Abstract
Background
Genome-wide association studies (GWAS) have identified a variant (rs9349379) at the phosphatase and actin regulator 1 (PHACTR1) locus that is associated with coronary artery disease (CAD). The same variant is also an expression quantitative trait locus (eQTL) for PHACTR1 in human coronary arteries (hCA). Here, we sought to characterize PHACTR1 splicing pattern in atherosclerosis-relevant human cells. We also explored how rs9349379 modulates the expression of the different PHACTR1 splicing isoforms.
Methods
We combined rapid amplification of cDNA ends (RACE) with next-generation long-read DNA sequencing to discover all PHACTR1 transcripts in many human tissues and cell types. We measured PHACTR1 transcripts by qPCR to identify transcript-specific eQTLs.
Results
We confirmed a brain-specific long transcript, a short transcript expressed in monocytes and four intermediate transcripts that are different due to alternative splicing of two in-frame exons. In contrast to a previous report, we confirmed that the PHACTR1 protein is present in vascular smooth muscle cells. In 158 hCA from our collection and the GTEx dataset, rs9349379 was only associated with the expression levels of the intermediate PHACTR1 transcripts.
Conclusions
Our comprehensive transcriptomic profiling of PHACTR1 indicates that this gene encodes six main transcripts. Five of them are expressed in hCA, where atherosclerotic plaques develop. In this tissue, genotypes at rs9349379 are associated with the expression of the intermediate transcripts, but not the immune-specific short transcript. This result suggests that rs9349379 may in part influence CAD by modulating the expression of intermediate PHACTR1 transcripts in endothelial or vascular smooth muscle cells found in hCA.
Electronic supplementary material
The online version of this article (10.1186/s12881-018-0616-7) contains supplementary material, which is available to authorized users.
Keywords: PHACTR1, Alternative splicing, Expression quantitative trait locus (eQTL), Myocardial infarction (MI), Coronary artery disease (CAD)
Background
Genome-wide association studies (GWAS) have identified a robust association between coronary artery disease (CAD) and the PHACTR1 locus on chromosome 6p24 [1]. This locus is highly pleiotropic as the same allele associated with increased risk of CAD is protective of migraine [2], cervical artery dissection [3], and fibromuscular dysplasia [4]. Previously, we fine-mapped the CAD association to the candidate causal variant rs9349379 located in an intron of PHACTR1 [5]. This SNP disrupts a MEF2 binding site and is an expression quantitative trait locus (eQTL) for PHACTR1 in human coronary arteries (hCA) [5]. The eQTL result was replicated in the Genotype-Tissue Expression (GTEx) dataset [6]. PHACTR1 encodes proteins of unclear biological functions: it was discovered in a yeast two-hybrid screen because it interacts with protein phosphatase-1 (PP1) and can also bind actin through its RPEL domains [7, 8]. Over-expression experiments have also implicated PHACTR1 in apoptosis [9], angiogenesis [10], cellular matrix remodelling [11], and cell motility [8]. Although these processes are important for atherogenesis and CAD, the precise role(s) that PHACTR1 plays at the site of atherosclerotic lesions in coronary arteries is unknown.
Gupta et al. used CRISPR-Cas9 genome editing experiments in endothelial cells (ECs) derived in vitro from induced pluripotent stem cells to test the effect of genotypes at rs9349379 on the expression of nearby genes [12]. In this cellular system, the regulatory element around rs9349379 did not modulate the expression of PHACTR1, but controlled the expression of the endothelin-1 (EDN1) gene located 600 kb upstream. EDN1 encodes ET-1, a 21 amino acids peptide with a potent vasoconstrictor effect in humans and clear implications for cardiovascular diseases. Paradoxically, rs9349379 is not an eQTL for EDN1 in our hCA samples nor in the GTEx dataset [5, 6]. Together, these results raise the possibility that rs9349379 might control the expression of both PHACTR1 and EDN1 in different cell types and/or upon different environmental stimuli (e.g. inflammation).
For these reasons, we continued to investigate the regulation of PHACTR1 expression in human tissues and cell types. Consistent with a recent study [13], we report here a long PHACTR1 transcript expressed in the brain and a short transcript expressed in the brain, the heart, and monocytes. We also confirmed four intermediate transcripts that are different due to alternative splicing of two exons. They are co-expressed in tissues and cells tested. The shorter intermediate transcripts have a higher expression in the brain, the heart, and monocytes, whereas the longer intermediate transcripts are more expressed in ECs and vascular smooth muscle cells (VSMCs). Finally, we tested genetic associations between genotypes at rs9349379 and the expression of PHACTR1 transcripts (or exons) in hCA samples. Our results strengthen the link between CAD-associated rs9349379 and PHACTR1 expression in non-immune cells found in hCA.
Methods
Cell lines and tissues
We purchased pooled human umbilical vein endothelial cells (HUVEC; C2519A), human aortic endothelial cells (HAEC; CC-2535), and human coronary artery endothelial cells (HCAEC; CC-2585; Lonza) and cultured them in EBM-2 media supplemented with EGM-2 or EGM2-MV SingleQuot kit. We purchased immortalized HAEC (teloHAEC; CRL-4052; ATCC) and cultured them in vascular cell basal medium supplemented with vascular endothelial cell growth kit and puromycin at 0.3 μg/mL. Human coronary artery smooth muscle cells (HCASMC) and human aortic smooth muscle cells (HASMC) were obtained from Dr. Tardif’s lab and cultured in medium 231 (M-231-500) supplemented with smooth muscle growth supplement (S-007-25; Gibco). Monocytes were obtained from Dr. Rioux’s lab and we cultured them in RPMI with 10% fetal bovine serum. RNA from tissues was extracted either with the Ribopure Kit (Ambion) or using EZ1-XL Advance and Qiagen EZ1 RNA Tissue Mini Kit. hCA were obtained from the “Réseau d’Échanges de Tissus et d’Échantillons Biologiques” (RÉTEB) biorepository at the Montreal Heart Institute and Quebec Heart and Lung Institute. We purchased adult brain total RNA (540005–41, lot#6048990) and adult heart total RNA (540011–41, lot#6056165; Agilent Technologies).
Rapid Amplification of cDNA Ends (RACE)
We extracted total RNA from HUVEC, teloHAEC, HCAEC, HAEC, HCASMC and HASMC P20 (with a passage (P) lower than 8 for primary cells unless specified) cells grown to confluence in 100 mm dishes with the RNeasy Plus Mini Kit (Qiagen). For the RACE experiments, we also added RNA from one hCA donor (#ICM167), adult brain, and adult heart. RNA integrity and concentration were measured by Agilent RNA 6000 Nano II assays (Agilent Technologies) on an Agilent 2100 Bioanalyzer. We reverse transcribed 1 μg of total RNA using a modified oligo (dT) primer and the SMARTScribe Reverse Transcriptase (Clontech). We specifically amplified cDNA of PHACTR1 using the SMARTer RACE 5′/3’ Kit (Clontech). The gene specific primers are in Additional file 1. The RACE products were purified by gel extraction with the NucleoSpin Gel and PCR Clean-Up Kit, cloned in pRACE vector with the In-Fusion HD Cloning Kit and transformed with Stellar Competent Cells (Clontech). After overnight incubation, plasmids were extracted from single bacterial colonies with QIAprep Spin miniprep kit (Qiagen). We sequenced PHACTR1 inserts by Sanger sequencing using M13 primers (Additional file 1).
Long-read Pacific Biosciences (PacBio) sequencing library preparation
We produced libraries using 1 μg of total RNA (HUVEC, HAEC, HCAEC, HASMC, HCASMC and brain) and 1 μL of 12 μM dT-Barcode (dT-BC), according to the manufacturer’s protocol (Procedure and checklist-Isoform sequencing using the SMARTer PCR cDNA synthesis kit (Clontech) and no size selection). Each library had its own dT-BC (see Additional file 1 for sequences). Briefly, we synthesized the first strand of cDNA with the following thermal profile: 72 °C for 3 min, slow ramp to 42 °C at 0.1 °C/sec and 42 °C for 2 min. We synthesized cDNA using the SMARTer PCR cDNA synthesis kit at 42 °C for 90 min. We performed a large-scale PCR reaction on each cDNA library with optimization for the numbers of cycle using the KAPA Hi-Fi PCR kit. As determined with a High Sensitivity DNA Chip (Agilent Technologies), we needed 11 to 13 PCR cycles to obtain 250 ng of each cDNA. Before capture, we washed cDNA with AMPure PB beads. To enrich for PHACTR1 transcripts, we designed biotinylated capture probes as Ultramers from IDT DNA according to the manufacturer’s protocol (full-length cDNA target sequence capture using SeqCap EZ libraries; see Additional file 1 for sequences). We used a total of 1.5 μg of cDNA (250 ng from each barcoded cDNA prep) to capture PHACTR1 transcripts. We used six different blocking oligos, corresponding to the reverse complement sequence of the dT-BC with a 3′ modification (3SpC3)(Additional file 1). After capture, we amplified the library with 14 cycles of PCR using Takara LA Taq DNA polymerase (Clontech). The long-read PacBio sequencing was performed on one SMRTcell with Pacbio RS II at the Genome Quebec/McGill Innovation Center.
Reverse transcription polymerase chain reaction (RT-PCR)
We extracted RNA from HUVEC, teloHAEC (treated with tumor necrosis factor (TNF)-α at 10 ng/ml for 4 h), monocytes, HCASMC, HASMC, HCAEC, and HAEC as described above. We analyzed unstimulated monocytes, as well as monocytes stimulated with 10 ng/ml of phorbol myristate acetate (PMA) for 48 h. Three hours later, some of the PMA-treated monocytes were also treated with lipopolysaccharides (LPS) (100 ng/ml) for one hour. We used the same adult brain total RNA and adult heart total RNA as for the RACE (see above). We measured RNA quality and concentration with Agilent RNA 6000 Nano II assays (Agilent Technologies) on an Agilent 2100 Bioanalyzer. We reverse transcribed 1 μg or less of total RNA (with RNA integrity number of 10 for all cells and above 7 for all tissues) using random primers and 1 U of the MultiScribe Reverse Transcriptase (Applied Biosystems) in a 20 μL reaction volume at 100 mM dNTPS and 20 U of RNase inhibitor with these three steps: 10 min at 25 °C, 120 min at 37 °C and 5 min at 85 °C. We mixed an equal volume of cDNA from each sample to create a pool of cDNA to use as positive control. We amplified fragments of PHACTR1 transcripts from these tissues and cell lines with GoTaq DNA polymerase (Promega) with the following thermal cycle: 95 °C for 2 min; 94 °C for 30 s, optimal temperature for 30 s and 72 °C for 1 min and 20 s (35 times); 72 °C for 5 min. We also amplified full PHACTR1 open reading frame (ORF) with SeqAmp DNA polymerase (Clontech) with the following thermal cycle: 94 °C for 1 min; 98 °C for 10 s, optimal temperature for 15 s and 68 °C for 3 min (30 times); 68 °C for 5 min. The sequences of the primers are in Additional file 1. We characterised the PCR products by agarose gel electrophoresis.
Quantitative polymerase chain reaction (qPCR)
We extracted DNA and RNA from 36 hCA samples from the RÉTEB. Of these 36 samples, 14 are new and 22 were previously analyzed [5]. Genotyping of PHACTR1-rs9349379 was performed at the Beaulieu-Saucier Pharmacogenomics Centre of the Montreal Heart Institute on the Illumina Infinium MEGA_Consortium_v2 BeadChip: nAA = 15, nAG = 13, nGG = 8. We measured RNA integrity and concentration with Agilent RNA 6000 Nano II assays (Agilent Technologies) on an Agilent 2100 Bioanalyzer. We reverse transcribed exactly 1 μg of total RNA as for the RT-PCR experiments (with RNA integrity number of 6 or above for all samples). We followed the MIQE guidelines to assess quality and reproducibility of our qPCR results [14]. We performed qPCR in triplicates for all samples using: 1.25 μL of cDNA (1/50 dilution), 5 μL of Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies) and 3.75 μL of primer pair mix at 0.8 μM on a CFX384 from Biorad or Eco Illumina qPCR system (Montreal Biotech). We used the following thermal profile: 10 min at 95 °C, and 40 cycles of 30 s at 95 °C, 30 s at 55 °C and 45 s at 72 °C. We carried out melting curve analyses after the amplification process to ensure the specificity of the amplified products. We also simultaneously performed qPCR reactions with no template controls for each gene to test the absence of non-specific products. Cq values were determined with the CFX Manager 3.1 (Bio-Rad) software and expression levels were normalized on the expression levels of the house-keeping genes TATA-box binding protein (TBP), hypoxanthine-guanine phosphoribosyltransferase (HPRT), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the ΔΔCt method. Based on geNORM principles for accurate normalization of qPCR data by geometric averaging of multiple internal control genes [15], mean M values of 0.5845 and 0.5356 (for two experiments) were generated from the GAPDH, HPRT1, and TBP genes. The primer sequences are in Additional file 1.
eQTL analyses using GTEx data
We downloaded from dbGaP genotypes at PHACTR1-rs9349379 and RNA-sequencing (RNA-seq) data from 122 hCA samples generated by the GTEx Project [6]. We used Trimmomatic (V0.36) on reads with the paired-end (PE) function to remove adapter sequences (TruSeq3-PE.fa:2:30:10) and to trim low-quality bases with the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. We then performed quality control checks using FastQC (V0.11.5). We aligned reads to the reference human genome sequence (GRCh37) using HISAT2 (V2.0.5) [16]. We selected reads that mapped to the PHACTR1 locus using BamTools (V2.4.1) with the parameters -region “6:12717037..13288073” -mapQuality “> = 30”, and used SortSam and BuildBamIndex tools from Picard (V1.130) to convert .sam files into .bam files and to build their index files. We quantified PHACTR1 expression with Stringtie (V1.3.3b) [16] with parameters -G -e -b; for these analyses we provided our own GTF file with genomic coordinates corresponding to the three PHACTR1 transcripts expressed in hCA (Additional file 2). Finally, we used Ballgown (V2.8.4) [16] to extract exon-level expression data for eQTL analyses with genotypes at PHACTR1-rs9349379.
Statistical analyses
We normalized PHACTR1 expression data (for full transcripts or exons) from our qPCR experiments or GTEx using inverse normal transformation. Using linear regression in R, we tested the association between PHACTR1 expression levels and genotypes at PHACTR1-rs9349379 (additive coding), correcting for sex, RNA integrity number (RIN), and the recruitment center (Montreal or Quebec city) when available.
Western blot
To detect PHACTR1 protein expression in VSMCs, 0.75 × 106 and 1 × 106 of cryopreserved HASMC and HCASMC cells, respectively, were pelleted and directly lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 0.1% SDS, 150 mM NaCl) containing protease inhibitor cocktail (Sigma), phosphatase inhibitor cocktail 2 and 3 (Sigma), and 1 mM PMSF. As controls, teloHAEC were transfected with 20 nM of scramble (Santa Cruz, sc-37,007) or PHACTR1 (Santa Cruz, sc-95,456) small interfering RNA (siRNA) in 2 mL of siRNA transfection medium (Santa Cruz, sc-36,868) for 4 h using siRNA Transfection Reagent (Santa Cruz, sc-29,528), and then grown for 48 h before cell lysis and protein extraction. Homogenized lysates were clarified by centrifugation to eliminate insoluble cell debris. We determined protein concentration with a Pierce BCA Protein Assay Kit (ThermoFisher) and prepared samples containing 30 μg of proteins in a 600 mM DTT reducing buffer that was denatured 5 min at 95 °C prior to loading on a 8% polyacrylamide gel (running buffer: 25 mM Tris base; 192 mM glycine; 0.1% SDS). We transferred proteins onto nitrocellulose membranes in Towbin buffer (25 mM Tris base; 192 mM glycine, 20% (v/v) methanol) and incubated them for 1.5 h in the blocking buffer (1× TBS, 0.1% Tween-20, 5% milk) at room temperature. We then incubated membranes with primary antibodies against PHACTR1 (1:1000; custom antibody generated by Biomatik) or GAPDH (1:10,000; Cell Signaling, #2118) at 4 °C overnight. For the secondary antibody, we incubated membranes with horseradish peroxidase-conjugated anti-rabbit IgG (1:10,000; GE Healthcare, NA934) at room temperature for 1 h. The immunoblotting signal was revealed with the SuperSignal west Pico PLUS chemiluminescent substrate (Thermo scientific, 34,580) and visualized with the ChemiDoc Touch system (Bio-Rad).
Results
Six PHACTR1 transcripts in human cells and tissues
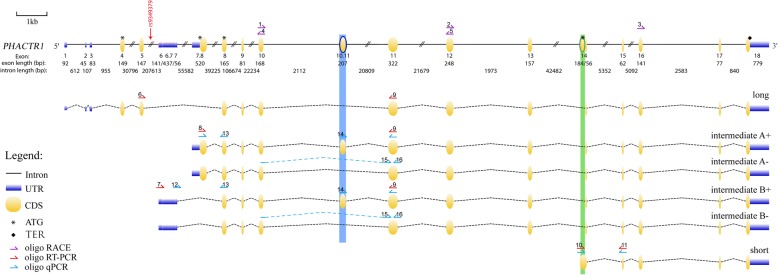
Public databases (e.g. ENSEMBL, Gencode, GTEx) report > 10 different PHACTR1 mRNA transcripts expressed in human samples. To confirm these results and comprehensively characterize the different PHACTR1 transcripts expressed in humans, we combined RACE and long-read next-generation (PacBio) DNA sequencing experiments in many cell types and tissues. These two complementary approaches generated highly concordant results that we also validated by sequencing of the whole ORF. Although we did identify a large number of PHACTR1 transcripts due to alternative splicing of non-coding 5′ and 3′ untranslated exons (Additional file 3), our transcriptomic profiling indicates that PHACTR1 can give rise to six main different transcripts that would encode six proteins based on their coding sequences (Fig. 1and Additional file 4). The long transcript (1743-bp) encodes a protein of 580 amino acids (Fig. 1). We confirmed four intermediate transcripts that are different due to the inclusion of the alternatively spliced exons 7.8 and 10.11 (Fig. 1). Both alternative exons are in-frame and add respectively 279- (exon 7.8) and 207-bp (exon 10.11) to the ORF. Intermediate transcripts A+ (1953-bp) and B+ (1674-bp) include exon 10.11 and encode proteins of 650 and 557 amino acids. Intermediate transcripts A- (1746-bp) and B- (1467-bp) exclude exon 10.11 and encode proteins of 581 and 488 amino acids. Finally, we confirmed a short transcript (435-bp) encoding a protein of 144 aa (Fig. 1). Its start codon is located in exon 14, 83-bp upstream of the 3′ splice site for the long and intermediate transcripts (Additional file 5).
Fig. 1.
Six PHACTR1 transcripts are expressed in human samples. We combined Rapid Amplification of cDNA Ends (RACE) and long-read DNA sequencing (Pacific Biosciences) to identify all PHACTR1 mRNA transcripts expressed in human samples. Although we identified multiple transcripts, they give rise to six main transcripts. The schematic diagram shows the PHACTR1 gene (top) and the six main transcripts below. The coronary artery disease-associated SNP rs9349379 (red arrow) is located in a large intron (207,613-bp) between exons 5 and 6. The start codons for the long, intermediate A, intermediate B and short transcripts are located, respectively, in exons 4, 7.8, 8, and 14. The stop codon for all six transcripts is in exon 18. In blue, we highlight the in-frame, 207-bp long, exon 10.11 that is included in intermediate transcripts A+ and B+. In green, we highlight part of exon 14 that is specific to the short transcript. We also indicated the different oligonucleotides used in our experiments; numbers referred to their sequences in Additional file 1. UTR, untranslated transcribed region; CDS, coding DNA sequence; ATG, start codon; TER, terminator codon. The figure was drawn to scale using GSDS2.0 [19]
PHACTR1 transcripts are differentially expressed
We used the sequence of the six PHACTR1 transcripts to design transcript-specific pairs of primers and carried out RT-PCR experiments on mRNA extracted from human brain, heart, monocytes, ECs, and VSMCs (Fig. 2). The long transcript, which never included exons 7.8 nor 10.11, was only expressed in the brain [7]. In contrast, both intermediate transcripts B were expressed in all samples tested, although we noted that the inclusion of exon 10.11 varied from one sample to the other. Intermediate transcript B+ was more abundant in ECs and VSMCs, whereas intermediate transcript B- was the major intermediate transcript expressed in the brain, the heart, and in monocytes (Fig. 2, third panel from top). Both intermediate transcripts A were expressed in the brain, the heart, untreated monocytes, and ECs. Similar to what we saw for the intermediate transcripts B, the inclusion of exon 10.11 in the intermediate transcripts A was variable from one tissue to the other (Fig. 2, second panel from top). We validated that the short transcript was expressed in monocytes [13]. Stimulation with PMA or LPS to differentiate monocytes into macrophages, which are of interest regarding atherosclerosis, did not induce the expression of the long transcript, but seems to weakly increase the expression of intermediate transcripts B (Fig. 2). We detected expression of the short transcript in the heart and the brain, although this is consistent with the presence of immune cells within these tissues.
Fig. 2.
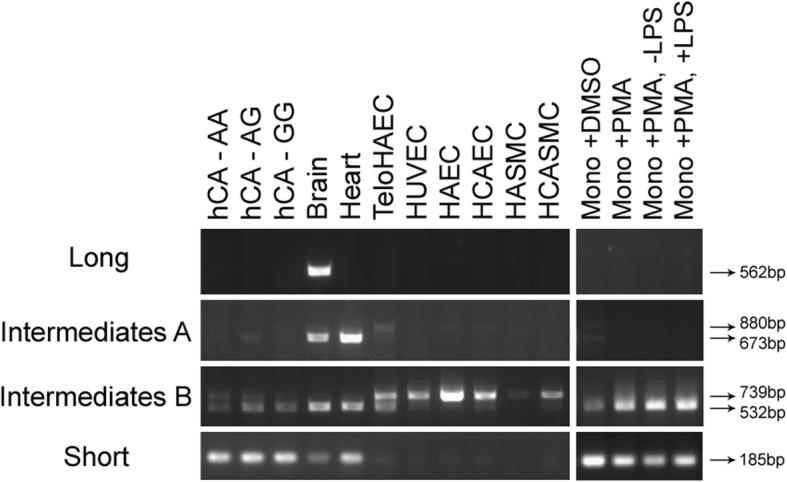
PHACTR1 transcripts differential expression in human tissues and cell types. We designed transcript-specific primers and amplified PHACTR1 by RT-PCR. The PCR fragment for the long transcript is 562-bp. We did not amplify a 769-bp fragment using this set of primers, indicating that exon 10.11 is absent from the long transcript. The PCR fragments for the intermediate transcripts A+, A-, B+ and B- are 880-bp, 673-bp, 739-bp and 532-bp, respectively. The PCR fragment for the short transcript is 185-bp. HUVEC: human umbilical vein endothelial cell; teloHAEC: immortalized human aortic endothelial cells; HCASMC: human coronary artery smooth muscle cells; HAEC: human aortic endothelial cells; HCAEC: human coronary artery endothelial cells; HASMC: human aortic smooth muscle cells; Mono: monocytes; PMA: phorbol myristate acetate; LPS: lipopolysaccharide
Although PHACTR1 transcripts had previously been detected in VSMCs, immunoblotting with a commercially available anti-PHACTR1 antibody failed to detect the corresponding protein [13]. We purchased and tested all commercially available anti-PHACTR1 antibodies, but none showed specificity in short interfering RNA (siRNA)-mediated knockdown experiments. For this reason, we developed our own polyclonal anti-PHACTR1 antibody (Materials and methods) and have shown its specificity using siRNA against PHACTR1 [17]. Using this antibody, we detected weak PHACTR1 protein expression in VSMCs from hCA (Fig. 3).
Fig. 3.
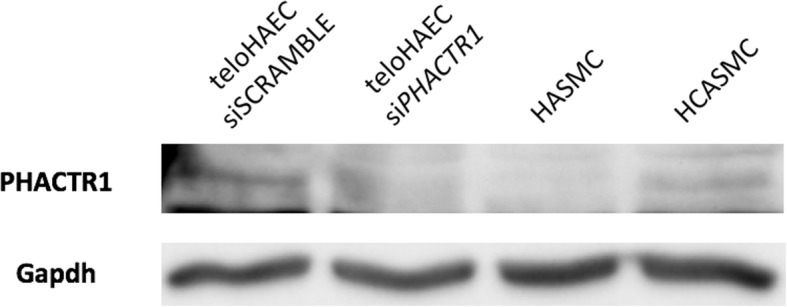
Presence of PHACTR1 protein in vascular smooth muscle cells. We tested for the presence of the PHACTR1 protein by immunoblotting using a custom anti-PHACTR1 antibody. The first and second lane, using teloHAEC protein extracts treated or not with a short interfering RNA (siRNA) against PHACTR1, serve as control to demonstrate the specificity of this anti-PHACTR1 antibody. Although we did not detect PHACTR1 in human aortic smooth muscle cells (HASMC), there is weak but specific band corresponding to PHACTR1 in human coronary artery smooth muscle cells (HCASMC). The expected 64 kDa PHACTR1 protein migrated with an apparent molecular weight of 74 kDa. GAPDH was used as loading control
CAD-associated rs9349379 is an eQTL for PHACTR1 intermediate transcripts A
Using primers that capture the long and intermediate PHACTR1 transcripts, we previously showed that genotypes at rs9349379 are associated with PHACTR1 expression levels in hCA [5]. That result was subsequently replicated in the GTEx database. Given that hCA are composed of different cell types (ECs, VSMCs, and immune cells) and that PHACTR1 transcripts are differentially expressed across tissues and cell types, we tested if rs9349379 is associated with the levels of all or specific PHACTR1 transcripts. We obtained DNA and RNA from 36 hCA, including 14 new samples not analyzed in our previous study [5]. We genotyped rs9349379 and measured by qPCR the levels of each of the five PHACTR1 transcripts expressed in hCA. We found a significant association between rs9349379 and intermediate transcripts A+ and B+, whereas the SNP was not significantly associated with intermediate transcripts A- and B- (Table 1 and Additional file 6). Although we detected strong expression of the short PHACTR1 transcript in hCA, genotypes at rs9349379 were not associated with its levels (Table 1 and Additional file 6).
Table 1.
rs9349379 is an eQTL for PHACTR1 intermediate transcripts A+ and B+ in human coronary arteries (hCA)
| Transcripts | Beta (SE) | P-value | Variance explained (%) |
|---|---|---|---|
| Intermediates A+ and A- |
−0.49 (0.21) | 0.026 | 7.8 |
| Intermediates B+ and B- |
−0.54 (0.17) | 0.0040 | 17.5 |
| Intermediates A+ and B+ |
−0.56 (0.19) | 0.0073 | 16.4 |
| Intermediates A- and B- |
−0.35 (0.21) | 0.10 | – |
| Short | −0.068 (0.20) | 0.73 | – |
We tested by linear regression the association between genotypes at rs9349379 (additive model) and normalized expression levels for the different PHACTR1 transcripts expressed in hCA. The long PHACTR1 transcript is not expressed in hCA. Effect sizes (Beta) and standard errors (SE) are in standard deviation units. The direction of the Beta is for the G-allele. We analyzed 36 hCA samples (NAA = 15, NAG = 13, NGG = 8). The frequency of the G-allele was 40%. The G-allele, associated with increased coronary artery disease risk, is associated with reduced PHACTR1 expression. When significant (P-value < 0.05), we provided the percentage of PHACTR1 expression variation explained by genotypes at rs9349379
Because our hCA sample size remains small, we attempted to replicate these eQTL results in the larger GTEx dataset with genotype and gene expression data available from 122 hCA. Since it is computationally challenging to assemble specific mRNA transcripts out of short-read RNA-seq results, we opted to test the association between rs9349379 and the expression levels of specific PHACTR1 exons. Overall, we found nominally significant associations (P-value < 0.05) with exons common to the intermediate and short transcripts (Table 2). However, the association between rs9349379 and the expression levels of the 5′ end of exon 14 that is specific to the short transcript (Additional file 5) was not significant (P-value = 0.47) (Table 2 and Additional file 6). Thus, using qPCR and RNA-seq gene expression data, we could not detect an eQTL effect between rs9349379 and the expression of the immune-specific short PHACTR1 transcript in 158 hCA samples.
Table 2.
Replication of rs9349379 eQTL effects in human coronary arteries (hCA) from GTEx
| Exon | Exon length (bp) | Beta (SE) | P-value | Variance explained (%) |
|---|---|---|---|---|
| 6 (5’UTR) | 141 | −0.084 (0.13) | 0.51 | – |
| 7 (5’UTR) | 56 | −0.11 (0.13) | 0.39 | – |
| 7.8 | 520 | −0.15 (0.12) | 0.22 | – |
| 8 | 165 | −0.31 (0.13) | 0.017 | 3.4 |
| 9 | 81 | −0.19 (0.13) | 0.14 | – |
| 10 | 168 | −0.35 (0.13) | 0.0065 | 3.1 |
| 10.11 | 207 | −0.30 (0.13) | 0.022 | 3.9 |
| 11 | 322 | −0.35 (0.13) | 0.0074 | 2.8 |
| 12 | 248 | −0.40 (0.13) | 0.0019 | 5.0 |
| 13 | 157 | −0.28 (0.13) | 0.029 | 3.0 |
| 14a | 240 | −0.17 (0.13) | 0.20 | – |
| 5′-14b | 184 | 0.095 (0.13) | 0.47 | – |
| 3′-14c | 56 | −0.25 (0.13) | 0.059 | – |
| 15 | 62 | −0.25 (0.13) | 0.058 | – |
| 16 | 141 | −0.27 (0.13) | 0.042 | 2.3 |
| 17 | 77 | −0.31 (0.13) | 0.017 | 3.5 |
| 18 (3’UTR) | 779 | −0.25 (0.13) | 0.055 | – |
We analysed by linear regression the association between genotypes at rs9349379 (additive model) and normalized levels of PHACTR1 exon expression. Expression levels were corrected for the length of the exons (Materials and methods). Effect sizes (Beta) and standard errors (SE) are in standard deviation units. The direction of the effect size is for the G-allele, associated with increased coronary artery disease risk. We analyzed data from 122 hCA samples (NAA = 48, NAG = 57, NGG = 17). The frequency of the G-allele was 37%. When significant (P-value < 0.05), we provided the percentage of PHACTR1 exon expression variation explained by genotypes at rs9349379. aThe full exon 14; bpart of exon 14 which is specific to the short PHACTR1 transcript; cpart of exon 14 which is present in all PHACTR1 transcripts (see Additional file 5 for details)
Discussion
Despite the recent report linking PHACTR1-rs9349379 with EDN1 in ECs [12], PHACTR1 itself remains a strong causal candidate gene at this CAD-associated locus because of the eQTL effect in hCA [5, 6]. Further, it was recently shown in an in vitro cell culture system that PHACTR1 contributes to VSMC calcification, a hallmark of CAD [18]. Using two different methods, we confirmed that the human PHACTR1 gene encodes six main transcripts that are differentially expressed. Although our results are largely consistent with a study by Reschen et al. [13], we note three important differences: First, we confirmed that there are six, and not three, PHACTR1 transcripts. Indeed, whereas Reschen and colleagues reported only one intermediate transcript (transcript B+ in Fig. 1), we found that four intermediate transcripts (with or without exons 7.8 and 10.11) are co-expressed in the heart (Fig. 2). Whether these four PHACTR1 isoforms, of 650, 581, 557 and 488 amino acids, have the same functions is currently unknown. Second, we confirmed using our own custom polyclonal antibody against PHACTR1 that the protein is present in VSMCs. In contrast, we have not been able to detect by immunoblotting the protein encoded by the short transcript. And third, our eQTL analyses in 158 hCA validated an effect of genotypes at rs9349379 on the expression of intermediate transcripts A+ and B+, but not the short immune-specific transcript. One possible interpretation of this result is that rs9349379 mediates in part its effect on CAD risk by regulating the expression or splicing of these PHACTR1 intermediate transcripts in ECs and/or VSMCs within hCA, and not through the short immune-specific PHACTR1 transcript. In support of this hypothesis, we also know that rs9349379 is associated with fibromuscular dysplasia [4], a disease without an inflammatory component. Precisely how, at the molecular level, genotypes at rs9349379 modulate cell type-specific transcriptional expression and/or splicing remains to be determined.
Our study presents with some limitations. As for every transcriptomic profiling experiments, weakly expressed transcripts might have missed detection. We tried to account for this by combining two complementary methods and profiling PHACTR1 transcripts in high quality mRNA prepared from many tissues and cell types. The number of available hCA samples for our eQTL analyses was small (N = 36). To increase statistical power, we also analyzed RNA-seq data from the GTEx Project (N = 122). Because short-reads cannot be used unambiguously to reconstruct specific transcripts, we decided to analyze the expression levels of specific exons. This analysis generated results that were highly concordant with our transcript-specific qPCR results. Finally, although we did not detect an eQTL effect for the short PHACTR1 transcript in hCA, it does not rule out a potential role for PHACTR1 in immune cells in the context of atherosclerosis progression and CAD. Indeed, the short PHACTR1 protein might play a critical atherosclerotic function in circulating monocytes, which would not have been captured in hCA samples. However, it is critical to confirm that the short PHACTR1 transcript can be translated into an active protein in human cells.
Conclusions
In this article, we provide a comprehensive survey of PHACTR1 transcripts in tissues and cells that are relevant to atherosclerosis initiation and progression. Importantly, we confirmed that the PHACTR1 protein is present in VSMCs and that the CAD-associated variant rs9349379 associates with the expression of specific PHACTR1 transcript in hCA. Gupta and colleagues have argued that the molecular link they identified between rs9349379 and EDN1 expression largely accounts for the association of this locus with several vascular diseases [12]. One natural extension of this model, which would be consistent with the pleiotropy observed as well as the PHACTR1 eQTL we detected in hCA, is that the same SNP influences the expression of two genes – EDN1 and PHACTR1 – which are expressed in CAD-relevant cells and could act together to contribute to vascular disease risk.
Additional files
List of primers used in this study. The numbers in the “Name” column refers to the primers shown in Fig. 1. All sequences are given in the 5′ to 3′ orientation. (PDF 43 kb)
Genomic coordinates of exons based on the reference human genome sequence GRCh37/hg19. (PDF 47 kb)
Summary of observed transcripts with their alternatively spliced 5′ and 3′ untranslated regions (UTRs). The blue rectangles on the left indicate transcripts that were detected in tissue samples whereas gray rectangles mean they were not detected. Introns are not shown for simplicity; black lines indicate that the exons were not present in the transcript. (PDF 75 kb)
Transcript and protein IDs of PHACTR1 gene (221,692; ENSG00000112137). (PDF 45 kb)
Zoom-in representation of PHACTR1 exon 14. The 5′-14 fraction of exon 14 is specific to the short immune-specific PHACTR1 transcript. We also illustrate the 3′ splice site used for the long and intermediate PHACTR1 transcripts. In blue is the intron between exons 13 and 14. The red arrow indicates a primer used to detect the short PHACTR1 transcript. The sequence of this primer #10 is in Additional file 1. (PDF 193 kb)
Associations between genotypes at rs9349379 and PHACTR1 expression levels in human coronary arteries. (A) By qPCR using transcript-specific primers, we measured the expression of PHACTR1 transcripts in 36 human coronary arteries (hCA)(NAA = 15, NAG = 13, NGG = 8). The long PHACTR1 transcript is not expressed in hCA. (B) We used GTEx data to test the associations between rs9349379 and the expression levels of PHACTR1 exons in 122 hCA (NAA = 48, NAG = 57, NGG = 17). Here, we only show results for five exons, but association results for all PHACTR1 exons are available in Table 2. Exon 10.11 corresponds to the alternatively spliced exon located between exons 10 and 11. 5′ exon 14 corresponds to part of exon 14 that is specific to the short PHACTR1 transcript. (PDF 350 kb)
Acknowledgments
We thank the Tardif and Rioux labs at the Montreal Heart Institute for providing HASMC, HCASMC, and monocytes. We thank Chantal St-Cyr from the “Réseau d’échange de tissus et d’échantillons biologiques (RÉTEB)” and Christine Racine from the “Biobanque du Centre de recherche de l’Institut Universitaire de Cardiologie et Pneumologie du Quebec (IUCPQ)” for providing human coronary artery samples. We also acknowledge the technical support of the Beaulieu-Saucier MHI Pharmacogenomic Center for RNA extraction and genotyping and the technical support of the Genome Quebec/McGill Innovation Center for Pacific Biosciences sequencing.
Funding
This work was supported by grants from the Canadian Institutes of Health Research (operating grant #136979), the Montreal Heart Institute Foundation, and the Canada Research Chair Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request. The GTEx dataset analyzed in the current study is available from dbGaP (https://www.ncbi.nlm.nih.gov/gap).
Abbreviations
- CAD
Coronary artery disease
- dT-BC
dT-Barcode
- ECs
Endothelial cells
- EDN1
Endothelin-1
- eQTL
Expression quantitative trait locus
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-wide association studies
- HAEC
Human aortic endothelial cells
- HASMC
Human aortic smooth muscle cells
- hCA
Human coronary arteries
- HCAEC
Human coronary artery endothelial cells
- HCASMC
Human coronary artery smooth muscle cells
- HPRT
Hypoxanthine-guanine phosphoribosyltransferase
- HUVEC
Human umbilical vein endothelial cells
- LPS
Lipopolysaccharides
- ORF
Open reading frame
- PacBio
Long-read Pacific Biosciences
- PHACTR1
Phosphatase and actin regulator 1
- PMA
Phorbol myristate acetate
- PP1
Protein phosphatase-1
- qPCR
Quantitative polymerase chain reaction
- RACE
Rapid amplification of cDNA ends
- RÉTEB
Réseau d’Échanges de Tissus et d’Échantillons Biologiques
- RIN
RNA integrity number
- RT-PCR
Reverse transcription polymerase chain reaction
- TBP
TATA-box binding protein
- teloHAEC
Immortalized HAEC
- TNFα
Tumor necrosis factor α
- VSMCs
Vascular smooth muscle cells
Authors’ contributions
VACF, MB and GL designed the study. VACF, MB and SL performed the experiments. KSL helped with long-read Pacific Biosciences analyses. VACF performed the statistical analyses. VACF, MB and GL wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Ethics approval and consent to participate
We obtained written consents from all donors of human coronary artery samples. The project was approved by the Montreal Heart Institute Ethics Committee (Project #2011–209).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12881-018-0616-7) contains supplementary material, which is available to authorized users.
Contributor Information
Valérie-Anne Codina-Fauteux, Email: Valerie-Anne.Codina-Fauteux@mhi-humangenetics.org.
Mélissa Beaudoin, Email: melissa.beaudoin@mhi-humangenetics.org.
Simon Lalonde, Email: Simon.Lalonde@mhi-humangenetics.org.
Ken Sin Lo, Email: ken.sin.lo@mhi-humangenetics.org.
Guillaume Lettre, Phone: 514-376-3330, Email: guillaume.lettre@umontreal.ca.
References
- 1.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freilinger T, Anttila V, de Vries B, Malik R, Kallela M, Terwindt GM, Pozo-Rosich P, Winsvold B, Nyholt DR, van Oosterhout WP, et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet. 2012;44(7):777–782. doi: 10.1038/ng.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, Pezzini A, Thijs V, Markus HS, Dichgans M, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47(1):78–83. doi: 10.1038/ng.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiando SR, Tucker NR, Castro-Vega LJ, Katz A, D'Escamard V, Treard C, Fraher D, Albuisson J, Kadian-Dodov D, Ye Z, et al. PHACTR1 is a genetic susceptibility locus for fibromuscular dysplasia supporting its complex genetic pattern of inheritance. PLoS Genet. 2016;12(10):e1006367. doi: 10.1371/journal.pgen.1006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaudoin M, Gupta RM, Won HH, Lo KS, Do R, Henderson CA, Lavoie-St-Amour C, Langlois S, Rivas D, Lehoux S, et al. Myocardial infarction-associated SNP at 6p24 interferes with MEF2 binding and associates with PHACTR1 expression levels in human coronary arteries. Arterioscler Thromb Vasc Biol. 2015;35(6):1472–1479. doi: 10.1161/ATVBAHA.115.305534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium GT. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen PB, Greenfield AT, Svenningsson P, Haspeslagh DC, Greengard P. Phactrs 1-4: a family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci U S A. 2004;101(18):7187–7192. doi: 10.1073/pnas.0401673101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiezlak M, Diring J, Abella J, Mouilleron S, Way M, McDonald NQ, Treisman R. G-actin regulates the shuttling and PP1 binding of the RPEL protein Phactr1 to control actomyosin assembly. J Cell Sci. 2012;125(Pt 23):5860–5872. doi: 10.1242/jcs.112078. [DOI] [PubMed] [Google Scholar]
- 9.Jarray R, Allain B, Borriello L, Biard D, Loukaci A, Larghero J, Hadj-Slimane R, Garbay C, Lepelletier Y, Raynaud F. Depletion of the novel protein PHACTR-1 from human endothelial cells abolishes tube formation and induces cell death receptor apoptosis. Biochimie. 2011;93(10):1668–1675. doi: 10.1016/j.biochi.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Allain B, Jarray R, Borriello L, Leforban B, Dufour S, Liu WQ, Pamonsinlapatham P, Bianco S, Larghero J, Hadj-Slimane R, et al. Neuropilin-1 regulates a new VEGF-induced gene, Phactr-1, which controls tubulogenesis and modulates lamellipodial dynamics in human endothelial cells. Cell Signal. 2012;24(1):214–223. doi: 10.1016/j.cellsig.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Jarray R, Pavoni S, Borriello L, Allain B, Lopez N, Bianco S, Liu W, Biard D, Demange L, Hermine O, et al. Disruption of phactr-1 pathway triggers pro-inflammatory and pro-atherogenic factors: new insights in atherosclerosis development. Biochimie. 2015;118:151–161. doi: 10.1016/j.biochi.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Gupta R, Hadaya J, Trehan A, Zekavat S, Roselli C, Klarin D, Emdin C, Hilvering C, Bianchi V, Mueller C, et al. A genetic variant associated with five vascular diseases is a distal regulator of Endothelin-1 gene expression. Cell. 2017;170:1–12. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reschen ME, Lin D, Chalisey A, Soilleux EJ, O'Callaghan CA. Genetic and environmental risk factors for atherosclerosis regulate transcription of phosphatase and actin regulating gene PHACTR1. Atherosclerosis. 2016;250:95–105. doi: 10.1016/j.atherosclerosis.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 15.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protocols. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalonde S, Stone OA, Lessard S, Lavertu A, Desjardins J, Beaudoin M, Rivas M, Stainier DYR, Lettre G. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS One. 2017;12(6):e0178700. doi: 10.1371/journal.pone.0178700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aherrahrou R, Aherrahrou Z, Schunkert H, Erdmann J. Coronary artery disease associated gene Phactr1 modulates severity of vascular calcification in vitro. Biochem Biophys Res Commun. 2017;491(2):396–402. doi: 10.1016/j.bbrc.2017.07.090. [DOI] [PubMed] [Google Scholar]
- 19.Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in this study. The numbers in the “Name” column refers to the primers shown in Fig. 1. All sequences are given in the 5′ to 3′ orientation. (PDF 43 kb)
Genomic coordinates of exons based on the reference human genome sequence GRCh37/hg19. (PDF 47 kb)
Summary of observed transcripts with their alternatively spliced 5′ and 3′ untranslated regions (UTRs). The blue rectangles on the left indicate transcripts that were detected in tissue samples whereas gray rectangles mean they were not detected. Introns are not shown for simplicity; black lines indicate that the exons were not present in the transcript. (PDF 75 kb)
Transcript and protein IDs of PHACTR1 gene (221,692; ENSG00000112137). (PDF 45 kb)
Zoom-in representation of PHACTR1 exon 14. The 5′-14 fraction of exon 14 is specific to the short immune-specific PHACTR1 transcript. We also illustrate the 3′ splice site used for the long and intermediate PHACTR1 transcripts. In blue is the intron between exons 13 and 14. The red arrow indicates a primer used to detect the short PHACTR1 transcript. The sequence of this primer #10 is in Additional file 1. (PDF 193 kb)
Associations between genotypes at rs9349379 and PHACTR1 expression levels in human coronary arteries. (A) By qPCR using transcript-specific primers, we measured the expression of PHACTR1 transcripts in 36 human coronary arteries (hCA)(NAA = 15, NAG = 13, NGG = 8). The long PHACTR1 transcript is not expressed in hCA. (B) We used GTEx data to test the associations between rs9349379 and the expression levels of PHACTR1 exons in 122 hCA (NAA = 48, NAG = 57, NGG = 17). Here, we only show results for five exons, but association results for all PHACTR1 exons are available in Table 2. Exon 10.11 corresponds to the alternatively spliced exon located between exons 10 and 11. 5′ exon 14 corresponds to part of exon 14 that is specific to the short PHACTR1 transcript. (PDF 350 kb)
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request. The GTEx dataset analyzed in the current study is available from dbGaP (https://www.ncbi.nlm.nih.gov/gap).