Abstract
Adoptive transfer of T cells engineered with a cancer-specific T cell receptor (TCR) has demonstrated clinical benefit. However, the risk for off-target toxicity of TCRs remains a concern. Here, we examined the cross-reactive profile of T cell clone (7B5) with a high functional sensitivity for the hematopoietic-restricted minor histocompatibility antigen HA-2 in the context of HLA-A*02:01. HA-2pos Epstein-Barr virus-transformed B lymphoblastic cell lines (EBV-LCLs) and primary acute myeloid leukemia samples, but not hematopoietic HA-2neg samples, are effectively recognized. However, we found unexpected off-target recognition of human fibroblasts and keratinocytes not expressing the HA-2 antigen. To uncover the origin of this off-target recognition, we performed an alanine scanning approach, identifying six out of nine positions to be important for peptide recognition. This indicates a low risk for broad cross-reactivity. However, using a combinatorial peptide library scanning approach, we identified a CDH13-derived peptide activating the 7B5 T cell clone. This was confirmed by recognition of CDH13-transduced EBV-LCLs and cell subsets endogenously expressing CDH13, such as proximal tubular epithelial cells. As such, we recommend the use of a combinatorial peptide library scan followed by screening against additional cell subsets to validate TCR specificity and detect off-target toxicity due to cross-reactivity directed against unrelated peptides before selecting candidate TCRs for clinical testing.
Keywords: off-target toxicity, adoptive T cell therapy, preclinical screening, SCT, TCR
Adoptive transfer of T cells engineered with a cancer-specific T cell receptor (TCR) has demonstrated clinical benefit. To validate TCR specificity and detect off-target toxicity before selecting candidate TCRs for clinical testing, Bijen et al. recommend the use of a combinatorial peptide library scan followed by screening against additional cell subsets.
Introduction
Immunotherapy has obtained a prominent role in the field of oncology and has proven valuable in the treatment of different types of tumors. A range of immunotherapies are under development, varying from chimeric antigen receptors (CARs),1 expanding tumor-infiltrating lymphocytes (TILs)2, 3 and T cell receptor (TCR)-transduced effector cells.4, 5 Various studies successfully make use of TCR-engineered T cells to enhance patients’ adaptive immune responses against malignancies, demonstrating potent anti-tumor reactivity.6, 7, 8, 9
No matter how promising therapies using TCR-engineered T cells may be, they come with side effects and risks. T cells are naturally checked for recognition of self-peptides in the context of self-human leukocyte antigen (HLA) and deleted in the thymus if this recognition is too strong, in a process called negative selection.10 This selection process is absent or non-optimal when TCRs are derived from donors with mismatched HLA typing, when they are isolated from humanized mouse models expressing only one HLA molecule and no human proteins, or when TCRs are structurally altered for affinity enhancement.11, 12
Affinity enhancement of TCRs can result in both on- and off-target toxicity.13 Recent examples of clinically tested TCRs directed against MAGE-A3 and MART-1 demonstrated severe toxicities.12, 14, 15, 16, 17 In the clinical trial testing a modified anti-MAGE-A3 TCR, derived from immunization of HLA-A*02:01 transgenic mice, two out of nine cancer patients developed fatal on-target neurological toxicity, due to recognition of a peptide derived from the same gene family that is expressed in the brain.15 In another trial, where an affinity-enhanced anti-MAGE-A3 TCR was tested in myeloma and melanoma patients, two patients died of off-target toxicity caused by recognition of a completely different peptide, resulting in severe myocardial damage.12, 17 These clinical cases show how difficult it is to predict the exact specificity and the resulting effects of TCRs that did not undergo optimal thymic selection. It is crucial to develop strategies to extensively validate the exact specificity of TCRs, particularly because TCR-engineered T cells are highly sensitive.14, 18
In this study, we aimed to develop a reliable screening pipeline for early detection of off-target toxicity due to the cross-reactivity of TCRs directed against unrelated peptides. In previous work, we have focused on the identification of therapeutic TCRs directed against hematopoietic restricted minor histocompatibility antigens (MiHAs) for the treatment of hematological malignancies. The HLA-A*02:01 restricted HA-2 peptide YIGEVLVSV is a well-known example of such an MiHA.19, 20 In recent experiments we isolated an HA-2-specific T cell clone, 7B5, using an in vitro HLA-mismatched setting. We validated its specificity for the HA-2 antigen in hematopoietic cell subsets. However, we found unexpected off-target recognition of human fibroblasts not expressing the HA-2 antigen, raising the questions what triggered this activation and what method is suitable to examine this. Thus, we investigated the off-target cross-reactive potential of this high-avidity T cell clone using different screening methods and characterized its fine specificity.
Currently, an amino acid scanning approach is the recommended method,21 which we compared with screening with a 9-mer combinatorial peptide library (CPL).22 To validate recognition of predicted cross-reactive target peptides by the 7B5 T cell clone, as discovered by the different screening approaches, we then screened the 7B5 T cell clone against several non-hematopoietic cell subsets endogenously expressing these peptides. As a control, we used an HA-2-specific T cell clone, HA2.27, previously isolated from a chronic myeloid leukemia patient who experienced a graft-versus-leukemia (GvL) response after HLA-matched stem cell transplantation (SCT) and subsequent donor lymphocyte infusion (DLI).23 The patient from whom these clones were isolated did not experience any harmful GvHD,24 and no ex vivo cross-reactivity was discovered.
We show that the amino acid scanning approach alone provided useful information about which amino acid positions in the peptide are important for TCR recognition, but its ability to elucidate the cross-reactivity profile of the 7B5 TCR was limited. Instead, we were able to detect off-target reactivity directed against a peptide derived from CDH13 using a CPL scanning approach. This was confirmed with CDH13-transduced HA-2neg Epstein-Barr virus-transformed B lymphoblastic cell lines (EBV-LCLs), and the potential in vivo consequences were deduced by screening against a broad range of different cell subsets.
Results
Identification of an HA-2-Specific T Cell Clone from the Allogeneic TCR Repertoire
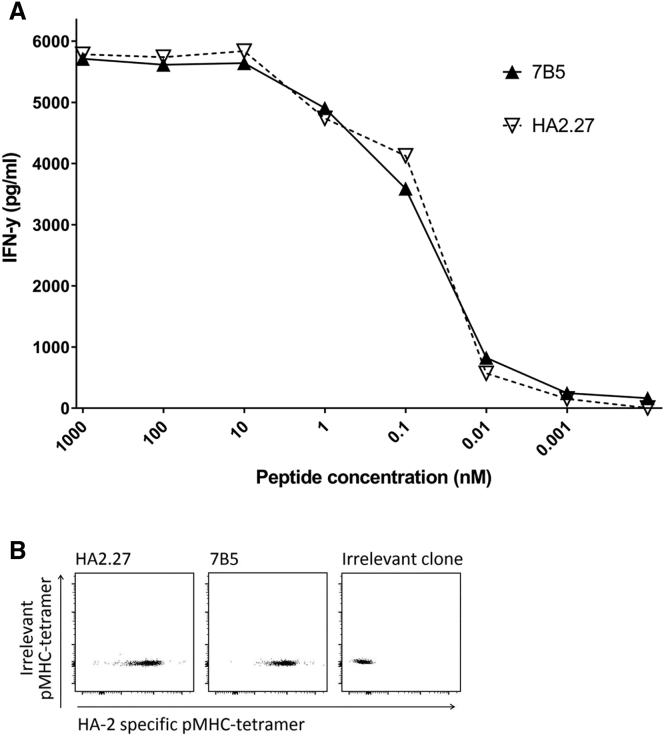
T cell clones from an HLA-A*02:01neg individual were isolated using peptide-major histocompatibility complex (pMHC)-tetramers composed of the HA-2 peptide bound to HLA-A*02:01. pMHC-tetramerpos cells were first enriched by magnetic-activated cell sorting (MACS) followed by single-cell sorting of pMHC-tetramerpos CD8pos T cells. T cell clones were selected that demonstrated reactivity against T2 cells loaded with the HA-2 peptide, but not against unloaded T2 cells. Among these T cell clones, clone 7B5 demonstrated the highest functional sensitivity when stimulated with titrated amounts of HA-2 peptide. This reactivity was comparable with the patient-derived control clone HA2.27, which was isolated from a beneficial GvL response after allogeneic SCT and subsequent DLI (Figure 1A).
Figure 1.
The 7B5 T Cell Clone Exhibits Similar Sensitivity to HA-2 Compared with Clone HA2.27 in a Peptide Stimulation Assay and pMHC-Tetramer Staining
(A) T cell clones 7B5 and HA2.27 (5,000/well) were cocultured with T2 cells (30,000/well) loaded with HA-2 peptide at different concentrations. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. (B) T cell clones were analyzed for binding to specific APC-labeled pMHC-tetramers. Depicted dot plots are gated for CD8pos lymphocytes.
Specific binding to the allophycocyanin (APC)-labeled HA-2 pMHC-tetramer was validated for both T cell clones. Both the 7B5 T cell clone and the HA2.27 T cell clone showed specific staining with the APC-labeled HA-2 pMHC-tetramer with similar intensity (Figure 1B).
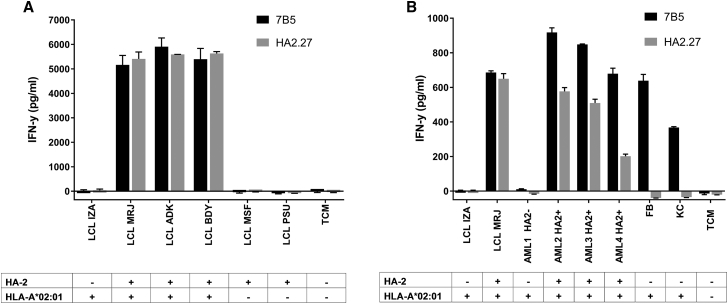
Expression of the MYO1G gene, encoding for the MiHA HA-2, is restricted to hematopoietic tissues.20 We confirmed this restricted expression in an in-house-generated microarray gene expression database (Figure S1).25 To investigate whether T cell clone 7B5 was able to selectively recognize cell subsets expressing HA-2, we tested the HA-2 reactive T cell clones against different hematopoietic cell subsets, starting with SNP-genotyped EBV-LCLs that were either positive or negative for the MiHA HA-2. Robust recognition of HLA-A*02:01pos HA-2pos EBV-LCLs, but not HLA-A*02:01pos HA-2neg or HLA-A*02:01neg EBV-LCLs, was detected, which was comparable with the HA2.27 T cell clone (Figure 2A). In addition, the anti-tumor reactivity of T cell clone 7B5 was tested against different HLA-A*02:01pos primary acute myeloid leukemia (AML) samples either positive or negative for the MiHA HA-2. HLA-A*02:01pos fibroblasts and HLA-A*02:01pos keratinocytes were included as HA-2neg controls. HA-2pos AML samples were similarly recognized by both the HA2.27 T cell clone and the 7B5 T cell clone, whereas the HA-2neg AML sample was not (Figure 2B). However, the HLA-A*02:01pos HA-2neg fibroblasts and keratinocytes that were not recognized by the HA2.27 T cell clone as expected were efficiently recognized by the 7B5 T cell clone (Figure 2B).
Figure 2.
The 7B5 T Cell Clone Recognizes HA-2-Expressing EBV-LCLs and Primary AML Samples
(A) T cell clones 7B5 and HA2.27 (5,000/well) were cocultured with several EBV-LCLs at a stimulator-to-responder (S:R) ratio of 6:1. Controls included EBV-LCLs from the patient (LCL MRJ, HLA-A*02:01pos HA-2pos) and donor (LCL IZA, HLA-A*02:01pos HA-2neg) we isolated T cell clone HA2.27 from, as well as HLA-A*02:01neg controls LCL-MSF and LCL-PSU. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. Depicted values are the average of duplicate measurements ± SD. (B) T cell clones 7B5 and HA2.27 (5,000/well) were cocultured with HLA-A*02:01pos HA-2neg primary AML sample (AML1), HLA-A*02:01pos HA-2pos primary AML samples (AML2,-3,-4), and HLA-A*02:01pos HA-2neg fibroblasts (FB) and keratinocytes (KC) at an S:R ratio of 6:1. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. Depicted values are the average of duplicate measurements ± SD.
All these results indicate that the 7B5 T cell clone is a high-avidity T cell clone, capable of targeting HA-2-expressing hematopoietic cell subsets in the context of HLA-A*02:01. Unexpectedly, the 7B5 T cell clone recognized HLA-A*02:01pos HA-2neg human fibroblasts and keratinocytes as well.
Analysis of Off-Target Cross-Reactivity of the 7B5 T Cell Clone
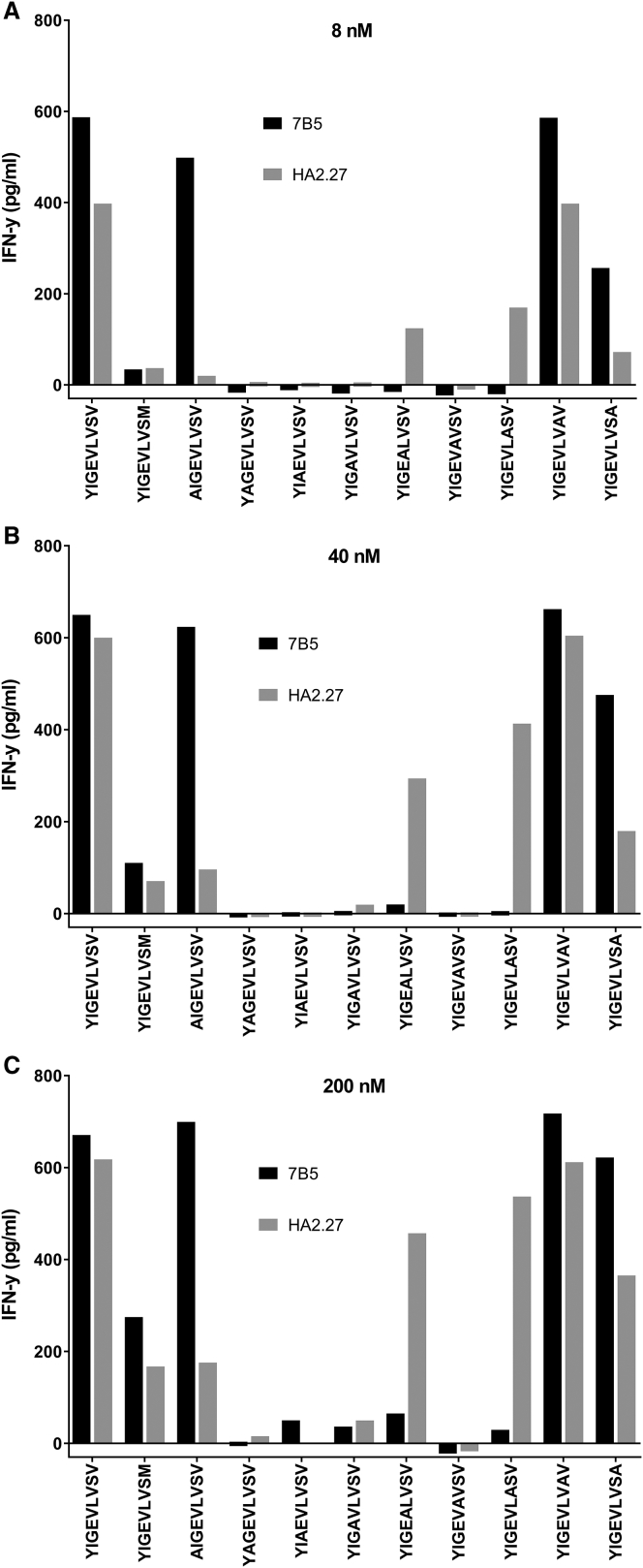
We showed that the 7B5 T cell clone and the HA2.27 T cell clone have a similar functional sensitivity for HA-2, but the 7B5 T cell clone demonstrated cross-reactivity against HA-2neg fibroblasts and keratinocytes. In order to assess what causes this off-target recognition of fibroblasts and keratinocytes, we used an alanine scanning approach. Both HA-2-reactive clones were tested against the immunogenic HA-2 sequence (YIGEVLVSV), the naturally occurring non-immunogenic counterpart (YIGEVLVSM), and newly synthesized HA-2 peptides in which every amino acid is sequentially replaced by an alanine (A). In Table 1 the different peptides are summarized. According to NetMHC4.0, none of the alanine substitutions interfere with HLA-A*02:01 binding to a relevant extent. Both HA-2 T cell clones were incubated overnight with peptide-pulsed T2 cells at different concentrations (8, 40, and 200 nM), and IFN-γ production was measured. When alanine substitution resulted in a substantial decrease in IFN-γ production compared with the immunogenic HA-2 sequence, this position was considered essential. The results, depicted in Figures 3A–3C, demonstrate that for T cell clone 7B5, positions 2, 3, 4, 5, 6, and 7 were important for HA-2-specific recognition. This recognition pattern was slightly different from the HA-2.27 clone, because for this clone, positions 1, 2, 3, 4, and 6 were important for HA-2-specific recognition.
Table 1.
HLA-A*02:01 Binding Properties of the Peptides Used for the Amino Acid Scanning Approach
| Sequence | netMHC4.0 (nM); Binding Level |
|---|---|
| YIGEVLVSV | 7.0; SB |
| YIGEVLVSM | 57.7; WB |
| AIGEVLVSV | 33.4; SB |
| YAGEVLVSV | 41.6; WB |
| YIAEVLVSV | 3.3; SB |
| YIGAVLVSV | 12.9; SB |
| YIGEALVSV | 7.6; SB |
| YIGEVAVSV | 14.9; SB |
| YIGEVLASV | 7.1; SB |
| YIGEVLVAV | 9.1; SB |
| YIGEVLVSA | 30.6; SB |
WB, weak binder; SB, strong binder.
Figure 3.
The Alanine Scanning Approach for T Cell Clones 7B5 and HA2.27
T cell clones 7B5 and HA2.27 (5,000/well) were cocultured with T2 cells (30,000/well), loaded with 8 (A), 40 (B), and 200 nM (C) peptides. After overnight incubation, IFN-γ concentrations were measured in the supernatant by ELISA. Where alanine substitution resulted in a substantial decrease in IFN-γ production compared with the immunogenic HA-2 sequence, the residue at this position was considered essential.
For the 7B5 T cell clone, an in silico search was carried out to identify protein sequences that contain the motif X-I-G-E-V-L-V-X-X. We searched for human self-protein-derived peptide sequences with this motif using the ScanProsite webtool. Besides the HA-2 peptide, this search resulted in only one other peptide sequence PIGEVLVSS derived from the human gene KLF6. However, this KLF6-derived sequence was not predicted to bind HLA-A*02:01 according to NetMHC4.0; thus, we concluded that there was a low risk of cross-reactivity, based on this amino acid scanning approach.
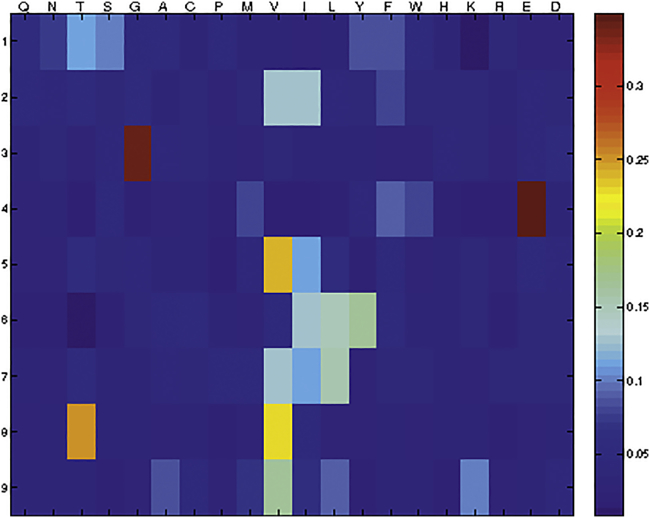
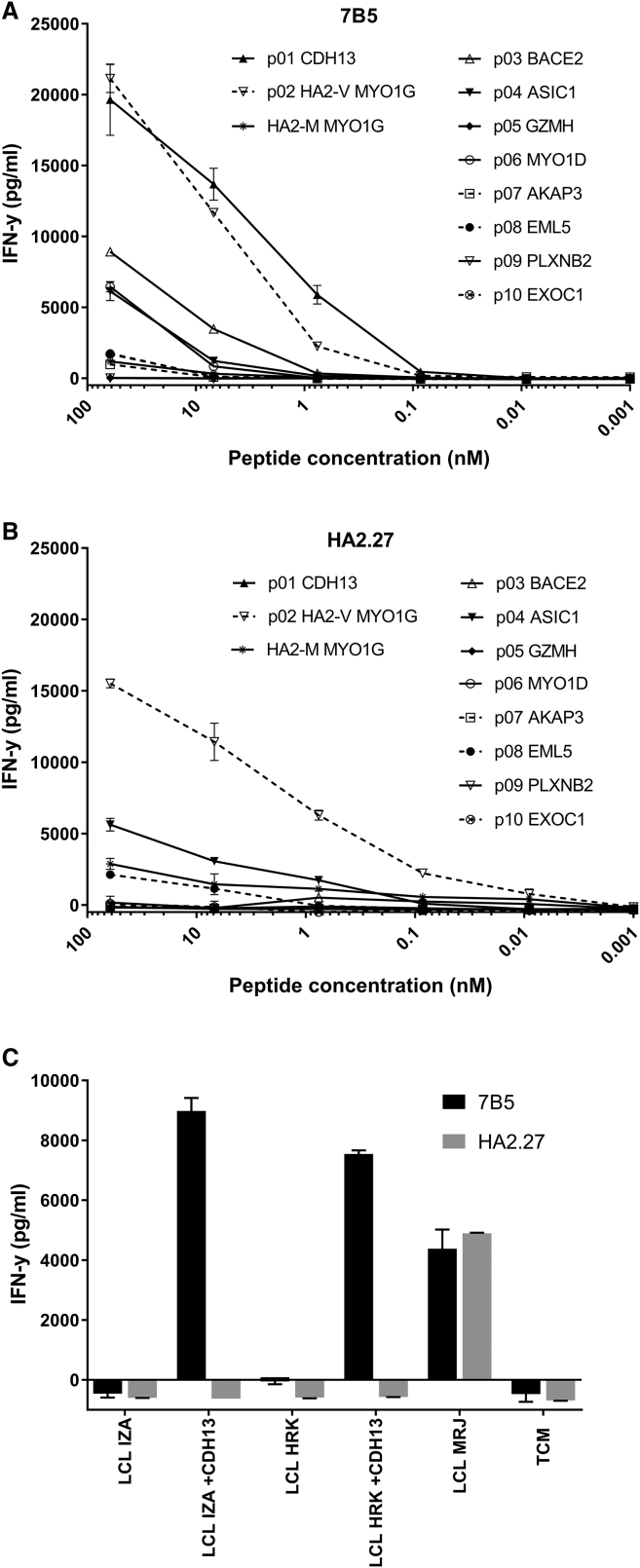
Next, we performed a 9-mer CPL scan. This CPL is composed of 180 different 9-mer peptide mixtures. In every peptide mixture one amino acid position has a fixed l-amino acid residue, but all other positions are degenerate, enabling incorporation of any 1 of 19 natural l-amino acids in all remaining 8 positions (cysteine is excluded). The 7B5 T cell clone was incubated overnight with T2 cells pulsed with the 180 different peptide mixtures, and IFN-γ production was measured. Results are depicted in a heatmap (Figure 4). Using the WSBC PI CPL webtool, we conducted a 9-mer CPL-driven search of the human self-protein database to produce a list of the top 100 peptide sequences ranked in order of likelihood of recognition (Table S1). After removing duplicate sequences, a list of 42 peptide sequences remained and were further refined on the basis of whether they were predicted to bind HLA-A*02:01 using NetMHC4.0. This resulted in a final list of 10 peptide sequences, which were subsequently synthesized (Table 2). The ability of the T cell clones 7B5 and HA2.27 to recognize these candidate peptides was determined by measuring the IFN-γ production after stimulation with T2 cells loaded with titrated concentrations of these peptides. Only one peptide, the CDH13-derived peptide (p01) SVGSVLLTV, was recognized with a similar sensitivity as the HA-2 peptide (p02) (Figure 5A). The patient-derived HA2.27 control clone did not recognize any other peptide but HA-2 (Figure 5B).
Figure 4.
A Heatmap Representation of a 9-Mer CPL Scan of the 7B5 T Cell Clone
A heatmap depicting the intensity of IFN-γ production by the 7B5 T cell clone (4,000/well) in response to T2 cells (20,000/well) loaded with the 180-peptide mixtures in a 9-mer CPL scan, indicating which amino acids are preferentially recognized at certain positions in the peptide sequence. CPL scan data are normalized in each row so that the values range from high (red) to low (blue); the maximum intensity is the largest of all red values in the rows. Amino acids are grouped according to their physicochemical properties as follows: polar, uncharged amines: Q, N; polar, uncharged alcohols: T, S; small: G, A, C; hydrophobic: A–H; aliphatic: V, I, L; aromatic: Y, F, W, H; large: F, W; charged basic: H, K, R; and charged acidic: E, D.
Table 2.
Selected Potential Cross-Reactive Human Self-Protein-Derived Peptide Targets of the 7B5 T Cell Clone
| Peptide No. | Peptide Sequence | Amino Acid Difference | netMHC4.0 (nM; Binding Level) | Gene | Protein Accession No. (NCBI) |
|---|---|---|---|---|---|
| p01 | SVGSVLLTV | 5 | 160.5; WB | CDH13 | NP_001207417 |
| p02 | YIGEVLVSV | – | 7.0; SB | MYO1G | NP_149043 |
| p03 | FVGEDLVTI | 5 | 42.6; WB | BACE2 | NP_036237 |
| p04 | YIGENILVL | 5 | 48.7; WB | ASIC1 | NP_001086 |
| p05 | TLQEVLLTV | 5 | 11.8; SB | GZMH | NP_219491 |
| p06 | FIGEVVVSV | 2 | 8.1; SB | MYO1D | NP_056009 |
| p07 | SVGEVLQSV | 3 | 116.2; WB | AKAP3 | NP_006413 |
| p08 | YIGQVLVTA | 3 | 253.2; WB | EML5 | NP_899243 |
| p09 | AQGEVQLTV | 5 | 144.1; WB | PLXNB2 | NP_036533 |
| p10 | TLGNVLVTV | 4 | 26.7; SB | EXOC1 | NP_001020095 |
The peptides in this table are selected based on their predicted recognition by the 7B5 TCR according to a CPL-driven search of the human self-protein database conducted using the WSBC PI CPL webtool, and subsequently filtered based on their ability to bind HLA-A*02:01 as predicted by NetMHC4.0. Peptides are ordered based on their predicted likelihood of recognition. WB, weak binder; SB, strong binder.
Figure 5.
The 7B5 T Cell Clone Recognizes the CDH13-Derived Peptide p01, Whereas T Cell Clone HA2.27 Does Not
T cell clones (A) 7B5 and (B) HA2.27 (5,000/well) were cocultured with T2 cells (30,000) loaded with the selected peptides at different concentrations. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. Depicted values are the average of duplicate measurements ± SD. p01, Cadherin-13 isoform 2; p02, Myosin-Ig MiHA HA-2 (V); p03, Beta-secretase 2 Isoform 4; p04, Amiloride-sensitive cation channel 2, neuronal isoform b; p05, granzyme H; p06, Myosin-Id; p07, A-kinase anchor protein 3; p08, Echinoderm microtubule-associated protein-like 5; p09, Plexin-B2 precursor; p10, Exocyst complex component 1 isoform 1; p11, Myosin-Ig AC HA-2 (M). (C) T cell clone 7B5 and HA-2 control clone (5,000/well) were cocultured with HA-2neg EBV-LCL IZA and HRK, untransduced or transduced with CDH13, at an S:R ratio of 6:1. The HA-2pos EBV-LCL MRJ was included as a positive control. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. Depicted values are the average of duplicate measurements ± SD.
In order to confirm processing and presentation of the CDH13-derived peptide in HLA-A*02:01, we transduced CDH13 into two different HLA-A*02:01pos HA-2neg EBV-LCLs (IZA and HRK). Recognition of the transduced and untransduced EBV-LCLs was evaluated by IFN-γ ELISA. The 7B5 T cell clone did not recognize untransduced HLA-A*02:01pos HA-2neg EBV-LCLs, similar to the HA2.27 T cell clone, but CDH13 transduced HLA-A*02:01pos HA-2neg EBV-LCLs were highly recognized by the 7B5 T cell clone, whereas the HA2.27 T cell clone was not able to recognize these transduced EBV-LCLs (Figure 5C). These results demonstrate that T cell clone 7B5 is cross-reactive against the CDH13-derived peptide.
Cross-Reactivity Directed against CDH13
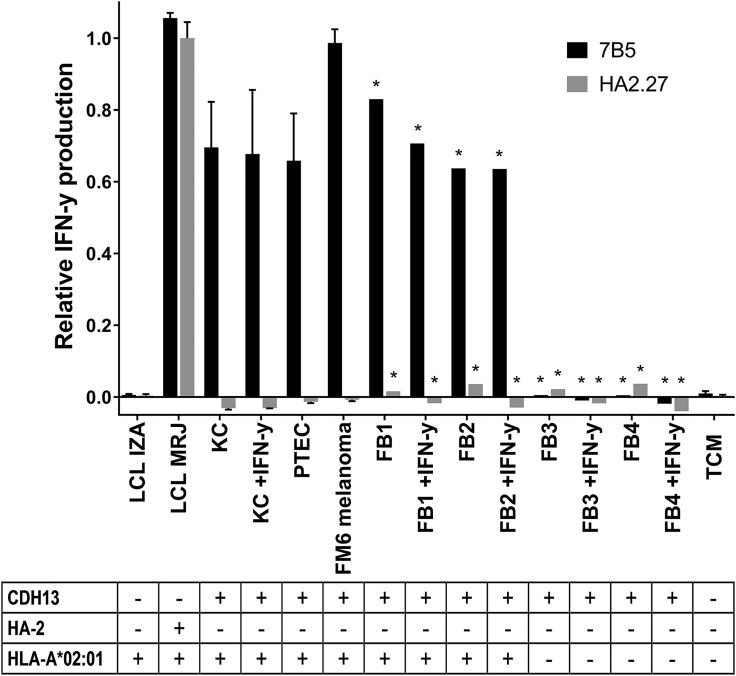
We showed that T cell clone 7B5 is cross-reactive toward a CDH13-derived peptide, which is presented by HLA-A*02:01. Next, we wanted to investigate the possible implications of this cross-reactivity in vivo. According to our gene expression microarray database,25 the CDH13 gene is expressed by a wide variety of healthy non-hematopoietic cell subsets, e.g., fibroblasts, keratinocytes, proximal tubular epithelial cells, and the melanoma cell line FM6 (Figure S2). We tested the 7B5 T cell clone against keratinocytes and fibroblasts derived from HLA-A*02:01pos donors, either untreated or pretreated with IFN-γ for 2 days to upregulate HLA expression, and HLA-A*02:01pos proximal tubular epithelial cells and melanoma cell line FM6. All cell subsets were recognized by the 7B5 T cell clone, but not by the HA2.27 T cell clone (Figure 6). To confirm that only HLA-A*02:01-restricted epitopes were recognized, we included HLA-A*02:01neg fibroblasts, either untreated or pretreated with IFN-γ for 2 days. The HLA-A*02:01neg fibroblasts were unable to activate the 7B5 T cell clone (Figure 6).
Figure 6.
The Endogenously Processed CDH13-Derived Peptide p01, Presented in HLA-A*02:01, Efficiently Activates the 7B5 T Cell Clone, but Not T Cell Clone HA2.27
T cell clones 7B5 and HA2.27 (5,000/well) were cocultured with different cell subsets endogenously expressing the CDH13 gene: keratinocytes and fibroblasts, untreated or pretreated with IFN-γ for 2 days to upregulate HLA expression, proximal tubular epithelial cells, and melanoma cell line FM6. To confirm that only HLA-A*02:01-restricted epitopes are recognized, we tested the 7B5 and HA2.27 T cell clone also against HLA-A*02:01neg fibroblasts, untreated or pretreated with IFN-γ. We also included the EBV-LCL controls from the patient (LCL MRJ, HLA-A*02:01pos HA-2pos) and donor (LCL IZA, HLA-A*02:01pos HA-2neg) we isolated the HA2.27 T cell clone from. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA. Average IFN-γ production by the HA2.27 T cell clone stimulated with LCL MRJ was set to 1. Depicted values are the average of duplicate measurements ± SD. Asterisks (*) indicate single measurements.
All these results demonstrate that the 7B5 T cell clone can activate in response to both the HA-2 antigen and the CDH13-derived peptide SVGSVLLTV, causing cross-reactivity restricted to HLA-A*02:01pos tissues.
Discussion
In this study, we detected off-target reactivity of the 7B5 TCR directed against a peptide derived from the gene CDH13 using a CPL scanning approach, which was confirmed by the ability of the 7B5 T cell clone to recognize CDH13-transduced HA-2neg EBV-LCLs and several cell subsets endogenously expressing CDH13. We show that the standard alanine scanning approach alone provided useful information about which amino acid positions in the peptide are important for TCR recognition, but its ability to elucidate cross-reactivity for the 7B5 TCR was limited.
The 7B5 T cell clone was isolated from an HLA-A*02:01-negative donor; thus, because of the HLA-mismatched experimental setting, thymic selection did not occur. The 7B5 T cell clone recognizes the hematopoietic restricted MiHA HA-2 in the context of HLA-A*02:01. The 7B5 T cell clone was shown to have a functional sensitivity that was similar to the patient-derived T cell clone HA2.27, because concentrations less than 1 nM HA-2 peptide loaded on T2 cells could still trigger IFN-γ production. Also, endogenously processed and presented HA-2 peptide was effectively recognized by the 7B5 T cell clone, as indicated by IFN-γ production upon incubation with HA-2pos EBV-LCLs and primary AML samples. Results were similar to the HA2.27 T cell clone, and no recognition of HA-2neg EBV-LCLs or primary AML samples was observed.
The results of the alanine scanning approach suggested a low risk for broad cross-reactivity of the 7B5 T cell clone, because exchanging the amino acid for an alanine at positions 2, 3, 4, 5, 6, or 7 resulted in complete loss of peptide recognition. These results do not exclude the possibility of cross-reactivity toward other specific peptides, but it dismisses the risk of a highly promiscuous TCR. A search for specific human self-protein-derived peptides with the motif X-I-G-E-V-L-V-X-X did not yield any potential off-target toxicity candidates. However, when we used the 9-mer CPL scan data to conduct a CPL-driven search of the human self-protein database, we found cross-reactivity directed against a peptide derived from the gene CDH13. This was confirmed by the ability of the 7B5 T cell clone to recognize HA-2neg EBV-LCLs transduced with the CDH13 gene and a range of tissues that endogenously express CDH13 including fibroblasts, keratinocytes, and proximal tubular epithelial cells (PTECs).
All of these results demonstrate that the 7B5 T cell clone can activate in response to healthy HLA-A*02:01pos tissues such as fibroblasts, which is likely to have severe clinical consequences if this TCR was engineered into T cells for adoptive therapy in patients. Therefore, we conclude that thorough validation of the specificity and characterization of cross-reactivity by combining several screening techniques should be a strict prerequisite for TCRs to be tested prior to their use in the clinic.
The alanine scanning approach was insufficient for detecting potentially dangerous cross-reactivities, whereas a CPL-driven search of the human self-protein database enabled the identification of a potentially dangerous reactivity directed toward a human self-protein-derived epitope. In the alanine scanning approach, only one amino acid is replaced at a time and the amino acids are only replaced with a single amino acid (alanine), and not with other amino acids possessing different properties. So it is no surprise that cross-reactivity against a peptide that differs from the MiHA peptide HA-2 in five amino acid positions cannot be discovered using this assay. Because the 9-mer CPL fixes only one amino acid at one position per peptide mixture, all possible sequences (except those with more than one cysteine) are represented in the assay, and cross-reactive peptides that differ substantially from the immunogenic target peptide can be identified. The downside of this approach is that every mixture consists of many peptides, lowering the concentration per unique peptide and increasing the competition for HLA binding within every mixture. These factors might influence the results and could explain why for some T cell clones that we tested with the 9-mer CPL, a sufficient peptide recognition signature could not be determined (data not shown). Also, the 9-mer CPL-driven search of the human self-protein database produces such a great number of potential candidates that a selection strategy is needed to keep further screening experiments manageable, risking unwanted exclusion of the target peptides you are searching for.
Although the 7B5 T cell clone, which is derived from an HLA-mismatched setting, showed a potentially dangerous cross-reactivity, this does not mean that all TCRs from an allogeneic setting are of lesser clinical interest compared with clones derived from HLA-matched settings, e.g., the patient-derived T cell clone HA2.27. All TCRs have an intrinsic cross-reactive capacity,26 which is necessary to cover all naturally occurring antigens that may be encountered.27 Thus, the relevant question is not whether a TCR is cross-reactive, but how to predict the clinical consequences of this cross-reactivity.28 Therefore, thorough screening for off-target toxicity must be included in the preclinical phases, to determine whether a TCR is safe for clinical application via adoptive transfer of TCR-engineered T cells. To further reduce potential risks, the use of a so-called suicide switch could also be considered,29 which enables immediate elimination of the transfused T cells upon signs of toxicity.
In summary, we demonstrate that a standard alanine scanning approach provided useful information about which positions in the peptide sequence are important for TCR recognition, but it did not predict off-target reactivity of the 7B5 TCR. However, we were able to detect off-target reactivity directed against a peptide derived from CDH13 using a CPL-driven search of the human self-protein database, which was confirmed using CDH13-transduced HA-2neg EBV-LCLs and screening against several different cell subsets that endogenously express CDH13. This example demonstrates the advantage and need of combining several screening techniques to characterize the cross-reactivity and specificity of TCRs.
Materials and Methods
Culture Conditions and Cells
All studies using human material were approved by the Leiden University Medical Center ethical review board. Peripheral blood was obtained from healthy individuals or patients after informed consent. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll gradient centrifugation.
T cells were cultured in T cell medium consisting of Iscove’s modified Dulbecco’s medium (IMDM) (Lonza) supplemented with 100 IU/mL IL-2 (Proleukin; Novartis Pharma), 5% fetal bovine serum (FBS; GIBCO, Life Technologies), 5% human serum, 2 mM L-glutamine (Lonza), and 1% penicillin/streptomycin (Lonza). T cells were stimulated using irradiated feeders in a 1:5 ratio in T cell medium supplemented with 0.8 μg/mL phytohemagglutinin (PHA; Biochrom AG) 8–16 days prior to experiments.
EBV-LCLs were generated using standard procedures and cultured in IMDM supplemented with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin.
Primary AML samples from patients at time of diagnosis were thawed and cultured overnight in IMDM supplemented with 10% fetal calf serum (FCS) before use in experiments.
Fibroblasts, keratinocytes, and other non-hematopoietic cell subsets were cultured either in the absence or presence of 200 IU/mL IFN-γ for 2 days and washed twice before use in experiments.
Isolation of the HA-2-Reactive T Cell Clone 7B5
To isolate HA-2-reactive T cell clones, we incubated 500 × 106 PBMCs from an HLA-A*02:01neg healthy donor with phycoerythrin (PE)-labeled pMHC-tetramers, composed of HA-2 9-mer peptide (YIGEVLVSV) bound in HLA-A*02:01, for 1 hr at 4°C. Cells were washed twice and incubated with anti-PE microbeads (Miltenyi Biotec) for 15 min at 4°C. PE-labeled cells were isolated on a large separator (LS) column (Miltenyi Biotec) according to the manufacturer’s instruction. Positively selected cells were stained with an anti-CD8 Alexa 700-labeled antibody (Invitrogen/Caltag) in combination with FITC-conjugated antibodies against CD4, CD14, and CD19 (BD Pharmingen). pMHC-tetramerpos CD8pos FITCneg T cells were single-cell sorted into round-bottom 96-well plates containing 5 × 104 irradiated feeders in 100 μL of T cell medium supplemented with 0.8 μg/mL PHA for expansion.
FACS Analysis
FACS acquisition was performed on an LSRII (BD Biosciences) and was analyzed using Diva Software (BD Biosciences). T cell clones were analyzed for binding to the HA-2-specific APC-labeled pMHC-tetramer, an irrelevant PE-labeled pMHC-tetramer, and an Alexa 700-conjugated antibody against CD8 (Invitrogen/Caltag). 10,000 T cells were first incubated with 2 μg/mL pMHC-tetramers for 15 min at 37°C and washed before anti-CD8 antibodies were added and incubated for an additional 15 min at 4°C.
Functional Analysis
For analysis of IFN-γ production, 5,000 T cells were cocultured with 30,000 target cells or T2 cells loaded with peptides in 384-well plates. Peptide loading was performed by incubating T2 cells for 30 min at 37°C and 5% CO2. T cells were washed twice before use. After overnight incubation, supernatants were harvested, and the concentration of IFN-γ was measured by ELISA (Sanquin Reagents).
To assess the peptide recognition signature, we used a previously described 9-mer CPL scan,22 consisting of 180 different peptide mixtures. In each peptide mixture all peptides have one fixed l-amino acid residue at one amino acid position, but all other positions are degenerate, enabling incorporation of any 1 of 19 natural l-amino acids in all remaining 8 positions (cysteine is excluded in non-fixed positions). For this assay, we pulsed 20,000 T2 cells with each of the 180 different peptide mixtures at a concentration of 100 μM for 2 hr at 37°C and 5% CO2 in 40 μL of T cell medium (IMDM supplemented with 100 IU/ml IL-2, 5% FBS, 5% human serum, 2 mM L-glutamine, and 1% penicillin/streptomycin). After peptide loading, 4,000 T cells in 20 μL of T cell medium were added per well. After overnight incubation, supernatants were harvested and IFN-γ production was measured by ELISA (Sanquin Reagents).
Analysis of the Peptide Recognition Signature
To analyze the data from the 9-mer CPL scan, we used the WSBC PI CPL webtool30 that applies a mathematical strategy to match the peptide recognition signature31 with the human proteome to predict the most likely human self-protein-derived peptide sequences that might be recognized by the tested TCR.26 We examined 100 peptides with the best log likelihood scores and removed duplicate sequences. The 10 peptide sequences that were predicted to bind HLA-A*02:01, according to NetMHC4.0, were synthesized.
Gene Transduction
The CDH13 gene was cloned into the modified MP71-retroviral backbone, together with the truncated nerve growth factor receptor (NGF-R; also known as CD271). The constructs were transfected into Phoenix-A cells using helper vector M57 and FuGENE HD reagent (Roche). Virus supernatant was harvested after 48 and 72 hr and stored at −80°C.
For transduction of EBV-LCLs, 24-well non-tissue culture plates were coated with 30 mg/mL RetroNectin (Takara) for 2 hr at room temperature and blocked with 2% human serum albumin (Sanquin Reagents) for 30 min. Viral supernatant was thawed, then 500 μL per well was added to the 24-well plate and spun down at 2,000 × g for 20 min at 4°C. 100,000 EBV-LCLs per well were added in 500 μL of fresh IMDM supplemented with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin and incubated overnight at 37°C and 5% CO2. CDH13pos EBV-LCLs were MACS purified using anti-CD271 PE-labeled antibody (BD Pharmingen) and anti-PE microbeads (Miltenyi Biotec). CDH13-transduced cells were isolated on an LS column following manufacturer’s instructions.
Author Contributions
H.M.B. designed the experiments, performed experiments, analyzed the data, and wrote the manuscript. D.M.v.d.S. performed the 9-mer CPL scan. A.K.W. performed experiments. R.S.H. prepared the virus supernatants. L.W. provided the 9-mer CPL and reviewed and edited the manuscript. J.H.F.F reviewed and edited the manuscript. M.H.M.H. acquired the funding, supervised the research, and reviewed and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This research was supported by Landsteiner Foundation for Blood Transfusion Research grant LSBR1331. We thank Marieke Griffioen for sharing the microarray gene expression database.
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.02.017.
Supplemental Information
References
- 1.Holzinger A., Barden M., Abken H. The growing world of CAR T cell trials: a systematic review. Cancer Immunol. Immunother. 2016;65:1433–1450. doi: 10.1007/s00262-016-1895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg S.A., Dudley M.E. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr. Opin. Immunol. 2009;21:233–240. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser M.J., Shapira-Frommer R., Schachter J. Tumor-infiltrating lymphocytes: clinical experience. Cancer J. 2015;21:465–469. doi: 10.1097/PPO.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 4.Thomas S., Stauss H.J., Morris E.C. Molecular immunology lessons from therapeutic T-cell receptor gene transfer. Immunology. 2010;129:170–177. doi: 10.1111/j.1365-2567.2009.03227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stromnes I.M., Schmitt T.M., Chapuis A.G., Hingorani S.R., Greenberg P.D. Re-adapting T cells for cancer therapy: from mouse models to clinical trials. Immunol. Rev. 2014;257:145–164. doi: 10.1111/imr.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins P.F., Kassim S.H., Tran T.L., Crystal J.S., Morgan R.A., Feldman S.A., Yang J.C., Dudley M.E., Wunderlich J.R., Sherry R.M. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 2015;21:1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rapoport A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., Badros A.Z., Garfall A., Weiss B., Finklestein J. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 2015;21:914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kageyama S., Ikeda H., Miyahara Y., Imai N., Ishihara M., Saito K., Sugino S., Ueda S., Ishikawa T., Kokura S. Adoptive transfer of MAGE-A4 T-cell receptor gene-transduced lymphocytes in patients with recurrent esophageal cancer. Clin. Cancer Res. 2015;21:2268–2277. doi: 10.1158/1078-0432.CCR-14-1559. [DOI] [PubMed] [Google Scholar]
- 9.Johnson L.A., Morgan R.A., Dudley M.E., Cassard L., Yang J.C., Hughes M.S., Kammula U.S., Royal R.E., Sherry R.M., Wunderlich J.R. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer E. Negative selection—clearing out the bad apples from the T-cell repertoire. Nat. Rev. Immunol. 2003;3:383–391. doi: 10.1038/nri1085. [DOI] [PubMed] [Google Scholar]
- 11.Kunert A., Obenaus M., Lamers C.H.J., Blankenstein T., Debets R. T-cell receptors for clinical therapy: in vitro assessment of toxicity risk. Clin. Cancer Res. 2017;23:6012–6020. doi: 10.1158/1078-0432.CCR-17-1012. [DOI] [PubMed] [Google Scholar]
- 12.Linette G.P., Stadtmauer E.A., Maus M.V., Rapoport A.P., Levine B.L., Emery L., Litzky L., Bagg A., Carreno B.M., Cimino P.J. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone J.D., Harris D.T., Kranz D.M. TCR affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr. Opin. Immunol. 2015;33:16–22. doi: 10.1016/j.coi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone J.D., Kranz D.M. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front. Immunol. 2013;4:244. doi: 10.3389/fimmu.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan R.A., Chinnasamy N., Abate-Daga D., Gros A., Robbins P.F., Zheng Z., Dudley M.E., Feldman S.A., Yang J.C., Sherry R.M. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 2013;36:133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Berg J.H., Gomez-Eerland R., van de Wiel B., Hulshoff L., van den Broek D., Bins A. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol. Ther. 2015;23:1541–1550. doi: 10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raman M.C., Rizkallah P.J., Simmons R., Donnellan Z., Dukes J., Bossi G., Le Provost G.S., Todorov P., Baston E., Hickman E. Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy. Sci. Rep. 2016;6:18851. doi: 10.1038/srep18851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn L., van der Steen D.M., Hagedoorn R.S., Hombrink P., Kester M.G., Schoonakker M.P., de Ridder D., van Veelen P.A., Falkenburg J.H., Heemskerk M.H. Generation of CD20-specific TCRs for TCR gene therapy of CD20low B-cell malignancies insusceptible to CD20-targeting antibodies. Oncotarget. 2016;7:77021–77037. doi: 10.18632/oncotarget.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voogt P.J., Goulmy E., Veenhof W.F., Hamilton M., Fibbe W.E., Van Rood J.J., Falkenburg J.H. Cellularly defined minor histocompatibility antigens are differentially expressed on human hematopoietic progenitor cells. J. Exp. Med. 1988;168:2337–2347. doi: 10.1084/jem.168.6.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulmy E. Human minor histocompatibility antigens. Curr. Opin. Immunol. 1996;8:75–81. doi: 10.1016/s0952-7915(96)80108-7. [DOI] [PubMed] [Google Scholar]
- 21.Hickman E.S., Lomax M.E., Jakobsen B.K. Antigen selection for enhanced affinity T-cell receptor-based cancer therapies. J. Biomol. Screen. 2016;21:769–785. doi: 10.1177/1087057116637837. [DOI] [PubMed] [Google Scholar]
- 22.Wooldridge L., Laugel B., Ekeruche J., Clement M., van den Berg H.A., Price D.A., Sewell A.K. CD8 controls T cell cross-reactivity. J. Immunol. 2010;185:4625–4632. doi: 10.4049/jimmunol.1001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heemskerk M.H., Hoogeboom M., de Paus R.A., Kester M.G., van der Hoorn M.A., Goulmy E., Willemze R., Falkenburg J.H. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 2003;102:3530–3540. doi: 10.1182/blood-2003-05-1524. [DOI] [PubMed] [Google Scholar]
- 24.Marijt W.A., Heemskerk M.H., Kloosterboer F.M., Goulmy E., Kester M.G., van der Hoorn M.A., van Luxemburg-Heys S.A., Hoogeboom M., Mutis T., Drijfhout J.W. Hematopoiesis-restricted minor histocompatibility antigens HA-1- or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc. Natl. Acad. Sci. USA. 2003;100:2742–2747. doi: 10.1073/pnas.0530192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pont M.J., Honders M.W., Kremer A.N., van Kooten C., Out C., Hiemstra P.S., de Boer H.C., Jager M.J., Schmelzer E., Vries R.G. Microarray gene expression analysis to evaluate cell type specific expression of targets relevant for immunotherapy of hematological malignancies. PLoS ONE. 2016;11:e0155165. doi: 10.1371/journal.pone.0155165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wooldridge L., Ekeruche-Makinde J., van den Berg H.A., Skowera A., Miles J.J., Tan M.P., Dolton G., Clement M., Llewellyn-Lacey S., Price D.A. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2012;287:1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degauque N., Brouard S., Soulillou J.P. Cross-reactivity of TCR repertoire: current concepts, challenges, and implication for allotransplantation. Front. Immunol. 2016;7:89. doi: 10.3389/fimmu.2016.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh N.K., Riley T.P., Baker S.C.B., Borrman T., Weng Z., Baker B.M. Emerging concepts in TCR specificity: rationalizing and (maybe) predicting outcomes. J. Immunol. 2017;199:2203–2213. doi: 10.4049/jimmunol.1700744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Stasi A., Tey S.K., Dotti G., Fujita Y., Kennedy-Nasser A., Martinez C., Straathof K., Liu E., Durett A.G., Grilley B. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szomolay B., Liu J., Brown P.E., Miles J.J., Clement M., Llewellyn-Lacey S., Dolton G., Ekeruche-Makinde J., Lissina A., Schauenburg A.J. Identification of human viral protein-derived ligands recognized by individual MHCI-restricted T-cell receptors. Immunol. Cell Biol. 2016;94:573–582. doi: 10.1038/icb.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wooldridge L. Individual MHCI-restricted T-cell receptors are characterized by a unique peptide recognition signature. Front. Immunol. 2013;4:199. doi: 10.3389/fimmu.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.