiency and higher viability of captured CTCs, the 3rd-generation Thermoresponsive NanoVelcro approach was developed to address such needs[102, 103]. Thermoresponsive polymer brushes (i.e., poly(N-isopropylacrylamide), PIPAAm) that were grafted onto SiNS gave the 3rd-gen NanoVelcro Chips (Figure 7) the ability to capture and release CTCs at 37 and 4°C, respectively. Our innovative technique of temperature-dependent conformational changes to polymer brushes could effectively transform the accessibility of the captured CTCs on the NanoVelcro chip, providing a more efficient CTC purification method with enhanced cell viability and molecular integrity. By strategically introducing biotin groups onto the polymer brushes, we facilitated their conjugation with anti-EpCAM, a CTC-capture agent. At 37°C, CTCs are captured on the NanoVelcro substrates due to their interactions with anti-EpCAM grafted on hydrophobic domains of the polymer brushes. On the other hand, at 4°C, CTC release occurrs due to conformational changes of the polymer brushes that induce internalization of anti-EpCAM. In addition to superior CTC-capture efficiency, thermoresponsive NanoVelcro demonstrated high recovery rates (>70%) and high recovered cell viability (>90%) at 4°C[103].
Figure 7. Operating mechanism of the thermoresponsive NanoVelcro CTC substrate.
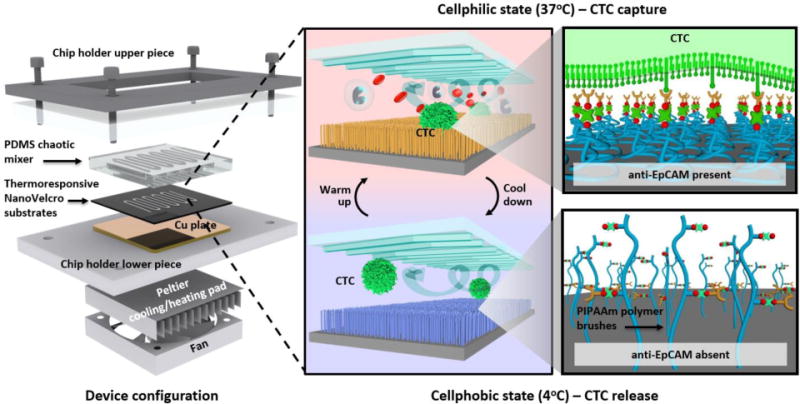
For capturing and releasing CTCs at 37 and 4°C, respectively. The temperature-dependent conformational changes of polymer brushes effectively alter the accessibility of anti-EpCAM on NanoVelcro substrates.
We continue to strive for rapid purification of CTCs from clinical blood samples, serving as a foundation for downstream CTC analysis for both molecular and functional aspects. Furthermore, obtaining viable CTCs for ex vivo expansion could be useful for a variety of applications, such as generating CTC-derived cancer lines and in vitro real-time monitoring of disease status. In the era of personalized and precision medicine, our device could assist the development of potential therapeutics for individual cancer patients.
There are other groups developing techniques of CTC capture and release using different thermoresponsive nanomaterials. Dr. Agarwal’s team at University of Miami[104] also utilized the thermoresponsive property of PIPAAm on a CTC separation slot filter. The filter is made of a parylene C membrane which binds CTCs using non-specific electrostatic interactions. When the temperature is lower than 32°C, PIPAAm demonstrates a hydrophilic property, allowing the non-specific electrostatic interactions between CTC membranes and the parylene C membrane of the filter. When the temperature is heated up over 37°C, PIPAAm becomes hydrophobic, thus releasing the electrostatically bound cells. Dr. Nagrath’s team at the University of Michigan[105] also used a similar approach by coating thermal-sensitive polymers combined with Graphene oxide (GO) to a chip. The chip demonstrated the ability to capture CTCs with anti-EpCAM at room temperature and release them at the lower critical solution temperature (LCST) of 13°C. Another thermoresponsive technique was carried out by Dr. Liu’s team at Beijing National Laboratory for Molecular Sciences, which combined a thermoresponsive nanostructured surface with hydrophobic interaction for its synergistic effect to capture and release CTCs[106]. In addition to thermoresponsive methods, other approaches, including photoresponsive[107] or electroresponsive[108] methods, have also been reported for capturing and releasing CTCs.
2.8. 4th-Gen NanoVelcro Chips
Although the 3rd-gen NanoVelcro Chip provided a stable and robust way to recover CTCs, we have also been exploring other methods to improve throughput and cell viability. Hence, we developed another method for CTC capture and release by utilizing surface chemistry with competitive binding. The 4th-gen NanoVelcro Chip (Sweet chip) [75] is fabricated with poly(3,4-ethylene-dioxythiophene)s (PEDOT)-based nanomaterial on the chip surface. In this platform, phenylboronic acid (PBA)-grafted PEDOT NanoVelcro combines the three-dimensional PEDOT nanosubstrate, and provides high specificity for CTC capture upon antibody conjugation. Furthermore, sorbitol competitive binding gently releases the captured cells. CTCs purified by this PEDOT NanoVelcro chip provide well-preserved RNA transcripts for the analysis of expression levels of several prostate cancer specific RNA biomarkers (e.g. AR-FL, AR-V7, KLK3, FOLH1 and SchLAP1), which may provide clinical insights into the disease. (Figure 8)
Figure 8. Illustration of 4th-gen NanoVelcro Chip (Sweet chip).
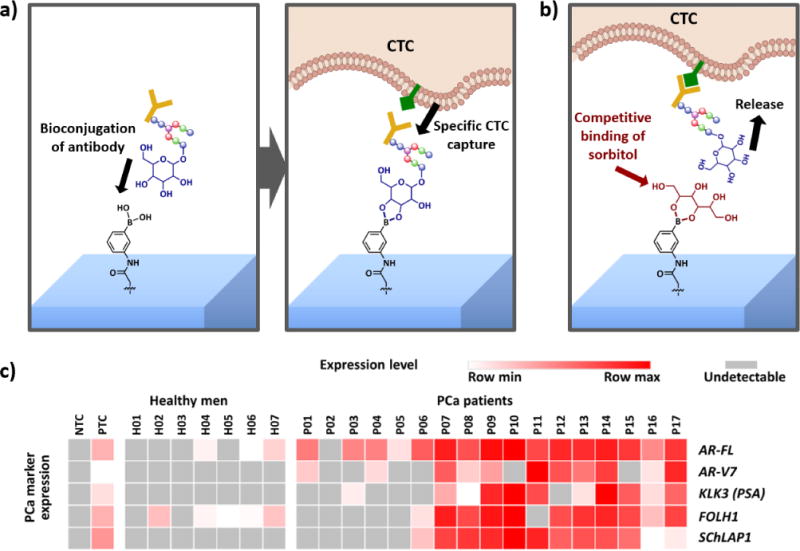
a) The mechanism for circulating tumor cell (CTC) capture is that the surface-grafted phenylboronic acid (PBA) conjugates with antibody, subsequently enabling specific CTC capture. b) The mechanism for CTC release is that the introduction of glycan with stronger affinity to PBA (i.e., sorbitol) results in competitive binding, allowing CTC release. c) RNA signature detection in 7 healthy men and 17 prostate cancer patients using 4th-gen NanoVelcro Chip. Expression of AR-FL, AR-V7, KLK3, FOLH1 and SchLAP1 is summarized in heatmap. PTC = positive control (RNA from 100 LNCaP cells + 100 22Rv1 cells in 1 million WBCs from a healthy man purified by PEDOT-NanoVelcro Chips); NTC = negative control (nuclease free water)[75].
There are other endeavors in the capture and release of viable CTCs using surface chemical reactions. Dr. Stott’s team at MGH utilized enzymatic reactions to release chip-bound CTCs by inducing cleavage of the bio-conjugation ligands of capture agents [109]. They also used small molecule Glutathione (GSH) to trigger ligand exchange to release CTCs [110]. Moreover, Dr. Liu’s team at Beijing National Laboratory for Molecular Sciences demonstrated the capture of CTCs by utilizing a dual responsive surface composed of poly(acrylamidophenylboronic acid) brush grated from aligned silicon nanowire array. This surface can capture and release CTCs by controlling pH and glucose concentration[111].
3. CTC enumeration with the 1st-gen NanoVelcro CTC Assay
3.1. Prostate cancer
With the proof-of-concept studies demonstrating the CTC capture performance of the 1st-gen NanoVelcro CTC Assay[90], a joint research team was formed by UCLA and Cedars-Sinai Medical Center to further test the use of this assay for prostate cancer CTC enumeration. We first focused on patients with advanced metastatic disease, in order to address the problem of CellSearch™ assay failing to detect CTCs in most metastatic castration-resistant prostate cancer patients [14]. The initial study [8] tested forty patients (8 with localized disease and 32 with metastatic disease). NanoVelcro CTC Assay was able to detect CTCs in all 40 patients, suggesting its consistent performance for CTC enumeration, particularly in the setting of advanced diseases. Serial enumerations were also performed to study the change of CTC count after initiation of therapies. Consistent with the observations made using CellSearch™ assay, the clinical responders had a statistically significant drop in CTC counts. Furthermore, in a patient who was able to provide serial blood specimens for as long as 460 days, we observed changes of CTC count corresponding to the patient’s initial response and subsequent failure during sipuleucel-t and nilutamide treatment. This initial observational study not only supported the use of 1st-gen NanoVelcro CTC Assay for CTC enumeration, but also suggested the potential clinical value of serial CTC measurements to monitor disease progression and response to treatment.
As the clinical studies continued to expand, the 1st-gen NanoVelcro CTC Assays also underwent improvements, including the introduction of a user-friendly system and protocols. The current version of the NanoVelcro CTC Enumeration Assay (Figure 9) has undergone extensive testing from calibration/interference tests with cancer cell lines. More than 100 prostate cancer patient samples have been tested for side-by-side comparison with CellSearch™ Assay. The research team noted the differences of the results of these two assays. The experience of the UCLA/CSMC team in combining NanoVelcro CTC Assay with higher resolution fluorescence microscopy suggested that certain immune cells, particularly neutrophils, might be falsely characterized as CTCs given the high CK false-positivity, resulting in the unusual high CTC readouts in the CellSearch™ Assay. To further improve the accuracy of CTC identification, the nature of CTCs as well as the mechanism of the assay will need to be further investigated. Clinical studies of larger scale are also needed to validate the performance of the assay and the clinical value of CTC enumeration in various disease settings. Meanwhile, this has led to the use of cytopathology standards and morphologic analysis in the NanoVelcro Assay, which not only helps minimize the false-positive events, but also results in the discovery of morphologic subsets of CTCs and their association with lethal visceral progression in prostate cancer.
Figure 9. The current version of NanoVelcro CTC Enumeration Assay.
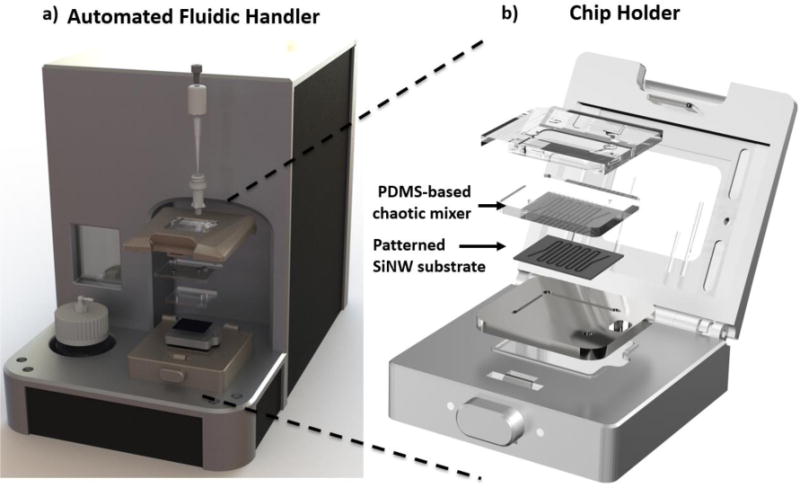
a) A fluidic handler designed to processe blood samples into NanoVelcro Chips. b) The mechanical click-on approach enables the NanoVelcro assay to assemble instantly.
3.2. Subclassification of prostate cancer CTCs
NanoVelcro CTC Assay combined with high-resolution fluorescence microscopy[73, 112] has allowed for better visualization and morphology analysis of the sub-cellular structures of CTCs. An observational study was conducted by UCLA/CSMC with serial blood samples collected from patients with different stages of prostate cancer. Mathematical modeling and unsupervised clustering of CTC nuclear size distribution was applied to identify three distinct subsets of CTCs. The CTC subsets were termed large-nuclear CTCs (lnCTCs, nuclear size > 15.0 μm), small-nuclear CTCs (snCTCs, nuclear size between 8.5 μm and 15.0 μm), and very-small-nuclear CTCs (vsnCTCs, nuclear size < 8.5 μm). vsnCTCs and snCTCs were more commonly seen in patients with metastatic disease. More importantly, we found that vsnCTCs predominantly occur in patients with visceral metastasis (i.e. metastasis to visceral organs such as the lung or liver). Moreover, patients with visceral metastasis had higher numbers of vsnCTCs compared to those without visceral lesions. Serial CTC analysis in this study also found several patients whose vsnCTCs emerged before clinical radiographic detection of visceral metastasis[113]. This observation has now been investigated by analyzing CTCs from serially collected blood specimens from larger patient cohorts focused on patients with metastatic prostate cancer who had progression under androgen receptor signaling inhibitors (ARSIs), such as abiraterone, enzalutamide, or other equivalent drugs. Initial analysis of another retrospective cohort consisting of 28 patients (13 had bone-only metastasis and 15 with visceral metastasis at the first CTC enumeration [113]) found vsnCTCs in all the patients with visceral metastasis. Six out of thirteen initially non-visceral metastatic patients developed visceral metastasis during continuous follow-up, and vsnCTCs were detected mostly within 150 days prior to clinical detection of visceral lesions by radiographic imaging; the only exception was a patient whose vsnCTCs were seen more than 400 days before radiographic detection of his visceral metastasis. In addition to the patients who developed visceral lesions, three other patients were found to have vsnCTCs but had not developed visceral metastasis by the time of this analysis, and will continue to be monitored for visceral metastasis in the future. It is worth noting that in this study, none of the vsnCTC-negative patients developed visceral metastasis. By incorporating with morphologic analysis (i.e., nuclear size) to CTC enumeration, it allows for further investigation of the association between emergence of vsnCTCs and visceral progression in patients with prostate cancer. (Figure 10)
Figure 10. 1stGen NanoVelcro CTC Device characterized very-small-nuclear CTCs (vsnCTCs) in visceral metastasis of prostate cancer.
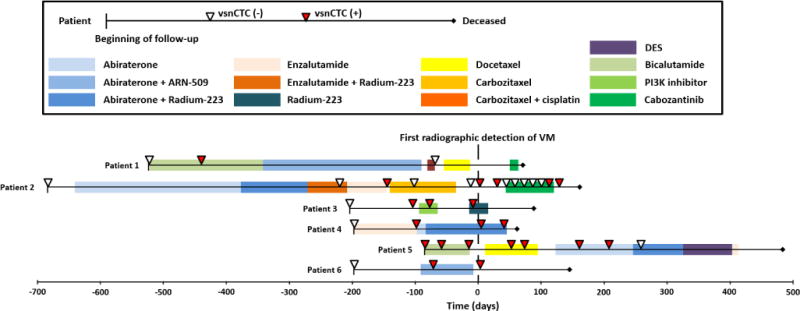
The correlation between presence of vsnCTCs and the presence of visceral metastasis is demonstrated. The presence of vsnCTCs in serial CTC enumerations before the detection of visceral metastasis suggests its predictive potential for visceral progression in prostate cancer patients.
3.3. Pancreatic cancer
In addition to prostate cancer, several research teams have also explored the use of NanoVelcro CTC Assay in other types of solid tumors. As one of the most difficult cancers to detect CTCs [114, 115], pancreatic ductal adenocarcinoma (PDAC) has attracted the attention of the research team at UCLA to investigate[116] the feasibility of applying the NanoVelcro Assay. The team hypothesized that NanoVelcro CTC Assay could detect CTCs in PDAC patients, especially when conventional image studies (e.g. CT scan, MRI) cannot reveal the tumors due to imaging thresholds, and that the CTC count could serve as an adjunctive biomarker for staging. With the initial success in cell line models, a prospective study was conducted to analyze CTCs using NanoVelcro assay from 100 consecutive, pre-treatment PDAC patients. Among the 36 patients without prior chemotherapy, a median of eight CTCs (IQR 3–12) were found in the 10 patients with occult metastatic disease, versus one CTC (IQR 0–2) in the 26 patients with localized disease (p\0.0001). With CTC enumeration from the patient’s venous blood, we can predict successfully resected tumors and occult metastatic disease among untreated patient with PDACs (Figure 11a.). Evaluation of CTC counts revealed the presence of CTCs in 54/72 patients with confirmed PDAC (sensitivity = 75.0%, specificity = 96.4%). Furthermore, a cut-off of >3 CTCs in 4-mL blood was able to distinguish patients with metastatic disease from those with local/regional PDAC. In comparison with CA19-9 as one of the conventional PDAC metastasis detection tools, CTC enumeration can clearly provide superior sensitivity, specificity, PPV, NPV and AUROC in distinguishing patients with occult metastatic disease from those with potentially curable and localized tumors (Figure 11b.). This study not only demonstrated that NanoVelcro assay can be readily adopted in different cancers, but also suggested a promising role of CTCs in early detection and occult cancer metastasis of PDAC. We envision that CTC enumeration can be utilized as an effective biomarker for clinical decision-making.
Figure 11. CTC circulating tumor cell, CA19-9 cancer antigen 19-9, ROC receiver operating characteristic curve, AUROC area under the receiver operating characteristic curve.
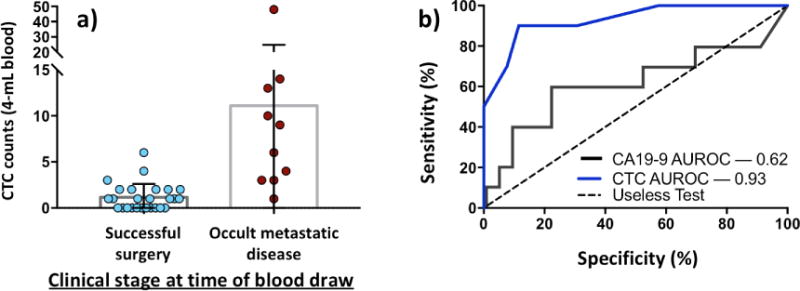
a) CTC count between successfully resected tumors and occult metastatic disease patients. The distinct difference in CTC number indicates that CTC enumeration can be utilized as a preoperative predictor of occult metastatic disease among untreated patients. b) The ROC curve of CTCs and CA19-9 in preoperative detection of patients with occult metastatic disease. AUROC difference between two methods (0.62 vs 0.93) shows effectiveness of CTC enumeration in occult metastatic disease detection.
3.4. Kidney cancer
Another example of the clinical applications of NanoVelcro CTC enumeration assay is the detection of CTCs in patients with renal cell carcinoma (RCC). With the clear cell subtype as the vast majority, RCC also encounters challenges in early detection. To capture CTCs from RCC patients, a new combination of capture antibodies (i.e., anti-CA9 and anti-CD147) were tested. A research team from the Fourth Military Medical University in China then led the effort to test the revised protocol in clinical samples [117]. The results showed CTCs in 72/76 clear cell RCC patients, with the CTC counts significantly higher than those in benign kidney tumors (12.8 ± 6.9 vs. 1.7 ± 1.7). No CTCs were detected in healthy volunteers. Moreover, CTC counts in patients with advanced disease (stages III and IV) were higher than counts in patients with early disease (stages I and II). Further analysis on CTC subsets found that the number of CTCs with vimentin expression correlated with the clinical stage of RCC, indicating a prognostic role of CTCs in RCC. Together these results demonstrated the versatility of NanoVelcro CTC Assay to be adopted in different cancers, and the role of CTCs in RCC as a tool for early detection and prognostication.
4. Molecular Analysis of CTCs using 2nd-gen and 3rd-gen NanoVelcro Chip
4.1. Mutational analysis of circulating melanoma cells (CMCs) and pancreatic CTCs
Utilizing the 2nd-gen NanoVelcro Chips, our first attempt to perform single-CTC mutational analysis was to detect a signature oncogenic mutation (i.e., BRAFV600E) in individual melanoma CTCs[96]. BRAFV600E is present in 50% of melanoma patients[118], and its presence predicts the treatment response to the specific inhibitors, including vemurafenib[119].
By utilizing melanoma-specific anti-CD146 as the capture agent, we isolated single CTCs from several stage IV melanoma patients with known BRAFV600E mutations by standard sequencing of tissue biopsy. After PCR-based WGA, the genomic materials from the isolated single CTCs were subjected to Sanger sequencing specifically looking for the BRAFV600E mutation. (Figure 12a) We not only demonstrated the detection of the oncogenic mutation in the CTCs, but also displayed a strong signal-to-noise ratio. This finding confirmed the purity of our isolated single CTC, which was different from the sequencing results of biopsied melanoma tissues, where melanoma cells inevitably mixed with normal cells surrounding the cancer. Our finding also suggested the feasibility of detecting BRAFV600E in melanoma CTC as a companion diagnostic tool for vemurafenib to avoid invasive tissue sampling.
Figure 12. a) Single-CMC WGA result and gel electrophoresis.
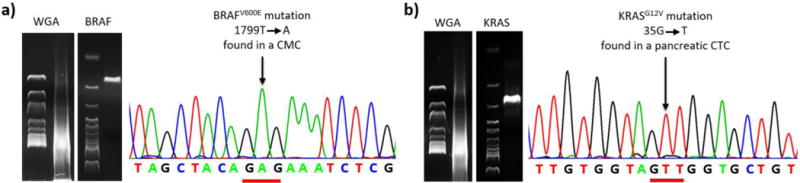
The CMCs exhibit the unique BRAFV600E mutation confirmed by Sanger sequencing [96]. b) KRASG12V mutation present in pancreatic CTCs [121].
Knowing that mutation can be detected in single-CTC sequencing, our team designed a study to assess the sensitivity of such detection utilizing pancreatic CTCs as a model system. In pancreatic cancer, KRAS oncogene mutations are present in more than 95% of the patients, and 98% of those mutations are located in codon 12[120]. Utilizing epithelial cancer specific anti-EpCAM as the capture agent, we identified pancreatic CTCs by their CEA+/CD45− characteristics under fluorescence microscopy. The dissected single CTCs were subjected to PCR-based WGA and Sanger sequencing.
Utilizing the 2nd-gen NanoVelcro Chips, we demonstrated that both KRASG12V (Figure 12b) and KRASG12D mutations [121] were detected in the pancreatic CTCs. 92% of the patients with known KRAS mutations were demonstrated to have the same mutation in their CTCs. We also revealed a low mutation detection rate in each CTC, indicating that PCR-based WGA was the most error-prone step in the CTC sequencing process. However, the sequencing of 10 CTCs from each patient was sufficient to compensate the false negative detection caused by random loss of allele in the WGA process.
4.2. Whole exome sequencing of CTCs
We further explored the feasibility of performing whole genome analysis on single-CTC DNA. In our first report, we performed whole exome sequencing on isolated single CTCs [95], and demonstrated the streamlined process of our platform. After PCR-based WGA, single-CTC DNA obtained from 2nd-gen NanoVelcro chips was sequenced by WES. WES was performed using a standard exome capture kit and Illumina HiSeq. 25 to 80% whole exome coverage was achieved with a mean depth of 29 to 48X. We observed no chromosomal loss in any CTCs, indicating no chromosomal loss during the extraction of single-CTC DNA. Limited by the coverage, we only compared the shared mutation between individual CTCs, CTCs and WBCs, and demonstrated the difference between CTCs and WBCs. This was the first report to demonstrate the feasibility of performing single-CTC WES to verify the difference between CTCs and WBCs at the whole genome level.
4.3. Whole genome sequencing (WGS) of CTCs
With the improvement of our processing technique and WGA method, we then sought to compare the single-CTC DNA with the paired tumor tissues from the same patient. In an index prostate cancer patient, we performed high-quality whole genome sequencing on isolated single CTCs as well as his primary prostate cancer and liver metastasis[16]. In the four sequenced single CTCs, we achieved high-quality sequencing with above 95% sequencing coverage with a standard 30X depth (Figure 13a). The data quality was comparable to the sequencing of the primary prostate tissue and the biopsied tissue from liver metastasis. 29% of the somatic single nucleotide variants (SSNVs) shared between the primary tumor and liver metastasis were found in the CTCs, and 86% of the mutations shared in CTCs were traced back to either the primary or metastatic tumor (Figure 13c). As the first team to perform WGS on CTCs, we also demonstrated the same rearrangements (in chr 3) in the tumor tissues and CTCs (Figure 13d) that were not present in the comparison WBCs or adjacent normal tissue (Figure 13b). Utilizing the high-quality WGS of single CTCs, we demonstrated the similarity between CTCs and tumor tissue at the whole genome level. Our report set the foundation for future studies to utilize isolated single CTCs as a surrogate source of tumor tissue to study cancer biology in vivo.
Figure 13. 2nd-gen NanoVelcro CTC Chips enabled high-quality WGS on single-CTCs.
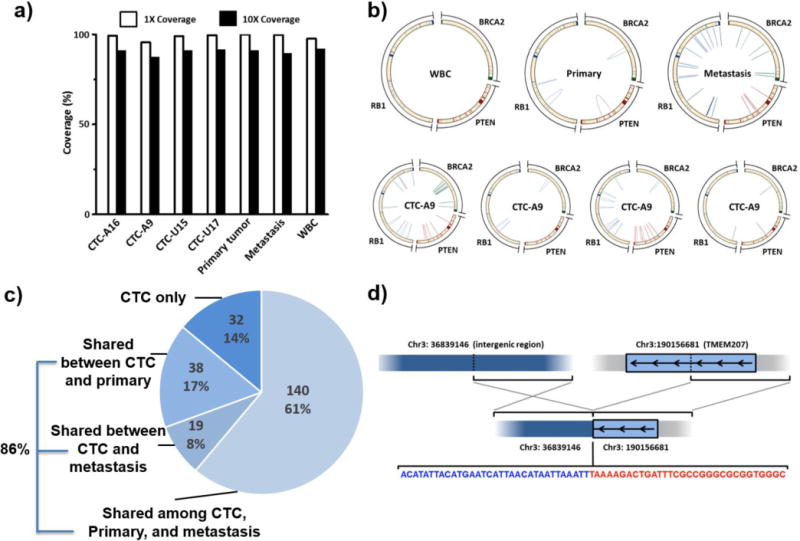
a) Above 95% coverage was demonstrated for single-CTC WGS[16]. b) Tumor suppressor gene rearrangements including RB1, BRCA2 and PTEN were detected in CTCs and tumors. No mutation was found in WBCs. c) We identified that more than 3 CTCs shared clonal somatic single nucleotide variants (SSNVs), which can be considered high confident mutations. 86.0% of CTC-clonal SSNVs are shared among either the primary tumor or metastatic tissue. d) TMEM207 inter-chromosomal rearrangement was detected in both tumor tissues and CTCs.
4.4. Molecular analysis of CTCs using 3rd-gen NanoVelcro Chip
In order to utilize the 3-gen Thermoresponsive NanoVelcro Chip for CTC capture and release, we first optimized performance by using artificial blood samples containing anti-EpCAM-positive H1975 non-small cell lung cancer (NSCLC) cell line. We performed 2 rounds of CTC capture and release to ensure higher purity, enabling point mutation analysis of EGFR (Figure 14a). Strong correlations with tumor tissues and molecular signatures of purified CTCs were found by Sanger sequencing of the 7 NSCLC patients’ CTCs. Additionally, we analyzed molecular signatures of serial CTCs for a NSCLC patient who was treated with EGFR inhibitor. We initially observed L858R mutation in the CTCs, which correlated to good drug response with EGFR inhibitor (i.e. Gefitinib). However, the emergence of T790M mutation was noticed after the treatment and the patient’s disease progressed, indicating possible drug resistance (Figure 14b-d). These results showed promising utility of CTCs for detecting resistance of targeted therapies.
Figure 14. CTC purification of non-small cell lung cancer (NSCLC) by using 3rd-Gen NanoVelcro CTC Chips and molecular analysis.
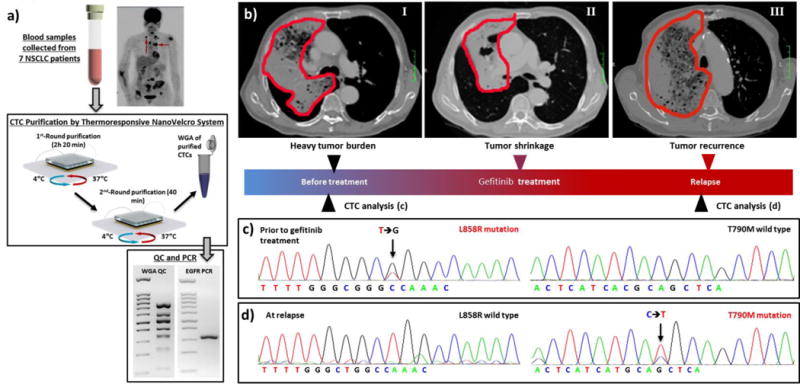
(a) CTC purification and EGFR mutation detection are demonstrated by 3rd-gen NanoVelcro Chips and electrophoresis. (b-d) Disease course and longitudinal mutational analysis of CTCs from a NSCLC patient. (b) Tumor regression and progression during gefitinib (1st-generation EGFR inhibitor) treatment course. (c) L8585R mutation was detected both in CTCs and tumor tissue before the patient was treated with gefitinib. (d) The secondary T790M mutation was detected in the CTCs after disease progression.
5. Technical summary for four generations of NanoVelcro CTC Assays
In summary, the four generations of NanoVelcro CTC Assays presented above introduced unique properties comparing to other existing systems. These four generations of NanoVelcro CTC Assays have similar operatiing mechanisms allowing the assays to achieve optimized CTC capture efficiency. First of all, the combination of embedded nanostructures and capture agents results in enhanced affinity between NanoVelcro substrates and CTCs. Second, increased contact frequency between NanoVelcro substrates and CTCs results from the chaotic flow created by the overlaid PDMS chaotic mixer. In addition, the reproducibility, scalability, and cost-effectiveness of producing these NanoVelcro CTC Assays can be guaranteed by employing micro-fabrication techniques for the two essential functional components (i.e., PDMS chaotic mixer and chip nanosubstrates). Other advantages include (i) Speed: Images of all CTCs are obtained via semi-automated microscopy in a short period of time allowing for rapid pathological review. A total of 4 hours is required to complete enumeration and isolation of individual CTCs with the 1st-gen NanoVelcro Enumeration Assay or 2nd gen NanoVelcro-LCM assay, which is significantly less than the time required for the FDA-approved CellSearch™ assay (4 hours for enumeration only). (ii) Flexibility: the four generations of assays demonstrated utilities for detecting and isolating CTCs with molecular intactness and cell viability; (iii) Sample utilization capacity: CTCs can be recovered from 2 mL blood samples (compared to 7.5 mL samples used by CellSearch™ Assay) given the high sensitivity of NanoVelcro Assays. Up to 5-mL of blood can be processed in one assay if needed, and high capacity can be achieved through multiple rounds performed in parallel. (iv) Cost efficiency: A CellSearch™ assay costs approximately $1,200. In comparison, a NanoVelcro CTC enumeration or isolation assay, including the materials, device fabrication, surface coating, and antibodies, can be performed at a much cheaper price; (v) Simple user interface: the user-friendly chip holder, the fluid handler (Figure 8) and computerized interface allow for easy setup and reduce variation among different users. Unique capacities to each generation of NanoVelcro Assay has been developed over the past decade including high-resolution fluorescent imaging for CTC morphological analysis, LCM technique and temperature-dependent purification for CTC isolation and enrichment. The NanoVelcro Assays have demonstrated the ability to address unmet needs in the field of oncology with their unique capacities, such as the stratification of heterogeneous CTC populations (1st-gen), molecular characterization of CTCs (2nd to 4th gens), and rapid CTC purification/enrichment (3rd and 4th gens).
5.1. Future scientific and clinical developments
With the rapid advancements in therapeutics and a strong drive towards personalization of cancer care, there is a clear, urgent, and unmet need for a liquid biopsy in the oncology clinic. This need is being addressed by many advancements in the field of nanotechnology-based diagnostics. Future research directions will focus on particular needs of nanotechnology-based CTC assays. These needs include (i) Exploring other possible capture/release mechanisms for CTC purification with physiologically compatible conditions for instant CTC isolation or purification that could provide high cell viability and molecular integrity for molecular profiling of the disease. (ii) Further understanding of the interactions between nanointerfaces and CTCs that could affect CTC-capture efficiency, molecular integrity and cell viability. (iii) Exploring the techniques of ex vivo expansion and rare-cell culture of purified CTCs for various research utilities such as drug susceptibility testing and xenograft models. (iv) Exploring the utilities of multi-omic analytical technologies to characterize CTC heterogeneity with single-cell resolution. (v) Characterization of other circulating rare cells such as circulating stromal cells and tumor-associated immune cells that harbor information about disease characteristics and tumor microenvironment.
5.2. Conclusion and outlook
The application of nanotechnology in clinical oncology has shown a promising future to address a myriad of unmet needs. As the current knowledge and understanding of cancer continues to evolve, cancer biologists and physician scientists look to characterize the dynamic biology of the disease. In such an evolving biological environment, oncologists are already familiar with handling temporal variation of data. Analysis of CTCs and other circulating biomarkers has shown promise for investigating the dynamic biology in the individual patient. From the molecular biological concepts of DNA, RNA and proteins, it is evident that there is a great deal of variability in the identities of patients and cancers. Utilizing a noninvasive means to explore these differences can help connect the laboratory and the clinic. Early successes in this field have served as motivation for continued work and inspired others to pursue interdisciplinary research in translational medicine.
Figure 6. Examination of circulating trophoblasts (cTBs) from maternal blood by using single-cell isolatable system, the 2nd-gen NanoVelcro Assay, to be characterized by its phenotype and genotype.
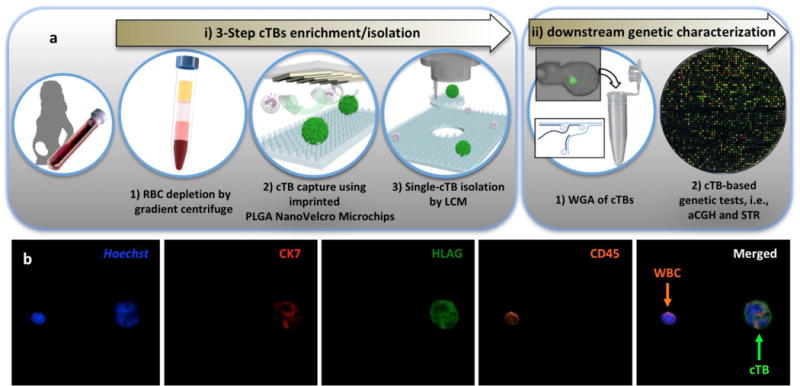
a) The workflow of rare cell enrichment and isolation. After centrifugation of the maternal blood, buffet coat (PBMC layer) was extracted to capture the target cells by NanoVelcro chips. Individual cell isolation is positioned by LCM to extract the targeted cell. The whole genome amplification (WGA) of target cells was conducted for further genetic analysis. b). Immunostaining of cTB by various specific markers. cTB is identified by specific markers, CK7 (red) and HLAG (green), to distinguish from other lymphocytes by CD45 (orange). Hoechst is utilized to stain cell nucleus. Adapted with permission from (Hou, et al., 2017) [100]. Copyright (2018) American Chemical Society.
Acknowledgments
This work was supported by National Institutes of Health, including the following grants R21CA151159, R33CA157396, P01CA168585, R33 CA174562, P01CA098912, U01 CA198900, R44 CA180482, and R01CA218356.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, Brewer DS, Kallio HM, Hognas G, Annala M, Kivinummi K, Goody V, Latimer C, O’Meara S, Dawson KJ, Isaacs W, Emmert-Buck MR, Nykter M, Foster C, Kote-Jarai Z, Easton D, Whitaker HC, I.P.U. Group. Neal DE, Cooper CS, Eeles RA, Visakorpi T, Campbell PJ, McDermott U, Wedge DC, Bova GS. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O’Donovan M, Malhotra S, di Pietro M, Ivakhno S, He M, Weaver JM, Lynch AG, Kingsbury Z, Ross M, Humphray S, Bentley D, Fitzgerald RC, C. Oesophageal Cancer, G. Molecular Stratification Study, C. Oesophageal Cancer, O.S.G. Molecular Stratification Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nature genetics. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, Zia A, Fox NS, Livingstone J, Shiah YJ, Wang J, Beck TA, Have CL, Chong T, Sam M, Johns J, Timms L, Buchner N, Wong A, Watson JD, Simmons TT, P’ng C, Zafarana G, Nguyen F, Luo X, Chu KC, Prokopec SD, Sykes J, Dal Pra A, Berlin A, Brown A, Chan-Seng-Yue MA, Yousif F, Denroche RE, Chong LC, Chen GM, Jung E, Fung C, Starmans MH, Chen H, Govind SK, Hawley J, D’Costa A, Pintilie M, Waggott D, Hach F, Lambin P, Muthuswamy LB, Cooper C, Eeles R, Neal D, Tetu B, Sahinalp C, Stein LD, Fleshner N, Shah SP, Collins CC, Hudson TJ, McPherson JD, van der Kwast T, Bristow RG. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nature genetics. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 6.Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, Pollett A, Gallinger S, McPherson J, Mullighan CG, Shibata D, Dick JE. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba PA., II Futreal, Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu YT, Zhao L, Shen Q, Garcia MA, Wu D, Hou S, Song M, Xu X, Ouyang WH, Ouyang WW, Lichterman J, Luo Z, Xuan X, Huang J, Chung LW, Rettig M, Tseng HR, Shao C, Posadas EM. NanoVelcro Chip for CTC enumeration in prostate cancer patients. Methods. 2013;64:144–152. doi: 10.1016/j.ymeth.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magbanua MJ, Sosa EV, Roy R, Eisenbud LE, Scott JH, Olshen A, Pinkel D, Rugo HS, Park JW. Genomic profiling of isolated circulating tumor cells from metastatic breast cancer patients. Cancer Res. 2013;73:30–40. doi: 10.1158/0008-5472.CAN-11-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 11.Green BJ, Saberi Safaei T, Mepham A, Labib M, Mohamadi RM, Kelley SO. Beyond the Capture of Circulating Tumor Cells: Next-Generation Devices and Materials. Angew Chem Int Ed Engl. 2016;55:1252–1265. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 12.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 13.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. The New England journal of medicine. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 14.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. The lancet oncology. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GS, Li T, Chabot MS, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nature biotechnology. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, Chung LW, Yang H, Li KC, Hou Y, Tseng HR, Hou S, Xu X, Wang J, Posadas EM. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, Xu L, An T, Ma Q, Wang Y, Wu M, Sun Y, Wang S, Li Z, Yang X, Yong J, Su XD, Lu Y, Bai F, Xie XS, Wang J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Brannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 21.Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Allen SG, Reka AK, Qian W, Han S, Zhao J, Bao L, Keshamouni VG, Merajver SD, Fu J. Nanoroughened adhesion-based capture of circulating tumor cells with heterogeneous expression and metastatic characteristics. BMC Cancer. 2016;16:614. doi: 10.1186/s12885-016-2638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob K, Sollier C, Jabado N. Circulating tumor cells: detection, molecular profiling and future prospects. Expert review of proteomics. 2007;4:741–756. doi: 10.1586/14789450.4.6.741. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, Xiao WZ, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earhart CM, Hughes CE, Gaster RS, Ooi CC, Wilson RJ, Zhou LY, Humke EW, Xu L, Wong DJ, Willingham SB, Schwartz EJ, Weissman IL, Jeffrey SS, Neal JW, Rohatgi R, Wakelee HA, Wang SX. Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips. Lab Chip. 2014;14:78–88. doi: 10.1039/c3lc50580d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. The New England journal of medicine. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M, Ionescu-Zanetti C. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay. Transl Oncol. 2013;6:528–+. doi: 10.1593/tlo.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. The VerIFAST: an integrated method for cell isolation and extracellular/intracellular staining. Lab Chip. 2013;13:391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 30.Winer-Jones JP, Vahidi B, Arquilevich N, Fang C, Ferguson S, Harkins D, Hill C, Klem E, Pagano PC, Peasley C, Romero J, Shartle R, Vasko RC, Strauss WM, Dempsey PW. Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell. PLoS One. 2014;9:e86717. doi: 10.1371/journal.pone.0086717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allan AL, Vantyghem SA, Tuck AB, Chambers AF, Chin-Yee IH, Keeney M. Detection and quantification of circulating tumor cells in mouse models of human breast cancer using immunomagnetic enrichment and multiparameter flow cytometry. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2005;65:4–14. doi: 10.1002/cyto.a.20132. [DOI] [PubMed] [Google Scholar]
- 32.He W, Wang H, Hartmann LC, Cheng JX, Low PS. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11760–11765. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Sensitive and High-Throughput Isolation of Rare Cells from Peripheral Blood with Ensemble-Decision Aliquot Ranking. Angew Chem Int Ed Engl. 2012 doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V, Feng Q, Chiu DT. An automated high-throughput counting method for screening circulating tumor cells in peripheral blood. Analytical chemistry. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecot CV, Bischoff FZ, Mayer JA, Wong KL, Pham T, Bottsford-Miller J, Stone RL, Lin YG, Jaladurgam P, Roh JW, Goodman BW, Merritt WM, Pircher TJ, Mikolajczyk SD, Nick AM, Celestino J, Eng C, Ellis LM, Deavers MT, Sood AK. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov. 2011;1:580–586. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27–29. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Wittner BS, Stojanov P, Brachtel E, Sgroi D, Kapur R, Shioda T, Ting DT, Ramaswamy S, Getz G, Iafrate AJ, Benes C, Toner M, Maheswaran S, Haber DA. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto DT, Luo X, Bardia A, Wittner BS, Ramaswamy S, Shioda T, Ting DT, Stott SL, Kapur R, Maheswaran S, Haber DA, Toner M. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nature methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew Chem Int Ed Engl. 2011;50:11769–11772. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castro CM, Ghazani AA, Chung J, Shao H, Issadore D, Yoon TJ, Weissleder R, Lee H. Miniaturized nuclear magnetic resonance platform for detection and profiling of circulating tumor cells. Lab Chip. 2014;14:14–23. doi: 10.1039/c3lc50621e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mach AJ, Kim JH, Arshi A, Hur SC, Di Carlo D. Automated cellular sample preparation using a Centrifuge-on-a-Chip. Lab on a Chip. 2011;11:2827–2834. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- 47.Dhar M, Wong J, Karimi A, Che J, Renier C, Matsumoto M, Triboulet M, Garon EB, Goldman JW, Rettig MB, Jeffrey SS, Kulkarni RP, Sollier E, Di Carlo D. High efficiency vortex trapping of circulating tumor cells. Biomicrofluidics. 2015;9:064116. doi: 10.1063/1.4937895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen JY, Tsai WS, Shao HJ, Wu JC, Lai JM, Lu SH, Hung TF, Yang CT, Wu LC, Chen JS, Lee WH, Chang YC. Sensitive and Specific Biomimetic Lipid Coated Microfluidics to Isolate Viable Circulating Tumor Cells and Microemboli for Cancer Detection. PLoS One. 2016;11:e0149633. doi: 10.1371/journal.pone.0149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohamadi RM, Besant JD, Mepham A, Green B, Mahmoudian L, Gibbs T, Ivanov I, Malvea A, Stojcic J, Allan AL, Lowes LE, Sargent EH, Nam RK, Kelley SO. Nanoparticle-mediated binning and profiling of heterogeneous circulating tumor cell subpopulations. Angew Chem Int Ed Engl. 2015;54:139–143. doi: 10.1002/anie.201409376. [DOI] [PubMed] [Google Scholar]
- 50.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. A rare-cell detector for cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Archives of pathology & laboratory medicine. 2009;133:1468–1471. doi: 10.1043/1543-2165-133.9.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, Ko AH, Korn WM, Schram E, Coward M, Yang X, Metzner T, Lamy R, Honnatti M, Yoshioka C, Kunken J, Petrova Y, Sok D, Nelson D, Kuhn P. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campton DE, Ramirez AB, Nordberg JJ, Drovetto N, Clein AC, Varshavskaya P, Friemel BH, Quarre S, Breman A, Dorschner M, Blau S, Blau CA, Sabath DE, Stilwell JL, Kaldjian EP. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360. doi: 10.1186/s12885-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schutze K, Capron F, Franco D, Pazzagli M, Vekemans M, Lacour B, Brechot C, Paterlini-Brechot P. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. The American journal of pathology. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 56.Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomedical microdevices. 2009;11:883–892. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 57.Tan SJ, Lakshmi RL, Chen P, Lim WT, Yobas L, Lim CT. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosensors & bioelectronics. 2010;26:1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 58.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. British journal of cancer. 2011;105:1338–1341. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, Le Moulec S, Andre F, Fizazi K, Soria JC, Vielh P. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. British journal of cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desitter I, Guerrouahen BS, Benali-Furet N, Wechsler J, Janne PA, Kuang Y, Yanagita M, Wang L, Berkowitz JA, Distel RJ, Cayre YE. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer research. 2011;31:427–441. [PubMed] [Google Scholar]
- 61.Gogoi P, Sepehri S, Zhou Y, Gorin MA, Paolillo C, Capoluongo E, Gleason K, Payne A, Boniface B, Cristofanilli M, Morgan TM, Fortina P, Pienta KJ, Handique K, Wang Y. Development of an Automated and Sensitive Microfluidic Device for Capturing and Characterizing Circulating Tumor Cells (CTCs) from Clinical Blood Samples. PLoS One. 2016;11:e0147400. doi: 10.1371/journal.pone.0147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta V, Jafferji I, Garza M, Melnikova VO, Hasegawa DK, Pethig R, Davis DW. ApoStream(), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gagnon ZR. Cellular dielectrophoresis: applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis. 2011;32:2466–2487. doi: 10.1002/elps.201100060. [DOI] [PubMed] [Google Scholar]
- 64.Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, Li Y, Burt DJ, Antonello J, Morrow CJ, Hodgkinson CL, Morris K, Priest L, Carter M, Miller C, Hughes A, Blackhall F, Dive C, Brady G. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nature Medicine. 2016;23:114. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 65.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 66.Riethdorf S, Pantel K. Advancing personalized cancer therapy by detection and characterization of circulating carcinoma cells. Annals of the New York Academy of Sciences. 2010;1210:66–77. doi: 10.1111/j.1749-6632.2010.05779.x. [DOI] [PubMed] [Google Scholar]
- 67.Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends in molecular medicine. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Yoon HJ, Kozminsky M, Nagrath S. Emerging role of nanomaterials in circulating tumor cell isolation and analysis. ACS Nano. 2014;8:1995–2017. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Asghar W, Demirci U, Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:347–387. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang P, Wang S. Designing fractal nanostructured biointerfaces for biomedical applications. Chemphyschem. 2014;15:1550–1561. doi: 10.1002/cphc.201301230. [DOI] [PubMed] [Google Scholar]
- 71.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed Engl. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Nanostructure embedded microchips for detection, isolation, and characterization of circulating tumor cells. Acc Chem Res. 2014;47:2941–2950. doi: 10.1021/ar5001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010;70:6128–6138. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen JF, Zhu Y, Lu YT, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posadas EM, Tseng HR. Clinical Applications of NanoVelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Theranostics. 2016;6:1425–1439. doi: 10.7150/thno.15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen M-Y, Chen J-F, Luo C-H, Lee S, Li C-H, Yang Y-L, Tsai Y-H, Ho B-C, Bao L, Jan YJ, Zhu Y, Cheng S, Feng FY, Chen P, Hou S, Agopian V, Hsiao Y-S, Tseng H-R, Posadas EM, Yu H-h. Glycan-Stimulation Enables Purification of Prostate Cancer Circulating Tumor Cells on PEDOT NanoVelcro Chips for RNA Biomarker Detection. Advanced Healthcare Materials. 2017 doi: 10.1002/adhm.201700701. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Functionalized conducting polymer nanodots for enhanced cell capturing: the synergistic effect of capture agents and nanostructures. Adv Mater. 2011;23:4788–4792. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 77.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 78.Hsiao YS, Luo SC, Hou S, Zhu B, Sekine J, Kuo CW, Chueh DY, Yu HH, Tseng HR, Chen P. 3D bioelectronic interface: capturing circulating tumor cells onto conducting polymer-based micro/nanorod arrays with chemical and topographical control. Small. 2014;10:3012–3017. doi: 10.1002/smll.201400429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fischer KE, Aleman BJ, Tao SL, Hugh Daniels R, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Biomimetic nanowire coatings for next generation adhesive drug delivery systems. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He R, Zhao L, Liu Y, Zhang N, Cheng B, He Z, Cai B, Li S, Liu W, Guo S, Chen Y, Xiong B, Zhao XZ. Biocompatible TiO2 nanoparticle-based cell immunoassay for circulating tumor cells capture and identification from cancer patients. Biomedical microdevices. 2013;15:617–626. doi: 10.1007/s10544-013-9781-9. [DOI] [PubMed] [Google Scholar]
- 81.Banerjee SS, Paul D, Bhansali SG, Aher ND, Jalota-Badhwar A, Khandare J. Enhancing Surface Interactions with Colon Cancer Cells on a Transferrin-Conjugated 3D Nanostructured Substrate. Small. 2012;8:1657–1663. doi: 10.1002/smll.201102354. [DOI] [PubMed] [Google Scholar]
- 82.Park GS, Kwon H, Kwak DW, Park SY, Kim M, Lee JH, Han H, Heo S, Li XS, Lee JH, Kim YH, Lee JG, Yang W, Cho HY, Kim SK, Kim K. Full Surface Embedding of Gold Clusters on Silicon Nanowires for Efficient Capture and Photothermal Therapy of Circulating Tumor Cells. Nano Letters. 2012;12:1638–1642. doi: 10.1021/nl2045759. [DOI] [PubMed] [Google Scholar]
- 83.Lee H, Jang Y, Seo J, Nam JM, Char K. Nanoparticle-Functionalized Polymer Platform for Controlling Metastatic Cancer Cell Adhesion, Shape, and Motility. ACS Nano. 2011;5:5444–5456. doi: 10.1021/nn202103z. [DOI] [PubMed] [Google Scholar]
- 84.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nano. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao W, Cui CH, Bose S, Guo D, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GS, Phillips JA, Dorfman DM, Karnik R, Karp JM. Bioinspired multivalent DNA network for capture and release of cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F, Jiang Y, Liu X, Meng J, Zhang P, Liu H, Yang G, Li G, Jiang L, Wan LJ, Hu JS, Wang S. Hierarchical Nanowire Arrays as Three-Dimensional Fractal Nanobiointerfaces for High Efficient Capture of Cancer Cells. Nano Lett. 2016;16:766–772. doi: 10.1021/acs.nanolett.5b04731. [DOI] [PubMed] [Google Scholar]
- 87.Hanson L, Zhao W, Lou HY, Lin ZC, Lee SW, Chowdary P, Cui Y, Cui B. Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat Nanotechnol. 2015;10:554–562. doi: 10.1038/nnano.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lou HY, Zhao W, Hanson L, Zeng C, Cui Y, Cui B. Dual-Functional Lipid Coating for the Nanopillar-Based Capture of Circulating Tumor Cells with High Purity and Efficiency. Langmuir. 2017;33:1097–1104. doi: 10.1021/acs.langmuir.6b03903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RHW, Macoska JA, Merajver SD, Fu J. Nanoroughened Surfaces for Efficient Capture of Circulating Tumor Cells without Using Capture Antibodies. ACS nano. 2013;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew Chem Int Ed Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science. 2002;295:647–651. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y, Hong H, Cai W. Tumor-Targeted Drug Delivery with Aptamers. Current medicinal chemistry. 2011;18:4185–4194. doi: 10.2174/092986711797189547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao L, Tang C, Xu L, Zhang Z, Li X, Hu H, Cheng S, Zhou W, Huang M, Fong A, Liu B, Tseng HR, Gao H, Liu Y, Fang X. Enhanced and Differential Capture of Circulating Tumor Cells from Lung Cancer Patients by Microfluidic Assays Using Aptamer Cocktail. Small (Weinheim an der Bergstrasse, Germany) 2016;12:1072–1081. doi: 10.1002/smll.201503188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang WH, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LW, Rettig M, Tseng HR, Posadas EM. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv Mater. 2013;25:2897–2902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew Chem Int Ed Engl. 2013;52:3379–3383. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fend F, Raffeld M. Laser capture microdissection in pathology. Journal of clinical pathology. 2000;53:666–672. doi: 10.1136/jcp.53.9.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvanen AC. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic acids research. 2003;31:e129. doi: 10.1093/nar/gng129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hou S, Chen JF, Song M, Zhu Y, Jan YJ, Chen SH, Weng TH, Ling DA, Chen SF, Ro T, Liang AJ, Lee T, Jin H, Li M, Liu L, Hsiao YS, Chen P, Yu HH, Tsai MS, Pisarska MD, Chen A, Chen LC, Tseng HR. Imprinted NanoVelcro Microchips for Isolation and Characterization of Circulating Fetal Trophoblasts: toward Noninvasive Prenatal Diagnostics. ACS Nano. 2017 doi: 10.1021/acsnano.7b03073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saracino GA, Cigognini D, Silva D, Caprini A, Gelain F. Nanomaterials design and tests for neural tissue engineering. Chem Soc Rev. 2013;42:225–262. doi: 10.1039/c2cs35065c. [DOI] [PubMed] [Google Scholar]
- 102.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo SC, Tseng HR, Yu HH. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv Mater. 2013;25:1547–1551. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR. Programming thermoresponsiveness of NanoVelcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano. 2015;9:62–70. doi: 10.1021/nn5056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ao Z, Parasido E, Rawal S, Williams A, Schlegel R, Liu S, Albanese C, Cote RJ, Agarwal A, Datar RH. Thermoresponsive release of viable microfiltrated Circulating Tumor Cells (CTCs) for precision medicine applications. Lab on a chip. 2015;15:4277–4282. doi: 10.1039/c5lc01024a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoon HJ, Shanker A, Wang Y, Kozminsky M, Jin Q, Palanisamy N, Burness ML, Azizi E, Simeone DM, Wicha MS, Kim J, Nagrath S. Tunable Thermal-Sensitive Polymer-Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv Mater. 2016;28:4891–4897. doi: 10.1002/adma.201600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L. Hydrophobic Interaction-Mediated Capture and Release of Cancer Cells on Thermoresponsive Nanostructured Surfaces. Advanced Materials. 2013;25:922–927. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 107.Lv SW, Wang J, Xie M, Lu NN, Li Z, Yan XW, Cai SL, Zhang PA, Dong WG, Huang WH. Photoresponsive immunomagnetic nanocarrier for capture and release of rare circulating tumor cells. Chemical Science. 2015;6:6432–6438. doi: 10.1039/c5sc01380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hsiao Y-S, Ho B-C, Yan H-X, Kuo C-W, Chueh D-Y, Yu H-h, Chen P. Integrated 3D conducting polymer-based bioelectronics for capture and release of circulating tumor cells. Journal of Materials Chemistry B. 2015;3:5103–5110. doi: 10.1039/c5tb00096c. [DOI] [PubMed] [Google Scholar]
- 109.Li W, Reátegui E, Park MH, Castleberry S, Deng JZ, Hsu B, Mayner S, Jensen AE, Sequist LV, Maheswaran S, Haber DA, Toner M, Stott SL, Hammond PT. Biodegradable nano-films for capture and non-invasive release of circulating tumor cells. Biomaterials. 2015;65:93–102. doi: 10.1016/j.biomaterials.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park MH, Reátegui E, Li W, Tessier SN, Wong KH, Jensen AE, Thapar V, Ting D, Toner M, Stott SL, Hammond PT. Enhanced Isolation and Release of Circulating Tumor Cells Using Nanoparticle Binding and Ligand Exchange in a Microfluidic Chip. Journal of the American Chemical Society. 2017;139:2741–2749. doi: 10.1021/jacs.6b12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu H, Li Y, Sun K, Fan J, Zhang P, Meng J, Wang S, Jiang L. Dual-Responsive Surfaces Modified with Phenylboronic Acid-Containing Polymer Brush To Reversibly Capture and Release Cancer Cells. Journal of the American Chemical Society. 2013;135:7603–7609. doi: 10.1021/ja401000m. [DOI] [PubMed] [Google Scholar]
- 112.Kamei K, Ohashi M, Gschweng E, Ho Q, Suh J, Tang J, For Yu ZT, Clark AT, Pyle AD, Teitell MA, Lee KB, Witte ON, Tseng HR. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen JF, Ho H, Lichterman J, Lu YT, Zhang Y, Garcia MA, Chen SF, Liang AJ, Hodara E, Zhau HE, Hou S, Ahmed RS, Luthringer DJ, Huang J, Li KC, Chung LW, Ke Z, Tseng HR, Posadas EM. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015 doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, Board R, Clack G, Hughes A, Blackhall F, Valle JW, Dive C. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. British journal of cancer. 2012;106:508–516. doi: 10.1038/bjc.2011.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kurihara T, Itoi T, Sofuni A, Itokawa F, Tsuchiya T, Tsuji S, Ishii K, Ikeuchi N, Tsuchida A, Kasuya K, Kawai T, Sakai Y, Moriyasu F. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. Journal of Hepato-Biliary-Pancreatic Surgery. 2008;15:189–195. doi: 10.1007/s00534-007-1250-5. [DOI] [PubMed] [Google Scholar]
- 116.Ankeny JS, Court CM, Hou S, Li Q, Song M, Wu D, Chen JF, Lee T, Lin M, Sho S, Rochefort MM, Girgis MD, Yao J, Wainberg ZA, Muthusamy VR, Watson RR, Donahue TR, Hines OJ, Reber HA, Graeber TG, Tseng HR, Tomlinson JS. Circulating Tumour Cells as a Biomarker for Diagnosis and Staging in Pancreatic Cancer. British journal of cancer. 2016 doi: 10.1038/bjc.2016.121. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu S, Tian Z, Zhang L, Hou S, Hu S, Wu J, Jing Y, Sun H, Yu F, Zhao L, Wang R, Tseng HR, Zhau HE, Chung LWK, Wu K, Wang H, Wu JB, Nie Y, Shao C. Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget. 2016;7:59877–59891. doi: 10.18632/oncotarget.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Current protocols in human genetics. 2008:11. doi: 10.1002/0471142905.hg1011s57. Chapter 10. Unit 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA, B.-S. Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends in biochemical sciences. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Court CM, Ankeny JS, Sho S, Hou S, Li Q, Hsieh C, Song M, Liao X, Rochefort MM, Wainberg ZA, Graeber TG, Tseng HR, Tomlinson JS. Reality of Single Circulating Tumor Cell Sequencing for Molecular Diagnostics in Pancreatic Cancer. J Mol Diagn. 2016;18:688–696. doi: 10.1016/j.jmoldx.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


