Abstract
Cadherin-6 (CDH6) is aberrantly expressed in cancer and closely associated with tumor progression. However, the functions of CDH6 in human osteosarcoma and the molecular mechanisms underlying CDH6 in osteosarcoma oncogenesis remain poorly understood. In this work, we assessed the role of CDH6 in human osteosarcoma and identified that the expression of CDH6 was closely related with the overall survival and poor prognosis of osteosarcoma patients. MicroRNAs (miRNAs) have been implicated as important epigenetic regulators during the progression of osteosarcoma. Using dual-luciferase reporter assays, we showed that miR-223-3p suppresses CDH6 expression by directly binding to the 3′ UTR of CDH6. miR-223-3p overexpression significantly inhibited cell invasion, migration, growth, and proliferation by suppressing the CDH6 expression in vivo and in vitro. Besides, CDH6 overexpression in the miR-223-3p-transfected osteosarcoma cells effectively rescued the inhibition of cell invasion, migration, growth, and proliferation mediated by miR-223-3p. Additionally, Kaplan-Meier analysis suggests that the expression of miR-223-3p predicts favorable clinical outcomes for osteosarcoma patients. Moreover, the expression of miR-223-3p was downregulated in osteosarcoma patients and was negatively associated with the expression of CDH6. Collectively, these data highlight that miR-223-3p/CDH6 axis is an important novel pleiotropic regulator and could early predict the metastatic potential in human osteosarcoma treatments.
Keywords: osteosarcoma, miR-223-3p, CDH6, therapy
Quanbo Ji et al. demonstrate that CDH6 is correlated with the overall survival and prognosis of osteosarcoma patients and identified as a novel direct target of miR-223-3p. miR-223-3p overexpression inhibited cell proliferation, invasion, and migration, indicating miR-223-3p and CDH6 could be novel prognostic biomarkers and therapeutic targets for treating osteosarcoma.
Introduction
Osteosarcoma has been implicated as the most common primary malignant tumors, affecting the children, adolescents, and young adults with highest incidence of complex karyotypes worldwide.1, 2, 3, 4 The survival rate of osteosarcoma patients with metastatic or recurrent disease is decreased to 10%–30%, leading to the limited effective options for successful treatments.5, 6 Recent advances in bioinformatics and scientific technologies have been developed to develop potential targets to treat osteosarcoma.7, 8, 9, 10, 11 However, the clinical outcomes have not yet been significantly improved for osteosarcoma patients. Therefore, the identification of new promising applicable therapeutic candidates and approaches for clinical management of osteosarcoma is urgent.
Cadherin-6 (CDH6), a type II cadherin and synaptic adhesion molecule, emerged as an important regulator involved in the kidney and CNS morphogenesis.12, 13, 14, 15, 16 Recent reports have shown that aberrant activation of CDH6 was found in cancer. CDH6 could be induced by transforming growth factor-β (TGF-β) during epithelial-to-mesenchymal transition (EMT) and strongly expressed in highly aggressive thyroid cancer cells,17, 18 suggesting CDH6 has key roles in aggressiveness of tumors. CDH6 influences active Rho distribution and promotes neural crest cell detachment via F-actin regulation during EMT.13 CDH6 could regulate cancer metastasis and EMT by restraining autophagy17 and promote mitochondrial network re-organization through a DRP1-mediated mechanism. In high-grade serous ovarian cancer, CDH6 is repressed by mutant p53 in the fallopian tube,19 whereas in nasopharyngeal carcinoma (NPC), CDH6 was revealed to be upregulated in LMP1-positive NPC tissues12 and was identified as a potential candidate of the epithelium-specific miR-203. Besides, CDH6 exerts as a node protein involved in the interplay of multiple signaling, including NF-κB and TGF-β in NPC. Although CDH6 has been implicated in cancer progression and development, its essential role in human osteosarcoma progression and oncogenesis remains unknown.
MicroRNAs (miRNAs), a family of endogenous small non-coding RNAs, have been described to be able to regulate target gene expression by directly binding to the 3′ UTR of the gene mRNA.20, 21, 22, 23, 24, 25, 26 Accumulating evidence suggests that miRNAs emerge as the posttranscriptional modulators in the regulation of biogenesis and cancer progression.27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43 Recently, studies have revealed that abnormal expression of miRNAs is involved in the impairment of normal function and osteosarcoma pathogenesis,5, 44, 45, 46, 47 such as miR-21,48, 49 miR-34a,50, 51 miR-143,52, 53 and miR-382.9 However, the molecular mechanisms by which miRNAs regulate CDH6 in human osteosarcoma progression and the specific roles of miRNAs in oncogenesis have not been fully clarified.
In the current study, we demonstrated that CDH6 was closely correlated with the overall survival and prognosis of osteosarcoma patients. In addition, CDH6 was identified as a novel direct target and functional modulator of miR-223-3p in osteosarcoma. We also demonstrated that miR-223-3p overexpression inhibited cell proliferation, invasion, and migration by using in vivo and in vitro approaches. Moreover, miR-223-3p expression was found to be downregulated in osteosarcoma patients and negatively associated with CDH6 expression. Thus, miR-223-3p and CDH6 may be novel prognostic biomarkers and common therapeutic targets for treating osteosarcoma.
Results
CDH6 Is Closely Correlated with Clinical Prognosis in Osteosarcoma Patients
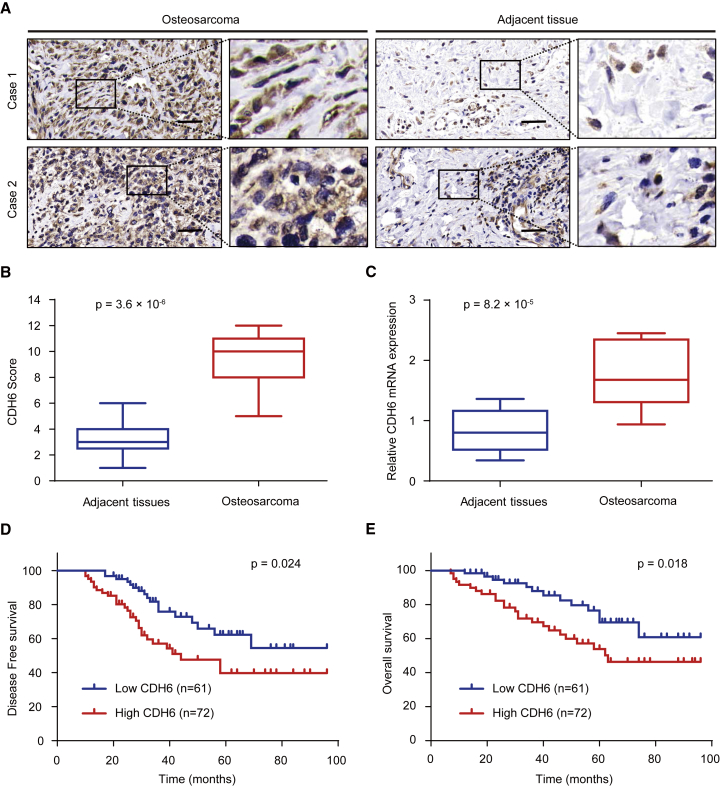
In order to investigate the exact role of CDH6 in human osteosarcoma, we performed histopathologic assay and examined the expression of CDH6 in 133 osteosarcoma samples and the adjacent noncancerous tissues (Figure 1A). The results showed that CDH6 gained a higher expression level in osteosarcoma tissues than the corresponding nontumor tissues (p = 3.6 × 10−6; Figure 1B). The qRT-PCR assay also indicated CDH6 expression was significantly upregulated in osteosarcoma samples (p = 8.2 × 10−5; Figure 1C), which was consistent with the results of histopathological assay. We then assessed the clinical parameters of CDH6 in osteosarcoma patients and investigated the correlation between clinicopathological characteristics and CDH6 expression. The data revealed that CDH6 expression was closely associated with histological stage and tumor size (Table 1). Moreover, on the basis of Kaplan-Meier survival analysis, we found the osteosarcoma patients with low expression of CDH6 had better disease-free survival (DFS) (p = 0.024) and overall survival (OS) (p = 0.018) than those with high CDH6 levels (Figures 1D and 1E). Taken together, these data indicated the important clinical significance of CDH6 in prognosis and metastasis of osteosarcoma patients.
Figure 1.
CDH6 Is Closely Correlated with Clinical Prognosis in Osteosarcoma Patients
(A) Representative immunohistochemistry images of CDH6 expression in human osteosarcoma tissues and adjacent tissues. The scale bars represent 50 μm. (B) CDH6 expression scores in osteosarcoma tissues and matched adjacent normal tissues (n = 133) were compared with the Mann-Whitney U test. (C) The mRNA expression levels of CDH6 expression scores in osteosarcoma tissues and matched adjacent normal tissues (n = 133) using qRT-PCR assay are shown. (D and E) Kaplan-Meier survival curves and log rank tests were used to compare (D) DFS and (E) OS of the osteosarcoma patients with low and high scores for CDH6.
Table 1.
Associations between CDH6 Expression and Clinicopathological Characteristics
| Characteristics | n | CDH6 Expression |
p Values | |
|---|---|---|---|---|
| High (n, %) | Low (n, %) | |||
| Gender | ||||
| Male | 76 | 46 (60.5%) | 30 (39.5%) | 0.088 |
| Female | 57 | 26 (45.6%) | 31 (54.4%) | |
| Tumor Size (cm) | ||||
| >7 | 69 | 49 (71.0%) | 20 (29.0%) | 4.985 × 10−5** |
| ≤7 | 64 | 23 (35.9%) | 41 (64.1%) | |
| Location | ||||
| Distal femur | 57 | 32 (56.1%) | 25 (43.9%) | 0.366 |
| Proximal tibia | 34 | 19 (55.9%) | 15 (44.1%) | |
| Proximal humerus | 26 | 10 (38.5%) | 16 (61.5%) | |
| Proximal femur | 11 | 8 (72.7%) | 3 (27.3%) | |
| Others | 5 | 3 (60.0%) | 2 (40.0%) | |
| TNM Stage | ||||
| I | 57 | 16 (28.1%) | 41 (71.9%) | 1.747 × 10−7** |
| II/III | 76 | 56 (73.7%) | 20 (26.3%) | |
| Relapse | ||||
| Yes | 26 | 24 (92.3%) | 2 (7.7%) | 1.331 × 10−5** |
| No | 107 | 48 (44.9%) | 59 (55.1%) | |
| Metastasis | ||||
| Lung | 49 | 35 (71.4%) | 14 (28.6%) | 0.005** |
| Others | 16 | 9 (56.3%) | 7 (43.8%) | |
| No | 68 | 28 (41.2%) | 40 (58.8%) | |
p values were calculated by Pearson’s chi-square test. **p < 0.01. TNM, tumor, node, metastasis.
miR-223-3p Suppresses CDH6 Expression by Directly Targeting Its 3′ UTR
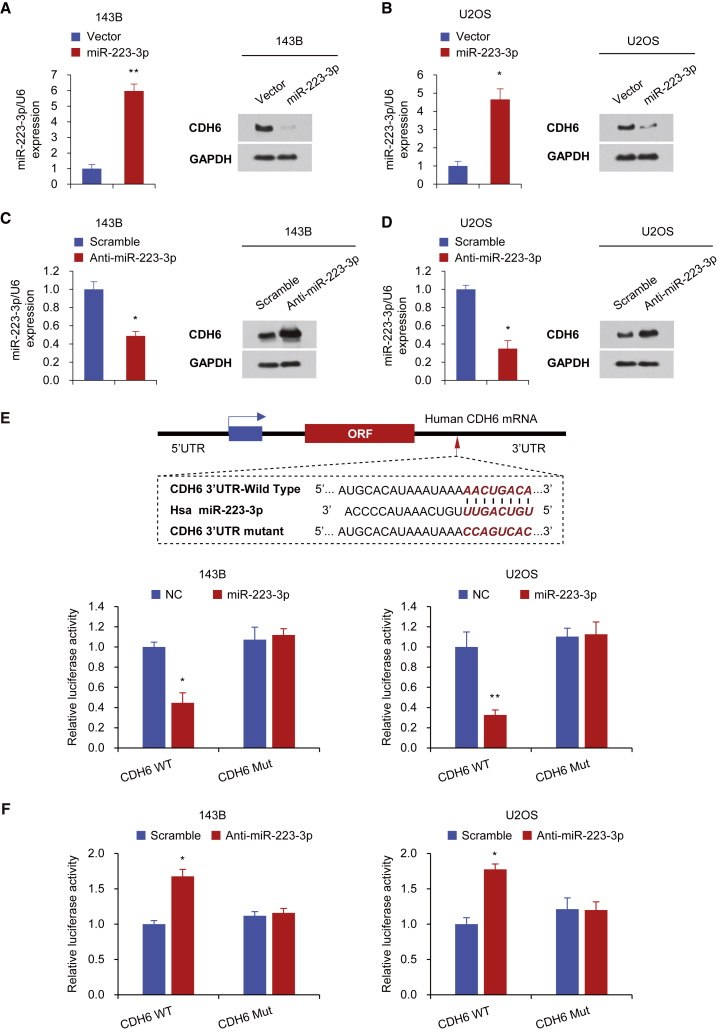
We next performed two target prediction programs, TargetScan and miRanda, to examine the potential CDH6-targeting miRNAs and predict the probable functional binding site. Several miRNA candidates that target CDH6 were selected, including miR-137, miR-203, miR-223-3p, and miR-373-3p. We then confirmed the effect of the above-mentioned miRNAs on the expression of CDH6 in human osteosarcoma cell lines using the western blot analysis (Figure S1). Consistent with the data previously found in cholangiocarcinoma,54 miR-203 also suppressed the expression of CDH6 in human osteosarcoma cells. Importantly, miR-223-3p had the most pronounced inhibitory effect on the expression of CDH6. Therefore, we then hypothesize that miR-223-3p might function as an important modulator in human osteosarcoma. Based on the western blot analysis, we demonstrated that miR-223-3p overexpression significantly suppressed the CDH6 production in osteosarcoma cell lines (Figures 2A, 2B, and S1). Besides, miR-223-3p inhibition promoted CDH6 expression in the same osteosarcoma cell lines (Figures 2C and 2D). Notably, miR-223-3p did not regulate the CDH6 mRNA expression level, indicating this regulation is posttranscriptional (Figure S2).
Figure 2.
miR-223-3p Suppresses CDH6 Expression by Directly Targeting Its 3′ UTR
(A–D) Immunoblot analysis of the indicated osteosarcoma cell lines transfected with miR-223-3p (A and B) or anti-miR-223-3p (C and D). The histograms on the left of the immunoblots show the corresponding miR-223-3p mRNA expression levels. (E) miRNA luciferase reporter assays in 143B and U2OS cells co-transfected with wild-type or mutated CDH6 reporters and miR-223-3p are shown. The top panel indicates wild-type and mutant forms of putative miR-223-3p target sequences in the 3′ UTR of CDH6. Bold and italicized fonts indicate putative miR-223-3p-binding sites in the 3′ UTR of human CDH6. Underlining indicates mutations introduced into the 3′ UTR of CDH6. (F) miRNA luciferase reporter assays in 143B and U2OS cells co-transfected with wild-type or mutated CDH6 reporters and anti-miR-223-3p are shown. Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05; **p < 0.01.
Next, to evaluate whether CDH6 is a specific and direct target of miR-223-3p, we transiently co-transfected 143B and U2OS cell lines with luciferase reporter constructs containing wild-type or mutated CDH6 3′ UTR and miR-223-3p or anti-miR-223-3p. The results showed that miR-223-3p overexpression inhibited the 3′ UTR luciferase reporter activity of CDH6 but did not have an effect on the luciferase activity of the reporter in which the miR-223-3p binding sites were mutated (Figure 2E). Moreover, inhibition of miR-223-3p expression promoted the CDH6 3′ UTR reporter luciferase activity (Figure 2F). Together, these results collectively revealed that miR-223-3p suppresses CDH6 expression by directly targeting the 3′ UTR of CDH6 in human osteosarcoma.
miR-223-3p Inhibits Osteosarcoma Cell Invasion, Migration, Growth, and Proliferation through the Suppression of CDH6 Production
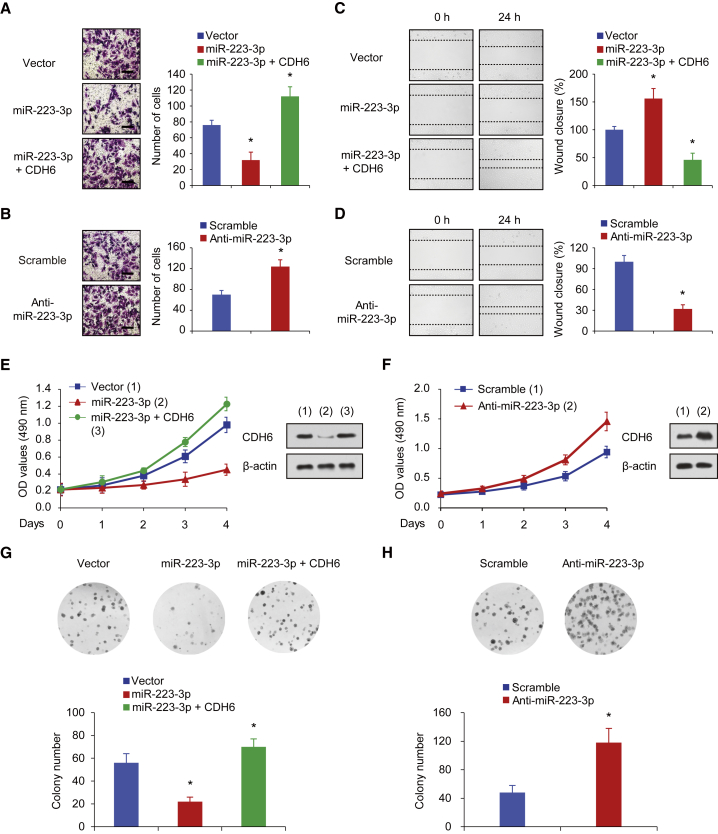
We next preformed Matrigel invasion assays to investigate whether miR-223-3p regulates invasive abilities of human osteosarcoma cell lines. The data showed that miR-223-3p overexpression significantly inhibited the invasion ability of human osteosarcoma cell lines, whereas CDH6 restoration rescued the miR-223-3p effects on osteosarcoma cells (Figure 3A). Similar results were also observed in wound healing assays. Briefly, miR-223-3p overexpression suppressed migratory ability of osteosarcoma cells, and CDH6 re-expression impaired the cell migration induced by miR-223-3p (Figure 3C). Furthermore, compared with a control treatment, miR-223-3p inhibition promoted the invasion and migration of human osteosarcoma cells (Figures 3B and 3D), which was in line with the findings above.
Figure 3.
miR-223-3p Inhibits Osteosarcoma Cell Invasion, Migration, Growth, and Proliferation through the Suppression of CDH6 Production
(A and B) Invasion of 143B cells transfected with miR-223-3p or miR-223-3p plus CDH6 (A) or miR-223-3p inhibitor (B) was evaluated using a Matrigel invasion chamber. The invaded cells were fixed and stained with crystal violet (A and B left images). The scale bar represents 100 μm. (C and D) Wound healing assays were conducted in 143B cells transfected with miR-223-3p or miR-223-3p and CDH6 (C) or miR-223-3p inhibitor (D). Cell migration was measured 24 hr after the cell layers were scratched. (E and F) 143B cells expressing miR-223-3p or miR-223-3p and CDH6 (E) and 143B cells transfected with miR-223-3p inhibitor (F) were cultured in regular medium. At the specified times, cell numbers were determined with the CCK-8 assay. The representative immunoblot shows CDH6 expression. (G and H) 143B cells transfected with miR-223-3p (G) or miR-223-3p inhibitor (H) were plated and assayed for colony formation. Representative images show colonies in plates (left panels). Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05; **p < 0.01.
Next, we assessed whether miR-223-3p could affect the cell proliferation phenotypes of osteosarcoma cell lines using cell growth and colony formation assays. Cells transfected with miR-223-3p were applied for cell growth analysis. Consistent with the findings above, overexpression of miR-223-3p inhibited the proliferative ability and colony formation (Figures 3E and 3G). Besides, CDH6 restoration reversed the effect of miR-223-3p on cell proliferation and colony formation (Figures 3E and 3G). Moreover, miR-223-3p inhibition promoted the osteosarcoma cell proliferation and colony formation (Figures 3F and 3H). Therefore, these findings demonstrated that miR-223-3p impaired cell proliferation though suppressing the production of CDH6, thus indicating that CDH6 is an important mediator of miR-223-3p function in human osteosarcoma metastasis.
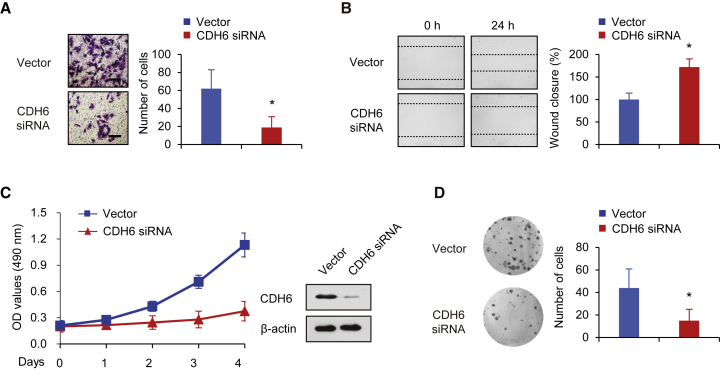
Inhibition of CDH6 Suppresses Osteosarcoma Cell Invasion, Migration, Growth, and Proliferation
To further confirm the role of CDH6 in osteosarcoma, we next preformed Matrigel invasion assays to investigate whether CDH6 inhibition could suppress invasive abilities of human osteosarcoma using CDH6-specific small interfering RNA (siRNA). The results indicated that CDH6 inhibition significantly inhibited the invasion ability of osteosarcoma (Figure 4A). Similar results were also observed in wound healing assays. Briefly, CDH6 inhibition suppressed osteosarcoma cells migratory ability (Figure 4B), which was consistent with the findings above. We next examined whether CDH6 inhibition could affect the cell proliferation phenotypes of osteosarcoma through cell growth and colony formation assays. In line with the findings above, inhibition of CDH6 suppressed the proliferative ability and colony formation (Figures 4C and 4D). Therefore, these data demonstrated that CDH6 inhibition impaired osteosarcoma cell invasion, migration, growth, and proliferation, suggesting that CDH6 plays an important role in human osteosarcoma.
Figure 4.
CDH6 Inhibition Suppresses Osteosarcoma Cell Invasion, Migration, Growth, and Proliferation
(A) Invasion of 143B cells transfected with CDH6 siRNA was evaluated using a Matrigel invasion chamber. The invaded cells were fixed and stained with crystal violet. The scale bar represents 100 μm. (B) Wound healing assays were conducted in 143B cells transfected with CDH6 siRNA. Cell migration was measured 24 hr after the cell layers were scratched. (C) 143B cells transfected with CDH6 siRNA were cultured in regular medium. At the specified times, cell numbers were determined with the CCK-8 assay. The representative immunoblot shows CDH6 expression. (D) 143B cells transfected with CDH6 siRNA were plated and assayed for colony formation. Representative images show colonies in plates (left panels). Each bar represents the mean ± SD of at least three independent experiments performed in triplicate. *p < 0.05; **p < 0.01.
miR-223-3p Inhibits Osteosarcoma Initiation and Metastasis
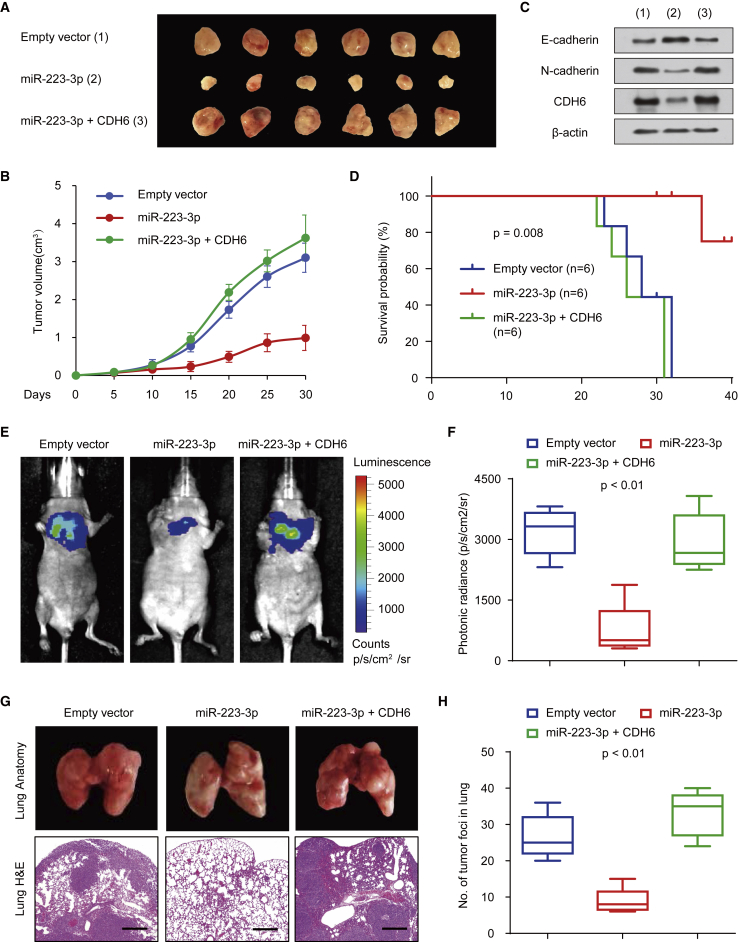
To determine the phenotype of miR-223-3p expression in vivo, we further investigated the effect of miR-223-3p on 143B cell growth in nude mice. We found that overexpression of miR-223-3p markedly suppressed the tumor growth in vivo (Figures 5A and 5B). Besides, the tumors in mice formed by miR-223-3p-overexpressing 143B cells gained downregulated expression of EMT markers (N-cadherin) and CDH6 (Figure 5C). In addition, tumors in mice inoculated with miR-223-3p plus CDH6-overexpressing 143B cells revealed a reversal effect of miR-223-3p on osteosarcoma tumor growth (Figures 5A and 5B). Moreover, according to the Kaplan-Meier survival analysis, the miR-223-3p-expressing group gained better survival probability than the control groups (p = 0.008; Figure 5D).
Figure 5.
miR-223-3p Inhibits Osteosarcoma Initiation and Metastasis
(A and B) Stable 143B cells overexpressing miR-223-3p and miR-223-3p and CDH6 were injected into nude mice (A). At the indicated times, tumors were measured with Vernier calipers (mean ± SD; n = 6) (B). (C) Immunoblot analysis of representative excised tumors in (A) is shown. (D) Kaplan-Meier survival curves and log rank tests were performed to evaluate the role of miR-223-3p and CDH6 treatments in vivo. (E and F) Bioluminescence imaging of metastasis of osteosarcoma cells in non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) mice at 28 days after intravenous injection of cells infected with PCDH-control, PCDH-miR-223-3p, or PCDH-miR-223-3p and CDH6 via the lateral tail vein is shown (E). The luminescence signal is represented by an overlaid false-color image with the signal intensity indicated by the scale (F). (G and H) Representative metastatic foci of lungs were subjected to anatomical (G) and histological (H) analyses. The data are shown as the mean ± SD (n = 6). *p < 0.05; **p < 0.01.
We then evaluated the effect of miR-223-3p on osteosarcoma metastasis. The data showed that, compared with the control group, miR-223-3p-expressing group revealed a more significant decrease in the lungs’ metastatic burden (Figure 5E). Besides, similar results were also found in the photonic radiance intensity of the lungs in the miR-223-3p-expressing group (Figure 5F). On the other hand, the miR-223-3p plus CDH6 group showed impaired expression of miR-223-3p (Figure 5F). We next performed the anatomic and histologic analysis of the lungs of mice. Indeed, the results confirmed the metastatic foci (Figures 5G and 5H). The miR-223-3p-expressing group displayed less tumor foci in the lungs than the control group, whereas the miR-223-3p plus CDH6 group showed the reversal effects of miR-223-3p on the metastatic foci. Taken together, these findings strongly demonstrated that miR-223-3p and CDH6 function as important regulators in osteosarcoma dissemination.
miR-223-3p Expression in Human Osteosarcoma Samples and the Correlation between miR-223-3p and CDH6
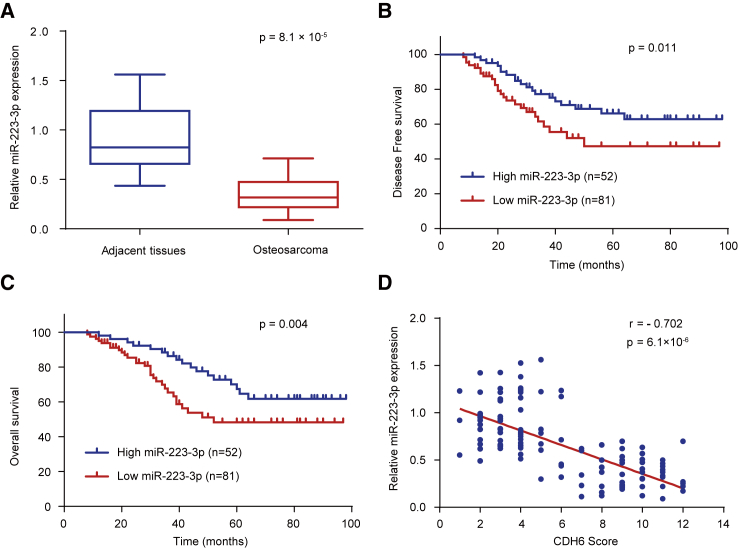
We further investigated the clinical significance of miR-223-3p in osteosarcoma patients. Using qRT-PCR assay, we detected the expression of miR-223-3p in 133 osteosarcoma samples and the matched adjacent nontumor tissues. According to the qRT-PCR results, the expression of miR-223-3p was significantly downregulated in osteosarcoma patients (p = 3.2 × 10−4; Figure 6A). We next evaluated the relationship between the expression of miR-223-3p and clinicopathological characteristics to investigate the clinical significance of miR-223-3p. The findings indicated that miR-223-3p expression level was closely associated with osteosarcoma histological stage and tumor size (Table 2). Besides, on the basis of the Kaplan-Meier survival analysis, patients with high expression levels of miR-223-3p had better DFS (p = 0.011) and OS (p = 0.004) than patients with low miR-223-3p expression levels, indicating that miR-223-3p might be a predictor of better human osteosarcoma clinical outcomes (Figures 6B and 6C). Furthermore, the expression of miR-223-3p was negatively correlated with the protein expression of CDH6 in the osteosarcoma samples (p = 6.1 × 10−6; r = −0.702; Figure 6D), which was consistent with miR-223-3p inhibition of CDH6 protein expression in cultured cells (Figures 2A and S1). Collectively, these results strongly suggested the important role of miR-223-3p and CDH6 in human osteosarcoma prognosis.
Figure 6.
miR-223-3p Expression in Human Osteosarcoma Samples and the Correlation between miR-223-3p and CDH6
(A) Expression of miR-223-3p in osteosarcoma tissues and matched adjacent normal tissues (n = 133) was compared using the Mann-Whitney U test. U6 small nuclear RNA was used as the internal control. (B and C) Kaplan-Meier survival curves and log rank tests were used to compare (B) DFS and (C) OS of osteosarcoma patients with low and high expression levels of miR-223-3p. (D) The relationship between miR-223-3p and CDH6 expression was assessed by Spearman’s rank correlation analysis in the osteosarcoma samples. The symbols represent individual samples.
Table 2.
Associations between miR-223-3p Expression and Clinicopathological Characteristics
| Characteristics | n | miR-223-3p Expression |
p Values | |
|---|---|---|---|---|
| High (n, %) | Low (n, %) | |||
| Gender | ||||
| Male | 76 | 29 (38.2%) | 47 (61.8%) | 0.798 |
| Female | 57 | 23 (40.4%) | 34 (59.6%) | |
| Tumor Size (cm) | ||||
| >7 | 69 | 19 (27.5%) | 50 (72.5%) | 0.005** |
| ≤7 | 64 | 33 (51.6%) | 31 (48.4%) | |
| Location | ||||
| Distal femur | 57 | 24 (42.1%) | 33 (57.9%) | 0.241 |
| Proximal tibia | 34 | 16 (47.1%) | 18 (52.9%) | |
| Proximal humerus | 26 | 6 (23.1%) | 20 (76.9%) | |
| Proximal femur | 11 | 3 (27.3%) | 8 (72.7%) | |
| Others | 5 | 3 (60.0%) | 2 (40.0%) | |
| TNM Stage | ||||
| I | 57 | 38 (66.7%) | 19 (33.3%) | 1.674 × 10−8** |
| II/III | 76 | 14 (18.4%) | 62 (81.6%) | |
| Relapse | ||||
| Yes | 26 | 3 (11.5%) | 23 (88.5%) | 0.001** |
| No | 107 | 49 (45.8%) | 58 (54.2%) | |
| Metastasis | ||||
| Lung | 49 | 7 (14.3%) | 42 (85.7%) | 2.660 × 10−5** |
| Others | 16 | 8 (47.1%) | 9 (52.9%) | |
| No | 68 | 38 (55.9%) | 30 (44.1%) | |
p values were calculated by Pearson’s chi-square test. **p < 0.01. TNM, tumor, node, metastasis.
Discussion
Currently, miRNAs have been implicated to be involved in almost every aspect of cellular functions and biological processes.32, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66 miRNAs are aberrantly expressed in various cancers and closely associated with invasion, proliferation, and metastasis.33, 41, 62, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76 Hence, one potential route to improve therapy for cancer patients is to understand miRNAs involved in tumor formation, which might be specific and effective for current therapies. Emerging evidence has shown that miR-223-3p acted as a novel microRNA regulator in biological behavior in various types of tumors.77, 78, 79 miR-223-3p promotes resistance to cetuximab and inhibits angiogenesis in head and neck squamous cell carcinoma.80 Besides, miR-223-3p suppresses ovarian cancer cell invasion and proliferation by targeting SOX11 expression.78
In this study, we also performed a software analysis of other putative targets of miR223-3p in human osteosarcoma using TargetScan and miRanda. The results revealed that several target genes potentially involved in osteosarcoma might be the potential target genes of miR-223-3p, including PAX5, CDH6, EYA3, SOX11, HDAC4, and TET3, among which only CDH6 and SOX11 were downregulated through miR-223-3p overexpression in osteosarcoma according to western blot analysis (Figure S3). To determine whether the forced expression of the target genes CDH6 and SOX11 have a similar role in osteosarcoma, we next preformed cell growth and wound healing assay to investigate whether SOX11 and CDH6 could rescue the miR-223-3p effects on cell growth and migration in osteosarcoma. Consequently, the results revealed that miR-223-3p inhibited the proliferative ability of osteosarcoma. However, CDH6 restoration, but not SOX11, could reverse the effect of miR-223-3p on cell proliferation in human osteosarcoma (Figures 3E and S4). Similar results were also observed in wound healing assays. Briefly, miR-223-3p overexpression suppressed migratory ability of osteosarcoma cells, and CDH6 re-expression, but not SOX11, could reverse the effect of the cell migration induced by miR-223-3p (Figures 3C and S5), thus suggesting that miR-223-3p impaired cell proliferation in osteosarcoma though suppressing the production of CDH6, but not SOX11. In addition, miR-223-3p also exerts its function as a tumor suppressor in bladder cancer and hepatocellular carcinoma.81, 82 However, the exact role of miR-223-3p in human osteosarcoma remains yet unelucidated. Here, in this present study, we identified that miR-223-3p exerts the tumor-suppressive effect on human osteosarcoma cell invasion, migration, growth, and proliferation by directly targeting CDH6. Besides, miR-223-3p expression in human osteosarcoma was markedly downregulated. Furthermore, osteosarcoma patients with low levels of miR-223-3p displayed worse DFS and OS, supporting that miR-223-3p functions as a novel prognostic and predictive role in human osteosarcoma.
CDH6, a member of the cadherin family and a membrane glycoprotein, regulates cell-to-cell adhesion and plays an important role in cell morphogenesis, contributing to cancer cell migration.17, 83 In this current study, we identify that the miR-223-3p/CDH6 axis regulates cell migration and invasion in osteosarcoma cells. EMT acts as a crucial cellular process in tumor metastasis and invasion. Studies have shown that CDH6 could promote EMT and cancer metastasis. In this study, we demonstrated CDH6 could regulate EMT in osteosarcoma, which was consistent with previous studies in other tumors. Besides, we also found that the expression of CDH6 directly increases with osteosarcoma progression. Importantly, CDH6 was correlated with osteosarcoma oncogenesis-associated cellular properties, such as motility and invasiveness, thus indicating that CDH6 might be potential attractive targets for therapeutic intervention of human osteosarcoma.
To date, emerging information has revealed that CDH6 knockdown alters tumor biological activity and cell motility.13 One recent study has shown that CDH6 was found to be upregulated in LMP1-positive NPC tissues and identified to be a potential target of miR-203,12 as was also demonstrated in osteosarcoma cells in our study. However, the exact function of CDH6 in osteosarcoma and the miRNAs in regulating CDH6 expression in osteosarcoma remains still unknown. Conceivably, inhibition of CDH6 might be a promising potential molecular therapeutic strategy for human osteosarcoma. In the present study, we identified that high levels of CDH6 expression were more frequent in human osteosarcoma tissues and patients with low levels of CDH6 had longer DFS and OS. Besides, miR-223-3p overexpression led to suppressed cell invasion, migration, growth, and proliferation in vitro and in vivo. Moreover, we performed clinical analysis data and found a negative association between the expression of miR-223-3p and CDH6 in human osteosarcoma samples, thereby suggesting that the miR-223-3p/CDH6 axis might be a novel and promising candidate for the prevention of metastasis and tumorigenesis in human osteosarcoma.
Taken together, our findings demonstrate that CDH6 exerts as an important independent biomarker that could predict human osteosarcoma clinical outcomes. miR-223-3p can inhibit cell invasion, migration, growth, and proliferation in osteosarcoma by directly targeting CDH6. CDH6 expression was upregulated in osteosarcoma patients and negatively correlated with that of miR-223-3p, thereby supporting that the miR-223-3p/CDH6 axis might be an ideal predictor and therapeutic candidate for human osteosarcoma clinical outcomes.
Materials and Methods
Plasmids and Reagents
The expression vector for CDH6 was generated by cloning the PCR product into a pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) or pCDH plasmid (System Biosciences, Mountain View, CA, USA). Mutant or wild-type promoter-containing luciferase reporters were constructed by the insertion of PCR-amplified promoter fragments from genomic DNA into the pGL4-Basic vector (Promega, Madison, WI, USA). The primer sequences are shown in Table S2. To introduce mutations into the seed sequences of predicted miR-223-3p target sites within the CDH6 3′ UTR, recombinant PCR was applied using the primers mentioned (Table S2). Lentiviruses were produced by co-transfection of HEK293T cells with recombinant lentiviral vectors and pPACK Packaging Plasmid Mix (System Biosciences, Mountain View, CA, USA) using MegaTran reagent (Origene, Rockville, MD, USA). After 48 hr transfection, the lentiviruses were collected and added to the medium of the target cells with 8 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA). Stable cell lines were selected for approximately 2 months with 1 μg/mL puromycin. Individual clones or pooled clones were screened by standard immunoblotting protocols and produced similar results. The expression vector for CDH6 siRNA was purchased from GenePharma (Shanghai, China).
Anti-CDH6 (MA1-06305), anti-PAX5 (PA1-109), anti-HDAC4 (PA1-863), and anti-TET3 (ab139311) were bought from Thermo Fisher Scientific (Rockford, IL, USA). Anti-EYA3 (ab95876), anti-SOX11 (ab134107), and anti-β-actin (ab8227) were purchased from Abcam (Cambridge, MA, USA).
Patients and Specimens
133 conventional osteosarcoma and adjacent noncancerous tissues used in the study were investigated on the basis of accepted radiological and pathological criteria. The follow-up information of the patients was updated every month. The overall survival was defined as the time elapsed from surgery to death. Specimens were divided into two portions: one portion was used for histopathologic assessment and the other portion was immediately snap frozen in liquid nitrogen and stored at −80°C until RNA extraction. The clinical and demographic characteristics are shown in Table S1. Clinical information was observed according to the patient records. This research was conducted with informed consent of the patients and approved by the Institutional Review Committee of the General Hospital of the People’s Liberation Army (Beijing, China).
Histopathologic Assessment
For histopathologic assays, tissues were fixed in 4% buffered paraformaldehyde for 48 hr and subsequently decalcified with buffered EDTA (20% EDTA [pH 7.4]). Tissues were then embedded in paraffin, sectioned, and stained with H&E. For immunohistochemistry (IHC) assays, briefly, the sections were pretreated with trypsin (0.05%) for 10 min before treatment with 3% (vol/vol) H2O2 for 15 min. Then the sections at room temperature for 1 hr were then blocked with 10% goat serum. After washing with PBS, anti-CDH6 antibody (1:25 dilution) was applied to the sections, and the sections were incubated at 4°C overnight. The sections were then washed with PBS for 15 min and incubated with biotinylated secondary antibody using a Histostain Plus kit (Invitrogen, Carlsbad, CA, USA). The sections were washed and incubated with 3, 3′-diaminobenzidine (DAB) substrate for 2 min.
The IHC staining was evaluated by two pathologists blinded to the origin of the specimen using light microscopy. H-score method that combines the values of immunoreaction intensity and the percentage of cells stained was applied to investigate the total immunohistochemical scoring as previously described.51 Briefly, H-score was achieved by multiplying the percentage of weakly stained cells (times 1), the percentage of moderately stained cells (times 2), and the percentage of strongly stained cells (times 3). Score ≤2.1 was defined as low score, and score between 2.1 and 3 was defined as high score.
Cell Culture and Transfection
143B and U2OS cell lines, which had been tested for mycoplasma contamination, were bought from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Stable cell lines overexpressing miR-223-3p and CDH6 were established by lentiviral transduction using a pCDH plasmid (System Biosciences, Mountain View, CA, USA). Cells were routinely cultured at 37°C in an atmosphere with 5% CO2 and in DMEM with high glucose supplemented with 100 μg/mL streptomycin, 10% fetal calf serum (FCS), and 100 IU/mL penicillin. About the transfection, cells were seeded with the indicated plasmids in 6- or 24-well plates using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) on the basis of the manufacturer’s instructions. The miRNA inhibitors (Ambion, Grand Island, NY, USA) were transfected at a concentration of 50 nM. The miRNA mimics were transfected into the cells using FuGENE HD (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
Luciferase Assay
The cells at 70% confluence were seeded in 24-well plates. The reporter constructs containing the mutant or wild-type CDH6 3′ UTR were co-transfected with miR-223-3p into cells using Lipofectamine 2000 reagent on the basis of the manufacturer’s protocol. The cells were collected after 48 hr and assessed for β-galactosidase and luciferase activities as previously described.84
Western Blotting
Total protein extracts were prepared for western blot analysis as previously described.85 The membranes were incubated with antibodies to CDH6 (1:200 dilution) and β-actin (1:500 dilution). The immunocomplexes were visualized via chemiluminescence using an ECL kit (Amersham Biosciences, Piscataway, NJ, USA).
RNA Extraction and qRT-PCR
Total RNA was extracted and reverse transcribed into cDNA using an RNeasy Mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. CFX Connect Real-Time PCR Detection System (Life Science Research, Hercules, CA, USA) was used to detect and quantify miRNA expression. The relative expression level of the miRNA was calculated using the comparative Ct method. Universal small nuclear RNA U6 (RNU6B) was used as the endogenous control for the miRNAs. Sequences of the primers used for qRT-PCR analysis are listed in Table S2.
Anchorage-Dependent and Anchorage-Independent Growth Assays
Cell proliferation was examined using a CCK-8 Kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. To analyze anchorage-independent growth, transfected cells were seeded in 96-well plates and examined at 0, 24, 48, 72, and 96 hr as previously described.84
Wound Healing Assays
Cells at 70% confluence were seeded in 6-well plates in culture medium for wound healing assays. After 24 hr, the confluent cellular monolayer was scratched with a fine pipette tip. For migration, using a microscope, the rate of wound closure was observed at the indicated times.
Cell Invasion Assays
Matrigel invasion chambers (BD Biosciences, San Jose, CA, USA) were performed to assess cell invasion according to the manufacturer’s instructions. Briefly, cells were placed on the upper surface of the Transwell inserts. After 24 hr, the invasive cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The number of invasive cells was counted in five randomly selected microscopic views and photographed.
Animal Experiments
Approximately 1.2 × 107 143B cells were injected into 6-week-old BALB/c mice. For the tumor growth model, cells labeled with firefly luciferase and stably transfected with the pCDH control vector, pCDH-miR-223-3p or PCDH-miR-223-3p, and CDH6 were subcutaneously injected. Tumor volume was examined according to the following formula: volume = (longest diameter × shortest diameter2) / 2. Tumor growth was calculated by caliper measurements. Excised tumors were weighed, and portions were frozen in liquid nitrogen or fixed in 4% paraformaldehyde for further study. The animals were imaged on day 35 using the IVIS200 imaging system (Xenogen, Alameda, CA, USA). The mice were then killed, and the lung was weighted and fixed with 4% paraformaldehyde for further study. For in vivo lung metastasis study, 1 × 106 143B cells labeled with firefly luciferase carrying indicated constructs were injected into the lateral tail vein of BALB/c female mice. The animal studies were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at the General Hospital of the People’s Liberation Army.
Statistical Analysis
The Cox regression model was used to perform univariate and multivariate analyses. The survival analysis was performed using the Kaplan-Meier method, and differences in survival curves were evaluated by the log rank test. The qRT-PCR data were investigated using one-way ANOVA with Tukey’s post hoc test. Correlation was determined through Pearson’s χ2 analysis using GraphPad PRISM 6 (GraphPad, San Diego, CA, USA). All statistical tests were two sided. Statistical calculations were applied using SPSS 17.0. The data are presented as the means ± SD. All in vitro experiments were performed in triplicate and were repeated three times. p < 0.05 was considered significant statistically.
Author Contributions
S.J. and Y.W. conceived the project and designed the experiments; Q.S., Q.J., X.X., Y.X., M.X., and W.B. performed experiments; Q.S., Q.J., and Y.W. wrote the manuscript; all authors analyzed data; and W.J.M. and S.B.G. supervised the project.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This study was financially supported by grants from National Programs for High Technology Research and Development (2015AA033701) and the National Natural Science Foundation (81672195, 81472589, 81672602, 81372834, and 81371976).
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.03.009.
Contributor Information
William J. Maloney, Email: wmaloney@stanford.edu.
Yan Wang, Email: yanwang301@126.com.
Supplemental Information
References
- 1.Gianferante D.M., Mirabello L., Savage S.A. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Gill J., Ahluwalia M.K., Geller D., Gorlick R. New targets and approaches in osteosarcoma. Pharmacol. Ther. 2013;137:89–99. doi: 10.1016/j.pharmthera.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Luetke A., Meyers P.A., Lewis I., Juergens H. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat. Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Dai H., Lv Y.F., Yan G.N., Meng G., Zhang X., Guo Q.N. RanBP9/TSSC3 complex cooperates to suppress anoikis resistance and metastasis via inhibiting Src-mediated Akt signaling in osteosarcoma. Cell Death Dis. 2016;7:e2572. doi: 10.1038/cddis.2016.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlanga P., Muñoz L., Piqueras M., Sirerol J.A., Sánchez-Izquierdo M.D., Hervás D., Hernández M., Llavador M., Machado I., Llombart-Bosch A. miR-200c and phospho-AKT as prognostic factors and mediators of osteosarcoma progression and lung metastasis. Mol. Oncol. 2016;10:1043–1053. doi: 10.1016/j.molonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S.N., Luo S., Liu C., Piao Z., Gou W., Wang Y., Guan W., Li Q., Zou H., Yang Z.Z. miR-491 inhibits osteosarcoma lung metastasis and chemoresistance by targeting αB-crystallin. Mol. Ther. 2017;25:2140–2149. doi: 10.1016/j.ymthe.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer S.V., Ranjan A., Elias H.K., Parrales A., Sasaki H., Roy B.C., Umar S., Tawfik O.W., Iwakuma T. Genome-wide RNAi screening identifies TMIGD3 isoform1 as a suppressor of NF-κB and osteosarcoma progression. Nat. Commun. 2016;7:13561. doi: 10.1038/ncomms13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuijjer M.L., Hogendoorn P.C., Cleton-Jansen A.M. Genome-wide analyses on high-grade osteosarcoma: making sense of a genomically most unstable tumor. Int. J. Cancer. 2013;133:2512–2521. doi: 10.1002/ijc.28124. [DOI] [PubMed] [Google Scholar]
- 9.Xu M., Jin H., Xu C.X., Sun B., Song Z.G., Bi W.Z., Wang Y. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol. Ther. 2015;23:89–98. doi: 10.1038/mt.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu K.P., Ma X.L., Zhang C.L. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 axis. Mol. Ther. 2017;25:2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H., Luo S., Wang Y., Liu C., Piao Z., Xu M., Guan W., Li Q., Zou H., Tan Q.Y. miR-135b stimulates osteosarcoma recurrence and lung metastasis via Notch and Wnt/β-catenin signaling. Mol. Ther. Nucleic Acids. 2017;8:111–122. doi: 10.1016/j.omtn.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo L.L., Zhang J., Liu L.Z., Zhou Q., Du S.J., Xin S.Y., Ning Z.P., Yang J., Yu H.B., Yue W.X. Cadherin 6 is activated by Epstein-Barr virus LMP1 to mediate EMT and metastasis as an interplay node of multiple pathways in nasopharyngeal carcinoma. Oncogenesis. 2017;6:402. doi: 10.1038/s41389-017-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Clay M.R., Halloran M.C. Cadherin 6 promotes neural crest cell detachment via F-actin regulation and influences active Rho distribution during epithelial-to-mesenchymal transition. Development. 2014;141:2506–2515. doi: 10.1242/dev.105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goeppert B., Ernst C., Baer C., Roessler S., Renner M., Mehrabi A., Hafezi M., Pathil A., Warth A., Stenzinger A. Cadherin-6 is a putative tumor suppressor and target of epigenetically dysregulated miR-429 in cholangiocarcinoma. Epigenetics. 2016;11:780–790. doi: 10.1080/15592294.2016.1227899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue Y.U., Asami J., Inoue T. Cadherin-6 gene regulatory patterns in the postnatal mouse brain. Mol. Cell. Neurosci. 2008;39:95–104. doi: 10.1016/j.mcn.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Kubota F., Murakami T., Mogi K., Yorifuji H. Cadherin-6 is required for zebrafish nephrogenesis during early development. Int. J. Dev. Biol. 2007;51:123–129. doi: 10.1387/ijdb.062200fk. [DOI] [PubMed] [Google Scholar]
- 17.Gugnoni M., Sancisi V., Gandolfi G., Manzotti G., Ragazzi M., Giordano D., Tamagnini I., Tigano M., Frasoldati A., Piana S., Ciarrocchi A. Cadherin-6 promotes EMT and cancer metastasis by restraining autophagy. Oncogene. 2017;36:667–677. doi: 10.1038/onc.2016.237. [DOI] [PubMed] [Google Scholar]
- 18.Sancisi V., Gandolfi G., Ragazzi M., Nicoli D., Tamagnini I., Piana S., Ciarrocchi A. Cadherin 6 is a new RUNX2 target in TGF-β signalling pathway. PLoS ONE. 2013;8:e75489. doi: 10.1371/journal.pone.0075489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karthikeyan S., Lantvit D.D., Chae D.H., Burdette J.E. Cadherin-6 type 2, K-cadherin (CDH6) is regulated by mutant p53 in the fallopian tube but is not expressed in the ovarian surface. Oncotarget. 2016;7:69871–69882. doi: 10.18632/oncotarget.11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aigner A. MicroRNAs (miRNAs) in cancer invasion and metastasis: therapeutic approaches based on metastasis-related miRNAs. J. Mol. Med. (Berl.) 2011;89:445–457. doi: 10.1007/s00109-010-0716-0. [DOI] [PubMed] [Google Scholar]
- 21.Venkatadri R., Muni T., Iyer A.K., Yakisich J.S., Azad N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Dis. 2016;7:e2104. doi: 10.1038/cddis.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vishnubalaji R., Hamam R., Abdulla M.H., Mohammed M.A., Kassem M., Al-Obeed O., Aldahmash A., Alajez N.M. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 2015;6:e1614. doi: 10.1038/cddis.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalbeth N., Pool B., Shaw O.M., Harper J.L., Tan P., Franklin C., House M.E., Cornish J., Naot D. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann. Rheum. Dis. 2015;74:786–790. doi: 10.1136/annrheumdis-2014-205409. [DOI] [PubMed] [Google Scholar]
- 24.Croci S., Zerbini A., Boiardi L., Muratore F., Bisagni A., Nicoli D., Farnetti E., Pazzola G., Cimino L., Moramarco A. MicroRNA markers of inflammation and remodelling in temporal arteries from patients with giant cell arteritis. Ann. Rheum. Dis. 2016;75:1527–1533. doi: 10.1136/annrheumdis-2015-207846. [DOI] [PubMed] [Google Scholar]
- 25.Nakamachi Y., Ohnuma K., Uto K., Noguchi Y., Saegusa J., Kawano S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann. Rheum. Dis. 2016;75:601–608. doi: 10.1136/annrheumdis-2014-206417. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q., Haupt S., Kreuzer J.T., Hammitzsch A., Proft F., Neumann C., Leipe J., Witt M., Schulze-Koops H., Skapenko A. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann. Rheum. Dis. 2015;74:1265–1274. doi: 10.1136/annrheumdis-2013-204377. [DOI] [PubMed] [Google Scholar]
- 27.Liang H., Liu S., Chen Y., Bai X., Liu L., Dong Y., Hu M., Su X., Chen Y., Huangfu L. miR-26a suppresses EMT by disrupting the Lin28B/let-7d axis: potential cross-talks among miRNAs in IPF. J. Mol. Med. (Berl.) 2016;94:655–665. doi: 10.1007/s00109-016-1381-8. [DOI] [PubMed] [Google Scholar]
- 28.Han R., Ji X., Rong R., Li Y., Yao W., Yuan J., Wu Q., Yang J., Yan W., Han L. MiR-449a regulates autophagy to inhibit silica-induced pulmonary fibrosis through targeting Bcl2. J. Mol. Med. (Berl.) 2016;94:1267–1279. doi: 10.1007/s00109-016-1441-0. [DOI] [PubMed] [Google Scholar]
- 29.Su Y., Wu H., Pavlosky A., Zou L.L., Deng X., Zhang Z.X., Jevnikar A.M. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsui M., Prakash T.P., Corey D.R. Argonaute 2-dependent regulation of gene expression by single-stranded miRNA mimics. Mol. Ther. 2016;24:946–955. doi: 10.1038/mt.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong P., Zhu X., Geng Q., Xia L., Sun X., Chen Y., Li W., Zhou Z., Zhan Y., Xu D. The microRNA-423-3p-Bim axis promotes cancer progression and activates oncogenic autophagy in gastric cancer. Mol. Ther. 2017;25:1027–1037. doi: 10.1016/j.ymthe.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pourshafie N., Lee P.R., Chen K.L., Harmison G.G., Bott L.C., Katsuno M., Sobue G., Burnett B.G., Fischbeck K.H., Rinaldi C. miR-298 counteracts mutant androgen receptor toxicity in spinal and bulbar muscular atrophy. Mol. Ther. 2016;24:937–945. doi: 10.1038/mt.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong H.A., Fatimy R.E., Onodera C., Wei Z., Yi M., Mohan A., Gowrisankaran S., Karmali P., Marcusson E., Wakimoto H. The Cancer Genome Atlas analysis predicts microRNA for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 2015;23:1234–1247. doi: 10.1038/mt.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang X., Teng Y., Samykutty A., Mu J., Deng Z., Zhang L., Cao P., Rong Y., Yan J., Miller D., Zhang H.G. Grapefruit-derived nanovectors delivering therapeutic miR17 through an intranasal route inhibit brain tumor progression. Mol. Ther. 2016;24:96–105. doi: 10.1038/mt.2015.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister E.L., Chase K.O., Sun H., Kennington L.A., Conroy F., Johnson E., Miller R., Borel F., Aronin N., Mueller C. Safe and efficient silencing with a Pol II, but not a Pol lII, promoter expressing an artificial miRNA targeting human Huntingtin. Mol. Ther. Nucleic Acids. 2017;7:324–334. doi: 10.1016/j.omtn.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam J.K., Chow M.Y., Zhang Y., Leung S.W. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X., Zhang H., Song Y., Zhou D., Wang J. Hemagglutinin-targeting artificial microRNAs expressed by adenovirus protect mice from different clades of H5N1 infection. Mol. Ther. Nucleic Acids. 2016;5:e311. doi: 10.1038/mtna.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staedel C., Varon C., Nguyen P.H., Vialet B., Chambonnier L., Rousseau B., Soubeyran I., Evrard S., Couillaud F., Darfeuille F. Inhibition of gastric tumor cell growth using seed-targeting LNA as specific, long-lasting microRNA inhibitors. Mol. Ther. Nucleic Acids. 2015;4:e246. doi: 10.1038/mtna.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, characterization, and lead selection of therapeutic miRNAs targeting Huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun C.C., Li S.J., Zhang F., Zhang Y.D., Zuo Z.Y., Xi Y.Y., Wang L., Li D.J. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol. Ther. Nucleic Acids. 2016;5:e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal S., Raynaud B., Weber M.J. The role of Ca2+ channels of the L-type in neurotransmitter plasticity of cultured sympathetic neurons. Brain Res. Mol. Brain Res. 1989;6:187–196. doi: 10.1016/0169-328x(89)90053-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Bei Y., Huang P., Zhou Q., Shi J., Sun Q., Zhong J., Li X., Kong X., Xiao J. Inhibition of miR-155 protects against LPS-induced cardiac dysfunction and apoptosis in mice. Mol. Ther. Nucleic Acids. 2016;5:e374. doi: 10.1038/mtna.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones K.B., Salah Z., Del Mare S., Galasso M., Gaudio E., Nuovo G.J., Lovat F., LeBlanc K., Palatini J., Randall R.L. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J., Song G., Tang Q., Yin J., Zou C., Zhao Z., Xie X., Xu H., Huang G., Wang J. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231–241. doi: 10.1038/onc.2016.194. [DOI] [PubMed] [Google Scholar]
- 46.Tsai H.C., Tzeng H.E., Huang C.Y., Huang Y.L., Tsai C.H., Wang S.W., Wang P.C., Chang A.C., Fong Y.C., Tang C.H. WISP-1 positively regulates angiogenesis by controlling VEGF-A expression in human osteosarcoma. Cell Death Dis. 2017;8:e2750. doi: 10.1038/cddis.2016.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Z., Gao Y., Shen J., Choy E., Cote G., Harmon D., Bernstein K., Lozano-Calderon S., Mankin H., Hornicek F.J. miR-15b modulates multidrug resistance in human osteosarcoma in vitro and in vivo. Mol. Oncol. 2017;11:151–166. doi: 10.1002/1878-0261.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua Y., Jin Z., Zhou F., Zhang Y.Q., Zhuang Y. The expression significance of serum MiR-21 in patients with osteosarcoma and its relationship with chemosensitivity. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2989–2994. [PubMed] [Google Scholar]
- 49.Yang C., Wang G., Yang J., Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am. J. Cancer Res. 2017;7:2009–2019. [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura S., Aryee D.N., Felicetti F., De Feo A., Mancarella C., Manara M.C., Picci P., Colombo M.P., Kovar H., Carè A., Scotlandi K. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-κB signaling. Oncogene. 2016;35:3944–3954. doi: 10.1038/onc.2015.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y., Tu M.J., Yu Y.F., Wang W.P., Chen Q.X., Qiu J.X., Yu A.X., Yu A.M. Combination therapy with bioengineered miR-34a prodrug and doxorubicin synergistically suppresses osteosarcoma growth. Biochem. Pharmacol. 2015;98:602–613. doi: 10.1016/j.bcp.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirahata M., Osaki M., Kanda Y., Sugimoto Y., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Kawai A., Ito H. PAI-1, a target gene of miR-143, regulates invasion and metastasis by upregulating MMP-13 expression of human osteosarcoma. Cancer Med. 2016;5:892–902. doi: 10.1002/cam4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H., Wang H., Liu H., Chen Y. Effect of miR-143 on the apoptosis of osteosarcoma cells. Int. J. Clin. Exp. Pathol. 2015;8:14241–14246. [PMC free article] [PubMed] [Google Scholar]
- 54.Li J., Gao B., Huang Z., Duan T., Li D., Zhang S., Zhao Y., Liu L., Wang Q., Chen Z., Cheng K. Prognostic significance of microRNA-203 in cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015;8:9512–9516. [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly E.J., Russell S.J. MicroRNAs and the regulation of vector tropism. Mol. Ther. 2009;17:409–416. doi: 10.1038/mt.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy M.S., Tayade C., Smith G.N. Maternal circulating microRNAs and pre-eclampsia: challenges for diagnostic potential. Mol. Diagn. Ther. 2017;21:23–30. doi: 10.1007/s40291-016-0233-0. [DOI] [PubMed] [Google Scholar]
- 57.Rasko J.E., Wong J.J. Nuclear microRNAs in normal hemopoiesis and cancer. J. Hematol. Oncol. 2017;10:8. doi: 10.1186/s13045-016-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J.Q., Papp G., Szodoray P., Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun. Rev. 2016;15:1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Krützfeldt J. Strategies to use microRNAs as therapeutic targets. Best Pract. Res. Clin. Endocrinol. Metab. 2016;30:551–561. doi: 10.1016/j.beem.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., Guo B., Guo P. Specific delivery of miRNA for high efficient inhibition of prostate cancer by RNA nanotechnology. Mol. Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guda S., Brendel C., Renella R., Du P., Bauer D.E., Canver M.C., Grenier J.K., Grimson A.W., Kamran S.C., Thornton J. miRNA-embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F induction. Mol. Ther. 2015;23:1465–1474. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C.L., Wu J.C., Chen G.Y., Yuan P.H., Tseng Y.W., Li K.C., Hwang S.M., Hu Y.C. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol. Ther. 2015;23:79–88. doi: 10.1038/mt.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang B., Yao K., Huuskes B.M., Shen H.H., Zhuang J., Godson C., Brennan E.P., Wilkinson-Berka J.L., Wise A.F., Ricardo S.D. Mesenchymal stem cells deliver exogenous microRNA-let7c via exosomes to attenuate renal fibrosis. Mol. Ther. 2016;24:1290–1301. doi: 10.1038/mt.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Q., Anderson C., Hanus J., Zhao F., Ma J., Yoshimura A., Wang S. Strand and cell type-specific function of microRNA-126 in angiogenesis. Mol. Ther. 2016;24:1823–1835. doi: 10.1038/mt.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi J.G., Bharaj P., Abraham S., Ma H., Yi G., Ye C., Dang Y., Manjunath N., Wu H., Shankar P. Multiplexing seven miRNA-based shRNAs to suppress HIV replication. Mol. Ther. 2015;23:310–320. doi: 10.1038/mt.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khella H.W.Z., Butz H., Ding Q., Rotondo F., Evans K.R., Kupchak P., Dharsee M., Latif A., Pasic M.D., Lianidou E. miR-221/222 are involved in response to sunitinib treatment in metastatic renal cell carcinoma. Mol. Ther. 2015;23:1748–1758. doi: 10.1038/mt.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D’Angelo B., Benedetti E., Cimini A., Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi R.U., Prieto-Vila M., Hironaka A., Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clin. Chem. Lab. Med. 2017;55:648–656. doi: 10.1515/cclm-2016-0708. [DOI] [PubMed] [Google Scholar]
- 69.Bertoli G., Cava C., Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int. J. Mol. Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mamoori A., Gopalan V., Smith R.A., Lam A.K. Modulatory roles of microRNAs in the regulation of different signalling pathways in large bowel cancer stem cells. Biol. Cell. 2016;108:51–64. doi: 10.1111/boc.201500062. [DOI] [PubMed] [Google Scholar]
- 71.Talekar M., Trivedi M., Shah P., Ouyang Q., Oka A., Gandham S., Amiji M.M. Combination wt-p53 and microRNA-125b transfection in a genetically engineered lung cancer model using dual CD44/EGFR-targeting nanoparticles. Mol. Ther. 2016;24:759–769. doi: 10.1038/mt.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsieh T.H., Hsu C.Y., Tsai C.F., Long C.Y., Wu C.H., Wu D.C., Lee J.N., Chang W.C., Tsai E.M. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol. Ther. 2015;23:656–666. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Tsuta K., Nokihara H., Tamura T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hiraki M., Nishimura J., Takahashi H., Wu X., Takahashi Y., Miyo M., Nishida N., Uemura M., Hata T., Takemasa I. Concurrent targeting of KRAS and AKT by miR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol. Ther. Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stiuso P., Potenza N., Lombardi A., Ferrandino I., Monaco A., Zappavigna S., Vanacore D., Mosca N., Castiello F., Porto S. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol. Ther. Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 76.Gallo Cantafio M.E., Nielsen B.S., Mignogna C., Arbitrio M., Botta C., Frandsen N.M., Rolfo C., Tagliaferri P., Tassone P., Di Martino M.T. Pharmacokinetics and pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol. Ther. Nucleic Acids. 2016;5 doi: 10.1038/mtna.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oksuz Z., Serin M.S., Kaplan E., Dogen A., Tezcan S., Aslan G., Emekdas G., Sezgin O., Altintas E., Tiftik E.N. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol. Biol. Rep. 2015;42:713–720. doi: 10.1007/s11033-014-3819-9. [DOI] [PubMed] [Google Scholar]
- 78.Fang G., Liu J., Wang Q., Huang X., Yang R., Pang Y., Yang M. MicroRNA-223-3p regulates ovarian cancer cell proliferation and invasion by targeting SOX11 expression. Int. J. Mol. Sci. 2017;18:E1208. doi: 10.3390/ijms18061208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo J., Cao R., Yu X., Xiao Z., Chen Z. MicroRNA-223-3p inhibits human bladder cancer cell migration and invasion. Tumour Biol. 2017;39 doi: 10.1177/1010428317691678. 1010428317691678. [DOI] [PubMed] [Google Scholar]
- 80.Bozec A., Zangari J., Butori-Pepino M., Ilie M., Lalvee S., Juhel T., Butori C., Brest P., Hofman P., Vouret-Craviari V. MiR-223-3p inhibits angiogenesis and promotes resistance to cetuximab in head and neck squamous cell carcinoma. Oncotarget. 2017;8:57174–57186. doi: 10.18632/oncotarget.19170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhi Y., Pan J., Shen W., He P., Zheng J., Zhou X., Lu G., Chen Z., Zhou Z. Ginkgolide B inhibits human bladder cancer cell migration and invasion through microRNA-223-3p. Cell. Physiol. Biochem. 2016;39:1787–1794. doi: 10.1159/000447878. [DOI] [PubMed] [Google Scholar]
- 82.Giray B.G., Emekdas G., Tezcan S., Ulger M., Serin M.S., Sezgin O., Altintas E., Tiftik E.N. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol. Biol. Rep. 2014;41:4513–4519. doi: 10.1007/s11033-014-3322-3. [DOI] [PubMed] [Google Scholar]
- 83.Li G., Passebosc-Faure K., Gentil-Perret A., Lambert C., Genin C., Tostain J. Cadherin-6 gene expression in conventional renal cell carcinoma: a useful marker to detect circulating tumor cells. Anticancer Res. 2005;25(1A):377–381. [PubMed] [Google Scholar]
- 84.Ji Q., Xu X., Xu Y., Fan Z., Kang L., Li L., Liang Y., Guo J., Hong T., Li Z. miR-105/Runx2 axis mediates FGF2-induced ADAMTS expression in osteoarthritis cartilage. J. Mol. Med. (Berl.) 2016;94:681–694. doi: 10.1007/s00109-016-1380-9. [DOI] [PubMed] [Google Scholar]
- 85.Xu X., Fan Z., Kang L., Han J., Jiang C., Zheng X., Zhu Z., Jiao H., Lin J., Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J. Clin. Invest. 2013;123:630–645. doi: 10.1172/JCI64265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.