Dear Editor,
Non-small cell lung cancer (NSCLC) accounts for over 75% cases of lung cancer, the leading cause of cancer deaths in the world. Most NSCLC patients are diagnosed at an advanced stage due to the inadequate screening program and late onset of clinical symptoms, leading to poor prognosis.1 Discovery of accurate and sensitive biomarkers is in urgent need. Recently, circular RNAs (circRNAs) are emerging as a novel biomarker because of their conservation, abundance, cell type-specific and tissue-specific expression, and their roles in disease progression.2,3
Increasing studies focus on molecular mechanisms of NSCLC with fusion genes, a hybrid of two otherwise-separated genes caused from aberrant chromosomal translocations. The fusion gene Echinoderm Microtubule-associated protein-Like 4 (EML4)-Anaplastic Lymphoma Kinase (ALK) is present in 4% to 5% of NSCLC cases and generates oncogenic activity by activating ALK kinase.4 Recently, mounting evidence demonstrates that fusion genes not only encode fusion proteins involved in tumorigenesis, but also generate non-coding RNAs contributing to tumor progression. For example, the circRNA generated by the MLL/AF9 fusion gene (f-circM9) in leukemia exerts pro-proliferative and pro-oncogenic activities,5 prompting us to investigate whether the EML4-ALK fusion gene produces circRNA with clinical relevance in NSCLC.
Unlike linear RNAs, circRNAs have a circular covalently-bonded structure, which endows circRNAs with higher tolerance to exonuclease digestion and prolonged lifetime in systemic circulation. CircRNAs are demonstrated to be enriched and stable in exosomes of peripheral blood.6 Compared to traditional biopsy biomarkers in tumor tissues, circRNAs in body fluids could be used as more convenient and non-invasive “liquid biopsy” biomarkers to detect tumor at early and late stages. Here, we report a novel circRNA named as F-circEA generated from the EML4-ALK fusion gene by back-splicing and identify its role in promoting tumor development. Notably, the existence of F-circEA in plasma suggests that F-circEA is a potentially novel “liquid biopsy” biomarker for diagnosis of EML4-ALK-positive NSCLC, guiding targeted therapy in clinic.
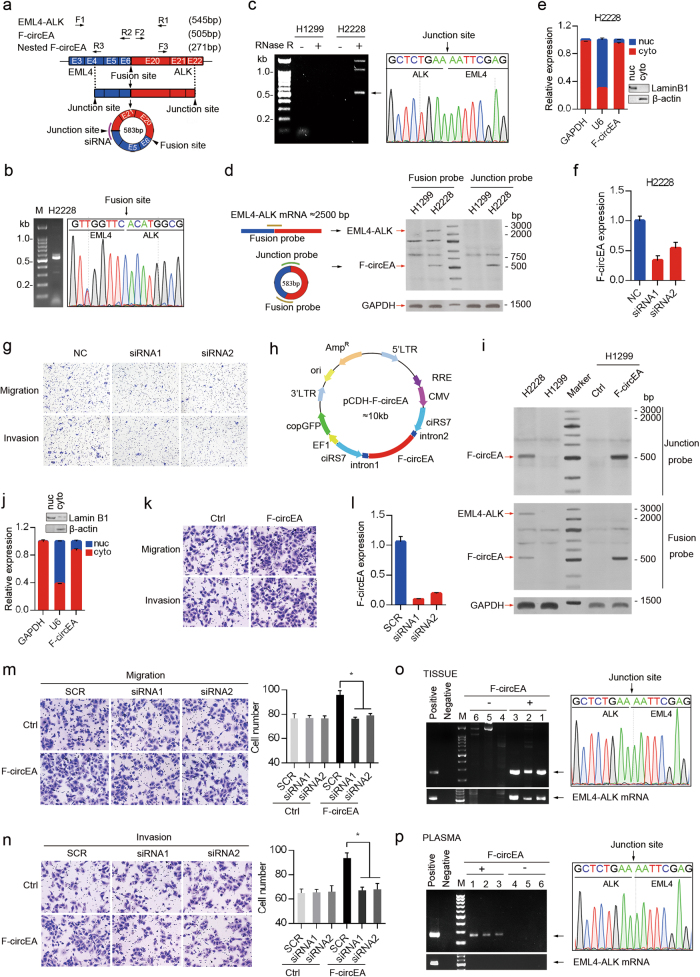
We firstly investigated the existence of endogenous F-circEA in H2228 cells harboring the EML4-ALK variant 3b translocation.7 The presence of the EML4-ALK fusion gene in H2228 cells was verified by sequencing the reverse transcription PCR (RT-PCR) products amplified with F1/R1 primers (Fig. 1a, b). Total RNAs were extracted from H2228 or H1299 cells (negative control without the fusion gene), and subjected to RNase-R digestion to remove linear RNA molecules. By using F2/R2 primers (Fig. 1a), potential circRNAs produced by the EML4-ALK fusion gene were specifically found in H2228 samples (Fig. 1c, left), and the one with an apparent length of ~0.55 kb was Sanger sequenced to confirm the back-splice junction between the 5′ head of EML4 exon 4 fragment and 3′ tail of ALK exon 22 fragment (Fig. 1c, right), different from that in H3321 cells with the EML4-ALK variant 1 translocation.5 Moreover, F-circEA was further confirmed by RNA hybridization assays with or without RNase-R digestion using 32P-labeled probes crossing the junction site and fusion site, respectively (Fig. 1d; Supplementary Information, Figure S1a). The back-splice junction does not fit the GT-AG pattern of a U2 intron and there is no obvious complementary structure in the adjacent introns, suggesting that F-circEA might be produced through a mechanism distinct from the canonical back-splicing pathway. We speculate the back-splice site might be an unconventional U12 intron, however, the exact mechanism needs experimental investigation. The subcellular fractionation and RT-qPCR analyses showed that F-circEA was mainly located in the cytoplasm of H2228 cells (Fig. 1e).
Fig. 1.
Identification and functional analysis of F-circEA in NSCLC. a Schematic representation of F-circEA generated from EML4-ALK fusion gene. The convergent primers (F1/R1) were used to detect EML4-ALK fusion site, and the divergent primer sets (F2/R2, F3/R3) were used to detect F-circEA. Violet curve represents siRNA location. b Agarose gel electrophoresis and Sanger sequencing of RT-PCR products from H2228 cells with F1/R1 primers, the arrow indicates EML4-ALK fusion site. c Identification of F-circEA in H2228 cells by RT-PCR with F2/R2 primers and Sanger sequencing. The arrow indicates F-circEA junction site. d Detection of F-circEA and EML4-ALK mRNA by RNA solution hybridization assay using 32P-labeled oligonucleotide probes crossing junction site and the fusion site. e Cell nucleus/cytoplasm fractionation and RT-qPCR analysis revealed the cytoplasmic distribution of F-circEA in H2228 cells. GAPDH mRNA and U6 snRNA represent cytoplasmic and nuclear RNA, respectively. Western blotting confirmed the efficiency of nucleus/cytoplasm isolation. Data are shown as the mean ± SD. f Knockdown efficiency of F-circEA siRNAs in H2228 cells analyzed by RT-qPCR. g F-circEA knockdown inhibited cell migration and invasion in H2228 cells. h Schematic representation of F-circEA-expressing plasmid with the reverse repeat of cirR-7 exons plus up-stream and down-stream flanking introns to facilitate RNA circularization. i RNA solution hybridization assays for F-circEA and EML4-ALK mRNA in H2228, H1299, empty vector-transfected (Ctrl-H1299), and F-circEA-overexpressing (F-circEA-H1299) H1299 cells. j Cell nucleus/cytoplasm fractionation and RT-qPCR analysis demonstrated that F-circEA was predominantly located in the cytoplasm of F-circEA-overexpressing H1299 cells. Data are shown as the mean ± SD. k Ectopic expression of F-circEA increased the migratory and invasive ability of H1299 cells. Results of statistical analysis are shown in the right panel. l RT-qPCR analysis showed effective knockdown of F-circEA by siRNA in F-circEA-overexpressing H1299 cells. The scramble siRNA (SCR) was used as negative control. m, n F-circEA knockdown attenuated the enhanced migratory and invasive ability of F-circEA-overexpressing H1299 cells. *P < 0.05. o, p Agarose gel electrophoresis and Sanger sequencing of RT-PCR products from tumor tissues (o) or plasma (p) of NSCLC patients with or without EML4-ALK fusion gene
As the EML4-ALK translocation is an oncogenic driver mutation associated with tumor proliferation, migration, and invasion,4 we planned to investigate the potential role of F-circEA in tumorigenesis. To this end, small interfering RNAs (siRNAs) targeting its back-splice junction were designed (Fig. 1a) and effectively knocked down the expression of endogenous F-circEA with minor influence on EML4-ALK mRNA level (Fig. 1f; Supplementary Information, Figure S1b). Transwell assays showed that F-circEA knockdown decreased cell migratory and invasion ability (Fig. 1g; Supplementary Information, Figure S1c), whereas it had little effect on cell proliferation and colony formation (Supplementary Information, Figure S1d and e). Moreover, these siRNAs had little effect on cell migration and invasion in H1299 cells without such fusion gene (Supplementary Information, Fig. S1f). To confirm these observations, the F-circEA expressing vector was constructed by cloning the circularizing sequence into the vector we made with reverse repeat of cirR-7 exons together with up-stream and down-stream flanking introns, which favored the formation of circular RNA (Fig. 1h). F-circEA was successfully expressed and correctly circularized in H1299 cells (Fig. 1i; Supplementary Information, Figure S1g and h), and was also predominantly located in the cytoplasm of H1299 and A549 cells (Fig. 1j; Supplementary Information, Figure S1i) that did not harbor the EML4-ALK translocation and therefore had no expression of endogenous fusion protein and corresponding circRNAs. Through Transwell assays and wound healing experiments, the cells expressing F-circEAs displayed higher migration and invasion ability than the cells expressing the empty vector in both H1299 (Fig. 1k; Supplementary Information, Figure S1j and k) and A549 cells (Supplementary Information, Figure S1l and m). Furthermore, the increased cell migration and invasion in F-circEA-expressing cells were attenuated upon the F-circEA silencing by siRNAs (Fig. 1l, m, n). Because F-circEA expression had little effect on cell proliferation in both H1299 and A549 cells, measured by MTT and colony formation assay (data not shown), F-circEA might participate in tumorigenesis through promoting cell migration and invasion.
To explore the clinical relevance of F-circEA, we investigated whether F-circEA existed in NSCLC patient samples. Two nested divergent primer sets (F2/R2, F3/R3) spanning the fusion site were designed to improve the specificity (Fig. 1a). Sanger sequencing of RT-PCR products clearly demonstrated that 5 of 6 patients with the EML4-ALK variant 3b translocation expressed the F-circEA with the back-splice junction in the tumors, whereas the patients without fusion gene or with the EML4-ALK variant 1 and variant 6 translocation did not (Fig. 1o; Supplementary Information, Figure S1n). Most importantly, F-circEA was specifically existed in the plasma of the patients with the EML4-ALK translocation (Fig. 1p; Supplementary Information, Figure S1o), suggesting that the detection of plasma F-circEA could be a specific and convenient approach to monitor the EML4-ALK translocation and guide the EML4-ALK-targeted NSCLC therapy, such as the use of ALK inhibitor crizotinib. In addition, EML4-ALK mRNA was detected in the NSCLC tumors, but not in plasma (Fig. 1o, p).
In summary, we report a fusion circRNA (F-circEA) produced from the EML4-ALK fusion gene mainly located in the cytoplasm. Moreover, F-circEA, independent of the EML4-ALK linear transcript and fusion protein, can promote cell migration and invasion, thus contributing to tumor development. These results add an extra layer of the regulation of tumorigenesis by the EML4-ALK fusion gene. Notably, the evidences that F-circEA specifically exists in the plasma of EML4-ALK-positive NSCLC patients suggest that F-circEA could be a novel “liquid biopsy” biomarker to monitor the EML4-ALK fusion gene in NSCLC. Taken together, our study not only expands the current knowledge regarding molecular mechanisms underlying the fusion gene-associated cancer progression, but also provides potential diagnostic and therapeutic implications.
Materials and Methods are available in Supplementary information, Data S1 and Table S1.
Electronic supplementary material
Supplementary information, Figure S1, Data S1 and Table S1
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFA0504304 and 2016YFA0502204 to Y.P.) and National Natural Science Foundation of China (81772960 and 81572739 to Y.P.).
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0033-7.
References
- 1.Gridelli C, et al. Nat. Rev. Dis. Prim. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 2.Chen LL. Nat. Rev. Mol. Cell Biol. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 3.Rybak-Wolf A, et al. Mol. Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, et al. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Guarnerio J, et al. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, et al. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivunen JP, et al. Clin. Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figure S1, Data S1 and Table S1