Abstract
Prostate cancer is screened by testing circulating levels of the prostate-specific antigen (PSA) biomarker, monitoring changes over time, or a digital rectal exam. Abnormal results often lead to prostate biopsy. Prostate cancer positive patients are stratified into very low-risk, low-risk, intermediate-risk, and high-risk, based on clinical classification parameters, to assess therapy options. However, there remains a gap in our knowledge and a compelling need for improved risk stratification to inform clinical decisions and reduce both over-diagnosis and over-treatment. Further, current strategies for clinical intervention do not distinguish clinically aggressive prostate cancer from indolent disease. This mini-review takes advantage of a large number of functionally characterized microRNAs (miRNA), epigenetic regulators of prostate cancer, that define prostate cancer cell activity, tumor stage, and circulate as biomarkers to monitor disease progression. Nanoparticles provide an effective platform for targeted delivery of miRNA inhibitors or mimics specifically to prostate tumor cells to inhibit cancer progression. Several prostate–specific transmembrane proteins expressed at elevated levels in prostate tumors are under investigation for targeting therapeutic agents to prostate cancer cells. Given that prostate cancer progresses slowly, circulating miRNAs can be monitored to identify tumor progression in indolent disease, allowing identification of miRNAs for nanoparticle intervention before the crucial point of transition to aggressive disease. Here, we describe clinically significant and non-invasive intervention nanoparticle strategies being used in clinical trials for drug and nucleic acid delivery. The advantages of mesoporous silica-based nanoparticles and a number of candidate miRNAs for inhibition of prostate cancer are discussed.
Keywords: mesoporous silica nanoparticles, microRNA, precision medicine, prostate cancer, targeted delivery
1 | INTRODUCTION
The goal of this review is to present the application of microRNA (miRNA) inhibitors and mimics delivered in silica-based nanoparticles specifically targeted to prostate cancer cells with the capability to inhibit tumor progression. This topic requires multidisciplinary expertise in miRNA biology, molecular mechanisms of cancer, nanoparticle synthesis and delivery, and physician-scientists treating patients. This review begins with a discussion of potential miRNA candidates for targeted intervention based on a wealth of studies from multiple laboratories that have established functional roles for miRNAs in prostate cancer cell and animal models. Having oncogenic and tumor suppressive activities, aberrant expression, or lack thereof, of these well characterized miRNAs accelerate aberrant cell growth, induce cell death, and regulate cancer related events, such as the epithelial-to-mesenchymal transition (EMT) and metastasis. Further, miRNAs functioning in the androgen signaling cascade are predictive of tumors unresponsive to androgen deprivation therapy (ADT). Importantly, candidate miRNAs for intervention can be monitored in body fluids, such as serum, plasma, and urine, and have been reported to distinguish patients with castration resistant prostate cancer (CRPC) or hormone sensitive disease from men with benign prostate hyperplasia (BPH). Thus, miRNA expression levels in a patient's body fluid or biopsy specimen will determine whether a miRNA inhibitor, mimic, or combination should be delivered to the developing tumor. We next present a general overview of targeted nanoparticle design and synthesis, detailing advantages, disadvantages, and clinical applicability. The benefits of mesoporous silica nanoparticles (MSN) compared to other nanoparticle formulations stems from their controllable pore size and the ability to covalently link a wide variety of molecules to their surface (e.g., targeting antibodies, fluorophores). The current use of preclinical mouse models provides the foundation for translation to clinical trials of nanoparticles loaded with tumor inhibiting miRNA cargo and targeted to prostate cancer cells. In summary, we describe a clinically significant and non-invasive intervention strategy using non-toxic, silica-based nanoparticles that carry nucleic acid cargo with antibody-targeting to prostate cancer cells capable of inhibiting disease progression.
2 | MICRORNAS CAN INFORM PROSTATE CANCER INTERVENTION DECISIONS
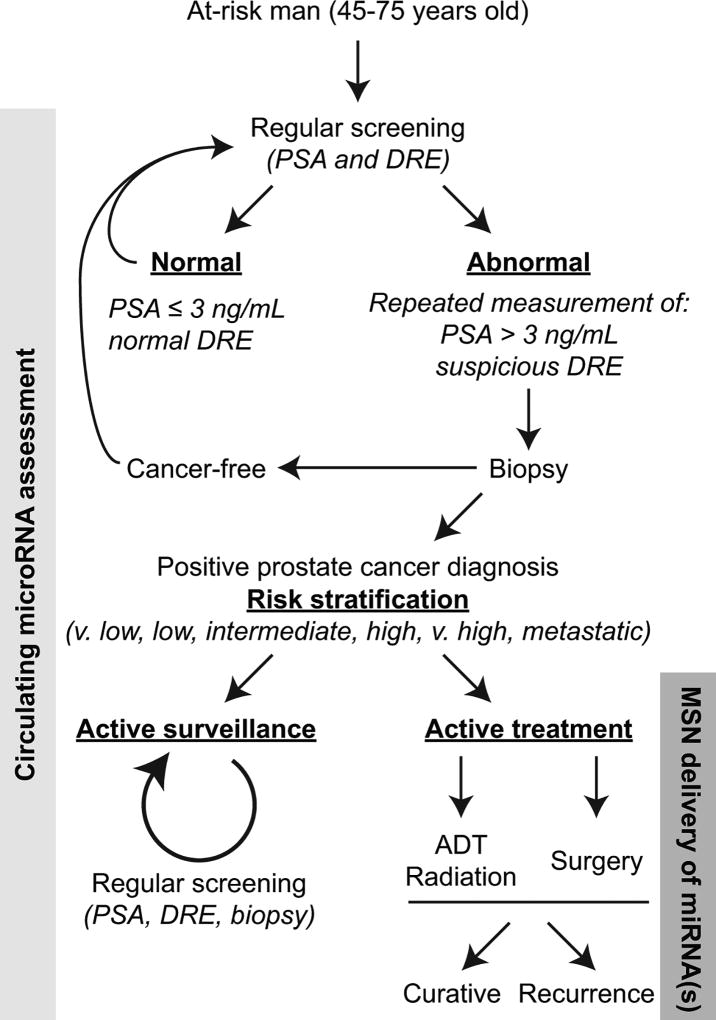
Prostate cancer is the most diagnosed and second leading cause of cancer-related death in men in the United States. More than 80% of men with aggressive prostate cancer develop skeletal complications leading to a dramatic reduction in quality of life (Siegel, Miller, & Jemal, 2017). Prostate cancer diagnosis utilizes multiple clinical parameters (PSA, Gleason score, digital rectal exam (DRE), family history, life expectancy—full NCCN Guideline recommendations available at NCCN.org). The decision as to when and how to treat early stage or indolent disease, as well as assessing treatment response and resistance to androgen deprivation, are still challenging questions (Udager & Tomlins, 2018). Presently, the only large scale randomized control trial that assessed the effectiveness between contemporary treatment modalities (radical prostatectomy, radiotherapy, active monitoring) in patients diagnosed with prostate cancer demonstrated no change in prostate cancer specific mortality after 10 years follow-up (Hamdy et al., 2016). Thus, current clinical risk-stratification algorithms are not sufficiently accurate nor incorporate molecular biomarkers to provide personalized treatment decisions for men facing a new diagnosis of prostate cancer. Very low- and low-risk patients are often over-treated or inappropriately offered active surveillance while intermediate- and high-risk patients are not treated at the appropriate time (early enough) to increase survival (Chang, Autio, & Roach, 2014). The problems in treating prostate cancer at any stage is compounded by the fact that both surgery and radiation treatment are associated with morbidities related to urinary, bowel, and sexual function that reduce patient quality of life (Donovan et al., 2016; Taylor et al., 2012). Further, invading tumor cells can enter a long period of dormancy resulting in disease recurrence and new metastases years after radiation therapy, prostatectomy, or other treatments (Summerer et al., 2013; Weilbaecher, Guise, & McCauley, 2011). The current literature clearly recognizes major concerns in managing prostate cancer patients, from over-treating indolent disease to the difficulties associated with biopsies and early detection of metastasis (Eggener et al., 2011; Narayan, Jiang, & Warlick, 2017; Welch, Schwartz, & Woloshin, 2005). It is these clinical concerns that can utilize circulating miRNA biomarkers (see Figure 1).
FIGURE 1.
Incorporating C-miRNAs and MSNs into prostate cancer screening and treatment. The flowchart of prostate cancer screening, diagnosis, and treatment is based on the 2017 NCCN Guideline recommendations (full version available at NCCN.org). We posit that assessment of prostate cancer associated miRNAs in circulation can complement current standard-of-care algorithms to provide personalized information on cancer cell-related activities. Targeted delivery of MSNs loaded with miRNA therapeutics specifically to prostate tumors may supplement ADT, radiation therapy, or surgery. PSA, prostate specific antigen; DRE, digital rectal exam; ADT, androgen-deprivation therapy; MSN, mesoporous silica nanoparticle
Non-invasive circulating biomarkers, such as PSA and nucleic acids, have practical applications within current clinical monitoring standards and can improve diagnostic and prognostic accuracy as well as track response to therapies. MicroRNAs are a class of small, non-coding RNA that are epigenetic regulators of gene and protein expression and control diverse biological processes (Berindan-Neagoe & Calin, 2014; Wilson & Doudna, 2013). Specifically, miRNAs bind mainly to the 3′ untranslated regions (UTRs) of messenger RNAs (mRNA) in a sequence-specific manner dependent upon the miRNA 5′ “seed” sequence of 6–8 nucleotides and block translation to functional proteins. With the ability to silence the expression of tens-to-hundreds of genes simultaneously, one miRNA can regulate entire biological networks. MicroRNAs have emerged as measurable biomarkers in the circulation and are well documented to be associated with tumor onset and progression (Garofalo, Leva, & Croce, 2014). Numerous miRNAs are well established markers of prostate cancer (Boll et al., 2013; Song et al., 2015; Stegeman et al., 2015; Tucci et al., 2012; Yang et al., 2015) and the associated metastatic bone disease (Browne, Taipaleenmaki, Stein, Stein, & Lian, 2014; Garofalo et al., 2014; Li et al., 2008; Shi, Liu, Duan, Shen, & Guo, 2010; Taipaleenmaki et al., 2015; Weilbaecher et al., 2011; Zhao, Luo, Ma, & Yu, 2015). These miRNAs can more precisely indicate cancer cell activities and thereby can better predict outcome for prostate cancer diagnosis and decisions for clinical intervention (Pettersson et al., 2017; Summerer et al., 2013). Circulating miRNAs (C-miRNA) are easily detected in human body fluids (Farina et al., 2017; Jackson, Grabowska, & Ratan, 2014; Mitchell et al., 2008; Nguyen et al., 2013) and are particularly advantageous when used in parallel to established clinical characteristics and biomarkers, such as rising PSA in the circulation, for monitoring patients under active surveillance or assessing treatment response.
A significant advance for prostate cancer intervention is the characterization of C-miRNAs as biomarkers for cancer subtypes and stages of disease progression (Selth, Tilley, & Butler, 2012; Sita-Lumsden, Dart, Waxman, & Bevan, 2013; Turchinovich, Weiz, & Burwinkel, 2012). It is possible to utilize established bioinformatics algorithms to analyze predicted mRNA targets to identify and validate cancer-associated function and biological relevance of candidate miRNAs (Mitra et al., 2014). Knowledge of predicted miRNA targets and cancer–related signaling pathways regulated by specific C-miRNA(s) in patient serum samples can lead to improved prostate cancer diagnosis and prognosis, as well as novel options for therapeutic interventions (Jackson et al., 2014; Nguyen et al., 2013; Summerer et al., 2013).
2.1 | Candidate microRNAs for intervention of prostate cancer
In cancer cells, miRNAs can be expressed as a consequence of initial transformation, gene mutations, chromosome abnormalities, and epigenetic silencing mechanisms, driving cancer-related phenotypes (Drusco & Croce, 2017; Fabris et al., 2016; Kotb et al., 2014; Peng & Croce, 2016; Pugliese, Palermo, Totaro, Bassi, & Pinto, 2016). Many miRNAs that are highly expressed in all cancers function as oncogenes, while other miRNAs, with important functions to support noncancerous prostate cells, act like tumor suppressors, and are often absent in aggressive cancer cells. MicroRNAs related to specific cancer events, including EMT, proliferation, androgen response, aggressive disease, and metastasis, are all viable therapeutic target miRNAs for specific delivery to prostate tumor cells.
The miRNAs presented in Table 1 have well established functional roles in prostate cancer initiation, progression, and metastasis. All six highlighted miRNAs have been reported as circulating biomarkers of prostate cancer and have differential expression between normal and tumorigenic prostate tissue. These miRNAs play integral roles in key biological processes in prostate cancer progression such as apoptosis, EMT, androgen resistance, and metastasis. For instance, miR-21 inhibits the TGF-β signaling pathway (Mishra et al., 2014) and promotes androgen independent prostate tumor growth (Ribas et al., 2009). Notably, elevated levels of miR-21 in resected primary tumor tissue is associated with prostate cancer recurrence after radical prostatectomy, demonstrating potential as a prognostic marker for biochemical recurrence (Jones, Grizzle, Wang, & Yates, 2013). The miRNA, miR-125b, is upregulated in prostate cancer cell and murine xenograft models, promoting tumorigenesis by directly targeting BAK1, p14ARF, p53, and PUMA, silencing protein expression and inhibiting apoptosis (Amir et al., 2013). Thus, providing a potential therapeutic target for management of patients with clinically aggressive prostate cancer.
TABLE 1.
Candidate miRNAs’ functional roles in prostate cancer progression
| miRNA | Function in prostate cancer | Reference |
|---|---|---|
| miR-21 | Oncogenic: interacts with AR; promotes androgen independent tumor growth, EMT, invasion and inhibits apoptosis | Ribas et al. (2009); Yaman Agaoglu et al. (2011) |
| miR-125b | Oncogenic: promotes cell proliferation, androgen independent tumor growth, and inhibits apoptosis | Mitchell et al, (2008); Amir et al. (2013) |
| miR-221 | Oncogenic: promotes cell cycle progression, survival and invasion; enhances colony formation and tumorigenesis | Mercatelli et al. (2008); Yaman Agaoglu et al. (2011) |
| miR-375 | Oncogenic: elevated expression correlates with higher Gleason score; promotes metastasis and inhibits apoptosis | Brase et al. (2011); Costa-Pinheiro et al. (2015); Wang et al. (2016) |
| miR-145-5p | Tumor suppressor: Interacts with AR; regulates androgen-dependent growth; inhibits cell proliferation, EMT, migration and invasion | Kojima et al. (2014); Shen et al. (2012) |
| miR-200b-3p | Tumor suppressor: inhibits EMT; inhibits chemo-sensitivity, proliferation, migration, invasion | Kojima et al. (2014); Kong et al. (2009) |
All miRNAs listed are found in prostate cancer cell lines, circulation of human PCa, human PCa tumors, and orthotopic tumors of mouse models.
One of the most described prostate cancer miRNAs, miR-221, blocks p27 protein expression, alleviating cell cycle checkpoints, and promotes aggressive proliferation (Galardi et al., 2007). Further, miR-221 has been reported to be elevated in circulation of patients with localized prostate cancer as compared to cancer-free men (Yaman Agaoglu et al., 2011). Simultaneously elevated levels of miR-21 and miR-221 in circulation are correlated with high sensitivity and specificity for detecting prostate cancer (80–90%) when comparing serum from men with BPH to prostate cancer patients (Gleason ≥ 6) (Kotb et al., 2014). Elevated expression of miR-375 in prostate tumor tissue has been correlated with higher Gleason score (>7) and more advanced pathological stage (Costa-Pinheiro et al., 2015). Concomitant elevated levels of miR-375 in serum and tissue from prostate cancer patients strongly supports its capacity to act as a biomarker for diagnosis, prognosis, and overall survival (Huang et al., 2015; Luu et al., 2017; Nguyen et al., 2013). Likewise, differential expression of miR-375, when combined with miR-143 and miR-145, in tumor and adjacent noncancerous tissue has been shown to have high sensitivity (>80%) and specificity (>90%) in cancer cell determination (Wach et al., 2012). These types of finding could effectively reduce unnecessary biopsies as a result of inappropriate diagnosis of prostate cancer.
In addition to oncogenic miRNAs, many miRNAs with tumor suppressor-like function in normal prostate tissue are downregulated in prostate cancer and have potential for novel clinical interventions. Prostate epithelial cells express high levels of miR-145, which targets a positive regulator of EMT, ZEB2, to prevent EMT and tumor aggressiveness (Ren et al., 2014). Further, levels of miR-145 in prostate tumors have an inverse correlation to clinical characteristics (Gleason score, clinical stage, tumor diameter, PSA). Low tumor miR-145 expression is significantly associated with reduced disease-free survival and increased risk for biochemical recurrence (Avgeris, Stravodimos, Fragoulis, & Scorilas, 2013). Moreover, miR-145-5p binds to androgen receptor mRNA and blocks transcription of androgen responsive genes (Larne et al., 2015). Numerous studies have also shown that EMT is negatively regulated by miR-200b-5p through inhibition of ZEB1 and ZEB2 (Burk et al., 2008; Gregory et al., 2008; Park, Gaur, Lengyel, & Peter, 2008).
Taken together, these many examples of miRNAs that support prostate cancer cell phenotypes at different stages of tumor progression support their use as viable targets for potential clinical intervention. The value of monitoring C-miRNAs in patients concurrent with standard-of-care clinical parameters for prostate cancer is an opportunity to define and appropriately treat aggressive prostate cancer. What remains necessary is a mechanism to specifically target miRNA-based therapeutics to prostate cancer cells.
3 | TYPES OF NANOPARTICLES IN TARGETED DELIVERY FORMULATIONS
Nanoparticles used in clinical applications are most often between 5 and 200 nm in diameter. Particle size is a major determining factor in circulation time, biodistribution, and cellular uptake (Mekaru, Lu, & Tamanoi, 2015; Yang & Yu, 2016). Other factors, such as surface chemistry, charge, shape, and biodegradability, are important in the fate of a nanoparticle delivery system (Barua & Mitragotri, 2014; Howard et al., 2014; Petros & DeSimone, 2010; Yang & Yu, 2016). The main advantage to using nanoparticles to deliver biomolecules is twofold. First, nanoparticles provide a concentrated reservoir of the molecule to be delivered, allowing a larger dose to be locally delivered at the site where it is needed in vivo. Second, they provide a scaffold for the addition of properties beneficial to the delivery process; that is, the nanoparticle can be chemically modified to add enhanced targeting, circulation, and cellular uptake capabilities without altering the molecule being delivered itself. A number of different types of nanoparticles currently in clinical trials are described below and summarized in Table 2.
TABLE 2.
Clinical trials using nanoparticles in molecular delivery
| Nanoparticle type | Product name | Clinical trial | Cancer/function |
|---|---|---|---|
| Lipid 80–210 nm | Anti-EGFR immunoliposomes | NCT01702129, NCT02833766 | Breast |
| Primary Use: Drug, siRNA, DNA delivery | |||
| MM-302 | NCT01304797, NCT02213744 NCT02735798 | Advanced breast metastasis | |
| SGT-53 | NCT02340156, NCT02354547 NCT00470613, NCT02340117 | Glioblastoma | |
| SGT-94 | NCT01517464 | Met. solid tumors | |
| MBP-426 | NCT00355888, NCT00964080 | Adv./met solid tumors | |
| Lipovaxin-MM | NCT01052142 | Met. melanoma | |
| 2B3-101 | NCT01386580, NCT01818713 | Recurrent glioma | |
| TKM 080301a | NCT01262235, NCT02191878 NCT01437007 | NET and ACC tumors | |
|
| |||
| Polymer 75–95 nm | BIND-014 | NCT02283320, NCT01812746 NCT01300533, NCT01792479 NCT02479178 | NSCLC (KRAS+ or squamous cell), solid tumor, Met. |
| Use: drug, siRNA, DNA delivery | |||
| CALAA-01 | NCT00689065 | Block tumor growth | |
|
| |||
| Iron oxide 100 nm Use: imaging | NCT01411904 | Identifies residual leukemia | |
|
| |||
| Silica 6–7 nm Use: imaging | C dots | NCT02106598 | Sentinel lymph nodes visualization |
Underline, closed; no underline, open and recruiting; Adv., advanced; Met., metastatic.
Refractory solid tumors or lymphomas; neuroendocrine tumors (NET) or adrenocortical carcinoma (ACC) tumors.
3.1 | Organic nanoparticles in clinical trials
The majority of organic nanoparticles are liposomes with polyethylene glycol (PEG) conjugated to their exteriors. PEG is added to reduce or eliminate protein adsorption, in turn leading to decreased aggregation, increased circulation and subsequent uptake time, and reduced clearance by immune cells (Anselmo & Mitragotri, 2016; Chen, Wu, & Huang, 2010; Eriguchi, Yanagie, Maruyama, & Fujisawa, 2004; Espelin, Leonard, Geretti, Wickham, & Hendriks, 2016; Mamot et al., 2005; Wicki et al., 2015). The liposomes are often cationic, which enhances cellular uptake. In clinical trials, these nanoparticles have been used to deliver genetic material, including p53 cDNA (Senzer et al., 2013) or siRNA (Northfelt et al., 2013). Organic nanoparticles can also be prepared from polymers (Davis, 2009; Hrkach et al., 2012). In one polymeric system, cyclodextin was modified with PEG and a human transferrin protein-targeting agent to encapsulate siRNA for delivery (Davis, 2009). Nanoparticles based on polylactic acid have also been used, modified with PEG and an antibody against prostate-specific membrane antigen (PSMA), for prostate cancer cell specific targeting (Von Hoff et al., 2016).
Although there has been progress in using organic nanoparticles in targeted molecular therapies, they have several drawbacks. For example, reliable scale-up is a concern for liposomal nanoparticles (Anselmo & Mitragotri, 2016), and the long-term stability of liposomal nanoparticles is uncertain. Polycationic polymer-based nanoparticles containing siRNA have been shown to be rapidly eliminated from circulation by the glomerular basement membrane (Zuckerman, Choi, Han, & Davis, 2012). Loading polymeric nanoparticles with nucleic acids often involve incorporating the genetic material during the polymerization process, which limits the amount that can be loaded. Post-synthesis loading of polymeric nanoparticles requires swelling the nanoparticles under appropriate conditions.
3.2 | Clinical trials using inorganic nanoparticles
Clinical trials using inorganic nanoparticles have emphasized biopsy and imaging application. Superparamagnetic iron oxide nanoparticles (SPIONs) have been modified to target CD34 receptors on leukemia cells to preferentially collect them during bone marrow sampling procedures (Lee et al., 2016). Similarly, HER2-conjugated SPIONs have been utilized for the detection of breast cancer (Adolphi et al.,2012), and PEGylated, monoclonal antibody-modified gold nanoparticles have been tested for in vivo imaging (Popovtzer et al., 2008). Dye-labeled core-shell silica nanoparticles, called “C dots,” were developed for sentinel lymph node mapping in the diagnosis and treatment of melanoma by modifying the particles with the PEGylated peptide cRGDY (Bradbury et al., 2013).
3.3 | Advantages of mesoporous silica nanoparticles in targeted therapies
Mesoporous silica nanoparticles (MSNs) are a promising platform for targeted molecular delivery due to their biocompatibility, large internal surface area, and pore volume with consequent high molecular loading capacity, and facile preparation and control of physical properties. Importantly, their rigid structure provides a permanent porosity. Both particle and pore size are easily tailored by altering synthesis conditions, which allows batch-to-batch reproducibility (Ngamcherdtrakul et al., 2015). As the chemical nature of MSNs is similar to silica glass, the same chemistry used to modify any glass material by silanization with an alkoxysilane can be applied, leading to a multitude of possible modifications. For example, triggered drug release systems have been synthesized by covalently linking the drug to the surface with cleavable bonds, or by functionalization of the MSN surface with a coating that will release the payload upon an environmental change, such as pH or temperature (Mamaeva, Sahlgren, & Lindén, 2013). Several other capping mechanisms are being explored in biomedical applications for MSNs and controlled release strategies have been shown to increase the efficacy and efficiency of drug therapies (Lee et al., 2016; Li, Barnes, Bosoy, Stoddart, & Zink, 2012).
MSNs contain amorphous silica, which is chemically distinct from crystalline forms of silica and is biocompatible. Highly cationic MSNs rapidly undergo hepatobiliary excretion (Souris et al., 2010); other MSNs are excreted in urine (He, Zhang, Gao, Li, & Shi, 2011; Lu, Li, Zink, & Tamanoi, 2012). In general, MSNs can accumulate in liver, spleen, and lungs, possibly due to phagocytosis by macrophages (Rosenholm, Mamaeva, Sahlgren, & Lindén, 2012). PEGylation reduces protein accumulation on the surface of MSNs, leading to reduced macrophage uptake and therefore decreased biodistribution in these organs (He et al., 2011; Rosenholm et al., 2012). One study comparing particle sizes between 80 and 360 nm found that smaller particles could avoid retention in the liver and spleen more easily than larger particles, and were consequently more slowly biodegraded (He et al., 2011). Numerous studies have found no pathological abnormalities of mice injected with MSNs (He et al., 2011; Lu et al., 2012; Steinbacher et al., 2010). Ultimately, silica-based nanoparticles with small sizes and long residence times are biodegraded to orthosilicic acid when concentration levels are below saturation, which is then excreted (Rosenholm et al., 2012).
3.4 | Loading genetic material into mesoporous silica nanoparticles
MSNs with larger pores (8–23 nm) and cationic surface modifications were shown to be the best for loading siRNA and DNA, rationalized on a geometric and size basis (Hartono et al., 2012; Kim et al., 2011; Na et al., 2012). However, MSNs that load the most material often bind it too tightly for effective release. While multilayer, enhanced loading of siRNA occurred in cationically modified MSNs with pore sizes of 15 nm, similarly modified MSNs with 8 nm pores proved to release a larger overall dose of siRNA (Steinbacher & Landry, 2014). The same study found that the degree of pore functionalization was also an important factor in optimizing the loading capacity and subsequent release of siRNA.
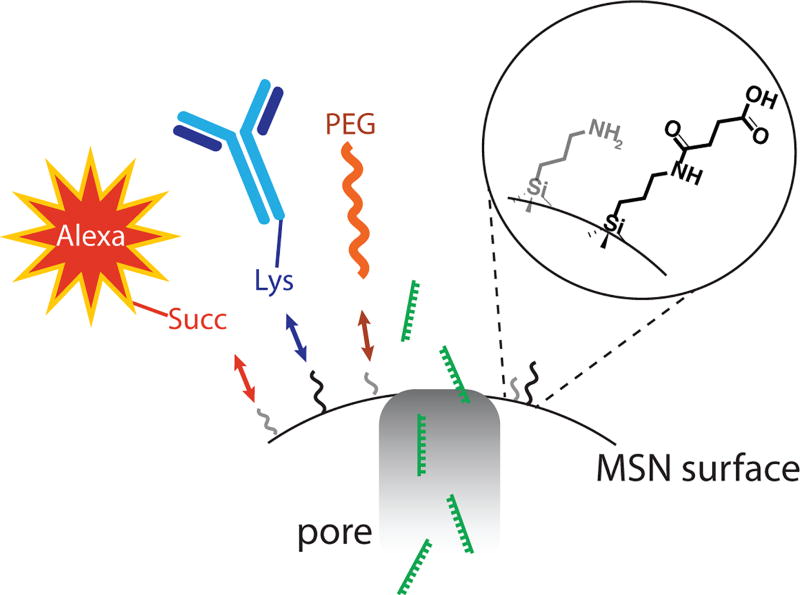
External functionalization of MSNs is equally important in successful delivery. MSNs can be functionalized with several surface groups such as a fluorophore for detection, an antibody for targeting, and PEG to increase circulation time and prevent unnecessary protein adsorption that might inhibit effective targeting (Figure 2). PEGylation of the surface of MSNs has been shown to increase particle delivery to tumor sites by 6.5% over non-PEGylated nanoparticles (Meng et al., 2015). “Smart” MSN systems that take advantage of the silica nanoparticle structure and ease of functionalization are also being developed. One example used the RGD peptide to improve cellular uptake and PEG to reduce intracellular aggregation (Yu, Qian, Uttamchandani, Li, & Yao, 2015); after filling the pores with the anti-cancer therapy doxorubicin, the “antagomir” anti-miRNA-122 was used to cap the pores through electrostatic interactions with surface amines. In PBS, no release was measured; however, within cells, protonation within the cellular endo-lysosome led to release of anti-miRNA-122 and doxorubicin following endosomal escape. Another “smart” system is nearing clinical trials. MSNs were coated with crosslinked PEI-PEG copolymer and coupled to the anti-HER2 mAb, Trastuzumab (Ngamcherdtrakul et al., 2015). In vivo studies found that multiple IV injections over three weeks significantly inhibited tumor growth and did not elicit an immune response.
FIGURE 2.
The MSN surface is readily functionalized. Cartoon showing a possible strategy for modification of MSN for targeted delivery of miRNA. The surface of MSNs contains both anime (NH2) and acid (COOH) moieties for easy modification. For delivery of miRNA(s) mimic or inhibitor (green), we propose that MSNs be modified with and fluorphore (Alexa–red), targeting antibody (blue), and PEG to prevent aggregation (orange). Succ, succinate; Lys, lysine; PEG, polyethylene glycol
4 | ONGOING STUDIES: MESOPOROUS SILICA NANOPARTICLES USED IN PROSTATE CANCER THERAPIES
Known transmembrane proteins with prostate cancer specific expression have been used to target miRNA delivery specifically to prostate tumor cells. For example, prostate-specific membrane antigen (PSMA) (Evans et al., 2016; Wu, Ding, et al., 2011; Wu, Zhang, et al., 2011) and prostate stem cell antigen (PSCA) (Barat et al., 2009) have been identified for targeted molecular delivery. Studies have successfully conjugated anti-PSMA and anti-PSCA to nanoparticles for targeting and these approaches are in clinical trials (Gao et al., 2012; Ruan, Yu, Cheng, Zhang, & Larre, 2012; Von Hoff et al., 2016). These capabilities and the advantageous physical properties of MSNs, particularly their large specific surface area and permanent, tailorable porosity, make MSNs a superior vehicle for targeted miRNA delivery to prostate cancer.
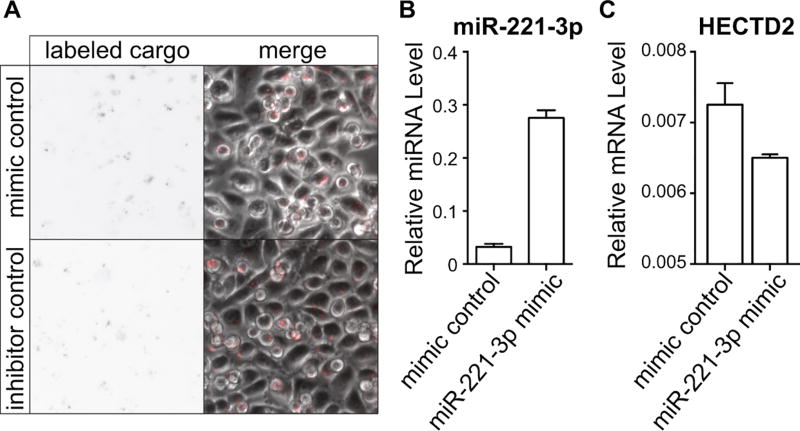
Preliminary data from our laboratories demonstrate that MSNs can be easily loaded with modified nucleic acid molecules designed to either inhibit or mimic miRNA activity. Importantly, MSNs loaded with either fluorophore-labeled miRIDIAN microRNA hairpin inhibitor or mimic negative control (Dharmacon) and added to the media of PC-3 cells are detected and retained within cells for at least 72 hr (Figure 3a). In these studies, miR-221-3p mimics were effectively delivered to PC3 cells in vitro by MSNs with qPCR levels of miR-221-3p increased more than 8-fold as compared to control (Figure 3b). As described in Table 1, miR-221-3p promotes prostate cancer cell survival, hyperplasia, and tumorigenesis (Mercatelli et al., 2008; Yaman Agaoglu et al., 2011). To confirm that the MSN-delivered functional miR-221-3p mimics, we interrogated the expression of HECTD2, a validated miR-221-3p target (Sun et al., 2014), and observed reduced mRNA levels following treatment with miR-221-3p mimic-loaded MSNs as compared to negative control (Figure 3c). We are currently preparing multifunctionalized MSNs, loaded with miRNA mimics or inhibitors and modified with anti-PSMA or anti-PSCA for targeting, a fluorophore for detection, and PEG for increased circulation time and reduced protein aggregation. These MSNs will be used in in vivo targeting and delivery studies on prostate cancer tumorigenesis and metastasis using xenograph mouse models.
FIGURE 3.
PSCA antibody-targeted MSNs deliver function miRNA cargo to prostate cancer cells. PEGylated MSNs covalently linked to an antibody against prostate stem cell antigen (PSCA) were loaded with either Dy547 labeled miRIDIAN microRNA hairpin inhibitor or mimic negative controls or miRIDIAN microRNA mimic against miR-221-3p (Dharmacon). (a) Epifluorescent (grayscale) and DIC images show uptake of Dy547-labed negative control molecules by PC3 cells. These nucleic acid molecules appear as dots in the grayscale image and are present in nearly all of the cells. (b and c) RNA was isolated 72 hr after treatment with MSNs loaded with mimic negative control or miR-221-3p mimic and qPCR levels of (b) miR-221-3p or (c) the validated target HECTD2. Relative miRNAs levels were normalized to U6 expression and relative mRNA levels normalized to GAPDH
5 | CLOSING REMARKS
There remains a compelling need in all cancers to utilize biomarkers that provide cellular and biochemical level information to supplement clinical parameters and expand the knowledgebase for cancer diagnosis, prognosis, and treatment strategy selection. This is an era where well characterized microRNAs are associated with distinct cancer-related cellular activities and signaling pathways that promote tumor progression. Thus, the opportunity is at hand for miRNAs detected in the circulation to serve as non-invasive biomarkers for risk assessment and stratification, early detection, prognosis outcomes and recurrence, and response to treatment by monitoring blood levels. Prostate cancer, which is often initially diagnosed as indolent, or slow growing, will transition into an aggressive and metastatic disease in a subset of men. However, effective identification of those patients that will most benefit from ADT, radiation therapy, and/or radical prostatectomy remains an unmet need. This problem lends itself to the application of monitoring specific miRNAs in the circulation that reflect stages of prostate tumor progression. As such, C-miRNA biomarkers can predict a change in tumor properties prior to detection by invasive procedures. In a related study, we have recently shown that a signature of multiple C-miRNAs defines women at clinical risk for breast cancer in blood sample years before cancer development (Farina et al., 2017). In prostate cancer, identifying a signature of miRNAs that can distinguish indolent disease, predict resistance to androgen-deprivation therapy, and warn of recurrence after treatment, would have a major impact on patient care.
The many studies that have identified miRNAs in surgically resected tumors and in body fluids confirm their significance as miRNAs that are targetable for intervention. The challenge now is developing a clinically safe protocol for delivery of miRNAs specifically to tumor cells. The advances made in biocompatible nanoparticles for targeted delivery to tumors cells has focused on fast growing and difficult to treat cancers as a priority for this approach to intervention. Because of the known resistance of tumor cells to androgen-deprivation and radiation therapy in prostate and other cancers, a population of resistant and dormant cells can become aggressive cancer cells. Further, the propensity of aggressive prostate cancer cells to metastasize to bone leads to a painful and devastating end stage of the disease. A nanoparticle delivery system that will specifically target any prostate cancer cell throughout the body, would be a major step forward for intervention of tumor progression and inhibition of metastasis and relapse. The advantages of using MSNs are multiple in assuring their effectiveness for inhibition of prostate cancer at any stage of the disease, as a result of the properties of MSNs that can readily be modified for individual needs, providing a tool for precision medicine. Among these are the control of MSN pore size to regulate release of cargo, miRNA mimic (s) and/or inhibitor (s), and the ability to track the particles in vivo with added fluorophores. For prostate cancer intervention, MSNs can accommodate attachment of an antibody to two highly specific prostate cancer cell surface markers. Mesoporous silica nanoparticle can then be targeted to these cell surface antigens and taken up into the tumor cell. The emerging use of nanoparticles in clinical trials for encapsulating miRNAs (or other cargo) with delivery to cancer cells are a future direction for developing therapies by a non-invasive procedure (peritoneal or venous injection) that can become an early preventive measure to limit cancer progression or as an adjuvant to other procedures to reduce cancer relapse.
Acknowledgments
Funding information
Congressionally Directed Medical Research Programs, Grant number: W81XWH-14-1-0468; National Cancer Institute, Grant number: P01 CA221769; Lake Champlain Cancer Research Organization, Grant number: 2015-031096
This publication was made possible by the University of Vermont Cancer Center's J Walter Juckett Award (JBL, CCL, GSS) and Postdoctoral Fellowship (NHF) from the Lake Champlain Cancer Research Organization and by support from the following grants: National Cancer Institute (P01 CA221769); Department of Defense (W81XWH-14-1-0468).
References
- Adolphi NL, Butler KS, Lovato DM, Tessier TE, Trujillo JE, Hathaway HJ, Flynn ER. Imaging of Her2-targeted magnetic nanoparticles for breast cancer detection: Comparison of SQUID-detected magnetic relaxometry and MRI. Contrast Media & Molecular Imaging. 2012;7(3):308–317. doi: 10.1002/cmmi.499. https://doi.org/10.1002/cmmi.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir S, Ma AH, Shi XB, Xue L, Kung HJ, Devere White RW. Oncomir miR-125b suppresses p14(ARF) to modulate p53-dependent and p53-independent apoptosis in prostate cancer. PLoS ONE. 2013;8(4):e61064. doi: 10.1371/journal.pone.0061064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioengineering & Translational Medicine. 2016;1(1):10–29. doi: 10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgeris M, Stravodimos K, Fragoulis EG, Scorilas A. The loss of the tumour-suppressor miR-145 results in the shorter disease-free survival of prostate cancer patients. British Journal of Cancer. 2013;108(12):2573–2581. doi: 10.1038/bjc.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat B, Sirk SJ, McCabe KE, Li J, Lepin EJ, Remenyi R, Wu AM. Cys-diabody quantum dot conjugates (immunoQdots) for cancer marker detection. Bioconjugate Chemistry. 2009;20(8):1474–1481. doi: 10.1021/bc800421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, Mitragotri S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: A review of current status and future prospects. Nano Today. 2014;9(2):223–243. doi: 10.1016/j.nantod.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berindan-Neagoe I, Calin GA. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clinical Cancer Research. 2014;20(24):6247–6253. doi: 10.1158/1078-0432.CCR-13-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll K, Reiche K, Kasack K, Morbt N, Kretzschmar AK, Tomm JM, Hackermuller J. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene. 2013;32(3):277–285. doi: 10.1038/onc.2012.55. [DOI] [PubMed] [Google Scholar]
- Bradbury MS, Phillips E, Montero PH, Cheal SM, Stambuk H, Durack JC, Patel S. Clinically-translated silica nanoparticles as dual-modality cancer-targeted probes for image-guided surgery and interventions. Integrative Biology. 2013;5(1):74–86. doi: 10.1039/c2ib20174g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. International Journal of Cancer Journal International Du Cancer. 2011;128(3):608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- Browne G, Taipaleenmaki H, Stein GS, Stein JL, Lian JB. MicroRNAs in the control of metastatic bone disease. Trends in Endocrinology and Metabolism. 2014;25(6):320–327. doi: 10.1016/j.tem.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AJ, Autio KA, Roach M, Scher HI., 3rd High-risk prostate cancer-classification and therapy. Nature Reviews Clinical Oncology. 2014;11(6):308–323. doi: 10.1038/nrclinonc.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu JJ, Huang L. Nanoparticles targeted with NGR motif deliver c-myc siRNA and doxorubicin for anticancer therapy. Molecular Therapy. 2010;18(4):828–834. doi: 10.1038/mt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Pinheiro P, Ramalho-Carvalho J, Vieira FQ, Torres-Ferreira J, Oliveira J, Goncalves CS, Jeronimo C. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clinical Epigenetics. 2015;7:42. doi: 10.1186/s13148-015-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: From concept to clinic. Molecular Pharmaceutics. 2009;6(3):659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- Donovan JL, Hamdy FC, Lane JA, Mason M, Metcalfe C, Walsh E, Protec TSG. Patient-reported outcomes after monitoring, surgery, or radiotherapy for prostate cancer. New England Journal of Medicine. 2016;375(15):1425–1437. doi: 10.1056/NEJMoa1606221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusco A, Croce CM. MicroRNAs and cancer: A long story for short rNAs. Advances in Cancer Research. 2017;135:1–24. doi: 10.1016/bs.acr.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Eggener SE, Scardino PT, Walsh PC, Han M, Partin AW, Trock BJ, Stephenson AJ. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. Journal of Urology. 2011;185(3):869–875. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriguchi M, Yanagie H, Maruyama K, Fujisawa T. Liposome preparations containing oxaliplatin. 2004002842 Google Patents. US Patent Application. 2004
- Espelin CW, Leonard SC, Geretti E, Wickham TJ, Hendriks BS. Dual HER2 targeting with trastuzumab and liposomal-encapsulated doxorubicin (MM-302) demonstrates synergistic antitumor activity in breast and gastric cancer. Cancer Research. 2016;76(6):1517–1527. doi: 10.1158/0008-5472.CAN-15-1518. [DOI] [PubMed] [Google Scholar]
- Evans JC, Malhotra M, Guo J, O'Shea JP, Hanrahan K, O'Neill A, O'Driscoll CM. Folate-targeted amphiphilic cyclodextrin. siRNA nanoparticles for prostate cancer therapy exhibit PSMA mediated uptake, therapeutic gene silencing in vitro and prolonged circulation in vivo. Nanomedicine. 2016;12(8):2341–2351. doi: 10.1016/j.nano.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S, Calin GA. The potential of microRNAs as prostate cancer biomarkers. European Urology. 2016;70(2):312–322. doi: 10.1016/j.eururo.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina NH, Ramsey JE, Cuke ME, Ahern TP, Shirley DJ, Stein JL, Wood ME. Development of a predictive miRNA signature for breast cancer risk among high-risk women. Oncotarget. 2017;8(68):112170–112183. doi: 10.18632/oncotarget.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. MiR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. The Journal of Biological Chemistry. 2007;282(32):23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- Gao X, Luo Y, Wang Y, Pang J, Liao C, Lu H, Fang Y. Prostate stem cell antigen-targeted nanoparticles with dual functional properties: In vivo imaging and cancer chemotherapy. International Journal of Nanomedicine. 2012;7:4037–4051. doi: 10.2147/IJN.S32804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Current Pharmaceutical Design. 2014;20(33):5328–5335. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, Protec TSG. 10-Year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. New England Journal of Medicine. 2016;375(15):1415–1424. doi: 10.1056/NEJMoa1606220. [DOI] [PubMed] [Google Scholar]
- Hartono SB, Gu W, Kleitz F, Liu J, He L, Middelberg AP, Qiao SZ. Poly-L-lysine functionalized large pore cubic mesostructured silica nanoparticles as biocompatible carriers for gene delivery. Acs Nano. 2012;6(3):2104–2117. doi: 10.1021/nn2039643. [DOI] [PubMed] [Google Scholar]
- He Q, Zhang Z, Gao F, Li Y, Shi J. In vivo biodistribution and urinary excretion of mesoporous silica nanoparticles: Effects of particle size and PEGylation. Small. 2011;7(2):271–280. doi: 10.1002/smll.201001459. [DOI] [PubMed] [Google Scholar]
- Howard M, Zern BJ, Anselmo AC, Shuvaev VV, Mitragotri S, Muzykantov V. Vascular targeting of nanocarriers: Perplexing aspects of the seemingly straightforward paradigm. ACS Nano. 2014;8(5):4100–4132. doi: 10.1021/nn500136z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrkach J, Von Hoff D, Ali MM, Andrianova E, Auer J, Campbell T, Horhota A. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Science Translational Medicine. 2012;4(128):128ra139–128ra 139. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang L. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. European Urology. 2015;67(1):33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BL, Grabowska A, Ratan HL. MicroRNA in prostate cancer: Functional importance and potential as circulating biomarkers. BMC Cancer. 2014;14:930. doi: 10.1186/1471-2407-14-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Grizzle W, Wang H, Yates C. MicroRNAs that affect prostate cancer: Emphasis on prostate cancer in African Americans. Biotechnic and Histochemistry. 2013;88(7):410–424. doi: 10.3109/10520295.2013.807069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-H, Na H-K, Kim Y-K, Ryoo S-R, Cho HS, Lee KE, Min D-H. Facile synthesis of monodispersed mesoporous silica nanoparticles with ultralarge pores and their application in gene delivery. ACS Nano. 2011;5(5):3568–3576. doi: 10.1021/nn103130q. [DOI] [PubMed] [Google Scholar]
- Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Seki N. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. Journal of Human Genetics. 2014;59(2):78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. MiR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27(8):1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotb S, Mosharafa A, Essawi M, Hassan H, Meshref A, Morsy A. Circulating miRNAs 21 and 221 as biomarkers for early diagnosis of prostate cancer. Tumour Biology. 2014;35(12):12613–12617. doi: 10.1007/s13277-014-2584-7. [DOI] [PubMed] [Google Scholar]
- Larne O, Hagman Z, Lilja H, Bjartell A, Edsjo A, Ceder Y. MiR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis. 2015;36(8):858–866. doi: 10.1093/carcin/bgv063. [DOI] [PubMed] [Google Scholar]
- Lee BY, Li Z, Clemens DL, Dillon BJ, Hwang AA, Zink JI, Horwitz MA. Redox-triggered release of moxifloxacin from mesoporous silica nanoparticles functionalized with disulfide snap-tops enhances efficacy against pneumonic tularemia in mice. Small. 2016;12(27):3690–3702. doi: 10.1002/smll.201600892. [DOI] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Stein GS. A microRNA signature for aBMP2-induced osteoblast lineage commitment program. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(37):13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Barnes JC, Bosoy A, Stoddart JF, Zink JI. Mesoporous silica nanoparticles in biomedical applications. Chemical Society Reviews. 2012;41(7):2590–2605. doi: 10.1039/c1cs15246g. [DOI] [PubMed] [Google Scholar]
- Lu J, Li Z, Zink JI, Tamanoi F. In vivo tumor suppression efficacy of mesoporous silica nanoparticles-based drug-delivery system: Enhanced efficacy by folate modification. Nanomedicine: Nanotechnology, Biology and Medicine. 2012;8(2):212–220. doi: 10.1016/j.nano.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu HN, Lin HY, Sorensen KD, Ogunwobi OO, Kumar N, Chornokur G, Di Pietro G. MiRNAs associated with prostate cancer risk and progression. BMC Urology. 2017;17(1):18. doi: 10.1186/s12894-017-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamaeva V, Sahlgren C, Lindén M. Mesoporous silica nanoparticles in medicine—Recent advances. Advanced Drug Delivery Reviews. 2013;65(5):689–702. doi: 10.1016/j.addr.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Mamot C, Drummond DC, Noble CO, Kallab V, Guo Z, Hong K, Park JW. Epidermal growth factor receptor-targeted immunoliposomes significantly enhance the efficacy of multiple anticancer drugs in vivo. Cancer Research. 2005;65(24):11631–11638. doi: 10.1158/0008-5472.CAN-05-1093. [DOI] [PubMed] [Google Scholar]
- Mekaru H, Lu J, Tamanoi F. Development of mesoporous silica-based nanoparticles with controlled release capability for cancer therapy. Advanced Drug Delivery Reviews. 2015;95:40–49. doi: 10.1016/j.addr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Wang M, Liu H, Liu X, Situ A, Wu B, Nel AE. Use of a lipid-coated mesoporous silica nanoparticle platform for synergistic gemcitabine and paclitaxel delivery to human pancreatic cancer in mice. ACS Nano. 2015;9(4):3540–3557. doi: 10.1021/acsnano.5b00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Ciafre SA. The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS ONE. 2008;3(12):e4029. doi: 10.1371/journal.pone.0004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Deng JJ, Gowda PS, Rao MK, Lin CL, Chen CL, Sun LZ. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014;33(31):4097–4106. doi: 10.1038/onc.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Edmonds MD, Sun J, Zhao M, Yu H, Eischen CM, Zhao Z. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014;20(9):1356–1368. doi: 10.1261/rna.042754.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na HK, Kim MH, Park K, Ryoo SR, Lee KE, Jeon H, Min DH. Efficient functional delivery of siRNA using mesoporous silica nanoparticles with ultralarge pores. Small. 2012;8(11):1752–1761. doi: 10.1002/smll.201200028. [DOI] [PubMed] [Google Scholar]
- Narayan V, Jiang S, Warlick CA. Early stage cancer in older adults: Prostate-avoiding overtreatment and undertreatment. Cancer Journal. 2017;23(4):238–241. doi: 10.1097/PPO.0000000000000273. [DOI] [PubMed] [Google Scholar]
- Ngamcherdtrakul W, Morry J, Gu S, Castro DJ, Goodyear SM, Sangvanich T, Beckman BL. Cationic polymer modified mesoporous silica nanoparticles for targeted siRNA delivery to HER2+ breast cancer. Advanced Functional Materials. 2015;25(18):2646–2659. doi: 10.1002/adfm.201404629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, Lee GS, Kantoff PW. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate. 2013;73(4):346–354. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northfelt DW, Hamburg SI, Borad MJ, Seetharam M, Curtis KK, Lee P, Gilbert MJ. ASCO Annual Meeting Proceedings. 2013;31(15_suppl):TPS2621. [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduction and Targeted Therapy. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nature Reviews. Drug Discovery. 2010;9(8):615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- Pettersson A, Gerke T, Fall K, Pawitan Y, Holmberg L, Giovannucci EL, Transdisciplinary Prostate Cancer P The ABC model of prostate cancer: A conceptual framework for the design and interpretation of prognostic studies. Cancer. 2017;123(9):1490–1496. doi: 10.1002/cncr.30582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovtzer R, Agrawal A, Kotov NA, Popovtzer A, Balter J, Carey TE, Kopelman R. Targeted gold nanoparticles enable molecular CT imaging of cancer. Nano Letters. 2008;8(12):4593–4596. doi: 10.1021/nl8029114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugliese D, Palermo G, Totaro A, Bassi PF, Pinto F. Clinical, pathological and molecular prognostic factors in prostate cancer decision-making process. Urologia. 2016;83(1):14–20. doi: 10.5301/uro.5000166. [DOI] [PubMed] [Google Scholar]
- Ren D, Wang M, Guo W, Huang S, Wang Z, Zhao X, Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell and Tissue Research. 2014;358(3):763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Lupold SE. MiR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69(18):7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenholm JM, Mamaeva V, Sahlgren C, Lindén M. Nanoparticles in targeted cancer therapy: Mesoporous silica nanoparticles entering preclinical development stage. Nanomedicine. 2012;7(1):111–120. doi: 10.2217/nnm.11.166. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Yu W, Cheng F, Zhang X, Larre S. Detection of prostate stem cell antigen expression in human prostate cancer using quantum-dot-based technology. Sensors (Basel) 2012;12(5):5461–5470. doi: 10.3390/s120505461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Tilley WD, Butler LM. Circulating microRNAs: Macro-utility as markers of prostate cancer? Endocrine-Related Cancer. 2012;19(4):R99–R113. doi: 10.1530/ERC-12-0010. [DOI] [PubMed] [Google Scholar]
- Senzer N, Nemunaitis J, Nemunaitis D, Bedell C, Edelman G, Barve M, Chang EH. Phase I study of a systemically delivered p53 nanoparticle in advanced solid tumors. Molecular Therapy. 2013;21(5):1096–1103. doi: 10.1038/mt.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Hruby GW, McKiernan JM, Gurvich I, Lipsky MJ, Benson MC, Santella RM. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72(13):1469–1477. doi: 10.1002/pros.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Liu D, Duan H, Shen B, Guo N. Metastasis-related miRNAs, active players in breast cancer invasion, and metastasis. Cancer Metastasis Reviews. 2010;29(4):785–799. doi: 10.1007/s10555-010-9265-9. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new biomarkers for prostate cancer. British Journal of Cancer. 2013;108(10):1925–1930. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Chen H, Wang T, Zhang W, Ru G, Lang J. Expression profile analysis of microRNAs in prostate cancer by next-generation sequencing. Prostate. 2015;75(5):500–516. doi: 10.1002/pros.22936. [DOI] [PubMed] [Google Scholar]
- Souris JS, Lee C-H, Cheng S-H, Chen C-T, Yang C-S, Ho J-A, Lo L-W. Surface charge-mediated rapid hepatobiliary excretion of mesoporous silica nanoparticles. Biomaterials. 2010;31(21):5564–5574. doi: 10.1016/j.biomaterials.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocrine-Related Cancer. 2015;22(2):265–276. doi: 10.1530/ERC-15-0013. [DOI] [PubMed] [Google Scholar]
- Steinbacher JL, Landry CC. Adsorption and Release of siRNA from porous silica. Langmuir. 2014;30(15):4396–4405. doi: 10.1021/la402850m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher JL, Lathrop SA, Cheng K, Hillegass JM, Butnor KJ, Kauppinen RA, Landry CC. Gd-labeled microparticles in MRI: In vivo imaging of microparticles after intraperitoneal injection. Small. 2010;6(23):2678–2682. doi: 10.1002/smll.201001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerer I, Niyazi M, Unger K, Pitea A, Zangen V, Hess J, Zitzelsberger H. Changes in circulating microRNAs after radiochemotherapy in head and neck cancer patients. Radiation Oncology. 2013;8:296. doi: 10.1186/1748-717X-8-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Kantoff PW. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2014;33(21):2790–2800. doi: 10.1038/onc.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmaki H, Browne G, Akech J, Zustin J, van Wijnen AJ, Stein JL, Lian JB. Targeting of Runx2 by miR-135 and miR-203 impairs progression of breast cancer and metastatic bone disease. Cancer Res. 2015;75(7):1433–1444. doi: 10.1158/0008-5472.CAN-14-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KL, Luta G, Miller AB, Church TR, Kelly SP, Muenz LR, Riley TL. Long-term disease-specific functioning among prostate cancer survivors and noncancer controls in the prostate, lung, colorectal, and ovarian cancer screening trial. Journal of Clinical Oncology. 2012;30(22):2768–2775. doi: 10.1200/JCO.2011.41.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Melino G. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(38):15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: The mystery of their origin and function. Trends in Biochemical Sciences. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Udager AM, Tomlins SA. Molecular biomarkers in the clinical management of prostate cancer. Cold Spring Harbor Perspectives in Medicine. 2018 doi: 10.1101/cshperspect.a030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Mita MM, Ramanathan RK, Weiss GJ, Mita AC, LoRusso PM, Sachdev JC. Phase I study of PSMA-targeted docetaxel-containing nanoparticle BIND-014 in patients with advanced solid tumors. Clinical Cancer Research. 2016;22(13):3157–3163. doi: 10.1158/1078-0432.CCR-15-2548. [DOI] [PubMed] [Google Scholar]
- Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Wullich B. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. International Journal of Cancer Journal International Du Cancer. 2012;130(3):611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lieberman R, Pan J, Zhang Q, Du M, Zhang P, Wang L. MiR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Molecular Cancer. 2016;15(1):70. doi: 10.1186/s12943-016-0556-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: A fatal attraction. Nature Reviews. Cancer. 2011;11(6):411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HG, Schwartz LM, Woloshin S. Prostate-specific antigen levels in the United States: Implications of various definitions for abnormal. Journal of the National Cancer Institute. 2005;97(15):1132–1137. doi: 10.1093/jnci/dji205. [DOI] [PubMed] [Google Scholar]
- Wicki A, Ritschard R, Loesch U, Deuster S, Rochlitz C, Mamot C. Large-scale manufacturing of GMP-compliant anti-EGFR targeted nanocarriers: Production of doxorubicin-loaded anti-EGFR-immunoliposomes for a first-in-man clinical trial. International Journal of Pharmaceutics. 2015;484(1–2):8–15. doi: 10.1016/j.ijpharm.2015.02.034. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual Review of Biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HX, Zhang SJ, Zhang JM, Liu G, Shi JL, Zhang LX, Bu WB. A hollow-core, magnetic, and mesoporous double-shell nanostructure: In situ decomposition/reduction synthesis, bioimaging, and drug-delivery properties. Advanced Functional Materials. 2011;21(10):1850–1862. [Google Scholar]
- Wu X, Ding B, Gao J, Wang H, Fan W, Wang X, Gao S. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. International Journal of Nanomedicine. 2011;6:1747–1756. doi: 10.2147/IJN.S23747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U. Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biology. 2011;32(3):583–588. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yu C. Advances in silica based nanoparticles for targeted cancer therapy. Nanomedicine: Nanotechnology, Biology and Medicine. 2016;12(2):317–332. doi: 10.1016/j.nano.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Yang J, Wei J, Wu Y, Wang Z, Guo Y, Lee P, Li X. Metformin induces ER stress-dependent apoptosis through miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis. 2015;4:e158. doi: 10.1038/oncsis.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Qian L, Uttamchandani M, Li L, Yao SQ. Single-vehicular delivery of antagomir and small molecules to inhibit miR-122 function in hepatocellular carcinoma cells by using “smart” mesoporous silica nanoparticles. Angewandte Chemie. 2015;127(36):10720–10724. doi: 10.1002/anie.201504913. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Luo F, Ma J, Yu X. Bone metastasis-related microRNAs: New targets for treatment? Current Cancer Drug Targets. 2015;15(8):716–725. doi: 10.2174/1568009615666150629102408. [DOI] [PubMed] [Google Scholar]
- Zuckerman JE, Choi CHJ, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proceedings of the National Academy of Sciences. 2012;109(8):3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]