Abstract
CRISPR-based gene editing is a powerful technology for engineering mammalian genomes. It holds the potential as a therapeutic, although much-needed in vivo delivery systems have yet to be established. Here, using the Cpf1-crRNA (CRISPR RNA) crystal structure as a guide, we synthesized a series of systematically truncated and chemically modified crRNAs, and identify positions that are amenable to modification while retaining gene-editing activity. Modified crRNAs were designed with the same modifications that provide protection against nucleases and enable wide distribution in vivo. We show crRNAs with chemically modified terminal nucleotides are exonuclease resistant while retaining gene-editing activity. Chemically modified or DNA-substituted nucleotides at select positions and up to 70% of the crRNA DNA specificity region are also well tolerated. In addition, gene-editing activity is maintained with phosphorothioate backbone substitutions in the crRNA DNA specificity region. Finally, we demonstrate that 42-mer synthetic crRNAs from the similar CRISPR-Cas9 system are taken up by cells, an attractive property for in vivo delivery. Our study is the first to show that chemically modified crRNAs of the CRISPR-Cpf1 system can functionally replace and mediate comparable gene editing to the natural crRNA, which holds the potential for enhancing both viral- and non-viral-mediated in vivo gene editing.
Keywords: synthetic CRISPR RNA, CRISPR, gene editing, gene targeting, Cpf1, scrRNA, RNA uptake
McMahon et al. show that AsCpf1 CRISPR RNAs containing chemical substitutions (scrRNA) known to provide protection against nucleases and enable wide distribution in vivo can functionally replace the natural CRISPR RNA (crRNA) to mediate efficient genome editing in human cells. Positions amenable or sensitive to chemical modifications were identified and characterized.
Introduction
The CRISPR systems are composed of an RNA-guided endonuclease that is essential for bacterial adaptive immunity.1, 2, 3 The class II CRISPR-associated (Cas) nucleases, most notably Cas9 and Cpf1 (CRISPR from Prevotella and Francisella 1), have been engineered to recognize and induce site-specific DNA double-stranded breaks (DSBs) in mammalian cells and other model organisms revolutionizing biomedical research.4, 5, 6, 7 In mammalian cells, the DSB is typically repaired by one of two major pathways. Mutagenic non-homologous end joining (NHEJ) can result in gene inactivation because of introduction of nucleotide insertions or deletions (indels) that disrupt a protein-encoding open reading frame. Alternatively, homologous recombination (HR) produced by introduction of a user-defined repair template can result in replacement of an initial DNA sequence surrounding the cleavage site with template-derived, precise nucleotide changes.
To induce a DSB, Cas9 protein complexes with two RNAs including the short CRISPR RNA (crRNA), which recognizes a target DNA sequence next to a protospacer adjacent motif (PAM), thereby providing nuclease site specificity and a second longer trans-activating crRNA (tracrRNA).4 A simplified system is typically used in mammalian cell applications in which the two RNAs are replaced by a single guide RNA (sgRNA), a 102-nt chimeric fusion of the crRNA and tracrRNA.4, 5 The two most commonly used Cas9 orthologs for mammalian genome editing are that from Streptococcus pyogenes (SpCas9),4, 5 which recognizes a DNA sequence next to an NGG PAM, or Staphylococcus aureus (SaCas9),8 which recognizes a DNA sequence next to an NNGRRT PAM. Both SpCas9 and SaCas9 have been shown to have comparable gene-editing activity.8
Like Cas9, Cpf1 can be programmed to cleave target DNA sequences, although strikingly it utilizes just a single 43-mer crRNA that recognizes target sequence next to a 5′ TTTN PAM7 and is smaller than SpCas9 (151 kDa compared with 160 kDa). Most notably, genome-wide off-target analysis has shown that Cpf1, from Acidaminococcus sp. BV3L6 (AsCpf1), has little to no off-target gene editing9, 10 as compared with what has been observed with SpCas9.11, 12
Over the past several years, many CRISPR-Cas gene-editing principles have been established in cell lines13, 14, 15 and animal models8, 16, 17, 18, 19 with therapeutic development still emerging.20, 21 Several challenges remain for therapeutic applications including delivery of Cas protein and RNA to target cells, controlling expression and persistence of system components, reducing off-target editing, and minimizing an immune response to the bacterial proteins.20, 21 The smaller size of AsCpf1 compared with SpCas9,7 its reduced off-target editing,9, 10 and its requirement for only a single RNA3, 7 make the AsCpf1 system attractive for therapeutic development.
We recently showed that a 29-mer synthetic crRNA (scrRNA), of the SpCas9 system, substituted with chemical modifications known to increase metabolic stability and affinity with complementary nucleic acid targets can mediate gene editing comparable with transcription-produced sgRNA, but with reduced off-target activity22 in human cell lines. Using the same experimental approach, we now demonstrate that truncated, 40-nt scrRNAs of the AsCpf1 CRISPR system can mediate genome editing in human cell lines. Using the Cpf1-crRNA crystal structure as a guide, we show that scrRNAs rationally substituted with phosphorothioate (PS) backbone modifications, 2′-fluoro (2′-F), 2′-O-Methyl (2′-O-Me), and S-constrained ethyl (cEt) sugar modifications, known to increase metabolic stability and binding affinity to RNA,23, 24 or replacement of RNA nucleotides with DNA, demonstrate highly efficient gene-editing activity comparable with the natural crRNA, and that scrRNAs with substitutions on the termini are resistant to exonuclease activity. Importantly, we show that scrRNAs can be active when added to culture media in the absence of agents or methodologies to enhance intracellular delivery, an essential feature for in vivo gene modification.
Results
crRNAs with Truncated DNA Specificity Region Retain Robust Gene-Editing Activity
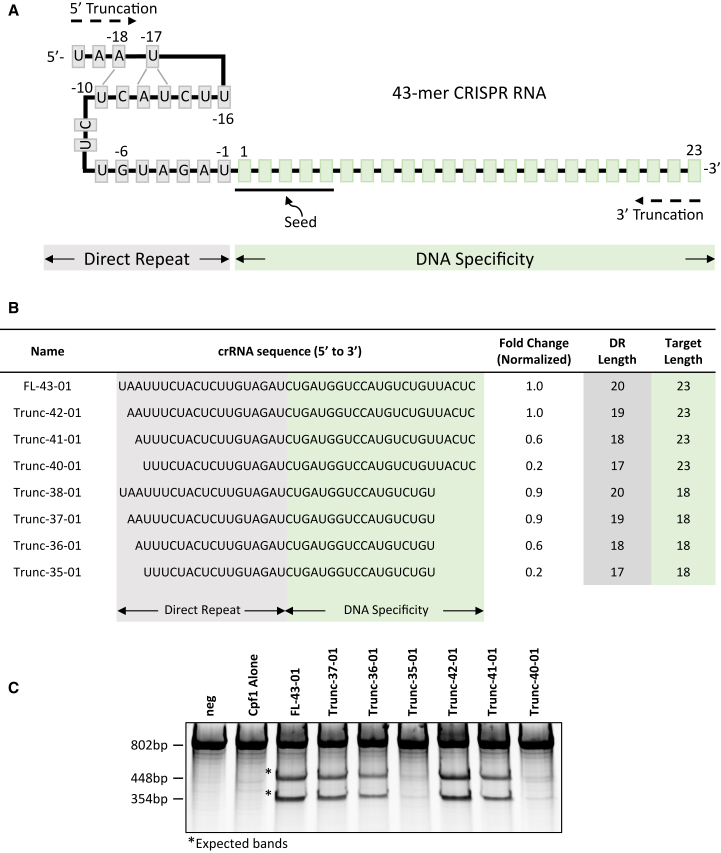
AsCpf1 is a class II type V effector and is composed of an RuvC nuclease domain and a second nuclease domain functionally analogous to the histidine and asparagine endonuclease domain (HNH) of Cas9 protein25 for cleavage of both the target and non-target DNA strands.3, 26 AsCpf1 utilizes a single 43-nt crRNA that provides specificity through the 3′ terminal 23 nt by direct Watson-Crick pairing with target DNA7 (Figure 1A). We sought to develop an optimal scrRNA for the AsCpf1 system, both for ease of chemical synthesis and to provide maximal in vivo stability. To do this, we first tested the minimal crRNA length required for AsCpf1 activity using human DNA methyltransferase 1 (DNMT1) as a target gene. As shown in Figure 1B (and Figure S1), we systematically designed crRNAs of variable length, with truncations from either the 5′, 3′, or both termini. HEK293T cells were transfected with plasmid encoding AsCpf1 protein and double-stranded DNA fragments encoding the variable-length crRNAs driven by the U6 promoter. Surveyor assay was performed 48 hr after transfection to quantify gene disruption mediated by AsCpf1-truncated crRNA. We observed that truncation by 1 nt on the 5′ terminus (Trunc-42-01; Figures 1B and 1C) did not affect gene disruption activity compared with the natural crRNA (FL-43-01; Figure 1B), but further removal of 1 additional nucleotide resulted in an approximately 40% loss in gene disruption activity compared with FL-43-01 (Trunc-41-01; Figures 1B and 1C). Three or more 5′ terminal nucleotide truncations resulted in near or complete loss of gene disruption activity (Trunc-40-01; Figure S1). Loss of activity with increasing 5′ nucleotide truncations was not unexpected because it has recently been reported that positions A(−18) and U(−16) of the crRNA are conserved in different CRISPR-Cpf1 systems7 and make specific base contacts required to form a pseudoknot structure that is critical for Cpf1 activity.25 Our data suggest that deletion of A(−18) as in Trunc-40-01 is negatively affecting the pseudoknot structure.
Figure 1.
Truncated crRNAs Mediate Gene Disruption Comparable with the Full-Length Natural crRNA
(A) Sequence recognition and structure of AsCpf1 43-mer crRNA with 5′ direct repeat indicated by nucleotides in gray box (all figures) and DNA specificity region indicated by nucleotides in light green box (all figures), including seed sequence (underlined). (B) Sequence of truncated DNMT1 targeting crRNAs where the fold change column is quantified from surveyor nuclease assay using genomic DNA isolated from transfected cells and normalized to full-length 43-mer crRNA (FL-43-01) as 1 (100%). Last two columns indicate the nucleotide length of the specified regions. Representative data from two or more replicate experiments. (C) Representative surveyor nuclease assay gel where “neg” indicates no transfection and AsCpf1 alone indicates transfection of the plasmid encoding the nuclease in the absence of crRNA. Asterisks (*) indicate expected bands.
In contrast, truncations on the 3′ end were well tolerated and did not compromise AsCpf1 activity. Greater than 90% of AsCpf1 activity was retained following reduction of the DNA specificity region from 23 to 18 nt (Trunc-38-01; Figure 1B). These findings are in agreement with recent work where it was shown that robust AsCpf1 activity was retained using crRNAs, where up to 6 nt were deleted from the 3′ end.9 We next designed a series of scrRNAs with deletions from both ends. Confirming truncations described above, a 37-mer crRNA (1-nt deletion from the 5′ end and 5-nt deletions from the 3′ end) retained robust activity, and 60% activity was retained for a slightly smaller 36-mer crRNA (Trunc-37-01 and Trunc-36-01; Figures 1B and 1C, respectively). Further 5′ end deletions completely abolished AsCpf1 activity (Trunc-35-01; Figures 1B and 1C; Figure S1). From this, we conclude that a minimal 36-mer crRNA (Trunc-36-01; Figures 1B and 2C) is sufficient for robust AsCpf1 gene disruption activity, although from this, we do not know how the length and chemical modifications will coordinate for optimal activity.
Figure 2.
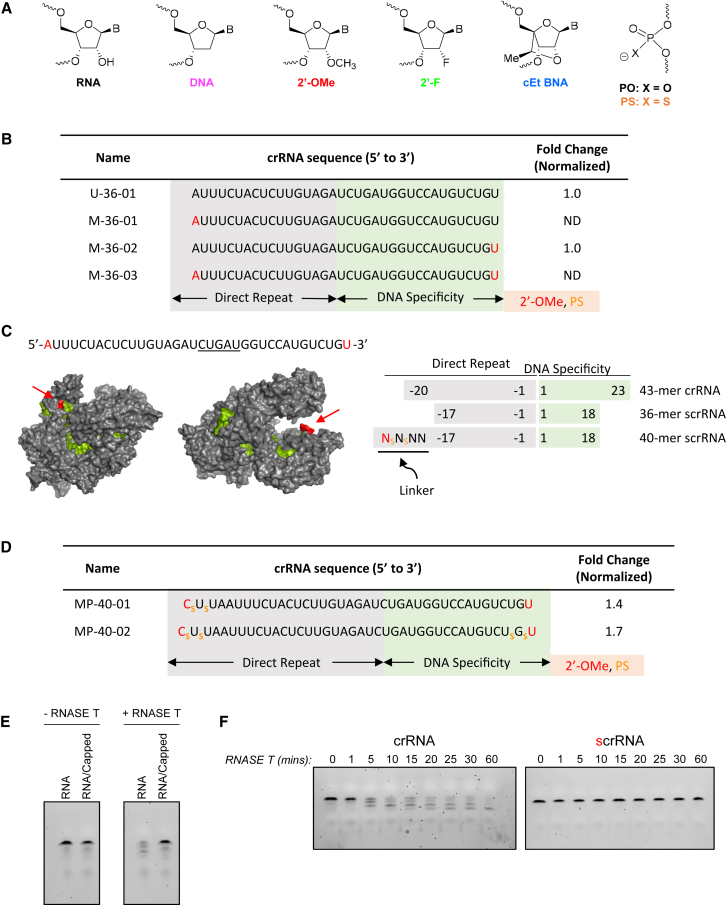
Synthetic crRNAs with Terminal 2′-O-Me Nucleotides Can Mediate Gene Disruption in Human Cells
(A) Structures of chemically modified nucleotides substituted in scrRNAs including phosphorothioate (PS), 2′-O-Methyl (2′-O-Me), 2′-Fluoro (2′-F), or S-constrained ethyl (cEt) substitutions. (B) Full sequence of scrRNAs targeting DNMT1 with 2′-O-Me-substituted nucleotides indicated in red. Data representative of gene disruption quantified from surveyor nuclease assay from two or more replicate experiments and normalized to U-36-01. (C) Surface representation of AsCpf1-crRNA-target DNA complex (from PDB: 5b43)25 with the red arrow indicating the 5′ and 3′ ends of the crRNA (left). (Right) Schematic representation of scrRNAs with variable length direct repeat and DNA specificity region. 43-mer crRNA represents the natural crRNA of the AsCpf1 system. “Linker” nucleotides are underlined, and chemical substitutions are indicated by color as in (B). (D) Full sequence of DNMT1 targeting scrRNAs and single-experiment gene disruption activity compared with U-36-01 (normalized as 1). (E) SYBRgold-stained TBE-urea denaturing gel for visualization of RNA degradation products after a 30-min co-incubation with RNase T. (F) Gel same as in (E) except with the addition of multiple time points up to 60 min.
Production and Activity of scrRNAs with 5′ and 3′ Terminal Chemical Modifications
RNA is highly unstable in vivo due to the presence of plasma and tissue ribonucleases. Yet, it has been demonstrated that specific chemical modifications to RNA can greatly increase stability, extending half-lives and enabling use of chemically modified RNA for therapeutic applications.23, 24, 27 One such modification, 2′-O-Me (Figure 2A) sugar substitution, forces RNA to adopt a structure that is more favorable for increasing Watson-Crick binding affinity in addition to providing nuclease resistance.28 Therefore, we wanted to test gene-editing activity of crRNAs substituted with 2′-O-Me-modified nucleotides at the termini. We hypothesized that we could improve the activity of the 36-mer scrRNA with a chemically modified nucleoside at position A(−18) by potentially stabilizing its interaction with U(−10) as shown in the crystal structure.25 We designed a series of 36-mer scrRNAs using the same DNMT1 targeting sequence, where either the 5′, 3′, or both terminal nucleotides were substituted with 2′-O-Me-modified nucleotides. We show that an scrRNA with a 3′ terminal 2′-O-Me-modified nucleotide (M-36-02; Figure 2B) had comparable gene disruption activity to the unmodified 36-mer crRNA (U-36-01; Figure 2B). On the other hand, chemical substitution with 2′-O-Me of the 5′ terminal nucleotide completely abolished activity (M-36-01 and M-36-03; Figure 2B). These data suggest that the critical and conserved interaction of A(−18) with U(−10) in the crRNA25 may be sensitive to chemical substitution and compromise AsCpf1 activity by affecting protein:RNA complexing and/or the RNA pseudoknot structure.
The previously published crystal structure of the crRNA and AsCpf1 complex revealed that A(−18) is just inside the complex and adjacent to an accessible “pocket or region,” and that crRNA position 18 is slightly exposed (Figures 1A and 2C, left). We hypothesized that: (1) extension of the 5′ end of the crRNA with a 3-nt linker, (2) substitution with chemically modified residues at both termini, and (3) addition of PS terminal linkages at both termini would not interfere with complex formation or activity but provide protection against nuclease degradation (Figure 2C, right). PS modifications (Figure 2A) in which one of the non-bridging oxygens is substituted with sulfur in the phosphate backbone is known to increase resistance to nuclease degradation.29 We synthesized and tested 40-mer scrRNAs (Figure 2C, right) with terminal 2′-O-Me nucleotides and two terminal PS substitutions (MP-40-01 and MP-40-02; Figure 2D), and demonstrate similar activity compared with natural crRNA restoring the activity that was lost with the slightly smaller scrRNA.
scrRNAs with Chemically Substituted Terminal Nucleotides Are Protected from Exonuclease Degradation
40-mer scrRNAs demonstrated a modest increase in gene-editing activity compared with the natural crRNA (∼1.4- to ∼1.7-fold). We hypothesized that one explanation for this increase in activity could be due to protection of the scrRNA from exonuclease activity resulting in a longer-lived scrRNA pool, compared with unmodified natural crRNA, available for complexing with AsCpf1 protein. We used a biochemical assay to test the susceptibility to exonuclease activity of scrRNAs with terminal PS linkages and 2′-O-Me chemically substituted nucleotides. In brief, crRNAs and scrRNAs were incubated with RNase T, a single-stranded RNA,30, 31 or DNA-specific exonuclease and reaction products run on a Tris/borate/EDTA (TBE)-urea polyacrylamide gel and stained using SYBRgold. As shown in Figure 2E, unmodified RNA was susceptible to RNase T activity as demonstrated by nearly complete degradation products detectable after 30 min of co-incubation, whereas an scrRNA capped with chemically substituted nucleic acids was completely resistant. A more detailed time course demonstrates that this activity is rapid as degradation products are detectable within 5 min of co-incubation for the unmodified RNA, but that even up to 1 hr the scrRNA is protected (Figure 2F). These data demonstrate that scrRNAs with terminal chemical modifications are resistant to exonucleases.
scrRNAs with PS Backbone Substitutions Retain High Levels of Gene-Editing Activity
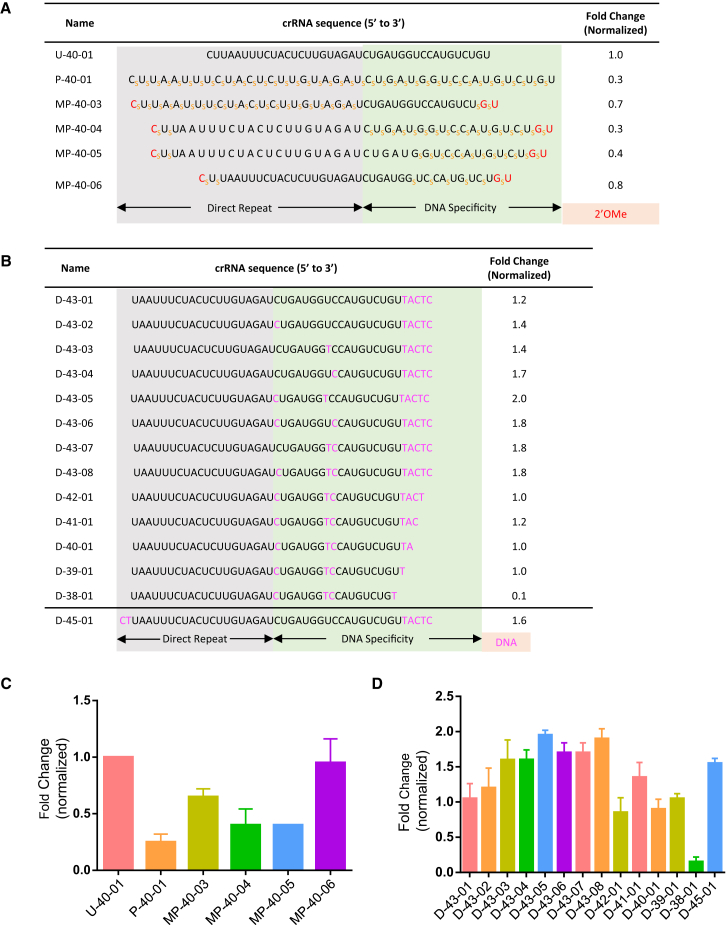
PS modification of small chemically modified oligonucleotides not only provides protection against exo-nuclease and endo-nuclease activity, but also increases binding affinity to serum proteins facilitating distribution to tissues and cellular uptake in the absence of special carriers or formulations.28 We previously established that scrRNAs of the SpCas9 system, with complete PS backbone substitution, can functionally replace unmodified crRNA.22 We applied the same strategy to AsCpf1 crRNA and show that unlike the SpCas9 system, there is an almost 70% loss of activity by a fully substituted PS backbone containing scrRNA (P-40-01; Figures 3A and 3C). This is likely due to structural changes in the scrRNA that disrupt scrRNA and AsCpf1 recognition of target DNA, although whether PS modification in the direct repeat or the DNA specificity region was primarily responsible for the reduction in gene disruption activity could not be determined with this scrRNA. Therefore, to determine which domain was more sensitive to PS modification, we tested two additional scrRNAs completely substituted in either domain (MP-40-03 and MP-40-04; Figures 3A and 3C). We observed a modest reduction (∼30%) in gene disruption activity by the direct repeat PS-modified scrRNA (MP-40-03), whereas complete PS substitution throughout the DNA specificity domain, including the seed sequence, significantly affected activity as shown by an almost 70% reduction in gene disruption activity (MP-40-04; Figures 3A and 3C). Next, to test whether the loss in activity was due to substitution in the seed sequence, we chemically synthesized an scrRNA with PS modification in the DNA specificity region up to this position (MP-40-05; Figure 3A) and observed that some activity was restored as demonstrated by a slight increase in gene disruption activity compared with MP-40-04. From the crystal structure, it was shown that the sugar-phosphate backbone of crRNA nucleotides from positions 1–8 make multiple contacts with the Wedge (WED) and REC1 domains of Cpf1,25 suggesting that PS substitutions in this region may not be well tolerated. Because of this, we further reduced the number of PS modifications and avoided positions that interact with AsCpf1 by chemically synthesizing an scrRNA with six alternating PS substitutions in the DNA specificity region, excluding the seed sequence, and show that gene disruption activity was restored to almost 80% that of unmodified crRNA (MP-40-06; Figures 3A and 3C). Taken together, PS substitutions are tolerated at crRNA positions that are non-interacting with AsCpf1.
Figure 3.
PS- or DNA-Substituted scrRNAs Can Mediate High Levels of Gene Disruption
(A) Complete DNMT1 target sequence with PS linkages indicated by orange subscript “S.” Gene disruption normalized to truncated and unmodified 40-mer crRNA, U-40-01 (= 1). Data represent gene disruption activity quantified from surveyor nuclease assay from two or more replicate experiments. (B) Complete DNMT1 target sequence with DNA nucleotides indicated in purple. Gene disruption activity is normalized to U-40-01 scrRNA (= 1). Representative data are from two or more replicates. (C) Graph shows fold change in gene disruption activity as indicated in last column of (A). (D) Graph shows fold change in gene disruption activity as indicated in last column of (B).
scrRNAs with DNA Substitutions Demonstrate Robust Gene-Editing Activity
AsCpf1 scrRNAs with the 3′ terminus truncated and substituted by chemically modified nucleotides demonstrated equivalent gene-editing activity compared with the natural crRNA. Because DNA is more stable than RNA, we next wanted to test the tolerability of DNA substitutions of the full-length 43-mer crRNA. We started with substitution of 3′ terminal RNA residues with DNA again using DNMT1 target sequence because the three-terminal target DNA and crRNA residues were disordered in the crystal structure, suggesting flexibility in this region.25 As shown in Figure 3B, D-43-01, a 43-mer scrRNA containing five 3′ terminal DNAs, retained comparable activity to the unmodified crRNA (Figure 3D).
It was previously shown that positions 1, 8, and 9 in the DNA specificity region of the crRNA tolerate mismatches,9 and hence we hypothesized that they could also be amenable to DNA substitution. Therefore, we tested a series of 43-mer scrRNAs with five 3′ terminal DNA nucleotide substitutions and with a combination of DNA substitutions at positions 1, 8, and 9 in the crRNA DNA specificity region. Surprisingly, we observed enhanced gene disruption (∼1.7-fold) by the RNA/DNA hybrid scrRNAs with single DNA substitutions at positions 1, 8, or 9 (D-43-02, D-43-03, D-43-04; Figures 3B and 3D). Combinations of two DNA substitutions at these positions further increased gene disruption activity by almost 2-fold compared with unmodified crRNA (D-43-05, D-43-06, D-43-07; Figures 3B and 3D), and this activity is retained when all three positions are substituted with DNA (D-43-08; Figures 3B and 3D). These data demonstrate that DNA can functionally replace RNA in multiple positions of the crRNA, but only up to a point as further increasing the number of DNA substitutions results in complete loss of activity (Figure S2). Next, we combined DNA substitution and 3′ truncation to make a series of scrRNAs and show that activity was comparable with unmodified crRNA with truncation by up to four bases (D-42-01, D-41-01, D-40-01, D-39-01; Figures 3B and 3D), but that a DNA containing crRNA with an 18-nt DNA specificity region (D-38-01; Figures 3B and 3D) lost almost 90% gene disruption activity. From the crystal structure, AsCpf1 Gln286 and crRNA nucleotide position 18 form a hydrogen bond/salt bridge, and a DNA nucleotide at this position may affect this interaction. Finally, we also demonstrate that a slightly longer 45-mer scrRNA, with DNA substitution at (1) the 5′ linker nucleotides (Figure 2C); (2) the 3′ terminal residues; and (3) the internal DNA specificity region positions 1, 8, and 9 is ∼1.6-fold more active than the natural crRNA (D-45-01; Figure 3B; Figure S2). Together, these data support further exploration and development of scrRNAs containing DNA substitutions.
scRNAs with 2′-F-Substituted Nucleotides in the DNA Specificity Region or Direct Repeat Have Opposing Effects on Gene Disruption Activity
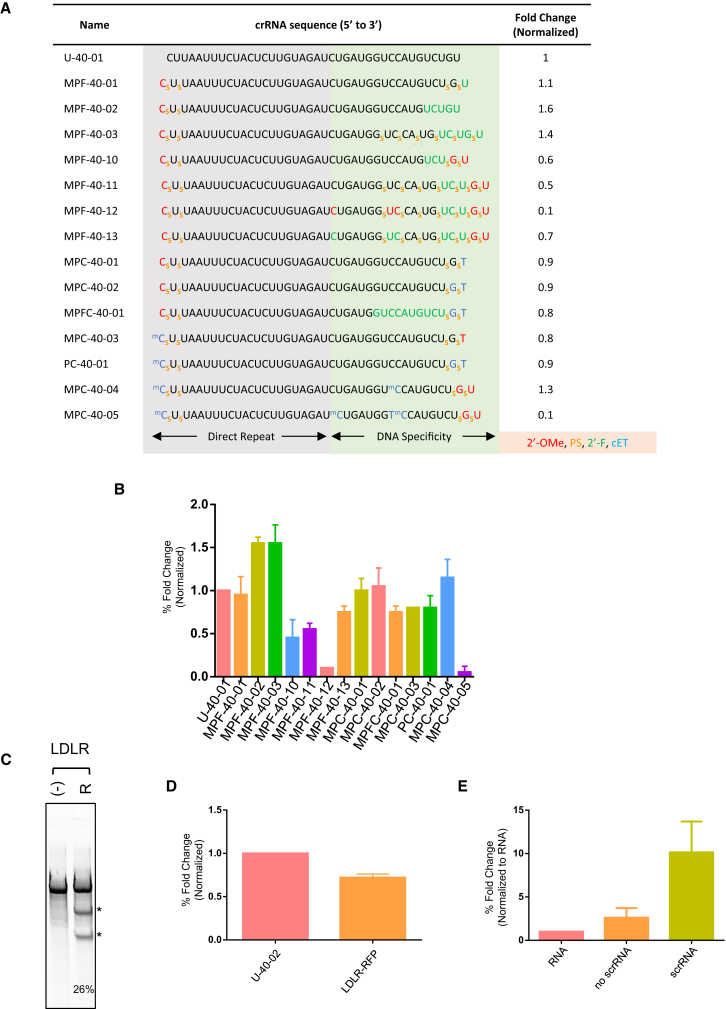
Recognizing that hybrid scrRNAs containing RNA and DNA bases are highly active, including crRNA substituted at internal positions of the DNA specificity region, we next tested whether additional modifications including replacement of the 2′-OH of the sugar with a 2′-F group (Figure 2A) were also active. We chose 2′-F chemically modified nucleic acids because they are known for increasing binding affinity to complementary DNA sequence, for improving metabolic stability, and have a small atomic radius.28 In fact, of the 2′-class of modifications, the 2′-F modification has been shown to impart the highest binding affinity for the complementary DNA target.28, 32 Using the same experimental approach and DNMT1 target sequence, we synthesized a series of scrRNAs with increasing substitution by 2′-F-modified nucleic acids at the 3′ terminus of the DNA specificity region and assessed gene disruption activity. We observed that an scrRNA with a single 3′ terminal 2′-F (MPF-40-01; Figures 4A and 4B; Figure S6) had gene disruption activity comparable with unmodified crRNA (U-40-01). scrRNAs with up to four 2′-F containing residues with or without alternating PS backbone substitutions in the DNA specificity region (MPF-40-02, MPF-40-03; Figures 4A and 4B) retained high levels of gene disruption activity (∼1.5-fold higher) compared with U-40-01 crRNA. Activity was reduced 40%–50% (compared with U-40-01) with scrRNA MPF-40-10 and MPF-40-11, each containing 3′ terminal 2′-O-Me substitutions in addition to internal 2′-F-substituted nucleotides. The combination of 2′-F and 2′-O-Me at the 3′ termini may structurally change the scrRNA so that it either has reduced complexing with AsCpf1 or change the conformation of the AsCpf1-scrRNA complex in an orientation not optimal for binding or cutting target DNA. In addition, we further tested substitution at positions 1, 8, and 9 of the DNA specificity region and found that 2′-F, but not 2′-O-Me, at these positions mediated gene editing comparable with unmodified crRNA (MPF-40-13, MPF 40-12; Figure 4A), confirming our findings with DNA at the same positions while also suggesting that activity is dependent on the type of chemically modified nucleic acid substituted.
Figure 4.
Additional Chemically Modified scrRNAs Mediate Comparable Gene Disruption to Natural crRNA
(A) Complete DNMT1 target sequence with 2′-F nucleotides indicated in green, 2′-O-Me in red, and cEt in blue. Gene disruption activity normalized to U-40-01 (= 1). Data represent gene disruption quantified from surveyor nuclease assay (representative gel in Figure S6) from two or more replicate experiments. (B) Graph shows fold change in gene disruption activity as indicated in last column of (A). (C) Representative surveyor nuclease assay gel for gene editing at the LDLR locus with expected bands indicated by an asterisk (*). (D) Graph shows fold change in LDLR gene disruption activity quantified from surveyor nuclease assay from two replicate experiments. (E) Graph shows quantification of TTR RNA levels measured in triplicate from three biological replicates and normalized to neg (no transfection). No scrRNA bar represents transfection of plasmids at 0 hr as indicated in Figure S7B, but no scrRNA. scrRNA bar represents transfection of plasmids at 0 hr, and addition of scrRNA at 24 hr as indicated in Figure S7B.
We also evaluated selective 2′-F substitutions to the direct repeat based on those positions that were either non-interacting or groove exposed from the crystal structure, presuming that those positions would be more amenable to substitution with chemically modified nucleic acids. To do this, we synthesized a series of scrRNAs with single 2′-F substitutions starting at position U(−16) to U(−1) (Figure S3A). All positions tested with 2′-F substitutions resulted in a 60%–70% reduction in gene disruption activity (Figure S3A).
scrRNAs Containing Bicyclic cEt Nucleic Acid Substitutions Retain High Levels of Gene Disruption Activity
We next tested gene disruption activity of scrRNAs containing replacement of the ribose sugar with the bicyclic nucleotide cEt (Figure 2A). Use of constrained bicyclic analogs like the cEt substitution, which links the 2′ and 4′ positions of the ribose sugar, improve nuclease resistance28 and increase binding to complementary sequences.33 As shown in Figure 4A, gene disruption activity mediated by a PS containing scrRNA with a single 5′ terminal 2′-O-Me containing nucleotide and a single 3′ terminal cEt-substituted nucleotide (MPC-40-01; Figures 4A and 4B) was comparable with unmodified crRNA (U-40-01). Additional 3′ terminal cEt substitution (positions 17 and 18) maintained gene disruption activity comparable with that of unmodified crRNA (MPC-40-02; Figures 4A and 4B). These data suggest that terminal 2′-O-Me and cEt substitutions are interchangeable. In addition, we designed an scrRNA with 10 2′-F-containing nucleotides in the DNA specificity region and demonstrate that gene disruption activity is comparable with that of U-40-01 (MPFC-40-01; Figures 4A and 4B). This is in contrast with the loss of activity we observed with DNA substitutions to the same positions (Figure S2). Also, just as we observed improved gene disruption activity with 5′ terminal 2′-O-Me-containing scrRNAs, replacement with cEt at this position is also well tolerated (MPC-40-03; Figures 4A and 4B). Moreover, cEt substitutions at both the 5′ and 3′ termini retained high levels of gene disruption activity (PC-40-01; Figure 4A) compared with unmodified and single-nucleotide-substituted termini scrRNAs, as expected because modified termini have increased resistance to exonuclease activity and, therefore, are likely more metabolically stable. We tested whether cEt substitution is tolerated at internal crRNA positions 1, 8, and 9, and show that cEt substitution at position 9 further improved gene disruption activity (MPC-40-04; Figures 4A and 4B), but increasing cEt substitution to positions 1 and 8 completely abolished activity (MPC-40-05; Figures 4A and 4B). It is likely that position 1, part of the seed sequence, is sensitive to substitution with locked nucleosides such as cEt because its constrained geometrical shape may affect the conformation of the scrRNA bound to target DNA in such a way that decreases the ability of AsCpf1 to induce a double-stranded break.34
cEt substitution is favored for increasing binding affinity between RNA:RNA interactions, so we additionally did a complete cEt walk of the direct repeat region assuming that it could stabilize the pseudoknot structure and possibly improve gene-editing activity. As shown in Figures S3B–S3D, scrRNAs with cEt substitution at A(−12), C(−11), U(−7), or U(−5) were as active or more active as unmodified crRNA. From the crystal structure, position A(−12) interacts with U(−17), and a cEt substitution could help stabilize the interaction and pseudoknot structure. It is not apparent from the crystal structure how cEt substitutions at the other positions can result in improved gene-editing activity, emphasizing the importance of our strategy to walk each position to better map positions amenable to chemical modification.
Validation of scrRNA Activity Using an Additional Gene Target
To demonstrate the applicability of the chemical designs to additional targets, we first examined gene disruption activity of unmodified crRNA against five additional genes including low-density lipoprotein receptor (LDLR), transthyretin (TTR), complement C5 (C5), empty spiracles homeobox 1 (EMX1), and glutamate ionotropic receptor N-methyl-D-aspartate (NMDA) type subunit 2B (GRIN2b). Gene disruption activity by unmodified crRNAs at their intended locus varied from no activity (crRNA to TTR; Figure S4) to almost 30% gene disruption activity (crRNA to LDLR; Figure 4C). Finding that not all potential AsCpf1 crRNAs can mediate gene disruption is consistent with what has been reported previously9 and because of this, we designed scrRNAs to LDLR. Because the natural crRNA and a chemically modified scrRNA had comparable activity (Figure 4D), we designed a series of LDLR-targeting scrRNAs using chemistries we already identified from DNMT1-targeting scrRNAs.
We show that gene disruption activity of LDLR-targeting scrRNAs containing DNA-substituted nucleic acids has comparable activity to unmodified crRNAs, including those substituted additionally with 2′-O-Me caps (MPD-43-01 and MPD-43-02; Figures S5A and S5B) and slightly longer scrRNAs MPD-45-01 and MPD-45-02 (Figure S5B), which is in agreement with our findings for the DNMT1-targeting scrRNAs. We also tested gene disruption activity of LDLR-targeting scrRNAs with additional substitutions and show that there is a near-complete loss of activity of MPF-40-14-containing 2′-O-Me-substituted nucleotides at DNA specificity region positions 1, 8, and 9, but that activity is restored (up to 60% compared with unmodified crRNA) by replacement of internal 2′-O-Me with 2′-F-substituted nucleic acids (MPF-40-15; Figures S5C and S5D). This again is in agreement with scrRNAs with the same chemical modifications targeting DNMT1 and suggests that these chemistries are applicable to multiple scrRNA sequences.
42-mer scrRNAs Are Cell Permeable in the Absence of Carrier Molecules
Chemical modification of the phosphodiester backbone and sugar moiety have been shown to increase in vivo half-life and to confer drug-like properties to oligonucleotides.23, 24 It has also been shown that chemically modified oligonucleotides, when injected systemically or infused into the cerebral spinal fluid, are rapidly distributed out of the plasma and taken up by cells in many tissues,28, 35, 36, 37, 38 or can be freely taken up by cells in culture.39, 40 To test the ability of scrRNAs to be freely taken up by cells, we used a two-RNA gene activation assay, similar to the synergistic activation mediator (SAM) that was previously described for the single-RNA SpCas9 CRISPR system.41 For SAM, assembly of a nuclease dead SpCas9 fused to a transcription factor assembles at the promoter region of a target gene in an sgRNA-dependent manner and induces gene transcription several orders of magnitude above basal levels.41 Just like SAM, our system assembles gene activation components at the promoter of the target gene, but in a scrRNA-dependent manner, as outlined in Figure S7A.
To test whether large chemically modified RNAs can be freely taken up by cells, we designed a 42-mer scrRNA that specifically recognizes a sequence within 100 bp of the transcription start site of TTR to activate its transcription following the experimental outline in Figure S7B. In brief, plasmids encoding MS2-tracrRNA, nuclease dead Cas9-VP64, and MS2-p65-HSF1 were transfected into HEK293T cells. Twenty-four hours after transfection, cells were washed two times, and the culture media were replaced with media containing 10 μM TTR-specific scrRNA and the cells incubated for a further 24 hr before isolating RNA and measuring TTR RNA levels by quantitative real-time PCR. The 42-mer TTR scrRNA, with chemistries similar to the AsCpf1 scrRNAs and sequence indicated in Figure S7C, activated gene transcription ∼10-fold compared with no RNA control (Figure 4E, scrRNA). These data demonstrate that when added to culture media, large chemically modified RNAs can be freely taken up by cells in culture in the absence of carrier molecules.
Discussion
In this report, we designed and tested scrRNAs of the AsCpf1 system and show that: (1) truncation of the 43-mer crRNA to a 36-mer or 37-mer crRNA retains high levels of gene-editing activity; (2) truncated 37-mer and 40-mer scrRNAs substituted with PS, 2′-O-Me, 2′-F, cEt, or DNA nucleotides at the terminal 5′ and 3′ ends or internal positions mediate high levels of gene editing; (3) terminally modified scrRNAs are resistant to exonuclease activity; and (4) 42-mer scrRNAs are freely taken up by cells in culture in the absence of carrier molecules. Our data suggest that the crRNA pseudoknot structure is sensitive to chemical modification, resulting in loss of biological function, but that terminal and many internal positions in the DNA specificity region, outside of the seed sequence, can be chemically substituted, and that these modifications help provide protection against ribonucleases. Based on the similarity in size and chemistry to RNAs currently being used for therapeutic applications,23, 24, 27 it is expected that the proposed scrRNAs will have greater in vivo stability and distribution compared with the natural crRNA, and a direct test of this in animal models is a next step.
Development of gene-editing technology has moved very rapidly, with CRISPR-Cas quickly becoming the most commonly used engineered nuclease in biomedical research, preferred over zinc-finger nucleases and transcription activator-like effector nucleases, because of its ease of use, multiplexing capabilities, and high success rate.42 It is transforming biomedical research by enabling manipulation of endogenous protein function through gene disruption, tagging, and/or correction.13, 14 Although still an emerging field, CRISPR technology as a potential therapeutic holds the promise for curing genetic diseases in patients who have no other treatment options. Proof-of-principle studies, including for those treating genetic diseases of the nervous system,21 initiated in cell-43, 44 and animal-based45, 46 models, have demonstrated not only efficacy at improving disease phenotype, but also feasibility of using CRISPR-Cas as a viable therapeutic strategy. In vivo delivery systems have included using recombinant adeno-associated virus (AAV) encoding CRISPR-Cas system components,8, 46 RNA-mediated delivery,47, 48 or lipid nanoparticles (LNPs) of sgRNA and SpCas9 mRNA.49 AAV-based therapeutics has experienced a rapid rise with several clinical examples demonstrating its safety and efficacy for gene delivery in patients50, 51 and more than 100 trials in early stages according to clinicaltrials.gov. But in the context of CRISPR systems, current viral delivery approaches do not allow for tunable alteration of gene expression, but do produce continuous, long-term activation of the nuclease. This activation enables high levels of gene editing at the intended target but without the ability to turn off expression of the nuclease, and also increases the likelihood of off-target gene editing.11, 12 RNA-mediated delivery of CRISPR-Cas components, such as ribonucleoprotein complexes (RNPs), has also gained momentum in the CRISPR field.47, 48 Because RNPs are present for only a short window of time compared with transcription produced CRISPR-Cas components, there may be a limit to the maximal on-target gene editing achievable, as well as limitations of systemic delivery.52, 53 LNP-mediated delivery of CRISPR components has shown success in the mouse liver49 and may prove to be advantageous over RNPs for broader delivery to target tissues.
Regardless of whether using a viral or non-viral delivery system, chemically modified crRNAs may enable tunable and enhanced in vivo gene editing. Synthetic, chemically modified oligonucleotides of similar size have been shown to be safe in rodents, non-human primates and humans, and when infused into the cerebral spinal fluid can be widely distributed throughout the CNS and taken up by both neurons and non-neurons.36, 38 In vivo delivery of AAV-encoded AsCpf1 and a gene-specific scrRNA could provide the gene-editing control needed as the duration of AsCpf1 nuclease activity is controllable by dosing of the scrRNA (single dose or multiple doses). We showed that scrRNAs are resistant to ribonuclease activity, so it is anticipated that RNPs complexed with scrRNAs will be more stable in vivo than complexes containing unmodified RNA. This is clearly an important next step. Finally, it was recently shown that LNP encapsulated SpCas9 mRNA and chemically modified sgRNA enhanced gene editing in the mouse liver compared with the natural unmodified RNA.49 Given that the same chemical modifications were introduced into our AsCpf1 crRNA, a similar enhancement is expected and should be tested.
Since the first reports describing use of SpCas9 for mammalian genome editing,4, 5, 6 many new CRISPR systems have been identified,3 as well as adaptation of the technology for new applications including using CRISPR systems for genome scale transcriptional activation or repression,41, 54 visualization of genomic loci,55, 56 targeting RNA (as in C2c2),57 and now for nucleic acid detection.58 Chemical modifications to the guide RNA can increase binding affinity to complementary sequence, provide protection from nuclease degradation, and increase distribution to target tissues, which if applied to these other CRISPR applications may be advantageous over unmodified RNA, expanding their use beyond simply providing specificity for mediating double-stranded breaks.
Using the crystal structure as a guide, we designed and tested an extensive set of AsCpf1 scrRNAs and identify several highly active and ribonuclease-resistant compounds. Sensitivity to chemical substitution at several crRNA positions was not expected based on the crystal structure, emphasizing the importance of the work done here to experimentally test and validate scrRNA activity. We cannot rule out that additional chemical modifications to the ones tested here may provide further benefits in vivo or that different chemistries may be required for maximizing scrRNA distribution and cellular uptake in different tissues. Nevertheless, using human cell lines, we have established a foundation for testing efficacy and safety of AsCpf1 scrRNAs in mice and for developing their use in viral and non-viral therapeutic strategies.
Materials and Methods
scrRNA Synthesis
scrRNAs were synthesized using a previously published protocol.59 In brief, scrRNAs were synthesized on a solid-phase DNA/RNA synthesizer using the tert-butyldimethylsilyl (2′-O-TBDMS) RNA phosphoramidites according to the reported protocols. 2′-F, 2′-O-Me, cEt, and 2′-O-MOE phosphoramidites with exocyclic amino groups protected with benzoyl (Bz for A and C) or isobutyryl (ibu for G) protecting groups were used for the synthesis of the RNA chimera. 0.12 M solution of the phosphoramidites in anhydrous acetonitrile was used for the synthesis. Oxidation of the internucleosidic phosphite to the phosphate was carried out using tert-butyl hydroperoxide/acetonitrile/water (10:87:3) with a 10-min oxidation time. PS linkages were introduced by sulfurization with a 0.1 M solution of xanthane hydride in 1:1 pyridine/CH3CN for a contact time of 3 min. The overall coupling efficiency of all modified phosphoramidites was more than 97%. Oligoribonucleotide-bearing solid supports were heated with aqueous ammonia/ethanol (3:1) solution at 55°C for 6 hr to deprotect the base labile-protecting groups. The 2′-O-TBDMS group was removed using a mixture of triethylamine trihydrofluoride/1-methyl-2-pyrrolidinone/triethyl amine mixture at 65°C for 6 hr. After deprotection, all the scrRNAs were isolated by HPLC on a strong anion exchange column and subsequently desalted using high-performance liquid chromatography (HPLC) on a reverse-phase column to yield scrRNAs. The scrRNAs were characterized by ion-pair-HPLC-mass spectrometry (MS) analysis.
Cell Lines and Transfection for Gene Disruption
HEK293T cells (ATCC), cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) and supplemented with 25 mM HEPES, antibiotic-antimycotic (10,000 U/mL penicillin, 10,000 μg/mL streptomycin, and 25 μg/mL Fungizone), and 10% FBS (Omega Scientific, Tarzana, CA) were transfected in six-well plates using 2 μg of plasmid encoding AsCpf1 protein [cloned by Gibson assembly of gene fragments (Integrated DNA Technologies, Coralville, IA, USA) into pcDNA3.1(-)] using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s instructions. Twenty-four hours after DNA transfection, cells were washed one time with culture media and transfected a second time with 3 μL of 100 μM scrRNA or unmodified RNA using RNAi max (Thermo Fisher Scientific, Waltham, MA) per the manufacturer’s instructions.
Gene Fragment Amplification
gBlock gene fragments (Integrated DNA Technology, San Diego, CA, USA), designed to encode the U6 promoter and DNMT1 minimal crRNA targeting sequence to be tested (Figure S7), were cloned into zero blunt PCR4 topo vector (Thermo Fisher Scientific, Waltham, MA, USA). Forward primer 5′-AAGGTCGGGCAGGAAGAG-3′ and variable reverse primers as indicated by underlined sequence in Figure S7 were used for PCR amplification using plasmid DNA isolated from a single colony and sequence verified as template (Q5 polymerase; New England Biolabs, Ipswich, MA, USA). PCR products were then gel purified (Zymo Research, Irvine, CA, USA) and 100 ng of product was used for co-transfection with 2 μg of plasmid encoding AsCpf1 protein using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Surveyor Nuclease Assay
The Surveyor nuclease assay was performed per manufacturer’s instructions (Integrated DNA Technology, San Diego, CA, USA). In brief, 24 hr after scrRNA transfection, genomic DNA was isolated using the Quick DNA miniprep kit (Zymo Research, Irvine, CA, USA) and 100 ng used as a template for PCR (all primer sequences in Figure S8) using Q5 polymerase (New England Biolabs, Ipswich, MA, USA) and DNMT1 primers: forward 5′-CTGGGACTCAGGCGGGTCAC-3′ and reverse 5′-CCTCACACAACAGCTTCATGTCAGC-3′ in a 50-μL reaction. Twelve microliters of the PCR was transferred to a new tube, melted to 95°C, and allowed to slowly anneal to room temperature using a T100 thermocycler (Bio-Rad, Irvine, CA, USA) and a stepwise temperature gradient program. Annealed products were then subjected to a 1-hr incubation at 42°C using the Surveyor nuclease kit (Integrated DNA Technologies, San Diego, CA, USA), products separated on a 10% TBE acrylamide gel (Bio-Rad, Irvine, CA, USA), stained with SYBRgold (Thermo Fisher Scientific) for visualization, and quantified using Image lab software using the following indel formula: indel (%) = 100 × [1 − (1 − fraction_cut)0.5]. Fraction_cut = (sum of the intensity of the cleaved product)/(sum of the intensity of cleaved product plus undigested product).
scrRNA Exonuclease Biochemical Assay
RNase T reactions were performed in 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, and 1 mM DTT (NEB Buffer 4) in a total volume of 100 μL. RNA and corresponding scrRNA were diluted in water to a concentration of 100 nM, and 3 μL was added to each 100-μL reaction (3 nM final). 3 U of RNase T (NEB) was added to each reaction, and 7 μL was removed at corresponding time points, added to 2X TBE-UREA sample buffer (Thermo Fisher Scientific), and heated to 95°C for 10 min. Reaction products were visualized by electrophoresis using 15% TBE-UREA denaturing polyacrylamide gels and stained with SYBRgold (Thermo Fisher Scientific).
Cell Lines and Transfection for Gene Activation
HEK293T cells (ATCC, Manassas, VA, USA), cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) and supplemented with 25 mM HEPES, antibiotic-antimycotic (10,000 U/mL penicillin, 10,000 μg/mL streptomycin, and 25 μg/mL Fungizone), and 10% FBS (Omega Scientific, Tarzana, CA, USA) were transfected in six-well plates using a total of 2 μg of plasmid encoding tracrRNA 2.0 driven by the U6 promoter (sequence 5′-GTTGGAACCATTCAAAACAGCATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGGCCAACATGAGGATCACCCATGTCTGCAGGGCCAAGTGGCACCGAGTCGGTGCTTT-3′), nuclease dead Cas9-VP64, or MS2-p65-HSF1 in a 1:1:1 molar ratio using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Twenty-four hours after DNA transfection, cells were washed two times and replaced with culture media containing 10 μM TTR-specific 42-mer scrRNA. RNA was isolated 24 hr later using the Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA, USA) and quantitative real-time PCR performed using GAPDH forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse primer 5′-GAAGATGGTGATGGGATTTC-3′, and probe 5′-CAGCTTCCCGTTCTCAGCCX-3′ and TTR forward primer 5′-CCCTGCTGAGCCCCTACTC-3′, reverse primer 5′-TCCCTCATTCCTTGGGATTG-3′, and probe 5′-ATTCCACCACGGCTGTCGTCAX-3′ and the Express One-Step Superscript qRT-PCR kit (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical Analysis
Statistics were performed with the Prism 6.0 statistical software package (GraphPad). Replicate error was assessed by calculating SD on the mean, where the replicates are greater than or equal to 2.
Author Contributions
M.A.M., T.P.P., D.W.C., C.F.B., and M.R. designed experiments; M.A.M. and M.R. conducted experiments; M.A.M., T.P.P., and M.R. contributed new reagents; M.A.M., T.P.P., C.F.B., and M.R. analyzed data; and M.A.M., T.P.P., D.W.C., C.F.B., and M.R. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Eric Swayze (Ionis Pharmaceuticals) for helpful input on scrRNA chemistry design and Beth Cisar (Ionis Pharmaceuticals) for review of the manuscript. This work was in part supported by grants R01-GM 074150 and R01-NS27036 from the NIH to D.W.C., who receives salary support from the Ludwig Institute for Cancer Research.
Footnotes
Supplemental Information includes nine figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.02.031.
Supplemental Information
References
- 1.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 3.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D., Kim J., Hur J.K., Been K.W., Yoon S.-H., Kim J.-S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon M.A., Rahdar M., Porteus M. Gene editing: not just for translation anymore. Nat. Methods. 2011;9:28–31. doi: 10.1038/nmeth.1811. [DOI] [PubMed] [Google Scholar]
- 14.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg S.H., Doudna J.A. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y., Cheong S.-A., Lee J.G., Lee S.-W., Lee M.S., Baek I.-J., Sung Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 19.Hur J.K., Kim K., Been K.W., Baek G., Ye S., Hur J.W., Ryu S.M., Lee Y.S., Kim J.S. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 2016;34:807–808. doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- 20.Cox D.B.T., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon M.A., Cleveland D.W. Gene therapy: gene-editing therapy for neurological disease. Nat. Rev. Neurol. 2017;13:7–9. doi: 10.1038/nrneurol.2016.190. [DOI] [PubMed] [Google Scholar]
- 22.Rahdar M., McMahon M.A., Prakash T.P., Swayze E.E., Bennett C.F., Cleveland D.W. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. USA. 2015;112:E7110–E7117. doi: 10.1073/pnas.1520883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A., Corey D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 24.Chiu Y.-L., Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 28.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 29.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 30.Deutscher M.P., Marlor C.W., Zaniewski R. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc. Natl. Acad. Sci. USA. 1984;81:4290–4293. doi: 10.1073/pnas.81.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deutscher M.P., Marlor C.W. Purification and characterization of Escherichia coli RNase T. J. Biol. Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- 32.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen X., Corey D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–1600. doi: 10.1093/nar/gkx1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 35.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 36.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.-H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 41.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 43.Park C.-Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 2015;13:234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- 44.Shin J.W., Kim K.-H., Chao M.J., Atwal R.S., Gillis T., MacDonald M.E., Gusella J.F., Lee J.M. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum. Mol. Genet. 2016;25:4566–4576. doi: 10.1093/hmg/ddw286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W., Lu Z., Li F., Wang W., Qian N., Duan J., Zhang Y., Wang F., Chen T. Efficient in vivo gene editing using ribonucleoproteins in skin stem cells of recessive dystrophic epidermolysis bullosa mouse model. Proc. Natl. Acad. Sci. USA. 2017;114:1660–1665. doi: 10.1073/pnas.1614775114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staahl B.T., Benekareddy M., Coulon-Bainier C., Banfal A.A., Floor S.N., Sabo J.K., Urnes C., Munares G.A., Ghosh A., Doudna J.A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H., Song C.-Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasi J., Rangarajan S., Walsh L., Lester W., Perry D., Madan B. Interim results from a phase1/2 AAV5-FVIII gene transfer in patients with severe hemophilia A. Res. Pract. Thromb. Haemost. 2017;1:1–15. [Google Scholar]
- 51.Shell R., Al-Zaidy S., Arnold W.D., Rodino-Klapac L., Prior T.W., Lowes L., Alfano L., Berry K., Church K., Kissel J.T. AVXS-101 phase 1 gene therapy clinical trial in SMA type 1: interim data demonstrates improvements in supportive care use. Eur. J. Paediatr. Neurol. 2017;21(Suppl 1):e14. [Google Scholar]
- 52.Lang A.E., Gill S., Patel N.K., Lozano A., Nutt J.G., Penn R., Brooks D.J., Hotton G., Moro E., Heywood P. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann. Neurol. 2006;59:459–466. doi: 10.1002/ana.20737. [DOI] [PubMed] [Google Scholar]
- 53.Bradley L.H., Fuqua J., Richardson A., Turchan-Cholewo J., Ai Y., Kelps K.A., Glass J.D., He X., Zhang Z., Grondin R. Dopamine neuron stimulating actions of a GDNF propeptide. PLoS One. 2010;5:e9752. doi: 10.1371/journal.pone.0009752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanenbaum M.E., Gilbert L.A., Qi L.S., Weissman J.S., Vale R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prakash T.P., Allerson C.R., Dande P., Vickers T.A., Sioufi N., Jarres R., Baker B.F., Swayze E.E., Griffey R.H., Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.