Abstract
Background
Shiga toxin producing E. coli (STEC) is an emerging zoonotic pathogen that can cause acute renal failure, especially in children. Clinical microbiology laboratories may fail to detect STEC and other diarrhoeic E. coli unless purposive rigorous screening procedures are followed using appropriate diagnostic technology; CHROMagar™STEC has rarely been used for isolation of African diarrhoeic E. coli hence characteristics of isolates on this medium are not yet fully understood. This study aimed to determine the prevalence and characteristics of STEC and other diarrhoeic E. coli isolated on CHROMagar™STEC from stool samples submitted to the microbiology laboratory of a South African public sector tertiary care hospital.
Results
In total, 733 stool samples were tested. Of these, 4.5% (33/733) possessed diarrhoeic E. coli. Of the diarrheic E. coli, 5/33 (15.2%) were STEC, 15/33 (45.5%) EAggEC, 6/33 (18.2%) atypical EPEC, 5/33 (15.2%) typical EPEC, and 1/33 (3%) DAEC. None of the STEC isolates had been identified by routine testing (based on using sorbitol media to test for E. coli O157: H7 strains and not the other STEC) in the laboratory. Of the 33 strains, 55% (95% CI = 40.8–72.7) showed resistance to ampicillin.
Conclusions
CHROMagar™STEC enabled detection of tellurite - resistant diarrhoeic E. coli that would be missed using routine methods. Further studies are needed to determine the proportion and characteristics of those which might have been missed using this approach.
Keywords: CHROMagar™STEC, Tellurite resistant diarrhoeic E. coli, STEC, Shiga toxin, Africa
Background
Shiga toxin-producing E. coli (STEC) are important causes of diarrhoea, haemorrhagic colitis, bloody diarrhoea, and haemolytic-uraemic syndrome (HUS) [1]. The use of sorbitol MacConkey to screen for STEC has limitations due to its inability to detect emerging sorbitol-fermenting non-O157 STEC and sorbitol-fermenting O157 strains [2]. Indeed, new chromogenic media, such as CHROMagar™STEC, have been developed to identify both O157 and non-O157 STEC with some targeting only specific STEC serotypes [3].
CHROMagar™STEC is a screening medium with a STEC recovery rate of 70%, which selects for tellurite-resistant STEC [3]. The STEC serotypes commonly associated with severe disease globally (O26, O45, O103, O111, O121, O145, and O157) [4] can readily be isolated on CHROMagar™STEC because they are commonly tellurite resistant [5]. Tellurite resistance is defined as growth at a minimum inhibitory concentration of 2.5 μg/ml to ≥20 μg/ml), [6] however the concentration of potassium tellurite in CHROMagar™STEC is proprietary. On this agar, the STEC serotypes that are commonly associated with severe human infections (O26, O45, O103, O111, O121, O145, and O157) [4, 7] form mauve colonies, other Enterobacteriaceae form blue or colourless colonies, while the growth of gram-positive bacteria is inhibited (http://www.chromagar.com/products-chromagar-stec-focus-on-stec-e-coli-51.html#.WNbR4vnyu01). On this media, tellurite-resistant E. coli pathotypes including STEC, form mauve coloured colonies [8]. The chemistry behind the formation of the mauve coloured colonies is proprietary information. Given that the routinely used culture method for STEC is based on sorbitol fermentation, and intended for E. coli O157: H7, it would miss sorbitol fermenting STEC.
Even though diarrhoea is not commonly treated using antibiotics, antibiograms of diarrhoeic E. coli can be utilised for sentinel surveillance of antimicrobial resistance [9]. Even though diarrhoeic E. coli infections are usually self-limiting, persistent and invasive infections, especially in the immunocompromised persons, may necessitate the use of antibiotics [10].
In an earlier study which evaluated STEC diagnostic technology on African isolates, an in-house-developed duplex real-time PCR assay for detection of stx1 and stx2 was validated and tested on diarrhoeic stool samples and then used as a reference standard to assess the performance of CHROMagar™STEC [11]. A related study used quantitative proteomics to show that mauve colonies on CHROMagar™STEC produced tellurite resistance proteins TerA, TerE, TerC, TerB, TerD, TerW, and TerZ [12]. This study purposed to determine the characteristics (pathotypes, serotypes, antibiogram, and sorbitol fermentation ability) of African isolates that grew on CHROMagar™STEC with and without TSB enrichment of stool samples submitted to the microbiology laboratory of a South African public sector tertiary hospital.
Methods
Sample collection and processing
This study is part of a larger study on STEC in human and non-human sources in Cape Town. At the National Health Laboratory Services (NHLS) laboratories at Groote Schuur Hospital (GSH), stool specimens are not routinely screened for diarrhoeic E. coli. Physicians are advised to contact the laboratory within seven days if there is a clinical suspicion of diarrhoeic E. coli infections such as STEC. Diarrhoea, in this study, was defined as more than three loose stools in less than 24 h. Freshly collected diarrhoeic stool specimens (in a screw-capped stool collection container and delivered in a temperature regulated box) submitted to the NHLS at GSH, Cape Town, between September 2014 and May 2015 for microbiological testing (as part of standard patient care) were screened for diarrhoeic E. coli. All the stool samples from diarrhoea samples were tested irrespective of the age of the patient as recommended by the Centre for Disease Control (CDC) [13]. In order to isolate tellurite resistant STEC and other diarrheic E. coli, samples were either inoculated directly onto CHROMagar™STEC (CHROMagar Microbiology, Paris, France) or subjected first to overnight enrichment in Tryptic Soy Broth (TSB; Oxoid, Basingstoke, U.K) before inoculation onto CHROMagar™STEC (Fig. 1). Mauve colonies, indicative of growth and therefore tellurite resistance, were visually identified and selected (a maximum of 5 per sample) for characterisation. Pathotypes and serotypes were determined as previously described [11].
Fig. 1.
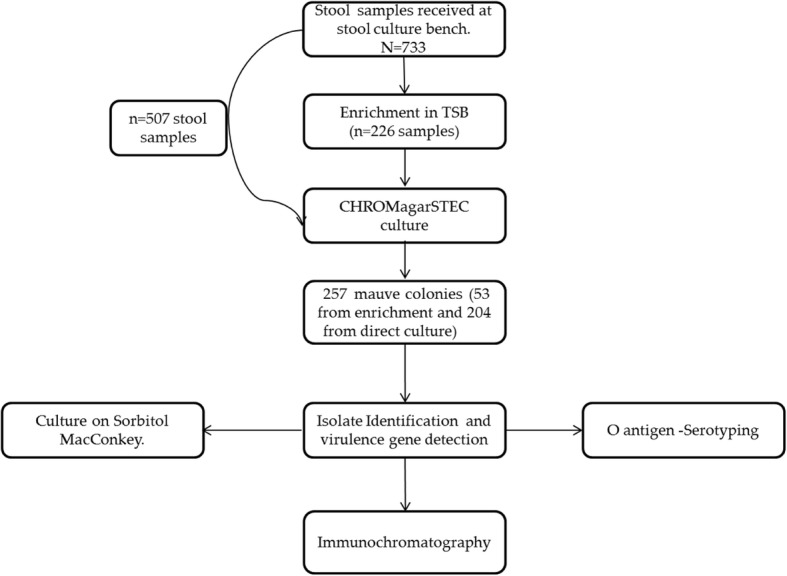
Summary of the methods used in this study
Briefly, to dertermine the pathotypes, end point PCR reactions targeting the fimbrial adhesion gene (daaC) for diffusely adherent E. coli (DAEC), the anti-aggregation protein transporter gene (aat) for enteroaggregative E. coli (EAggEC), heat-stable (ST) and heat-labile (LT) enterotoxin genes of enterotoxigenic E. coli (ETEC), the invasive plasmid antigen gene (ipa) for enteroinvasive E. coli (EIEC), and the intimin coding gene (eae) for enteropathogenic E. coli (EPEC) were done as previously described [14]. An in-house developed duplex real-time PCR assay was used to screen for stx1 and stx2. The stx + E. coli isolates were tested for Shiga toxin production using the immunocard STAT® EHEC (Meridian Biosciences Inc., Cincinnati, Ohio, United States). Subsequently, clinical data were reviewed for all diarrhea cases that carried STEC to assess for presentation of typical haemolytic uremic syndrome (tHUS).
To determine the serotypes, serology was done to detect the somatic O- antigens using reagents supplied by the Statens Serum Institut, Copenhagen, Denmark and the tube agglutination method as previously described [15]. Serology to detect the H-antigens was not done.
Culture characteristics on sorbitol MacConkey
Briefly, mauve isolates identified as E. coli using the VITEK 2 system (bioMérieux, USA) were cultured on Sorbitol MacConkey and incubated for 18 h at 35 °C. Colourless colonies were considered to be non-sorbitol fermenting. Antimicrobial susceptibility testing.
Isolates were tested for susceptibility to 19 antibiotics following the Clinical Laboratory Standards Institute guidelines (CLSI, 2015). Since VITEK offers a limited MIC range, we further re-tested these isolates using the broth microdilution based GNX2F sensititre MIC plate (Thermo-scientific, USA) to confirm the MICs observed using VITEK 2 systems. MIC values from broth microdilution were used as the final reference for classification as resistant (R), intermediate (I) or susceptible (S). E. coli ATCC 25922 was used as the control strain. The antibiotics included in the broth microdilution panel on the GNX2F sensititre plate were: amikacin (AMK), aztreonam (AZT), cefepime (FEP), cefotaxime (CTX), ceftazidime (CAZ), ciprofloxacin (CIP), colistin (COL), doripenem (DOR), doxycycline (DOX), ertapenem (ETP), gentamicin (CIN), imipenem (IMP), levofloxacin (LEV), meropenem (MEM), minocycline (MIN), piperacillin-tazobactam (TZP), polymixin B (POL), ticarcillin-clavulanic acid (TIM), tigecycline (TGC), tobramycin (TOB), and trimethoprim – sulfamethoxazole (SXT).
MIC data were entered into WHONET 5.6 software. Resistance to third-generation cephalosporins and or carbapenems was confirmed using the ESB1F sensititre MIC plates (Thermo-scientific, USA).
Data analysis
Data on cultural characteristics, serotypes, and pathotypes was entered into Microsoft Office Excel 2016 (Microsoft Corporation, USA) software, coded and exported to STATA version 12 for analysis. Further analysis was done using the R-statistical package [16]. Results were entered in WHONET 5.6 (World Health Organization, Geneva) and interpreted in accordance with current Clinical Laboratory Standards Guidelines [17].
Results
The mean age of the patients whose stool samples possessed diarrheic E. coli was 20.6 (± 17.4) years (Table 1). Of the 733 specimens, 507 were directly inoculated on CHROMagar™STEC while 226 were first enriched in TSB before inoculation on CHROMagar™STEC. Of 733 specimens screened on CHROMagar™STEC, 257 (35%) yielded mauve colonies. Mauve colonies were obtained from 40% (204/507) of the directly inoculated specimens, and 24% (53/226) of stool samples that were enriched first in tryptic soy broth (p = 0.001). Of the mauve colonies obtained from direct culture, 5.9% (12/204)were diarrhoeic E. coli while 39.6% (21/53) of the mauve colonies obtained following enrichment in TSB were diarrhoeic E. coli.
Table 1.
Growth and Virulence properties of diarrhoeic E. coli that formed Mauve colonies on CHROMagar™STEC
| Isolate No. | HUS | Bloody diarrhoea | Serotype | Virulence genes. | Pathotype | Immunoassay | TSB enrichment | Sorbitol |
|---|---|---|---|---|---|---|---|---|
| 271 | – | O55 | eae,bfp | Typical EPEC | – | – | ||
| 724 | – | O55 | eae,bfp | Typical EPEC | + | – | ||
| 722.1 | – | O55 | eae,bfp | Typical EPEC | + | – | ||
| 279 | – | O55 | eae, bfp | Typical EPEC | – | – | ||
| 344 | – | O55 | eae, bfp | Typical EPEC | + | – | ||
| 345.1 | – | O55 | eae, bfp | Typical EPEC | + | + | ||
| 291 | – | O182 | eae | Atypical EPEC | – | + | ||
| 286 | – | O111 | eae | Atypical EPEC | + | + | ||
| 600 | + | Not Identified | eae | Atypical EPEC | + | + | ||
| 19 | + | O26 | eae | Atypical EPEC | – | – | ||
| 284 | – | O25 | eae | Atypical EPEC | – | + | ||
| 15.2 | + | O26 | eae | Atypical EPEC | – | – | ||
| 63.2 | – | O1 | daaC | DAEC | – | + | ||
| 689 | – | O175 | aat | EAggEC | + | – | ||
| 326 | – | O16 | aat | EAggEC | + | – | ||
| 231 | – | O104 | aat | EAggEC | – | – | ||
| 229.1 | – | O104 | aat | EAggEC | – | – | ||
| 473 | – | O33 | aat | EAggEC | + | + | ||
| 371.1 | + | O25 | aat | EAggEC | + | – | ||
| 336 | – | O175 | aat | EAggEC | + | + | ||
| 229 | – | O104 | aat | EAggEC | – | – | ||
| 733 | – | O104 | aat | EAggEC | + | – | ||
| 502 | – | O9 | aat | EAggEC | + | + | ||
| 688 | – | O3 | aat | EAggEC | + | + | ||
| 480 | – | O8 | aat | EAggEC | + | + | ||
| 250 | – | O175 | aat | EAggEC | – | + | ||
| 207 | – | O104 | aat | EAggEC | – | + | ||
| 696 | – | O175 | aat | EAggEC | + | + | ||
| 598 | – | – | O186 | stx 1 , eae | STEC | – | + | + |
| 29.5 | – | – | O186 | eae, stx 1 | STEC | – | + | + |
| 602 | – | – | O101 | eae, stx 1 , stx 2 | STEC | – | + | – |
| 232.1 | – | – | Not identified | stx 1 | STEC | Shiga toxin 1 | + | + |
| 73 | – | Not identified | stx 1 | STEC | Shiga toxin 1 | + |
EAggEC-enteroaggregative E. coli, STEC-Shiga-toxin producing E. coli, EPEC-enteropathogenic E. coli, DAEC-diffusely adherent E. coli. Fimbrial adhesion gene (daaC), the anti-aggregation protein transporter gene (aat), heat-stable (ST) and heat-labile (LT) enterotoxin genes, the intimin coding gene (eae) and the bundle-forming pili gene (bfp), Shiga toxin 1 (stx1) and 2 (stx2)
Tellurite resistant diarrhoeic E. coli were therefore obtained from 5% (33/733) of the stool specimens.
The growth and virulence attributes of the isolates that formed mauve colonies on CHROMagar™STEC are shown in Table 1. Of the 33 diarrhoeic E. coli, 64% (21/33) were obtained following enrichment in TSB while 36% (12/33) were obtained without enrichment (p = 0.004). Of these, 15.2% (5/33) were STEC, 45.5% (15/33) were EAggEC, 18.2% (6/33) were atypical EPEC, 15.2% (5/33) were typical EPEC, and 3% (1/33) were DAEC. No EIEC or ETEC were detected.
Among the diarrhoea cases that carried STEC, co-infection with other bacterial pathogens (as evidenced from the routine microbiology testing results) was not detected; and none presented with HUS.
A total of 16 different serotypes were identified, of which serotypes O104 (5/33, 15.2%) and O55 (6/33, 18.2%) were most common. All the E. coli isolates that belonged to serotypes O104 and O55 were EAggEC and EPEC respectively.
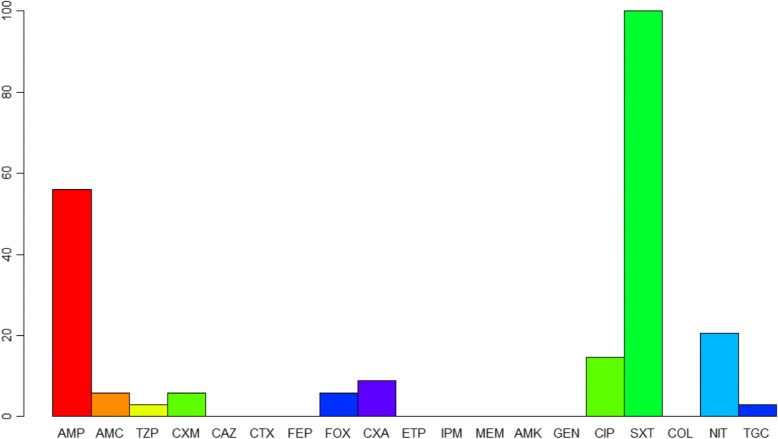
Of the thirty-three diarrhoeic E. coli, 54.5% (18/33) were resistant to AMP, 3% (1/33) to TZP, 6.1% (2/33) to CXM, 3% (1/33) to FOX, 21.2% (7/33) to NIT, 3% (1/33) to TGC and 3% (1/33) to ciprofloxacin (Fig. 2). All the isolates were resistant to SXT.
Fig. 2.
Antimicrobial susceptibility patterns of diarrheic E. coli isolated on CHROMagar™STEC; AMP-ampicillin, AMC-amoxicillin clavulanate, CXM-cefuroxime, CAZ-ceftazidime, CTX-cefotaxime, FEP-cefepime, FOX-cefoxitin, CXA-cefuroxime axetil, ETP- ertapenem, IPM-imipenem, MEM-meropenem, AMK-amikacin, GEN-gentamicin, CIP-ciprofloxacin, SXT- trimethoprim-sulfamethoxazole, COL-colistin, NIT-nitrofurantoin, TGC-tigecycline
Only three isolates were multidrug-resistant, one of which (EAggEC, serotype O25) was resistant to six antibiotics while the atypical (serotype O182) and typical EPEC (serotype O55) were resistant to three and four antibiotics respectively. The commonest resistance profile was resistance to only ampicillin (Table 2). All the STEC were susceptible to all the antibiotics except SXT.
Table 2.
Resistance profiles of Diarrhoeic E. coli obtained with and without enrichment
| Number of isolates | Resistance Profile |
|---|---|
| 18 | AMP |
| 1 | AMP AMC CXA |
| 1 | AMP CXM CXA FOX |
| 1 | AMP AMC TZP CXM CXA FOX |
AMP ampicillin, AMC amoxicillin clavulanate, CXA cefuroxime axetil, CXM cefuroxime, FOX cefoxitin, TZP tazobactam piperacillin, EAggEC enteroaggregative E. coli, EPEC enteropathogenic E. coli
Of the 33 tellurite-resistant diarrhoeic E. coli, 45.5% (15/33) were non – sorbitol fermenting. Of the 15, seven were EPEC (46.7%) while seven (46.7%) were EAggEC. Of the five STEC isolates, only one was non-sorbitol fermenting.
Discussion
The main findings of this study were; (1) Tellurite resistant diarrhoeic E. coli were obtained from 5% (33/733) of the stool specimens and included; five STEC, twelve EPEC, one DAEC, and fifteen EAggEC strains, (2) All the diarrhoeic E. coli isolated in this study were resistant to trimethoprim-sulphamethoxazole while 55% (18/33) were resistant to ampicillin.
Detection of diarrhoeic E. coli in stool
The inhibitory action of potassium tellurite on coliforms was first reported by Fleming in 1940 [18]. Tellurite-containing media have been routinely used to screen for STEC [19], but not the other E. coli pathotypes. However, Hirvonen et al., 2012 reported the formation of mauve colonies by other tellurite resistant diarrhoeic E. coli on this medium [8]. Of the 733 stool samples processed in this study, 33 (5%) contained tellurite-resistant diarrhoeic E. coli. However, tellurite-containing media have limited ability to select for STEC. In a study conducted on STEC from human, animals, and food in Austria, the prevalence of tellurite resistance amongst STEC was 26% - therefore 74% of the STEC would not be detected by the tellurite-containing medium. [20]
Tellurite resistance and antimicrobial resistance are often observed in highly pathogenic diarrhoeic E. coli strains [20]. Commercial selective media such as CHROMagar™STEC and Sorbitol MacConkey with cefixime and potassium - tellurite use this property to select STEC O157: H7 and other pathogenic STEC. There was a relatively low prevalence (5%, 33/733) of tellurite-resistant diarrhoeic E. coli in stool samples submitted to NHLS, GSH. We were unable to determine the prevalence of tellurite susceptible STEC in this study; this is because these would not grow on CHROMagar™STEC.
STEC
Five STEC were detected in this study. All the five were detected after enrichment in TSB, and none was obtained by directly streaking on CHROMagar™STEC. Since the other diarrheic E. coli pathotypes were detected with and without enrichment, it can, therefore, be speculated that TSB enrichment is more beneficial for STEC isolation relative to the other diarrheic E. coli pathotypes. Overall, the number of diarrheic E. coli obtained after enrichment in TSB was significantly higher than the number obtained without (p = 0.004). This may suggest that enrichment in TSB may be a beneficial step as regards recovering STEC (and other pathotypes) from a stool sample. The STEC diagnostic strategy routinely employed by the NHLS clinical laboratory at the GSH targets only O157 STEC and is based on the non-sorbitol fermenting attribute of STEC O157: H7. Therefore, this strategy would miss the non-O157 STEC and sorbitol-fermenting E. coli O157. Of the five STEC reported in this study, only one was non-sorbitol fermenting and belonged to serotype O101. The five non- O157 STEC that we report in this study would have been missed (four because they were sorbitol fermenting and one because it was not O157) given that the routinely used method is based on absence of sorbitol fermentation and targets only E. coli O157:H7. Only two of the five STEC tested positive for Shiga toxin production by immunoassay because the other three did not express the stx genes.
The relatively low rate of STEC isolation in this study could be due to several reasons: (1) The stx primers may not have been suitable for detection of all the stx gene variants [21]. (2) The tellurite susceptible STEC could not be isolated on CHROMagar™STEC. (3) This study had a short sampling frame and not all children presenting with diarrhoea may have had a stool specimen taken. (4) There could have been a low prevalence of STEC in stool samples processed, especially given that the stool specimens were not from patients presenting with bloody diarrhoea, or HUS. There are higher chances of recovering STEC from the stool of patients with bloody diarrhoea or HUS as opposed to those without [22].
In South Africa, as in other African countries, there may be a lower prevalence of STEC as compared to America and Europe. This may be related to the type of diet given to ruminants in America and Europe that favour the proliferation of STEC in cattle-the primary ruminant reservoirs for STEC [23].
It is important to detect STEC in stool because the use of antibiotics such as the quinolones leads to bacterial lysis and toxin release which increases the chances of HUS in infected patients [23]. Four of the five STEC detected in this study carried the stx1 genes. Possession of stx2 genes has been associated with the more severe form of illness [24]. Also, we detected eae in three of the five STEC, while two did not possess the eae gene. The eae gene which is located in the locus of enterocyte effacement (LEE) codes for the intimin protein which is necessary for the formation of attaching-effacing lesions in the intestinal tract [25]. STEC that carry eae (LEE-positive STEC) have been shown to cause more severe disease.
Per the manufacturer of CHROMagar™STEC, the commonly encountered STEC serotypes should form mauve coloured colonies on this medium. However, the serotypes were categorised as “common” (O157, O26, O45, O145, O111, O121, and O103) based on outbreaks that occurred in developed countries. In this study, STEC belonged to serotypes O101 and O186. These serotypes were not detected in an earlier study conducted at the NICD (2006 to 2009) which screened 2378 diarrhoeic E. coli isolates. In that study, STEC in stool had not been purposively targeted (and thus CHROMagar™STEC was not used) but was an incidental finding among EPEC (the strains carried the eae gene) that had been sent to the central public health laboratory for serotyping. The 14 STEC identified in that study belonged to serotypes O4, O5, O21, O26, O84, O111, and O157 [26]. Other related studies reported STEC serotypes O8 and O9 in pigs in South Africa [27]. We did not detect E. coli O157: H7 despite high sensitivity of CHROMagar™STEC for this serotype [3].
Clinical relevance of STEC carriage
On review of the clinical records of the five patients that carried STEC, none presented with typical Hemolytic Uremic Syndrome (HUS). However, the assessment of clinical relevance of STEC carriage was not possible due to the protracted time lag between the onset of sickness in the community and reporting to primary health care and eventual referral to tertiary health care. Earlier research has shown that only a small percentage of acute diarrhea cases in South Africa report to the health centres [28].
EAggEC, EPEC, and DAEC
At the GSH, stool from diarrhoea patients is not routinely screened for diarrhoeic E. coli other than non-sorbitol fermenting O157. Of all the 33 diarrhoeic E. coli strains isolated on CHROMagar™STEC, 45% (15/33) were EAggEC, 18% (6/33) were atypical EPEC, 15% (5/33) were typical EPEC, and 3% (1/33) were DAEC. No enteroinvasive E. coli (E. coli) were detected. This finding is in agreement with an earlier study which showed that EAggEC were most likely to be cultured from stool since they cause a more persistent form of diarrhea [14].
Serotypes
Of the 16 diarrhoeic E. coli serotypes that were isolated using CHROMagar™STEC in this study, only serotypes O111, O104 and O26 were previously reported to be detectable on this medium by studies conducted in Europe [3, 8]. In this study, we report the detection of the other tellurite resistant serotypes including O16, O175, O182, O186, O25, O3, O33, O175, O8, O9, O55, and O101. The dominant tellurite resistant diarrhoeic E. coli serotypes we identified in this study were O104 (15%), and O55 (18%). The cluster of six serotype O55 EPEC had clearly distinct antimicrobial resistance patterns and so was not an outbreak cluster. Since the cluster of five serotype O104 EAggEC strains were noted within a collection period of 38 days and had the same antimicrobial susceptibility pattern, they are possibly an outbreak cluster. However, we were unable to perform an epidemiological investigation to support this hypothesis.
Antibiograms
Even though diarrhoea is not normally treated with antibiotics (except if accompanied with invasive disease), E. coli is used for sentinel surveillance of antimicrobial resistance. The STEC detected in this study were only resistant to trimethoprim-sulphamethoxazole (SXT). This finding is similar to a 2011 study that was conducted in Kenya that reported a high level of resistance to SXT among STEC [29]. EAggEC, EPEC, and DAEC in this study showed resistance to SXT (100%, 28/28) and ampicillin (64%, 18/28). These findings are similar to reports from Kenya which showed a high prevalence of resistance to sulfamethoxazole among intestinal E. coli [30]. There are increasing reports of resistance to multiple antibiotics among EAggEC [31]. We identified multidrug resistance in one EAggEC (resistant to six antibiotics), one atypical EPEC strain (resistant to four antibiotics), and one typical EPEC strain (resistant to three antibiotics).
Sorbitol fermentation
Of the 33 diarrhoeic E. coli isolated on CHROMagar™STEC, 15 (45%) were non-sorbitol fermenting. A similar study conducted in Tanzania reported a 14% prevalence of non-sorbitol fermenting E. coli in 1049 human stool and non-human samples [32]. This is higher than the 2% (15/733) prevalence of non-sorbitol fermenting E. coli reported in this study. The high number of non-sorbitol fermenting EAggEC can be explained by the fact that serotype O104 is one of the prevalent non-sorbitol fermenting serotypes globally [3].
Limitations of this study
CHROMagar™STEC only permits the growth of tellurite resistant STEC and not the tellurite susceptible strains. This study did not have a long sampling timeframe, and not all children that presented with diarrhoea may have had stool specimens taken. Only diarrhoeic E. coli that possessed virulence genes were characterised; therefore, we might have missed strains that lost the virulence genes.
Conclusions
Given that clinical laboratories in Africa largely rely on screening for sorbitol negative STEC O157, the use of CHROMagar™STEC (with enrichment) would enable the detection of non-O157 STEC and other diarrhoeic E. coli pathotypes. However, more research is needed to figure out the extent of tellurite susceptible STEC which would be missed on use of this medium.
Additionally, further research is needed to characterise resistance to SXT and AMP in this region and to establish the clinical relevance of isolating STEC in the absence of a HUS or bloody diarrhea outbreak situation.
Acknowledgements
We acknowledge the Centre for Enteric Diseases of the National Institute for Infectious Disease, for serotyping the diarrheic E. coli.
Funding
This study was funded by the National Research Foundation, South Africa, RISE ACP scholarship and the AfDB-HEST program. The funding bodies had no role in the design of the study, collection, analysis, interpretation of data, and in writing of the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- aat
Anti – aggregation protein transporter gene
- AMK
Amikacin
- ATCC
American type culture collection
- AZT
Aztreonam
- bfp
bundle forming pili gene
- CAZ
Ceftazidime
- CDC
Centre for disease control
- CIN
Gentamicin
- CIP
Ciprofloxacin
- CLSI
Clinical Laboratory Standards Institute
- COL
Colistin
- CTX
Cefotaxime
- daaC
fimbrial adhesion gene
- DAEC
Diffusely adherent Escherichia coli
- DOR
Doripenem
- DOX
Doxycyclin
- eae
intimin gene
- EAggEC
Enteroaggregative Escherichia coli
- EIEC
Enteroinvasive Escherichia coli
- EPEC
Enteropathogenic Escherichia coli
- ETEC
Enterotoxigenic Escherichia coli
- ETP
Ertapenem
- F
Female
- FEP
Cefepime
- GSH
Groote schuur hospital
- HUS
Haemolytic uremic syndrome
- IMP
Imipenem
- ipa
invasive plasmid antigen
- LEV
Levofloxacin
- LT
Heat labile enterotoxin gene
- M
Male
- MEM
Meropenem
- MIC
Minimum inhibitory concentration
- MIN
Minicycline
- NHLS
National health laboratory services
- PCR
Polymerase chain reaction
- POL
Polymixin
- ST
Heat stable enterotoxin gene
- STEC
Shiga toxin producing Escherichia coli
- Stx
Shiga toxin gene
- SXT
Trimethoprim - sulfamethoxazole
- Ter
Tellurium resistance gene
- TGC
Tigecycline
- TIM
Ticarcillin – clavulanic acid
- TOB
Tobramycin
- TSB
Tryptic soy broth
- TZP
Tazobactam - piperacillin
Authors’ contributions
JBK conducted laboratory testing and participated in the writing of the manuscript. KHK and MPN supervised the laboratory testing and participated in the writing of the manuscript. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Written consent was obtained from all study participants or their guardians (for those under the age of 18 years). Ethics approval to conduct this study was obtained from Higher Research and Ethics Committee of the University of Cape Town, Faculty of Health Sciences (HREC 014/2015).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
John Bosco Kalule, Email: kaluleb@gmail.com.
Karen H. Keddy, Email: karenk@nicd.ac.za
Mark P. Nicol, Email: Mark.Nicol@uct.ac.za
References
- 1.Ardissino G, Salardi S, Colombo E, Testa S, Borsa-Ghiringhelli N, Paglialonga F, Paracchini V, Tel F, Possenti I, Belingheri M, et al. Epidemiology of haemolytic uremic syndrome in children. Data from the north Italian HUS network. Eur J Pediatr. 2016;175(4):465–473. doi: 10.1007/s00431-015-2642-1. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2006;43(12):1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 3.Gouali M, Ruckly C, Carle I, Lejay-Collin M, Weill FX. Evaluation of CHROMagar STEC and STEC O104 chromogenic agar media for detection of Shiga toxin-producing Escherichia coli in stool specimens. J Clin Microbiol. 2013;51(3):894–900. doi: 10.1128/JCM.03121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.USDA Foodsafety and Inspection Services: Risk Profile for Pathogenic Non-O157 Shiga Toxin-Producing Escherichia coli (non-O157 STEC). In.: Office of Public Health Science,Office of Policy and Program Development Food Safety and Inspection Service United States Department of Agriculture; 2011: 8–12.
- 5.Kase JA, Maounounen-Laasri A, Son I, Lin A, Hammack TS. Comparison of eight different agars for the recovery of clinically relevant non-O157 Shiga toxin-producing Escherichia coli from baby spinach, cilantro, alfalfa sprouts and raw milk. Food Microbiology. 2015;46(0):280–287. doi: 10.1016/j.fm.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Kerangart S, Douellou T, Delannoy S, Fach P, Beutin L, Sergentet-Thevenot D, Cournoyer B, Loukiadis E. Variable tellurite resistance profiles of clinically-relevant Shiga toxin-producing Escherichia coli (STEC) influence their recovery from foodstuffs. Food Microbiol. 2016;59:32–42. doi: 10.1016/j.fm.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Anses.: Opinion of the French Food Safety Agency on the advisability of revising the definition of pathogenic STEC, specified in AFSSA's Opinion of 15 July 2008. In.: French Agency for Food, Environmental and Occupational Health and Safety ; 2010. Anses, Maisons-Alfort, France.
- 8.Hirvonen JJ, Siitonen A, Kaukoranta SS. Usability and performance of CHROMagar STEC medium in detection of Shiga toxin-producing Escherichia coli strains. J Clin Microbiol. 2012;50(11):3586–3590. doi: 10.1128/JCM.01754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO: Integrated Surveillance of Antimicrobial Resistance. In: Integrated Surveillance of Antimicrobial Resistance. Geneva, Switzerland: World Health Organisation; 2013: 8–10.
- 10.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalule JB, Keddy KH, Smith A, Nicol MP, Robberts L. Development of a real-time PCR assay and comparison to CHROMagarTM STEC to screen for Shiga toxin-producing Escherichia coli in stool, cape town, South Africa. AJLM. 2017;6(1) 10.4102/ajlm.v6i1.609. [DOI] [PMC free article] [PubMed]
- 12.Kalule JB, Fortuin S, Calder B, Robberts L, Keddy KH, Nel AJM, Garnett S, Nicol M, Warner DF, Soares NC et al: Proteomic comparison of three clinical diarrhoeagenic drug-resistant Escherichia coli isolates grown on CHROMagarSTEC media J Proteomics 2017. https://doi: 10.1016/j.jprot.2017.09.003. [DOI] [PubMed]
- 13.National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance. [http://www.cdc.gov/nationalsurveillance/ecoli-surveillance.html].
- 14.Tau NP, Meidany P, Smith AM, Sooka A, Keddy KH, Group for Enteric R, Meningeal Disease Surveillance in South A: Escherichia coli O104 associated with human diarrhea, South Africa, 2004-2011. Emerg Infect Dis 2012, 18(8):1314–1317. https://doi.org/10.3201%2Feid1808.111616 [DOI] [PMC free article] [PubMed]
- 15.Orskov I, Orskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41(3):667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R: A language and environment for statistical Computing R Foundation for Statistical Computing. [ http://www.R-project.org/.]
- 17.CLSI: Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement (M100-S25). In. Wayne,PA: CLSI; 2016.
- 18.Fleming A, Young MY. The inhibitory action of potassium tellurite on coliform bacteria. J Pathol Bacteriol. 1940;51(1):29–35. doi: 10.1002/path.1700510106. [DOI] [Google Scholar]
- 19.Zadik PM, Chapman PA, Siddons CA. Use of tellurite for the selection of verocytotoxigenic Escherichia coli O157. J Med Microbiol. 1993;39(2):155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]
- 20.Orth D, Grif K, Dierich MP, Wurzner R. Variability in tellurite resistance and the ter gene cluster among Shiga toxin-producing Escherichia coli isolated from humans, animals and food. Res Microbiol. 2007;158(2):105–111. doi: 10.1016/j.resmic.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins C, Lawson AJ, Cheasty T, Willshaw GA. Assessment of a real-time PCR for the detection and characterization of verocytotoxigenic Escherichia coli. J Med Microbiol. 2012;61(Pt 8):1082–1085. doi: 10.1099/jmm.0.041517-0. [DOI] [PubMed] [Google Scholar]
- 22.Werber D, Fruth A, Buchholz U, Prager R, Kramer MH, Ammon A, Tschape H. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur J Clin Microbiol Infect Dis. 2003;22(12):726–730. doi: 10.1007/s10096-003-1025-0. [DOI] [PubMed] [Google Scholar]
- 23.Boukhors K, Pradel N, Girardeau JP, Livrelli V, Ou Said AM, Contrepois M, Martin C. Effect of diet on Shiga toxin-producing Escherichia coli (STEC) growth and survival in rumen and abomasum fluids. Vet Res. 2002;33(4):405–412. doi: 10.1051/vetres:2002026. [DOI] [PubMed] [Google Scholar]
- 24.Melton-Celsa AR. Shiga toxin (Stx) classification, structure, and function. Microbiology Spectrum. 2014;2(4) https://doi.org/10.1128%2Fmicrobiolspec.ehec-0024-2013. [DOI] [PMC free article] [PubMed]
- 25.McDaniel TK, Kaper JB. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23(2):399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 26.Smith AM, Tau NP, Sooka A, Keddy KH, Group for Enteric R, Meningeal Disease Surveillance in South A: Surveillance for enterohaemorrhagic Escherichia coli associated with human diarrhoea in South Africa, 2006-2009. J Med Microbiol 2011, 60(Pt 5):681–683. https://doi.org/10.1099%2Fjmm.0.022947-0 [DOI] [PubMed]
- 27.Henton MM, Engelbrecht MM. Escherichia coli serotypes in pigs in South Africa. Onderstepoort J Vet Res. 1997;64(3):175–187. [PubMed] [Google Scholar]
- 28.Okeke IN. Diarrheagenic Escherichia coli in sub-Saharan Africa: status, uncertainties and necessities. Journal of infection in developing countries. 2009;3(11):817–842. doi: 10.3855/jidc.586. [DOI] [PubMed] [Google Scholar]
- 29.Sang WK, Oundo V, Schnabel D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J Infect Dev Ctries. 2012;6(7):572–578. doi: 10.3855/jidc.2196. [DOI] [PubMed] [Google Scholar]
- 30.Chiller TM, Polyak CS, Brooks JT, Williamson J, Ochieng B, Shi YP, Ouma P, Greene C, Hamel M, Vulule J, et al. Daily trimethoprim-sulfamethoxazole prophylaxis rapidly induces corresponding resistance among intestinal Escherichia coli of HIV-infected adults in Kenya. J Int Assoc Physicians AIDS Care (Chic) 2009;8(3):165–169. doi: 10.1177/1545109709333112. [DOI] [PubMed] [Google Scholar]
- 31.Okeke IN, Nataro JP. Enteroaggregative Escherichia coli. Lancet Infect Dis. 2001;1(5):304–313. doi: 10.1016/S1473-3099(01)00144-X. [DOI] [PubMed] [Google Scholar]
- 32.Lupindu AM, Olsen JE, Ngowi HA, Msoffe PL, Mtambo MM, Scheutz F, Dalsgaard A: Occurrence and characterization of Shiga toxin-producing Escherichia coli O157:H7 and other non-sorbitol-fermenting E. coli in cattle and humans in urban areas of Morogoro, Tanzania. Vector borne and zoonotic diseases (Larchmont, NY) 2014, 14(7):503–510. https://doi.org/10.1089%2Fvbz.2013.1502 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.