Abstract
Dysregulated mRNA translation and aberrant energy metabolism are frequent in cancer. Considering that mRNA translation is an energy demanding process, cancer cells must produce sufficient ATP to meet energy demand of hyperactive translational machinery. In recent years, the mammalian/mechanistic target of rapamycin (mTOR) emerged as a central regulatory node which coordinates energy consumption by the translation apparatus and ATP production in mitochondria. Aberrant mTOR signaling underpins the vast majority of cancers whereby increased mTOR activity is thought to be a major determinant of both malignant translatomes and metabolomes. Nonetheless, the role of mTOR and other related signaling nodes (e.g. AMPK) in orchestrating protein synthesis and cancer energetics is only recently being unraveled. In this review, we discuss recent findings that provide insights into the molecular underpinnings of coordination of translational and metabolic programs of cancer cells, and potential strategies to translate these findings into clinical treatments.
Keywords: mRNA translation, energy metabolism, mTOR, cancer
Introduction
The dysregulation of mRNA translation is a prominent characteristic of cancer cells (Bhat et al., 2015; Pelletier et al., 2015). Elevated protein synthesis is required to support neoplastic growth, which consequently results in a high energy demand (Buttgereit and Brand, 1995; Morita et al., 2015; Rolfe and Brown, 1997). As mRNA translation is one of the most energetically demanding process in the cell, cancer cells require sufficient ATP production to maintain elevated mRNA translation rates (Buttgereit and Brand, 1995; Topisirovic and Sonenberg, 2011). Protein synthesis occurs in four major steps: initiation, elongation, termination and ribosome recycling (Hershey et al., 2012). Each step of mRNA translation comprises a complex interplay between mRNA, ribosomes, transfer RNAs and auxiliary proteins also known as translation factors that function together in a highly orchestrated manner to generate newly synthesized proteins (Hershey et al., 2012) (Figure 1).
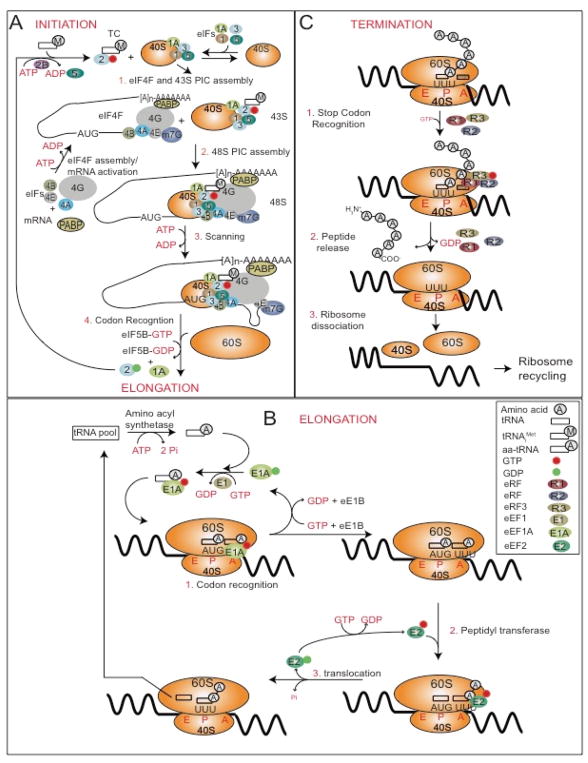
Figure 1. Energy consumption by the eukaryotic translational machinery.
In eukaryotes, protein synthesis occurs in four major steps: initiation, elongation, termination and ribosome recycling. (A) Step 1: Initiation. Initiation requires the assembly of the 43S pre-initiation complex (PIC) and eIF4F (1). The 5′ capped mRNA is activated in an ATP-dependent manner by eIF4F. The 48S PIC is assembled by association of 43S PIC and the eIF4F complexes (2). As a part of eIF4F, eIF4A unwinds 5′UTR in an ATP-dependent manner while the 5′UTR is scanned in the 5′-->3′ direction (3). Recognition of the translation initiation codon triggers hydrolysis of GTP from the ternary complex (TC) resulting in TC release (4). This is followed by the dissociation of other initiation factors (eIFs). eIF5B accelerates the release of eIFs and the joining of the 60S ribosomal subunit which is accompanied by the hydrolysis of an additional GTP (4). (B) Step 2: Elongation. Aminoacyl-tRNAs (aa-tRNA) are recruited by elongation factor (eEF) 1A. The anticodon of the incoming aa-tRNA is matched against the mRNA codon positioned in the A site resulting in the hydrolysis of GTP which is stimulated by eIF1B leading to the release of eEF1A (1). The growing polypeptide chain is covalently linked to the new amino acid, leaving an empty tRNA in the P site (2). As the mRNA moves one codon forward, the empty tRNA from the P site is displaced to the E site as the peptidyl tRNA is translocated into the P site which is facilitated by eEF2 and requires GTP hydrolysis. tRNAs are aminoacylated by aminoacyl tRNA synthetase, which requires hydrolysis of ATP to AMP (3). These steps are repeated until the ribosome encounters an in-frame stop codon. (C) Step 3: Termination. An in-frame stop codon is positioned in the A site (1). Release factors (eRFs) 1, 2 and 3 assemble with GTP forming a complex near the A site (1). Upon recognition of the stop codon by eRF1 and eRF2, GTP hydrolysis is triggered by eRF3 resulting in the release of the polypeptide chain (2). eRFs are released followed by the dissociation of the 40S, 60S ribosomal subunits and mRNA (3). The ribosomal subunits are then recycled. Abbreviations: eIF, eukaryotic initiation factor, eRF, eukaryotic release factor, eEF, eukaryotic elongation factor, PIC, preinitiation complex, TC, ternary complex, PABP, poly(A) binding protein, tRNAiMet, initiator tRNA, M7G, 7-methylguanylate cap.
Translation and energy demand
During the initiation of mRNA translation in both eukaryotes and prokaryotes, a number of initiation factors recruit the initiator tRNA (tRNAiMet in eukaryotes) and mRNA to the small ribosomal subunit (Hinnebusch, 2014; Voigts-Hoffmann et al., 2012). The initiator tRNA is positioned in the P site of the small ribosomal subunit upon recognition of the start codon which is followed by joining of the large ribosomal subunit to form the translationally competent ribosome (Hinnebusch, 2014; Voigts-Hoffmann et al., 2012). Although the general mechanisms of translation in prokaryotes and eukaryotes exhibit some resemblance, eukaryotes utilize many more initiation factors, larger ribosomal complexes and more energy. In eukaryotes, cap-dependent initiation of translation of most cellular mRNAs occurs by the scanning mechanism which requires the formation of a 43S pre- initiation complex (PIC) consisting of eukaryotic translation initiation factors (eIF) 1, 3, 5, the 40S ribosomal subunit and the ternary complex (TC) which comprises GTP bound eIF2 and tRNAiMet (Hinnebusch, 2014). The eIF4F cap-binding complex, which contains eIF4A DEAD box helicase, eIF4E cap-binding protein, and eIF4G which acts as a scaffold, recruits 43S PIC to the ribosome via the interaction of eIF4G and eIF3, which leads to the 48S PIC assembly (Hinnebusch, 2014). This stimulates 43S PIC scanning of the mRNA 5′ UTR towards the start codon (Hinnebusch, 2014). 43S PIC scanning requires the removal of secondary structure present in the 5′UTR which is achieved by eIF4A and requires ATP hydrolysis (Rogers et al., 1999; Svitkin et al., 2001). Recognition of the start codon triggers hydrolysis of GTP from the TC resulting in its release followed by the dissociation of other eIFs and joining of the 60S ribosomal subunit, which is stimulated by initiation factor 5B, resulting in the hydrolysis of an additional GTP (Hinnebusch, 2014). In prokaryotes, initiation of protein synthesis is much simpler whereby the recruitment of the initiator tRNA (N-formyl methionine tRNA) to the 30S ribosomal subunit does not require the eIF4F complex assembly nor complex scanning mechanisms (Wintermeyer and Gualerzi, 1983). This makes initiation in eukaryotes more energetically expensive, inasmuch as it requires two GTP for the recycling of the TC and the formation of the 80S monosome and one ATP for mRNA activation, as compared to one GTP utilized by prokaryotes when forming the 70S monosome (Laursen et al., 2005; Voigts-Hoffmann et al., 2012). In contrast to initiation, elongation of mRNA translation is well conserved between eukaryotes and prokaryotes (Rodnina and Wintermeyer, 2009). Eukaryotic elongation factor 1 (eEF1) consists of eEF1A and eEF1B, wherein eEF1A (homologous to EF-Tu in prokaryotes) delivers amino-acyl tRNAs (Voigts-Hoffmann et al., 2012) to the A site of the ribosome (Carvalho et al., 1984). Upon proper codon recognition, rapid hydrolysis of eEF1A-bound GTP induced by eEF1B causes the release of eEF1 resulting in the formation of a new peptide bond (Voorhees et al., 2010). Secondary to peptide-bond formation, eEF2 (EF-G in prokaryotes), facilitates the translocation of the ribosome to free the A-site, whereas uncharged tRNA is transferred to the E-site which requires hydrolysis of another GTP molecule (Stark et al., 2000; Taylor et al., 2007). The tRNA itself is recycled by the amino acyl synthetase complex that requires hydrolysis of ATP to AMP, which is equivalent to two ATP molecules (Han et al., 2003) (Figure 1). This makes elongation the most energetically demanding step of mRNA translation requiring a total of two ATPs and two GTPs (Ibba and Soll, 1999) (Figure 1). The termination step of mRNA translation requires hydrolysis of GTP for the release of the nascent polypeptide, however the mechanism is quite different between prokaryotes and eukaryotes. In eukaryotes, release factors are likely part of the ribosome complex (Pisareva et al., 2006) where eukaryotic release factor 1 (eRF1) triggers peptidyl-tRNA hydrolysis while eRF3 accelerates this process. Upon recognition of the stop codon eRF1 stimulates GTP hydrolysis by eRF3 which facilitates the release of the nascent polypeptide (Alkalaeva et al., 2006) (Figure 1). In prokaryotes, release factors are recruited to the ribosome upon recognition of the stop codon and promote release of the nascent polypeptide followed by GTP hydrolysis (Zavialov and Ehrenberg, 2003). Collectively, these data indicate that mRNA translation imposes a significant energetic burden which cells must resolve by orchestrating protein synthesis rates and ATP production.
mTOR dictates mRNA translation and metabolic reprograming of cancer cells
As noted above, the complexity of the initiation increased more dramatically throughout evolution as compared to the other phases of mRNA translation (Malys and McCarthy, 2011). This, in conjunction with high energy demand of elongation, suggests that initiation represents the rate-limiting step of protein synthesis wherein the most of regulation takes place to prevent extensive energy consumption by the translational apparatus (Chu et al., 2016; Sonenberg and Hinnebusch, 2009). Initiation is therefore tightly regulated, which is in a large part achieved by the mammalian/mechanistic target of rapamycin (mTOR) (Figure 2). mTOR adjusts global protein synthesis rates as well as the composition of the translatome in response to a number of environmental stimuli and intracellular cues to support cellular growth and proliferation (Efeyan et al., 2015; Morita et al., 2015; Saxton and Sabatini, 2017). More recently, the findings pointing out the importance of the mTOR-dependent regulation of the elongation step of translation in the context of energy homeostasis are starting to emerge (Leprivier et al., 2015; Proud, 2015). Accordingly, frequent hyperactivation of mTOR in cancer is thought to underpin dysregulation of translation and energy metabolism which fuel neoplastic growth (Bhat et al., 2015).
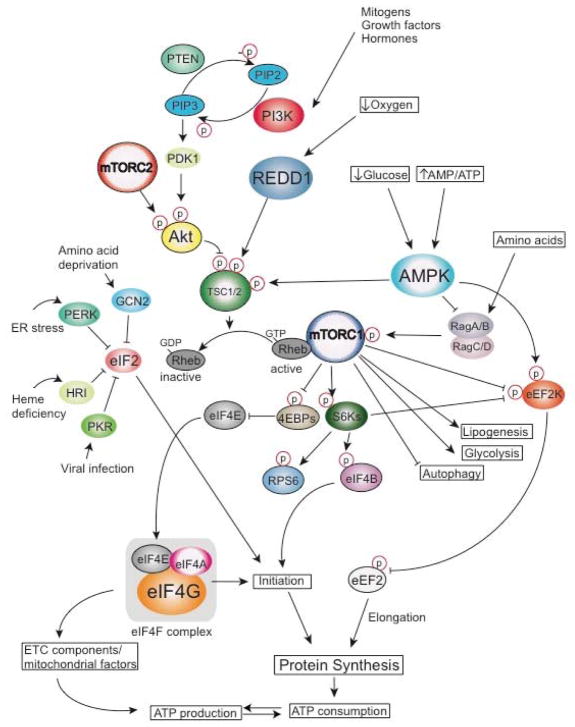
Figure 2. Simplified scheme of signaling pathways that coordinate energy production and protein synthesis.
The mammalian/mechanistic target of rapamycin (mTOR) pathway emerged as a pivotal regulator of protein synthesis and energy metabolism. It is present in at least two functionally and structurally distinct complexes mTORC1 and mTORC2. mTORC1 integrates a number of signals via various upstream pathways. For instance, hormones and growth factors (e.g. insulin and IGFs) which activate receptor tyrosine kinases (e.g. insulin receptor) lead to activation of PI3K which via AKT inactivates TSC1/2 complex. TSC1/2 complex acts as a GAP (GTPase-activating protein) towards the Ras homologue enriched in brain (RHEB) GTPase, which converts RHEB-GTP to its inactive RHEB-GDP form thus preventing activation of mTORC1. In addition, nutrients and in particular amino acids activate mTORC1 via RAG GTPases, while the effects of oxygen tension and energy status in the cell on mTORC1 activity are mediated by REDD1 and AMPK, respectively. mTORC1 stimulates translation by modulating the activity of its downstream effectors including S6Ks, 4EBPs and eEF2K. mTOR simultaneously perturbs other metabolic processes including induction of lipogenesis and glycolysis, and suppression of autophagy. Increase in energy consumption under conditions wherein mTOR is activated is compensated by the perturbations in the translatome that allow selective increase in translation of the nuclear-encoded mRNAs that encode proteins that bolster mitochondrial number and functions. In addition to mTOR, a number of stress conditions including amino acid deprivation, ER stress, heme deficiency and viral infection translation is downregulated via eIF2α kinases which phosphorylate eIF2α and impedes the recycling of ternary complex. Emerging results suggest that the activity of AMPK, mTOR and/or eIF2α phosphorylation may be co-regulated. Abbreviations: PDK1, 3-phosphoinositide-dependent protein kinase-1, RAG, Ras-related GTP-binding protein, RPS6, ribosomal protein S6, PIP3, Phosphatidylinositol (3,4,5)-trisphosphate, PIP2, Phosphatidylinositol 4,5-bisphosphate, eIF, eukaryotic initiation factor, eEF, eukaryotic elongation factor.
mTOR is a serine/threonine kinase that plays a critical role in regulating cell growth and proliferation (Figure 2) (Saxton and Sabatini, 2017). In mammals, mTOR exists in at least two different complexes: mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Saxton and Sabatini, 2017). Both complexes share the catalytic subunit mTOR, GTPase β-subunit like protein GβL (also known as mLST8, which is orthologous to LST8 in S. cerevisiae) and negative regulator DEPTOR (disheveled, Egl-10, pleckstrin [DEP] domain containing mTOR interacting protein), whereas RAPTOR (regulatory-associated protein of TOR) and RICTOR (rapamycin-insensitive companion of TOR); mSIN1 (mammalian stress-activated protein kinase (SAPK)-interacting protein) and protor (Proline-rich protein 5, also known as PRR5) are mTORC1- and mTORC2-specific components, respectively (Saxton and Sabatini, 2017). mTORC1 is the better studied of the two complexes and it senses alteration in nutrients (e.g. amino acids, glucose), oxygen levels, growth factors (e.g. IGFs), and hormones (e.g. insulin) via RAG GTPases and the Phosphoinositide 3-kinase (PI3K)/AKT/tuberous sclerosis complex (TSC)/Ras-homologue enriched in brain (RHEB) pathway (Ben-Sahra and Manning, 2017; Bond, 2016; Saxton and Sabatini, 2017; Wolfson and Sabatini, 2017) (Figure 2). Moreover, energy depletion resulting in increased AMP/ATP ratio and hypoxia both reduce mTORC1 activity via AMP-activated protein kinase (AMPK) (Bolster et al., 2002; Hardie et al., 2012; Inoki et al., 2006) and are regulated in development and DNA damage responses (REDD1), respectively (Brugarolas et al., 2004). mTORC1 mainly induces anabolic processes such as protein and lipid synthesis, stimulates glycolysis and inhibits autophagy, thereby promoting cell growth and proliferation (Duvel et al., 2010; Saxton and Sabatini, 2017) (Figure 2). In most cell lines, mTORC1 also appears to be more sensitive to naturally occurring allosteric inhibitor rapamycin than mTORC2, at least during acute treatment (Sarbassov et al., 2006). Upstream regulators of mTORC2 are largely unknown, and its functions are chiefly mediated by AGC kinases [e.g. AKT, protein kinase C (PKC) and serum and glucocorticoid-regulated kinase 1 (SGK1)] which regulate cytoskeletal organization and survival (Destefano and Jacinto, 2013; Su and Jacinto, 2011). Moreover, mTORC2 has been shown to associate with ribosomes where it is thought to regulate stability of newly synthesized polypeptides (Zinzalla et al., 2011) and regulate lipid and glucose metabolism (Hagiwara et al., 2012; Lamming and Sabatini, 2013; Masui et al., 2013).
Downstream effectors of mTOR-dependent orchestration of cancer translatome and energetics
Since other mTORC1 effectors and their impact on translation and metabolism are covered in several excellent recent reviews (Ben-Sahra and Manning, 2017; Bond, 2016; Saxton and Sabatini, 2017; Thoreen, 2017), we will focus on recent findings highlighting the mTORC1/4E-BP and mTORC1/S6K/eEF2K axis as major nodes which orchestrate energy metabolism and protein synthesis.
The mTORC1/4E-BP axis
mTORC1 phosphorylates and inactivates translational suppressors 4E-binding proteins (4E-BP1-3 in humans) (Burnett et al., 1998; Gingras et al., 1999; Hara et al., 1997; Roux and Topisirovic, 2012; von Manteuffel et al., 1996). 4E-BPs bind to mRNA 5′ cap binding protein eIF4E thereby impeding assembly of the eIF4F complex and recruitment of the mRNA to the ribosome (Pause et al., 1994). This leads not only to a decrease in global protein synthesis rates, but also to a selective increase in synthesis of a subset of proteins including those with mitochondrial functions [e.g. electron transport chain (ETC) components, NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 (NDUF6), ATP synthase subunit O (ATP5O), and ATP synthase subunit D (ATP5D)] (Larsson et al., 2012; Morita et al., 2013). The unifying features of the vast majority of these mRNAs that encode proteins with mitochondrial function is that they harbor short 5′UTRs (<40 nucleotides), which appears to render translation of these transcripts exceptionally sensitive to changes in levels and/or activity of eIF4E, but not eIF4A component of the eIF4F complex (Elfakess et al., 2011; Gandin et al., 2016b; Sinvani et al., 2015). Notably, eIF1 prevents translation from 5′mRNA cap proximal start codons by inducing leaky scanning and favoring the open, scanning competent state of the 43S preinitiation complex (Hinnebusch, 2014; Hinnebusch et al., 2016). Recently it has been shown that a subset of mRNAs with short 5′UTRs, including those containing the Translation Initiator of Short 5′ UTR (TISU) element, efficiently initiate from 5′ mRNA cap-proximal start codons (Elfakess et al., 2011). Although the precise mechanism of this cap-dependent but scanning free process of translation initiation remains largely unknown, it appears that it encompasses of interactions between eIF1 and eIF4G (Sinvani et al., 2015) as well as eIF1A-directed association of ribosomal protein S3 (RPS3) and RP10a with TISU element (Haimov et al., 2017). This selective increase in translation of factors which are involved in mitochondrial functions enhances ATP production. Therefore, translational reprograming caused by mTORC1 activation increases synthesis of ETC components and other factors with mitochondrial functions (e.g. TFAM), which stimulates energy production required to fuel protein synthesis as well as other anabolic processes in the cells, ultimately stimulating cell proliferation and growth (Gandin et al., 2016b).
The mTORC1/S6K/eEF2K axis
In addition to 4E-BPs, eEF2 kinase (eEF2K) appears to play a major role in coordinating protein synthesis rates and cancer energetics (Ryazanov, 2002) (Figure 2). eEF2K phosphorylates eEF2 on Thr56 in humans and inhibits its ribosome association, thereby preventing ribosome translocation and attenuating elongation (Carlberg et al., 1990). In turn, mTORC1 increases elongation rates via phosphorylating and inactivating eEF2K (at Ser 366 in humans), through the action of ribosomal protein S6 kinases (S6Ks) (Wang et al., 2001). mTORC1 has also been shown to directly phosphorylate eEF2K (Ser 78 and 359 in humans), which leads to its inactivation (Browne and Proud, 2004; Smith and Proud, 2008). Under physiological conditions, energy depletion in the muscle stimulates release of calcium and consequently increases eEF2K association with calmodulin (Kenney et al., 2014). This results in the activation of eEF2K and subsequent reduction in ATP consumption by the translation machinery (Kenney et al., 2014). Understanding of the biological consequences of eEF2K phosphorylation in cancer however is still largely incomplete. In general, protein synthesis correlates with proliferation, and therefore it is expected that increased elongation rates upon eEF2K inhibition stimulate neoplastic growth. Indeed, in a mouse model of intestinal carcinogenesis caused by the adenomatous polyposis coli (APC) tumor suppressor loss, ablation of eEF2K drives oncogenic mTOR signaling, suggesting that eEF2K may exert tumor suppressive properties (Faller et al., 2015). In stark contrast, in a variety of cancer cell lines and xenograft models, eEF2K exerts tumor protective properties, in particular under conditions wherein nutrients are limiting (Leprivier et al., 2013). In this context, eEF2K engenders decrease in protein synthesis thereby conserving energy when nutrients are limiting, which occurs when tumors outstrip their vasculature (Kenney et al., 2014; Leprivier et al., 2013; Leprivier et al., 2015). These findings suggest that whereas the loss of eEF2K activity bolsters tumor initiation and early carcinogenesis, the increase in eEF2K may maintain energy homeostasis and prevent energy crisis under conditions when energy resources are compromised.
The role of AMPK in orchestrating protein synthesis and cancer energetics
AMPK acts as a central sensor of energy status in the cell (Hardie et al., 2012). Inadequate energy state leads to AMPK activation and consequent engagement of mechanisms which reduce anabolic processes such as lipogenesis and protein synthesis and induce autophagy, oxidation of fatty acids and other catabolic processes to conserve energy (Hardie and Pan, 2002; Hardie et al., 2012; Li et al., 2011; Shaw et al., 2004; Woods et al., 2003). AMPK is a heterotrimeric enzyme, composed of catalytic α, and regulatory β and γ subunits, which is traditionally thought to be activated by an increase in the AMP/ATP ratio, whereby AMP (or ADP) associates with γ subunit, leading to the phosphorylation of the activation loop (Thr172 in human protein) by a number of kinases, most notably LKB1 (Hardie et al., 2016). More recently, however, it has been shown that glucose withdrawal activates AMPK prior to increase in AMP/ATP levels by a mechanism that likely involves glycolytic enzyme aldolase (Zhang et al., 2017). Upon activation, AMPK reduces catabolic processes including protein and lipid synthesis which is chiefly mediated by inactivation of mTORC1 and acetyl-CoA carboxylase (ACC), respectively (Inoki et al., 2003; Zang et al., 2004; Zannella et al., 2011). AMPK has been shown to inhibit mTORC1 by a multitude of mechanisms including via TSC1/2, which dampens ATP consumption by translational machinery (Hong-Brown et al., 2012; Zannella et al., 2011) (Figure 2). In addition, it has been shown that AMPK can directly phosphorylate translational regulators such as eEF2K (Ser398 in humans), leading to its activation and a reduction in protein synthesis (Browne et al., 2004) (Figure 2). Recent studies wherein energy stress was induced by anti-diabetic biguanides (i.e. metformin and phenformin), which represses complex I of ETC and subsequently reduces mitochondrial ATP production (Andrzejewski et al., 2014; Bridges et al., 2014; Owen et al., 2000), revealed that the LKB1/AMPK/mTORC1 axis may play an essential role in preventing energy crisis and death of cancer cells (Algire et al., 2010; Shackelford et al., 2013). To this end, biguanides exhibited extensive cytotoxicity in cells which do not possess functional LKB1 and are thus incapable of reducing protein synthesis and other anabolic processes in response to energy stress (Algire et al., 2010; Shackelford et al., 2013). In contrast, biguanides induced only minimal cytotoxic effect in LKB1-proficient cells (Algire et al., 2010; Shackelford et al., 2013). Notwithstanding that AMPK regulates a number of anabolic and catabolic processes which are involved in adaptation to energy stress (Hardie et al., 2012), it is reasonable to postulate that an energy crisis in LKB1-deficient cells is at least in part caused by their inability to suppress protein synthesis. Consistently, it has been demonstrated that cancer cells adapt to nutrient stress by engaging AMPK-eEF2K axis and reducing mRNA translation (Leprivier et al., 2013).
eIF2α kinases and coordination of cancer energetics and protein synthesis
In addition to the eIF4F complex assembly, ternary complex (TC) recycling is an additional rate limiting step of translation initiation (Hinnebusch, 2014). As mentioned above, TC consists of eIF2 bound to GTP along with initiator tRNA (tRNAiMet). tRNAiMet delivery to the P site of the ribosome is accompanied by the hydrolysis of GTP, which results in the release of the eIF2:GDP complex. Next, eIF2:GDP is recycled to eIF2:GTP via the action of a multi-subunit guanine nucleotide exchange factor (GEF) eIF2B for the next round of initiation (Hinnebusch, 2014; Wortham and Proud, 2015) (Figure 1). The regulatory eIF2α subunit of eIF2 (also containing β and γ subunits) undergoes phosphorylation at position S51 (in mouse; S52 in human) in response to various stimuli including amino acids deprivation, ER stress, viral infection or heme deficiency, via General Control Nonderepressible 2 (GCN2), protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), protein kinase R (PKR) and heme-regulated inhibitor kinase (HRI) kinases, respectively (Hinnebusch, 2014) (Figure 2). In turn, PPP1R15 family members growth arrest and DNA damage-inducible protein 34 (GADD34) or the constitutive reverter of eIF2α phosphorylation (CReP), in collaboration with protein phosphatase 1 (PP1), dephosphorylate eIF2α (Jousse et al., 2003; Novoa et al., 2001). Phosphorylation of eIF2α leads to enhanced association of eIF2 with eIF2B, which blocks eIF2B GEF activity thus limiting TC availability (Hinnebusch, 2014). This leads to downregulation of translation of most cellular mRNAs, which is accompanied by translational upregulation of mRNAs which harbor inhibitory upstream open reading frames (uORFs) in their 5′UTR, including activating transcription factor 4 (ATF4), CCAAT-enhancer-binding protein homologous protein (CHOP) and GADD34 (Hinnebusch et al., 2016). While GADD34 induces dephosphorylation of eIF2α to resolve acute stress response (Brush et al., 2003; Novoa et al., 2001), ATF4 and CHOP are transcription factors that play a major role in metabolic regulation including governing glucose and glutathione metabolism and stimulating expression of amino acid transporters (Huggins et al., 2015; Krokowski et al., 2013; Wan et al., 2014). Moreover, it has been shown that aberrant ATF4 and CHOP expression leads to oxidative stress and increase in protein synthesis, which results in cell death and can be averted by suppressing translation via e.g. depletion of ribosomal proteins (Han et al., 2013). Accordingly, eIF2α phosphorylation which limits protein synthesis, has been shown to protect cancer cells from cell death upon glucose and amino acids depletion (Muaddi et al., 2010; Ye et al., 2010). In recent years a number of cross-talk mechanism between eIF2α phosphorylation, ATF4, mTORC1 and AMPK have been described (Ben-Sahra et al., 2016; Gandin et al., 2016a; Liu et al., 2006; Mounir et al., 2011; Park et al., 2017; Wengrod et al., 2015). This suggests that mTORC1, AMPK and the eIF2α/ATF4 axis may constitute central nodes of a master regulatory network that adjusts protein synthesis rates to cellular energy status.
Potential application of eIF4A inhibitors to target mechanisms coordinating protein synthesis and energy metabolism in neoplasia
Considering that uncoupling protein synthesis and cancer energetics appears to be detrimental to the survival of neoplastic cells, there is a heightened interest to develop therapeutic modalities to disrupt orchestration of mRNA translation and energy metabolism in the clinic (Erazo et al., 2016; Guichard et al., 2015; Powles et al., 2016). Considering the central role of mTOR in metabolic control, it is not surprising that mTOR inhibitors induce profound metabolic changes in the cell including reduction of protein, nucleotide and lipid synthesis and glycolysis (Duvel et al., 2010). Moreover, mTOR inhibitors also perturb metabolism at the organismal level (Gonzalez and Hall, 2017). A number of mTOR inhibitors have been discovered and/or developed in the last four decades. Rapamycin, a macrolide antibiotic produced by S. hygroscopicus which exerts antifungal and immunosuppressive properties (Houchens et al., 1983; Martel et al., 1977) acts as an allosteric inhibitor of mTOR and also exhibits anti-neoplastic properties (Garcia-Echeverria, 2010). Rapamycin binds to FKBP–rapamycin-binding (FRB) domain of mTOR in complex with FK-506-binding Protein 12 (FRBP12) and allosterically interferes with mTORC1 function, whereas it has only marginal effect on mTORC2, at least during the acute treatment (Aylett et al., 2016; Saxton and Sabatini, 2017). mTORC2 however appears to be rapamycin-sensitive over prolonged treatment in some cell lines and in vivo (Sarbassov et al., 2006). Rapamycin and its synthetic analogs (rapalogs) showed promising anti-neoplastic properties in a number of pre-clinical models which led to clinical trials for oncological indications and resulted in FDA approval for several indications including kidney and breast cancers (Basho et al., 2017; Chan et al., 2005; Hudes, 2007). Nonetheless, the anti-neoplastic efficacies of rapalogs were less than expected (Benjamin et al., 2011; Faes et al., 2017). This in part was attributed to the inhibition of the S6K-insulin receptor substrate 1 (IRS1)-PI3K-AKT feedback loop, which results in AKT activation and incomplete suppression of some mTORC1 outputs including 4E-BP phosphorylation (Dowling et al., 2010; Faes et al., 2017). Later findings spearheaded the development of the second generation of mTORC1 inhibitors which target the ATP binding pocket of mTOR (e.g. torin1, INK128) and thus inhibit both mTORC1 and mTORC2 (Benjamin et al., 2011). More recently, a third generation mTOR inhibitor (e.g. rapalink) which combines allosteric effects and targeting of the active site was developed (Rodrik-Outmezguine et al., 2016). Notwithstanding that the second and third generation of mTOR inhibitors exert more potent anti-proliferative effects than rapamycin in pre-clinical models, their effects, at least in the preclinical models, appear to be cytostatic, but not cytotoxic (Faes et al., 2017). This can at least in part be explained by the effects of mTOR inhibitors on the translatome. mTOR inhibitors simultaneously suppress translation of the short 5′UTR mRNAs which encode proteins with mitochondrial function (e.g. components of the ETC), as well as those which contain long 5′UTR and encode for proteins which maintain mitochondrial integrity [e.g. B-cell lymphoma 2 (BCL2) family members] (Gandin et al., 2016b). This decreases mitochondrial energy production which is compensated by reduced energy consumption by protein synthesis machinery. Moreover, mTOR inhibitors stimulate fusion of mitochondria and enhance removal of damaged mitochondria by autophagy (Gandin et al., 2016b; Morita et al., 2017). Together, this is expected to result in metabolic dormancy and cytostatic but not cytotoxic effects. In turn, inhibition of eIF4A results in selective inhibition of translation of mRNAs with long 5′UTR which are enriched in genes encoding for proteins which protect mitochondrial integrity (e.g. BCL-2 family members), but not those with short 5′UTRs encoding proteins with essential mitochondrial functions (ETC component complexes) (Gandin et al., 2016b). In addition, eIF4A inhibitors slightly increase mTORC1 signaling and thus do not induce autophagy (Galicia-Vazquez et al., 2012; Gandin et al., 2016b). This eIF4A inhibitor-induced combination of translational reprograming and suppression of autophagy is paralleled by mitochondrial depolarization and cell death (Gandin et al., 2016b). These findings suggest that eIF4A, but not mTOR inhibitors disrupt coordination between translational machinery and energy metabolism thereby resulting in a cytotoxic effect. Dramatic differences in translational and metabolic programs of non-transformed and cancer cells are thought to provide sufficient therapeutic window to employ eIF4A inhibitors in the clinic. Indeed, recent studies have shown that eIF4A inhibitors at doses that eradicate cancer cells exert only minimal toxicity in non-transformed cells and mice (Cencic et al., 2009; Cencic et al., 2013; Nasr et al., 2013).
Concluding remarks
Protein synthesis is one of the most energetically expensive biological processes (Buttgereit and Brand, 1995). In cancer cells where mRNA translation is commonly hyperactive, increased protein synthesis rates require elevated ATP production (Buttgereit and Brand, 1995; Morita et al., 2015; Rolfe and Brown, 1997). Emerging data indicate that this is achieved via the interplay between AMPK, mTORC and eIF2α phosphorylation. Notably, translational programs governed by the latter factors appear to be further integrated with other levels of regulation of gene expression including transcription. To this end, mTORC1 was reported to upregulate expression of mitochondrial genes involved in oxidative phosphorylation via the transcriptional factor Ying-Yang 1 (YY1) (Cunningham et al., 2007), and more recently to be directly involved in transcription of metabolic genes (Audet-Walsh et al., 2017; Chaveroux et al., 2013). The cross talk between energy production and protein synthesis is further corroborated by the findings that p62, a positive regulator of a glutamate transporter, may enable cancer cells with hyperactivated mTOR signaling to limit mitochondrial dysfunction by maintaining intercellular pools of glutathione (Lam et al., 2017). Additional components of the translational machinery have also been proposed to be implicated in metabolic regulation. For example, eIF6, which is involved in 60S ribosome biogenesis and subunit joining (Brina et al., 2015a), increases synthesis of transcriptional factors which regulate lipogenesis and glycolysis (Brina et al., 2015b), whereas eIF3, a large multiprotein complex that participates in the recruitment of mRNA to the ribosome, appears to control translation of mRNAs encoding mitochondrial factors (Shah et al., 2016). In conclusion, emerging findings highlight several mechanisms that underpin cross-talk between protein synthesis and cancer energetics, and suggest that future research is required to delineate cellular networks which orchestrate mRNA translation and energy metabolism of cancer cells, which may eventually result in novel therapeutic avenues to treat neoplasia.
Acknowledgments
We apologize to those authors whose work was not cited due to space constraints. We thank the members of our lab for critical reading of the manuscript. IT is a Junior 2 scholar of the Fonds de Recherche du Québec - Santé (FRQS). Research in our laboratory is funded in part by grants from Prostate Cancer Canada, Cancer Research Society, Canadian Cancer Society Research Institute, Canadian Institutes for Health Research, Terry Fox Research Institute and National Institutes of Health.
References
- Algire C, Amrein L, Bazile M, David S, Zakikhani M, Pollak M. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene. 2010 doi: 10.1038/onc.2010.483. [DOI] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125(6):1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Andrzejewski S, Gravel SP, Pollak M, St-Pierre J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014;2:12. doi: 10.1186/2049-3002-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audet-Walsh E, Dufour CR, Yee T, Zouanat FZ, Yan M, Kalloghlian G, Vernier M, Caron M, Bourque G, Scarlata E, Hamel L, Brimo F, Aprikian AG, Lapointe J, Chevalier S, Giguere V. Nuclear mTOR acts as a transcriptional integrator of the androgen signaling pathway in prostate cancer. Genes & development. 2017 doi: 10.1101/gad.299958.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. Architecture of human mTOR complex 1. Science (New York, NY) 2016;351(6268):48–52. doi: 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- Basho RK, Gilcrease M, Murthy RK, Helgason T, Karp DD, Meric-Bernstam F, Hess KR, Herbrich SM, Valero V, Albarracin C, Litton JK, Chavez-MacGregor M, Ibrahim NK, Murray JL, 3rd, Koenig KB, Hong D, Subbiah V, Kurzrock R, Janku F, Moulder SL. Targeting the PI3K/AKT/mTOR Pathway for the Treatment of Mesenchymal Triple-Negative Breast Cancer: Evidence From a Phase 1 Trial of mTOR Inhibition in Combination With Liposomal Doxorubicin and Bevacizumab. JAMA oncology. 2017;3(4):509–515. doi: 10.1001/jamaoncol.2016.5281. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science (New York, NY) 2016;351(6274):728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Current opinion in cell biology. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015 doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. The Journal of biological chemistry. 2002;277(27):23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- Bond P. Regulation of mTORC1 by growth factors, energy status, amino acids and mechanical stimuli at a glance. Journal of the International Society of Sports Nutrition. 2016;13:8. doi: 10.1186/s12970-016-0118-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges HR, Jones AJ, Pollak MN, Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. The Biochemical journal. 2014;462(3):475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brina D, Miluzio A, Ricciardi S, Biffo S. eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochimica et biophysica acta. 2015a;1849(7):830–835. doi: 10.1016/j.bbagrm.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Brina D, Miluzio A, Ricciardi S, Clarke K, Davidsen PK, Viero G, Tebaldi T, Offenhauser N, Rozman J, Rathkolb B, Neschen S, Klingenspor M, Wolf E, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Quattrone A, Falciani F, Biffo S. eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat Commun. 2015b;6:8261. doi: 10.1038/ncomms9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. The Journal of biological chemistry. 2004;279(13):12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Molecular and cellular biology. 2004;24(7):2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & development. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Molecular and cellular biology. 2003;23(4):1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95(4):1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. The Biochemical journal. 1995;312(Pt 1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlberg U, Nilsson A, Nygard O. Functional properties of phosphorylated elongation factor 2. European journal of biochemistry. 1990;191(3):639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- Carvalho MD, Carvalho JF, Merrick WC. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Archives of biochemistry and biophysics. 1984;234(2):603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, Porco JA, Jr, Pelletier J. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PloS one. 2009;4(4):e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Robert F, Galicia-Vazquez G, Malina A, Ravindar K, Somaiah R, Pierre P, Tanaka J, Deslongchamps P, Pelletier J. Modifying chemotherapy response by targeted inhibition of eukaryotic initiation factor 4A. Blood Cancer J. 2013;3:e128. doi: 10.1038/bcj.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S, Scheulen ME, Johnston S, Mross K, Cardoso F, Dittrich C, Eiermann W, Hess D, Morant R, Semiglazov V, Borner M, Salzberg M, Ostapenko V, Illiger HJ, Behringer D, Bardy-Bouxin N, Boni J, Kong S, Cincotta M, Moore L. Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(23):5314–5322. doi: 10.1200/JCO.2005.66.130. [DOI] [PubMed] [Google Scholar]
- Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, Sonenberg N, Giguere V. Molecular and genetic crosstalks between mTOR and ERRalpha are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell metabolism. 2013;17(4):586–598. doi: 10.1016/j.cmet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Chu J, Cargnello M, Topisirovic I, Pelletier J. Translation Initiation Factors: Reprogramming Protein Synthesis in Cancer. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Destefano MA, Jacinto E. Regulation of insulin receptor substrate-1 by mTORC2 (mammalian target of rapamycin complex 2) Biochemical Society transactions. 2013;41(4):896–901. doi: 10.1042/BST20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science (New York, NY) 2010;328(5982):1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517(7534):302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39(17):7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erazo T, Lorente M, Lopez-Plana A, Munoz-Guardiola P, Fernandez-Nogueira P, Garcia-Martinez JA, Bragado P, Fuster G, Salazar M, Espadaler J, Hernandez-Losa J, Bayascas JR, Cortal M, Vidal L, Gascon P, Gomez-Ferreria M, Alfon J, Velasco G, Domenech C, Lizcano JM. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(10):2508–2519. doi: 10.1158/1078-0432.CCR-15-1808. [DOI] [PubMed] [Google Scholar]
- Faes S, Demartines N, Dormond O. Resistance to mTORC1 Inhibitors in Cancer Therapy: From Kinase Mutations to Intratumoral Heterogeneity of Kinase Activity. Oxidative medicine and cellular longevity. 2017;2017:1726078. doi: 10.1155/2017/1726078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, Dudek KM, Casey HA, Scopelliti A, Cordero JB, Vidal M, Pende M, Ryazanov AG, Sonenberg N, Meyuhas O, Hall MN, Bushell M, Willis AE, Sansom OJ. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517(7535):497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 2012;18(7):1373–1384. doi: 10.1261/rna.033209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Cargnello M, Gyenis L, McLaughlan S, Cai Y, Tenkerian C, Morita M, Balanathan P, Jean-Jean O, Stambolic V, Trost M, Furic L, Larose L, Koromilas AE, Asano K, Litchfield D, Larsson O, Topisirovic I. mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat Commun. 2016a;7:11127. doi: 10.1038/ncomms11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, Furic L, Pollak M, Porco JA, Jr, St-Pierre J, Pelletier J, Larsson O, Topisirovic I. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016b;26(5):636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C. Allosteric and ATP-competitive kinase inhibitors of mTOR for cancer treatment. Bioorganic & medicinal chemistry letters. 2010;20(15):4308–4312. doi: 10.1016/j.bmcl.2010.05.099. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13(11):1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. The EMBO journal. 2017;36(4):397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard SM, Curwen J, Bihani T, D’Cruz CM, Yates JW, Grondine M, Howard Z, Davies BR, Bigley G, Klinowska T, Pike KG, Pass M, Chresta CM, Polanska UM, McEwen R, Delpuech O, Green S, Cosulich SC. AZD2014, an Inhibitor of mTORC1 and mTORC2, Is Highly Effective in ER+ Breast Cancer When Administered Using Intermittent or Continuous Schedules. Molecular cancer therapeutics. 2015;14(11):2508–2518. doi: 10.1158/1535-7163.MCT-15-0365. [DOI] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell metabolism. 2012;15(5):725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin-Ben-Harush A, Vardy A, Dikstein R. Efficient and Accurate Translation Initiation Directed by TISU Involves RPS3 and RPS10e Binding and Differential Eukaryotic Initiation Factor 1A Regulation. Molecular and cellular biology. 2017;37(15) doi: 10.1128/MCB.00150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature cell biology. 2013;15(5):481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Kim JY, Kim S. Molecular network and functional implications of macromolecular tRNA synthetase complex. Biochemical and biophysical research communications. 2003;303(4):985–993. doi: 10.1016/s0006-291x(03)00485-6. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. The Journal of biological chemistry. 1997;272(42):26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Pan DA. Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochemical Society transactions. 2002;30(Pt 6):1064–1070. doi: 10.1042/bst0301064. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends in Cell Biology. 2016;26(3):190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Sonenberg N, Mathews MB. Principles of Translational Control: An Overview. Cold Spring Harbor Perspectives in Biology. 2012;4(12):a011528. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science (New York, NY) 2016;352(6292):1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong-Brown LQ, Brown CR, Kazi AA, Navaratnarajah M, Lang CH. Rag GTPases and AMPK/TSC2/Rheb mediate the differential regulation of mTORC1 signaling in response to alcohol and leucine. American journal of physiology Cell physiology. 2012;302(10):C1557–1565. doi: 10.1152/ajpcell.00407.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchens DP, Ovejera AA, Riblet SM, Slagel DE. Human brain tumor xenografts in nude mice as a chemotherapy model. European journal of cancer & clinical oncology. 1983;19(6):799–805. doi: 10.1016/0277-5379(83)90012-3. [DOI] [PubMed] [Google Scholar]
- Hudes GR. mTOR as a target for therapy of renal cancer. Clinical advances in hematology & oncology : H&O. 2007;5(10):772–774. [PubMed] [Google Scholar]
- Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, Kasoji M, Haines DC, Quinones OA, Johnson PF. C/EBPgamma Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF4. Molecular and cellular biology. 2015;36(5):693–713. doi: 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D. Quality control mechanisms during translation. Science (New York, NY) 1999;286(5446):1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & development. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, Ron D. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. The Journal of cell biology. 2003;163(4):767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney JW, Moore CE, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Advances in biological regulation. 2014;55:15–27. doi: 10.1016/j.jbior.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Krokowski D, Han J, Saikia M, Majumder M, Yuan CL, Guan BJ, Bevilacqua E, Bussolati O, Broer S, Arvan P, Tchorzewski M, Snider MD, Puchowicz M, Croniger CM, Kimball SR, Pan T, Koromilas AE, Kaufman RJ, Hatzoglou M. A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. The Journal of biological chemistry. 2013;288(24):17202–17213. doi: 10.1074/jbc.M113.466920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HC, Baglini CV, Lope AL, Parkhitko AA, Liu HJ, Alesi N, Malinowska IA, Ebrahimi-Fakhari D, Saffari A, Yu JJ, Pereira A, Khabibullin D, Ogorek B, Nijmeh J, Kavanagh T, Handen A, Chan SY, Asara JM, Oldham WM, Diaz-Meco MT, Moscat J, Sahin M, Priolo C, Henske EP. p62/SQSTM1 Cooperates with Hyperactive mTORC1 to Regulate Glutathione Production, Maintain Mitochondrial Integrity, and Promote Tumorigenesis. Cancer research. 2017;77(12):3255–3267. doi: 10.1158/0008-5472.CAN-16-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Sabatini DM. A Central role for mTOR in lipid homeostasis. Cell metabolism. 2013;18(4):465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A. 2012;109(23):8977–8982. doi: 10.1073/pnas.1201689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiology and molecular biology reviews : MMBR. 2005;69(1):101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Prakash Somasekharan S, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH. The eEF2 Kinase Confers Resistance to Nutrient Deprivation by Blocking Translation Elongation. Cell. 2013;153(5):1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochimica et biophysica acta. 2015;1849(7):845–860. doi: 10.1016/j.bbagrm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, Gao B, Wierzbicki M, Verbeuren TJ, Shaw RJ, Cohen RA, Zang M. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell metabolism. 2011;13(4):376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Molecular cell. 2006;21(4):521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malys N, McCarthy JEG. Translation initiation: variations in the mechanism can be anticipated. Cellular and Molecular Life Sciences. 2011;68(6):991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Canadian journal of physiology and pharmacology. 1977;55(1):48–51. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, Campos C, Zhu S, Yang H, Yong WH, Cloughesy TF, Mellinghoff IK, Cavenee WK, Shaw RJ, Mischel PS. mTOR Complex 2 Controls Glycolytic Metabolism in Glioblastoma through FoxO Acetylation and Upregulation of c-Myc. Cell metabolism. 2013;18(5):726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, Sonenberg N. mTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell metabolism. 2013;18(5):698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Hulea L, Larsson O, Pollak M, St-Pierre J, Topisirovic I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14(4):473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, St-Pierre J, Larsson O, Topisirovic I, Vali H, McBride HM, Bergeron JJ, Sonenberg N. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Molecular cell. 2017;67(6):922–935. e925. doi: 10.1016/j.molcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, Hatzoglou M, Koromilas AE. Akt determines cell fate through inhibition of the PERK-eIF2alpha phosphorylation pathway. Sci Signal. 2011;4(192):ra62. doi: 10.1126/scisignal.2001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muaddi H, Majumder M, Peidis P, Papadakis AI, Holcik M, Scheuner D, Kaufman RJ, Hatzoglou M, Koromilas AE. Phosphorylation of eIF2alpha at serine 51 is an important determinant of cell survival and adaptation to glucose deficiency. Molecular biology of the cell. 2010;21(18):3220–3231. doi: 10.1091/mbc.E10-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr Z, Robert F, Porco JA, Jr, Muller WJ, Pelletier J. eIF4F suppression in breast cancer affects maintenance and progression. Oncogene. 2013;32(7):861–871. doi: 10.1038/onc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. The Journal of cell biology. 2001;153(5):1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. The Biochemical journal. 2000;348(Pt 3):607–614. [PMC free article] [PubMed] [Google Scholar]
- Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell reports. 2017;19(6):1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F Translation Initiation Complex: A Critical Nexus for Cancer Development. Cancer research. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. The Journal of biological chemistry. 2006;281(52):40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Powles T, Lackner MR, Oudard S, Escudier B, Ralph C, Brown JE, Hawkins RE, Castellano D, Rini BI, Staehler MD, Ravaud A, Lin W, O’Keeffe B, Wang Y, Lu S, Spoerke JM, Huw LY, Byrtek M, Zhu R, Ware JA, Motzer RJ. Randomized Open-Label Phase II Trial of Apitolisib (GDC-0980), a Novel Inhibitor of the PI3K/Mammalian Target of Rapamycin Pathway, Versus Everolimus in Patients With Metastatic Renal Cell Carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(14):1660–1668. doi: 10.1200/JCO.2015.64.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Regulation and roles of elongation factor 2 kinase. Biochemical Society transactions. 2015;43(3):328–332. doi: 10.1042/BST20140323. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Current opinion in cell biology. 2009;21(3):435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, Barratt DG, Cosulich S, Klinowska T, Rosen N, Shokat KM. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534(7606):272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. The Journal of biological chemistry. 1999;274(18):12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77(3):731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol. 2012;4(11) doi: 10.1101/cshperspect.a012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazanov AG. Elongation factor-2 kinase and its newly discovered relatives. FEBS letters. 2002;514(1):26–29. doi: 10.1016/s0014-5793(02)02299-8. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Molecular cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, Shaw RJ. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer cell. 2013;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Su D, Scheliga JS, Pluskal T, Boronat S, Motamedchaboki K, Campos AR, Qi F, Hidalgo E, Yanagida M, Wolf DA. A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell reports. 2016;16(7):1891–1902. doi: 10.1016/j.celrep.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer cell. 2004;6(1):91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. Translational Tolerance of Mitochondrial Genes to Metabolic Energy Stress Involves TISU and eIF1-eIF4GI Cooperation in Start Codon Selection. Cell metabolism. 2015;21(3):479–492. doi: 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Smith EM, Proud CG. cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle- and amino acid-dependent manner. The EMBO journal. 2008;27(7):1005–1016. doi: 10.1038/emboj.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100(3):301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Su B, Jacinto E. Mammalian TOR signaling to the AGC kinases. Critical reviews in biochemistry and molecular biology. 2011;46(6):527–547. doi: 10.3109/10409238.2011.618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7(3):382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. The EMBO journal. 2007;26(9):2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC. The molecular basis of mTORC1-regulated translation. Biochemical Society transactions. 2017;45(1):213–221. doi: 10.1042/BST20160072. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. mRNA Translation and Energy Metabolism in Cancer: The Role of the MAPK and mTORC1 Pathways. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- Voigts-Hoffmann F, Klinge S, Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Current Opinion in Structural Biology. 2012;22(6):768–777. doi: 10.1016/j.sbi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci U S A. 1996;93(9):4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science (New York, NY) 2010;330(6005):835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XS, Lu XH, Xiao YC, Lin Y, Zhu H, Ding T, Yang Y, Huang Y, Zhang Y, Liu YL, Xu ZM, Xiao J, Li XK. ATF4- and CHOP-dependent induction of FGF21 through endoplasmic reticulum stress. BioMed research international. 2014;2014:807874. doi: 10.1155/2014/807874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. The EMBO journal. 2001;20(16):4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengrod J, Wang D, Weiss S, Zhong H, Osman I, Gardner LB. Phosphorylation of eIF2alpha triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci Signal. 2015;8(367):ra27. doi: 10.1126/scisignal.aaa0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, Gualerzi C. Effect of Escherichia coli initiation factors on the kinetics of N-Acphe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry. 1983;22(3):690–694. doi: 10.1021/bi00272a025. [DOI] [PubMed] [Google Scholar]
- Wolfson RL, Sabatini DM. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell metabolism. 2017;26(2):301–309. doi: 10.1016/j.cmet.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13(22):2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Wortham NC, Proud CG. eIF2B: recent structural and functional insights into a key regulator of translation. Biochemical Society transactions. 2015;43(6):1234–1240. doi: 10.1042/BST20150164. [DOI] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. The EMBO journal. 2010;29(12):2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang M, Zuccollo A, Hou X, Nagata D, Walsh K, Herscovitz H, Brecher P, Ruderman NB, Cohen RA. AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. The Journal of biological chemistry. 2004;279(46):47898–47905. doi: 10.1074/jbc.M408149200. [DOI] [PubMed] [Google Scholar]
- Zannella VE, Cojocari D, Hilgendorf S, Vellanki RN, Chung S, Wouters BG, Koritzinsky M. AMPK regulates metabolism and survival in response to ionizing radiation. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2011;99(3):293–299. doi: 10.1016/j.radonc.2011.05.049. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114(1):113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng JW, Zhu M, Wu YQ, Li TY, Ye Z, Lin SY, Yin H, Piao HL, Hardie DG, Lin SC. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548(7665):112–116. doi: 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144(5):757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]