Abstract
Background
Atopic dermatitis (AD) is a common inflammatory skin disease. A subset of AD is susceptible to disseminated herpes simplex virus (HSV) infection, a complication termed as eczema herpeticum(ADEH+). The immune mechanisms causing ADEH+ remain elusive. Using RNA-sequencing, we recently found that ankyrin repeat domain 1 (ANKRD1) was significantly induced in human peripheral blood mononuclear cells (PBMCs) upon HSV-1 stimulation; and its induction in ADEH+ was significantly reduced as compared to AD patients without a history of EH (ADEH−).
Objective
To validate ANKRD1 gene expression in non-atopic (NA), ADEH− and ADEH+ subjects; to delineate the biological function of ANKRD1 and the signaling pathway(s) involved.
Methods
Purification of human PBMCs, monocytes, B cells, dendritic cells, T cells and NK cells; RNA extraction and qRT-PCR; small interfering RNA technique; co-immunoprecipitation; and western-blot assays were used.
Results
ANKRD1 was significantly reduced in PBMCs from ADEH+ patients after HSV-1 stimulation as compared to PBMCs from ADEH−. We found that the induction of ANKRD1 by HSV-1 and multiple pathogen pattern recognition receptor (PRR) agonists are mediated by inflammatory cytokines. Silencing ANKRD1 gene expression in APCs led to increased viral load and reduced IFNb1 and IL-29 production. Using co-immunoprecipitation methods, we demonstrated that ANKRD1 formed protein complexes with IRF3 and IRF7, which are important transcription factors regulating PRRs’ signaling transduction. Over-expression of ANKRD1 enhanced the IRF3-mediated signaling pathways.
Conclusion
ANKRD1 is involved in IRF3 mediated anti-viral innate immune signaling pathways. Its reduced expression in ADEH+ subjects may contribute to the pathogenesis of ADEH+.
Keywords: herpes simplex virus, Ankyrin repeat domain 1, innate immunity, atopic dermatitis, eczema herpeticum, IRF3, NFκB1, IFNb1, IL-29
Introduction
Atopic dermatitis (AD) is the most common chronic skin inflammatory disease worldwide, affecting up to 25 % of children and 10% of adults.1 A subset of AD patients are susceptible to disseminated skin viral infection including herpes simplex virus (HSV), molluscum contagiosum and vaccinia virus.2, 3 The most common viral complication in AD is eczema herpeticum (ADEH+), which is mainly caused by HSV-1.2 ADEH+ subjects are invaluable resources for mechanistic investigations of the interplay between host immune defense and HSV-1 in humans. Based on this perspective, the National Institute of Allergy and Infectious Diseases (NIAID) funded the Atopic Dermatitis Research Network (ADRN) to investigate ADEH+ with comprehensive mechanistic studies.
We previously reported a study comparing transcriptomes of peripheral blood mononuclear cells (PBMCs) from atopic dermatitis without a history of eczema herpeticum (ADEH−) versus ADEH+ subjects.4 This study found ankyrin repeat domain 1 (ANKRD1) was one of the most down-regulated genes in ADEH+.4 ANKRD1, also known as c-193 and Cardiac Ankyrin Repeat Protein (CARP), is a pleiotropic protein containing four ankyrin repeat domains, a nuclear localization signal, a sequence which is rich in proline, glutamic acid, serine and threonine, and multiple phosphorylation consensus sites.5 ANKRD1 was discovered as a nuclear DNA-binding protein induced by inflammatory cytokines in human dermal vascular endothelial cells.5 Subsequently, ANKRD1 was found to be abundantly expressed in cardiac myocytes and skeletal muscle tissues, where they localized to the I band of sarcomere through binding to titin and myopalladin.6–8 ANKRD1 can also enter the nuclear of cardiac myocytes to regulate cardiomyogenesis as a transcriptional regulator.9 Up-regulation and gene mutations of ANKRD1 are associated with cardiomyopathy.10–12 Additionally, induction of ANKRD1 has been found in ovarian cancer, injured podocytes, cutaneous wounds, transforming growth factor-β induced mouse mammary epithelial cells, hepatitis C infected hepatocytes and papilloma virus infected keratinocytes.13–18 Although ANKRD1 has been extensively studied, its induction by HSV-1 in human immune cells and the biological significance of this response has not been investigated.
In the current study, we first confirmed our previous RNA-seq result using an increased number of subjects demonstrating that ANKRD1 transcripts were significantly down-regulated in HSV-1 stimulated PBMCs from ADEH+ subjects as compared to PBMCs cells from ADEH− subjects. We then explored the regulation of ANKRD1 expression in PBMCs, its cell source and its biological function in host anti-viral responses. The results of our study support a functional role for reduced ANKRD1 expression in ADEH+ pathogenesis.
Materials and Methods
Human subjects
Individuals ranging in age from 6 to 65 years participated in the study. They included 21 controls without atopy (non-atopic, NA), 19 ADEH−, and 20 ADEH+. The groups were stratified based on age and gender. None of the ADEH+ subjects had acute HSV-1 infection. All human subjects were examined for serum HSV-1 IgG and HSV-2 IgG values. The demographic characteristics of the 60 subjects is shown in supplemental table 1. An additional 10 healthy adults were recruited to participate in the mechanistic investigation. The institutional review board at National Jewish Health approved the study and all subjects provided written informed consent to participate.
Virus, PRR agonists, recombinant cytokines, neutralization antibodies and plasmids
HSV-1 virus stock (VR-733) was purchased from ATCC (Manassas, VA). PRR agonists CpG (ODN2395), CL264 (cat#: tlrl-c264e), LPS (cat#: tlrl-b5lps), Pam3CSK4 (cat#: tlrl-pms), PolydA:dT (cat#: tlrl-patc), PolyI:C-LMW (cat#:tlrl-picwlv), polyI:C-TLR3 agonist (cat#: tlrl-picw), and SS40 (cat#: tlrl-lrna40) were purchased from InvivoGen (San Diego, CA). Recombinant human TNFα (cat# 210-TA-020/CF), IL-1β (cat# 201-LB-005), IFNγ, IL-4, IL-22, GM-CSF, monoclonal mouse IgG1 Clone#11711 (Cat# MAB002), human IL-1β/IL-1F2 antibody (MAB601), human TNFαR1/TNFRSF1A antibody (MAB625), and anti-human IFNγR1 antibody (MAB6731) were bought from R&D systems, Inc. (Minneapolis, MN). Recombinant human IFNα (cat#:11101-1) and mouse anti-human IFNα (cat#: 21112-1) were purchased from PBL Biomedical Laboratories (Piscataway, NJ). Myc-DDK1-tagged RELA (RC220780), Myc-DDK1-tagged NFKB1 (RC208384), Myc-DDK1-tagged IRF7 (RC217934), Myc-DDK1-tagged ANKRD1 (RC205609) and Myc-DDK1-tagged STING were purchased from OriGene (Rockville, MD). Dr. Hong-Bing Shu (Wuhan University, China) kindly provided pCMV-flag-IRF3 and pCMV-flag-MyD88. PRK-neo-HA-ANKRD1 was generated in our lab by insertion of an encoding cDNA fragment in frame into PRK-neo-HA vector (a kind gift from Dr. Hong-Bing Shu).
Peripheral blood mononuclear cell isolation and purification of different cell types
Human PBMCs were isolated using Ficoll-Hypaque® density gradient centrifugation of heparinized venous blood from donors. The PBMCs were subjected to sequential isolation of T cells, NK cells and monocytes using anti-CD3, anti-CD56 and anti-CD14 microbeads according to manufacturer’s guidelines (Miltenyi Biotec Inc., San Diego, CA). Then the rest of the cells were further separated using a human B cell isolation kit II purchased from Miltenyi Biotec Inc. to obtain B cells and dendritic cells. In some experiments, we used antigen presenting cells (APCs, i.e. a mix of B cells, dendritic cells and monocytes) isolated from PBMCs by depletion of T cells and NK cells.
Cell treatment
PBMCs and other types of cells isolated from PBMCs were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin (50 I.U./ml) and streptomycin (50 μg/ml). For 21 NA subjects, 20 ADEH+ and 19 ADEH− subjects, one million PBMCs in 200μl culture media were stimulated with sham or HSV-1 at a multiplicity of infection (MOI) of 0.1 for 21 hours. For mechanistic studies, PBMCs and other types of cells were suspended in complete RPMI 1640 at 1×106 per ml, seeded in 96 well plates, and stimulated with HSV-1 at MOI of 0, 0.01 and 0.1 and various PRR agonists and cytokines for 24 hours. For experiments with neutralizing antibodies, PBMCs were first incubated with antibody at 5ug/ml for 2 hours and then exposed to medium or toll-like receptor 9 agonist CpG ODN 2395 at 50μM or 200μM for an additional 24 hours. The cells were harvested at the end of treatment for RNA extraction and real-time PCR.
Small interfering RNAs (siRNAs) silencing experiment
ANKRD1 and negative non-targeting scrambled siRNA duplexes were purchased from Dhamarcon (Lafayette, CO). The ON-TARGETplus Non-targeting pool sequences are: 5′-UGGUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAUGUUGUGUGA-3′, 5′-UGGUUUA CAUGUUUUCHGA-3′, 5′-UGGUUUACAUGUUUUCCUA-3′. The ON-TARGETplus ANKRD1 siRNA-SMARTpool target sequences are: 5′-CUACAAGACCUCUCGCAUA-3′, 5′-GAACCAAAGCAAUAUUCGA-3′, 5′-CGAAUUCCGUGAUAUGCUU-3′, 5′-GCUAUAAG AUGAUCCGACU-3′. APC cells were plated in 96 well plates at 1×105 per well and transfected with 0.5 pmol of siRNA duplexes per well using lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Twenty-four hours after transfection, HSV-1 was added to the cells at MOI of 0, 0.01 and 0.1 for an additional 24 hours. The cells were then harvested for RNA extraction and qRT-PCR.
RNA extraction and real-time PCR analysis
Total RNA was extracted from cells using RNeasy Mini Kits (Qiagen, Valencia, CA) according to the manufacturer’s guidelines. RNA was reverse transcribed into cDNA using SuperScript®III Reverse Transcriptase (Invitrogen). Real time PCR was conducted in an ABI Prism 7000 sequence detector (Applied Biosystems, Foster City, CA) as previously described.19 The primers for ANKRD1 (Hs00923599_m1), IL-29(Hs00601677_g1), IFNb1 (Hs01077958_s1), HPRT1 (Hs02800695_m1) and 18S (Hs99999901_s1) were purchased from Applied Biosystems. The primer sequences for transcripts of HSV-1 were prepared as previously described.20 Quantities of all target genes in test samples were normalized to the corresponding HPRT1 and 18S levels.
Over-expression, Co-immunoprecipitation and western-blotting
We purchased 293 FT cells from Invitrogen and cells were maintained in Dulbecco’s Modification of Eagle’s Medium (DMEM) with 4.5g/L glucose, 10% of fetal bovine serum and penicillin/streptomycin (50 units/ml/50 μg/ml) (Life Technologies, Carlsbad, CA ). For over-expression experiments, the cells were seeded in 24 well dishes at 1×105/well at the day before transfection. The following day, pCMV6-AN-HA, HA-ANKDR1, IRF3-Flag at 0.4 μg/well or indicated amounts were transfected into cells. The cells were harvested after 24 hours of transfection, or after stimulated with Poly I:C LMW for additional 8 hours. Western blotting and qRT-PCR assays were performed using these samples. For co-immunoprecipitation experiments, the cells were seeded in 100 mm diameter of petri dishes at 80–90% of confluence the day before transfection. The following day, plasmids of Flag-tagged or Myc-DDK1-tagged IRF3, IRF7, NFkB1, RELA, STING and MyD88 (10 μg of each) were co-transfected with 10 μg HA-tagged ANKRD1 using Lipofectamine 2000 according to the manufacturer’s guidelines (Invitrogen). After overnight incubation, the cells were harvested and lysed in 1 ml of protein lysis buffer (20mM Tris-HCl (pH 7.4), 150mM NaCl, 1mM EDTA,1% Triton-X100) supplemented with protease inhibitor (Sigma-Aldrich). Protein lysates were incubated with 0.5 μg of anti-HA antibody and 30 μl of protein A/G plus agarose (Santa Cruz Biotechnology) in 1.5 ml Eppendorf tubes and rotated at 4°C for one hour. The beads were then washed three times using protein lysis buffer with 0.5M NaCl. After wash, 30 μl of the 2xLaemmli sample buffer (Bio-Rad) was added into the beads and then boiled for 5 minutes. The supernatants from each sample were then subjected to western-blotting for detection of either Flag/DDK1 tagged protein or HA-tagged protein. Both anti-Flag antibody and anti-HA antibody were purchased from Sigma-Aldrich. A standard western-blot protocol was used.
Statistical analysis
Comparisons between NA, ADEH−, and ADEH+ subjects in ANKRD1 gene expression were analyzed using a general linear mixed model, adjusting for gender. Log transformation was performed on the normalized gene expression values and least squares means were used for the group comparisons between NA vs. ADEH−, NA vs. ADEH+, and ADEH− vs. ADEH+. No adjustments for multiple comparisons were made. SAS version 9.4 software (SAS Institute, Inc., Cary, NC) was used for all analyses. Comparisons of expression levels were performed using analysis of variance (ANOVA) techniques and independent sample t tests, as appropriate. Differences were considered significant at p<0.05.
Results
The induction of ANKRD1 by HSV-1 in PBMCs from ADEH+ is decreased
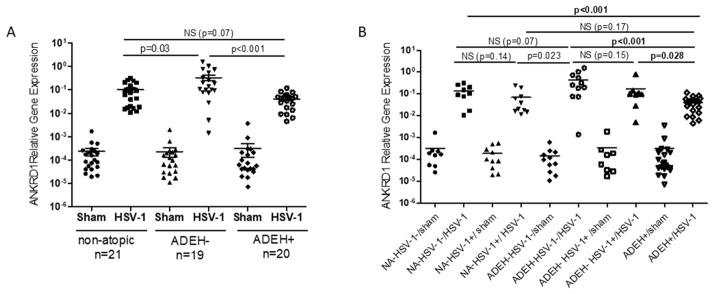
In a previous publication using a small number of patients and RNA sequencing, we reported that the levels of RNA transcripts of ANKRD1 in PBMCs induced by HSV-1 infection were significantly lower in ADEH+ (n=5) versus ADEH− (n=6) subjects.4 To validate this observation, we investigated ANKRD1 expression in the presence and absence of HSV-1 stimulation in 21 NA subjects, 19 ADEH− subjects and 20 ADEH+ subjects. As shown in Figure 1, ANKRD1 transcripts were expressed at either extremely low levels or were undetectable in PBMCs without HSV-1 exposure. Upon HSV-1 stimulation, ANKRD1 transcription was significantly induced in PBMCs in all three groups; however, the level in ADEH+ PBMCs was significantly lower than HSV-1 stimulated ADEH− PBMCs (p<0.001).
Figure 1. ANKRD1 is significantly down-regulated in HSV-1 stimulated ADEH+ PBMCs.
(A) PBMCs from normal (n=21), ADEHminus; (n=19) and ADEH+ (n=20) were stimulated with sham and HSV-1 (Multiplicity of infection: 0.1) overnight. ANKRD1 mRNA levels were evaluated by qRT-PCR. (B) ANKRD1 mRNA levels in human subjects were grouped based on their serological HSV-1 IgG positivity. NS indicates not significant.
To exclude the confounding factor of previous HSV-1 exposure that may affect responses of PBMCs to HSV-1, we further analyzed the data based on HSV-1 IgG serological positivity. 10 NA, 8 ADEH− and 20 ADEH+ were HSV-1 IgG positive. Among them, one NA, two ADEH− and one ADEH+ also were HSV-2 IgG positive (Supplemental table 1). Two NA subjects with HSV-2 IgG positive alone were excluded in the analyses. As shown in Fig 1B, ANKRD1 transcription levels in ADEH+ group (0.041 ± 0.006, n=20) were still significantly lower than HSV-1 IgG positive ADEH− group (0.169 ± 0.087, n=8) (p=0.028), while there were no significant differences between HSV-1 IgG positive subjects and HSV-1 IgG negative subjects for both of NA group (p= 0.14) and ADEH− group (p=0.15).
ANKRD1 transcription is significantly induced by PRR agonists and cytokines
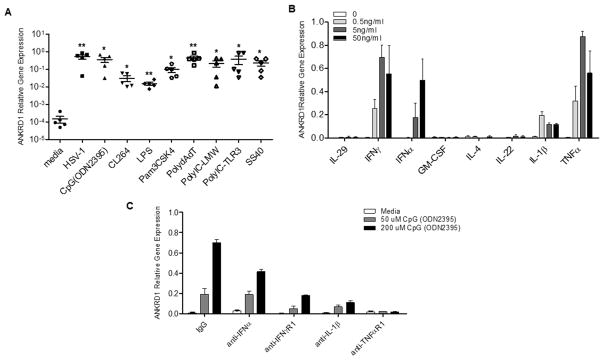
Since we found that ANKRD1 transcription could be induced in PBMCs by HSV-1 virus, and host innate immune cells recognize invading viruses through various pattern recognition receptors,21 we determined whether Pam3CSK4 (TLR2 agonist), CpG (TLR9 agonist), Poly dA: dT (cytoplasmic DNA sensor agonist), LPS (TLR4 agonist), CL264 (TLR7 agonist), Poly I:C-LMW (cytoplasmic RNA sensor), and Poly I:C –TLR3, SS40 (TLR8 agonist) could induce ANKRD1 gene expression. As shown in Figure 2A, all these agonists up-regulated ANKRD1 gene expression in PBMCs. In order to determine whether the up-regulation of ANKRD1 was a direct effect of PRRs’ signal activation or a secondary effect mediated by cytokine production after PRRs’ activation, we tested a panel of human recombinant cytokines, and found that ANKRD1 gene expression could be up-regulated in human PBMCs by IFNα, IFNγ, TNFα and IL-1β, but not IL-29, IL-4, IL-22 and GM-CSF (Figure 2B). We then used neutralizing antibodies to block the activity of IFNα, IFNγ, TNFα and IL-1β. We found that PBMCs pretreated with anti-IFNα, anti-IFNγR1, anti-IL-1β and anti-TNFR1, but not the control IgG, had reduced ANKRD1 gene expression upon CpG stimulation, demonstrating that CpG–induced ANKRD1 is a secondary response of CpG-induced cytokine production (Figure 2C).
Figure 2. The induction of ANKRD1 by PRR agonists is the secondary effect of cytokines.
(A) PBMCs from 5 normal subjects were treated with HSV-1 and various PRR agonists for 21 hours. ANKRD1 mRNA expression levels were evaluated by qRT-PCR. * indicates p<0.05; ** indicates p<0.01. (B) PBMCs from normal subjects (n=3) were treated with various cytokines at indicated concentrations. ANKRD1 mRNA expression levels were evaluated by qRT-PCR. Data shown as mean ±SE. (C) PBMCs from normal subjects (n=3) were pretreated with 5μg/ml of indicated neutralizing antibodies for 2 hours, the cells were then treated with indicated doses of ODN2395 for an additional 21 hours. ANKRD1 mRNA expression levels were evaluated by qRT-PCR. Data shown as mean ±SE.
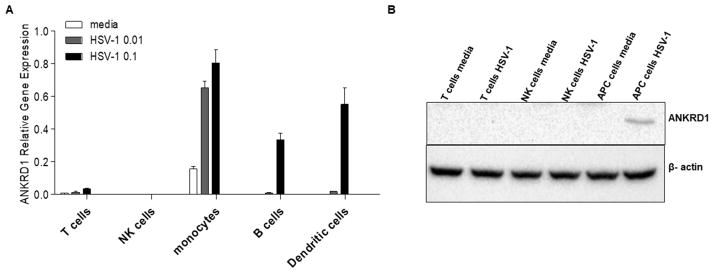
ANKRD1 transcription is predominantly induced in primary human antigen-presenting cells by HSV-1
To determine which cell types in PBMCs express ANKRD1, we purified T cells, NK cells, B cells, monocytes and dendritic cells, and stimulated these cells with HSV-1 of 0, 0.01 and 0.1 MOI for 24 hours. ANKRD1 transcription levels were significantly induced in APCs, i.e. B cells, monocytes and dendritic cells, but not in T cells and NK cells (Figure 3A). Additionally, we detected increased protein expression of ANKRD1 in HSV-1 stimulated mixed APCs, but couldn’t detect ANKRD1 protein in both sham and HSV-1 treated T cells and NK cells, as well as sham treated APCs (Fig 3B). The induction of ANKRD1 mRNA by HSV-1 was only found in primary APCs, but not in Epstein-Barr virus transformed B cell lines or the U937 and THP-1 human monocyte cell lines (data not shown). Additionally, ANKRD1 was not induced by IFNα, IFNγ, TNFα and IL-1β in these cell lines (data not shown). These data suggest that the induction of ANKRD1 expression by HSV-1 and cytokines may occur specifically in primary APC cells but not transformed cells or monocyte cell lines.
Figure 3. ANKRD1 is predominantly expressed and induced in APCs by HSV-1.
(A) Primary B cells, dendritic cells, monocytes, T cells and NK cells were purified from PBMCs isolated from normal subjects (n=3). ANKRD1 mRNA expression levels were evaluated by qRT-PCR. Data shown as mean ± SE. (B) ANKRD1 protein was detected by western blotting in T cells, NK cells and APCs with and without HSV-1 stimulation for 24 hours.
Gene Knockdown of ANKRD1 impairs innate immune responses against HSV-1
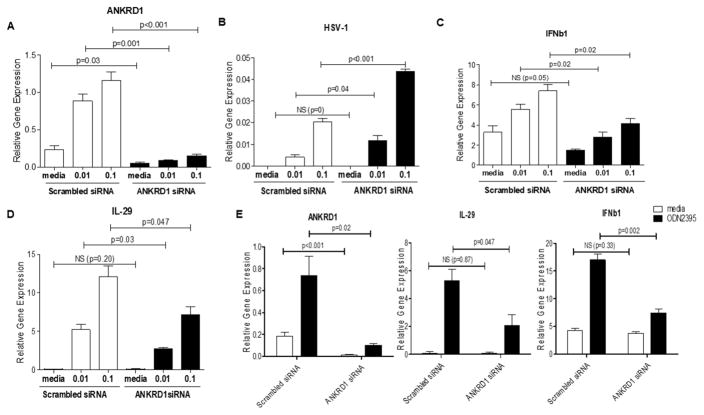
Since HSV-1 predominantly up-regulates ANKRD1 transcription in APCs, we hypothesized that it may be involved in anti-viral innate immune responses. To test this hypothesis, we silenced the expression of ANKRD1 in human APCs, then stimulated cells with various doses of HSV-1. We then evaluated HSV-1 viral load, as well as HSV-1 induced type I and type III interferons (IFNs), because type I (IFNα/IFNβ) and type III IFNs (IL-29/IL-28A) are the most important cytokines that protect host cells from HSV infection.22–24 As shown in Figure 4 (A–D), silencing ANKRD1 led to significantly increased HSV-1 viral loads and decreased IL-29 and IFNb1 mRNA levels as compared to cells transfected with scrambled siRNA. We also found that silencing ANKRD1 transcription in primary dendritic cells leads to decreased IL-29 and IFNb1 mRNA levels in response to CpG stimulation (Figure 4E). These results suggest that ANKRD1 is involved in anti-viral innate immune signaling pathways, specifically the signaling pathways leading to the production of type I and type III IFNs.
Figure 4. Knock-down of ANKRD1 enhances HSV-1 viral loads and reduces production of type I and type III IFNs in APCs.
Primary APCs from normal subjects (n=3) were isolated by depletion of T cells and NK cells. The cells were then transfected with scrambled siRNA duplexes and ANKRD1 siRNA duplexes. After 24 hours of incubation, HSV-1 at indicated doses was added to the cultures for an additional 24 hours of incubation. The cells were then harvested for RNA extraction and qRT-PCR. (A) ANKRD1, (B) HSV-1, (C) IFNb1 and (D) IL-29 were evaluated by qRT-PCR. (E). Primary dendritic cells from normal subjects (n=3) were isolated and transfected with siRNA duplexes for 24 hours. The cells were then stimulated with ODN 2395 for an additional 24 hours. ANKRD1, IFNb1 and IL-29 were evaluated by qRT-PCR. Data represented as mean ±SE. The data are representative of three independent experiments.
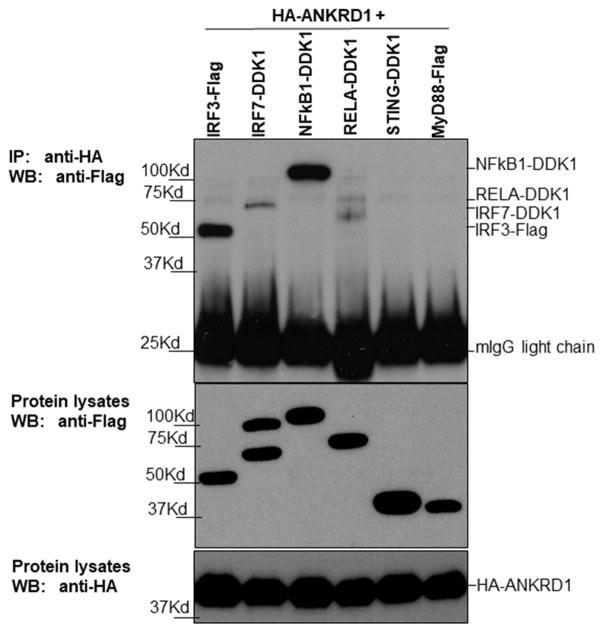
ANKRD1 forms protein complexes with IRF3
Since the current study found that ANKRD1 expression can be induced by multiple PRR agonists and cytokines (Figure 2A&B), we hypothesized that it may interact with some master regulators in PRR signaling transduction pathways. PRRs mediate their signaling through three major master regulators: IRF3, IRF7, and NFκB.21, 25 Therefore, we examined whether ANKRD1 could form protein complexes with IRF3 and IRF7, as well as the two subunits of NFκB: RELA and NFκB1. Two cytoplasmic adaptor protein STING-Flag and MyD88-Flag were used as controls. As shown in Figure 5, IRF3-DDK1, IRF7-DDK1 and NFkB1-DDK1 were all pulled down by HA-ANKRD1; but not RELA-DDK1, STING-Flag and MyD88-Flag. Additionally, IRF3-DDK1 and NFkB1-DDK1 had quantitatively more protein pulled down by HA-ANKRD1 although the amount of protein in the lysates was similar, suggesting that ANKRD1 may preferentially complex with IRF3 and NFkB1 rather than IRF7. Our data supports the previous report that ANKRD1 interacts with NFκB1.26 Additionally, we identified IRF3 and IRF7 as new binding partners for ANKRD1.
Figure 5. ANKRD1 can be pulled-down by IRF3, IRF7 and NFkB1.
The indicated expression plasmids were transfected into 293 FT cells and incubated overnight. The cells were then harvested and co-IP experiments were performed. Flag/DDK1-tagged protein expression and HA-tagged ANKRD1 protein expression are shown in IP samples and lysates. Data presented as one of three independent experiments.
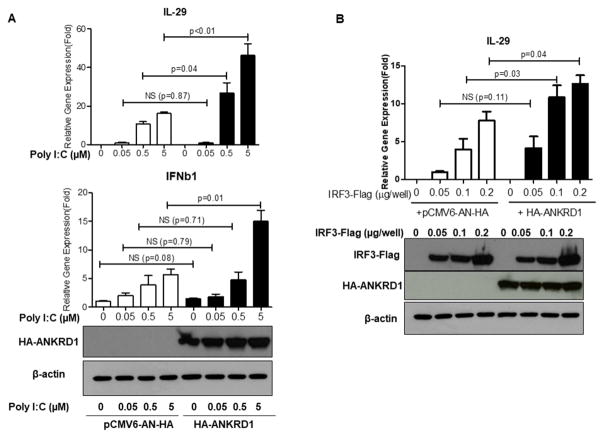
Over-expression of ANKRD1 enhances the IRF3 signaling pathway
Since ANKRD1 forms protein complexes with IRF3 and IRF7, and knockdown of ANKRD1 impairs innate immune responses against HSV-1 in APCs, we then investigated whether over-expression of ANKRD1 affects IFNb1 and IL-29 induction. HA-ANKRD1 and pCMV6-AN-HA empty vector were transiently transfected into 293FT cells, then stimulated with different concentrations of poly I:C LMW, an agonist that activates the IRF3/IRF7 signaling pathway. As shown in Figure 6A, both IFNB1 and IL-29 were significantly enhanced in HA-ANKRD1 overexpressed cells as compared to the control cells transfected with pCMV6-AN-HA empty vector upon poly I:C LMW stimulation. We also transfected IRF3-Flag with or without co-transfection of HA-ANKRD1. We found that HA-ANKRD1 by itself didn’t induce IL-29, however, it enhanced IRF3-Flag –induced IL-29 in 293T cells (Fig 6B). These data provide further support that ANKRD1 plays a role in the IRF3 signaling pathway by enhancing induction of type I and type III IFNs.
Figure 6. ANKRD1 enhances IRF3 signaling pathway.
(A) PRK control plasmid and HA-ANKRD1 plasmids were transfected into 293 FT cells. The following day cells were treated with poly I:C LMW at indicated concentrations for 8 hours. The cells were then harvested and IL-29 and IFNb1 gene expression were evaluated by qRT-PCR. Western-blot assay confirmed that HA-ANKRD1was over-expressed. (B) Different amounts of IRF3-Flag plasmids with or without HA-ANKRD1 plasmids were transfected into 293FT cells, the following day the cells were harvested and IL-29 gene expression was evaluated by qRT-PCR. Western-blot assay confirmed that IRF3-Flag and HA-ANKRD1 were over-expressed. Data represented as mean ±SE. The data are representative of four independent experiments.
Discussion
ADEH+ is a devastating complication of AD, as it can be complicated by keratoconjunctivitis, viremia, meningitis and encephalitis.27 It has been postulated that the increased propensity of subjects with AD to EH may be caused by reduced antimicrobial peptides and skin barrier proteins due to the influence of Th2 cytokines.28, 29 Additionally, decreased IFN responses have been reported to play a role in ADEH+ etiology.4, 30 To date, the molecular biomarkers that distinguish ADEH+ from ADEH− have not been identified. In the current study, we identified that ANKRD1 was significantly decreased in ADEH+ PBMCs upon HSV-1 stimulation as compared to ADEH− PBMCs (Fig 1). 42% of ADEH− subjects in our study (n=8) were serologically HSV-1 IgG positive, suggesting that they have had prior HSV-1 exposure. After we stratified subjects based on their HSV-1 IgG positivity, the induction of ANKRD1 was still significantly greater in ADEH− subjects as compared to ADEH+ subjects (Fig 1 B). This result demonstrates that reduced ANKRD1 in PBMCs from ADEH+ subjects, in comparison with ADEH−, is not caused by differences on HSV-1 exposure history, supporting the concept that there are fundamental differences between ADEH− and ADEH+.
The current study is the first to demonstrate that ANKRD1 is induced by HSV-1 virus, PRRs and IFNs in human PBMCs. Although previous studies reported that ANKRD1 could be induced by IL-1α, TNFα and LPS in endothelial cells, by HCV in hepatocytes and by HPV in keratinocytes, 5, 16, 17 the current study examined a more comprehensive spectrum of stimuli on ANKRD1 induction in human PBMCs as compared to prior studies. We further demonstrate that the induction of ANKRD1 by PRRs may be a secondary effect of PRR induced cytokine production, as cytokines’ neutralizing antibodies can reduce ANKRD1 induction by CpG (Fig 2C).
ANKRD1 has been extensively studied in cardiac myocyte biology and pathology where it is expressed at the highest level. Although ANKRD1 knockout mice are born normal,31 up-regulation of ANKRD1 has been found in cardiac tissues caused by cardiac overload, hypertension and heart failure.32–34 Induction of ANKRD1 has also been reported in various physiological and pathological conditions in different tissues and cells. Since ANKRD1 is significantly up-regulated in skin wounds, it has been suggested to play a role in angiogenesis and tissue remodeling through regulation of MMP13 gene expression in wound repair.15, 35, 36 ANKRD1 up-regulation has also been found in ovarian cancer cells. Inhibition of ANKRD1 in ovarian cancer cells promotes cisplatin-induced apoptosis.37 Additionally, Matsuura et al reported that ANKRD1 was up-regulated in renal podocytes by LPS-induced injury and its up-regulation was associated with proteinuria in lupus nephritis.14
In the case of viral infections, Kaczkowski et al found ANKRD1 was up-regulated by HPV11, HPV16 and HPV45 in the HaCat keratinocyte cell line, but they didn’t investigate the biological significance of this response.17 Than et al found that ANKRD1 was induced significantly by HCV in hepatocytes. Their study suggested that ANKRD1 might been involved in facilitating HCV entry.16 Although previous studies couldn’t definitively address the biological function of ANKRD1 protein, these studies revealed a common phenomenon that ANKRD1 was up-regulated by various different types of stress and insults, suggesting that ANKRD1 acts as a danger sensor. In the current study, we found ANKRD1 was significantly induced in human primary APCs by HSV-1, demonstrating that up-regulation of ANKRD1 is a response to HSV-1 infection. Thus, our current study provides an additional example to support the notion that ANKRD1 is a danger sensor.
In the context of cardiomyogenesis, ANKRD1 has been found to interact with several transcription factors, including Y-box transcription factor 1 (YB-1), androgen receptor (AR), p50 subunit of NF-κB and p53.6, 26, 38, 39 Functionally, ANKRD1 works as a repressor to suppress the transcriptional activity of YB-1, AR and p50, while it works as a co-activator to p53. Our current study demonstrates that ANKRD1 mRNA expression is at either very low or undetectable levels in PBMCs in different individuals when they are unstimulated. Its expression is significantly induced by the stimulation of PRR induced cytokines including IFNs (Figure 2). This suggests that up-regulation of ANKRD1 in PBMCs is a response to the activation of viral recognition pathways, leading us to hypothesize that it may function in viral–induced host cell innate immunity. HSV-1 viruses are known to activate innate immune cells to produce type I/type III IFNs through multiple PRR signaling pathways.16, 40–44 The IFNs then prepare host cells to enter an “anti-viral state” by producing large amounts of anti-viral proteins to inhibit the replication of invading viruses. Viral recognition pathways of TLR3, RIG-1/MDA5 and cGAS/STING signaling pathways all use the transcription factor IRF3 to transduce signaling cascade to the production of type I and type III IFNs. In some cases, the transcription factor IRF7 co-operates with IRF3 to promote IFN production.25 We therefore investigated whether IRF3 and IRF7 were the binding partners of ANKRD1. As expected, ANKRD1 indeed interacts with IRF3 and IRF7, but it mainly binds with IRF3 (Fig 5). Functionally, we found that silencing ANKRD1 led to enhanced HSV-1 viral load and reduced production of the anti-viral cytokines, IFNb1 and IL-29, in response to HSV-1 stimulation (Figure 4 A–D). When ANKRD1 is over-expressed, it enhances poly I:C induced IL-29 and IFNb1 transcription; co-expression of ANKRD1 and IRF3 also enhances IRF3-induced IL-29 (Fig 6). These data suggest that ANKRD1 works in a positive feedback loop to regulate viral recognition pathways, i.e. after upregulation by PRRs, it promotes optimal type I/III IFN production by working as a co-activator of IRF3 in viral recognition pathways.
We also detected the interaction of ANKRD1 and NFκB1 (p50). However, we found silencing ANKRD1 led to reduced IFN transcription by the TLR9 agonist CpG stimulation (Fig 4E). TLR9 activated IFN responses is mainly transduced by NFκB.21 Our results therefore suggest that ANKRD1 acts as a coactivator to NF-κB in APCs. This contradicts a previous study by Liu et al, who found ANKRD1 inhibited TNF-α induced NF-κB activity in myoblasts.26 This discrepancy may be explained by cell type specific responses although further work is needed to sort out these differences.
In summary, our study has identified ANKRD1 as a novel biomarker that is differentially expressed between ADEH+ subjects and ADEH− subjects. We demonstrate it interacts with IRF3 and IRF7, and enhances IRF3 mediated signaling pathways. These findings suggest a novel antiviral pathway that is subverted in AD subjects prone to eczema herpeticum.
Supplementary Material
Key Messages.
Induction of ANKRD1 by HSV-1 is reduced in ADEH+ subjects.
ANKRD1 expression induced by HSV-1 and pattern recognition receptor agonists is secondary to cytokines.
ANKRD1 is involved in the IRF3 signaling pathways.
Acknowledgments
This work was funded by NIH/NIAID through the Atopic Dermatitis Research Network U19 AI117673. Lianghua Bin is also supported by National Natural Science Foundation of China (No: 81371716) and 111 project (B16021). The authors acknowledge the CTRC nurses for their hard work in recruiting human subjects for this study. CTRC is supported in part by the Colorado Clinical and Translational Science Award/Colorado Clinical &Translational Sciences Institute grant UL1 RR025780 from National Center for Research Resources/NIH and UL1 TR0001082 from NIH/National Center for Advancing Translational Sciences. Additionally, the authors wish to acknowledge The Edelstein Family Foundation for their generous support of our work.
ABBREVIATIONS
- AD
Atopic dermatitis
- ADEH+
Atopic dermatitis with a history of eczema herpeticum
- ADEH−
Atopic dermatitis without a history of eczema herpeticum
- ANKRD1
Ankyrin repeat domain 1
- APC
Antigen presenting cell
- HA
Hemagglutinin
- HSV-1
Herpes simplex virus 1
- IFN
Interferon
- MOI
Multiplicity of infection
- NA
non-atopic
- PBMCs
Peripheral blood mononuclear cells
- Poly I:C-LMW
Polyinosinic-polycytidylic acid low molecular weight
- Poly dA:dT
Poly(deoxyadenylic-deoxythymidylic) acid sodium salt
- PRR
Pathogen pattern recognition receptor
- qRT-PCR
Quantitative reverse transcriptase polymerase chain reaction
- SE
Standard error
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387:1109–22. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- 2.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–9. 9 e1–7. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung DY. Why is eczema herpeticum unexpectedly rare? Antiviral Res. 2013;98:153–7. doi: 10.1016/j.antiviral.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bin L, Edwards MG, Heiser R, Streib JE, Richers B, Hall CF, et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol. 2014;134:848–55. doi: 10.1016/j.jaci.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu W, Burns DK, Swerlick RA, Presky DH. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J Biol Chem. 1995;270:10236–45. doi: 10.1074/jbc.270.17.10236. [DOI] [PubMed] [Google Scholar]
- 6.Zou Y, Evans S, Chen J, Kuo HC, Harvey RP, Chien KR. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]
- 7.Miller MK, Bang ML, Witt CC, Labeit D, Trombitas C, Watanabe K, et al. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol. 2003;333:951–64. doi: 10.1016/j.jmb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Bang ML, Mudry RE, McElhinny AS, Trombitas K, Geach AJ, Yamasaki R, et al. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol. 2001;153:413–27. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeyaseelan R, Poizat C, Baker RK, Abdishoo S, Isterabadi LB, Lyons GE, et al. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem. 1997;272:22800–8. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- 10.Moulik M, Vatta M, Witt SH, Arola AM, Murphy RT, McKenna WJ, et al. ANKRD1, the gene encoding cardiac ankyrin repeat protein, is a novel dilated cardiomyopathy gene. J Am Coll Cardiol. 2009;54:325–33. doi: 10.1016/j.jacc.2009.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling SSM, Chen YT, Wang J, Richards AM, Liew OW. Ankyrin Repeat Domain 1 Protein: A Functionally Pleiotropic Protein with Cardiac Biomarker Potential. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18071362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duboscq-Bidot L, Charron P, Ruppert V, Fauchier L, Richter A, Tavazzi L, et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur Heart J. 2009;30:2128–36. doi: 10.1093/eurheartj/ehp225. [DOI] [PubMed] [Google Scholar]
- 13.Scurr LL, Guminski AD, Chiew YE, Balleine RL, Sharma R, Lei Y, et al. Ankyrin repeat domain 1, ANKRD1, a novel determinant of cisplatin sensitivity expressed in ovarian cancer. Clin Cancer Res. 2008;14:6924–32. doi: 10.1158/1078-0432.CCR-07-5189. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura K, Uesugi N, Hijiya N, Uchida T, Moriyama M. Upregulated expression of cardiac ankyrin-repeated protein in renal podocytes is associated with proteinuria severity in lupus nephritis. Hum Pathol. 2007;38:410–9. doi: 10.1016/j.humpath.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y, Reitmaier B, Regenbogen J, Slowey RM, Opalenik SR, Wolf E, et al. CARP, a cardiac ankyrin repeat protein, is up-regulated during wound healing and induces angiogenesis in experimental granulation tissue. Am J Pathol. 2005;166:303–12. doi: 10.1016/S0002-9440(10)62254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Than TT, Tran GV, Son K, Park EM, Kim S, Lim YS, et al. Ankyrin Repeat Domain 1 is Up-regulated During Hepatitis C Virus Infection and Regulates Hepatitis C Virus Entry. Sci Rep. 2016;6:20819. doi: 10.1038/srep20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaczkowski B, Rossing M, Andersen DK, Dreher A, Morevati M, Visser MA, et al. Integrative analyses reveal novel strategies in HPV11,-16 and -45 early infection. Sci Rep. 2012;2:515. doi: 10.1038/srep00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, et al. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 19.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–9. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 20.Bin L, Howell MD, Kim BE, Streib JE, Hall CF, Leung DY. Specificity protein 1 is pivotal in the skin’s antiviral response. J Allergy Clin Immunol. 2011;127:430–8. e1–2. doi: 10.1016/j.jaci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–40. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–9. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deonarain R, Alcami A, Alexiou M, Dallman MJ, Gewert DR, Porter AC. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J Virol. 2000;74:3404–9. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J Exp Med. 1999;189:663–72. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Liu S, Chen ZJ. SnapShot: pathways of antiviral innate immunity. Cell. 2010;140:436–e2. doi: 10.1016/j.cell.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XH, Bauman WA, Cardozo C. ANKRD1 modulates inflammatory responses in C2C12 myoblasts through feedback inhibition of NF-kappaB signaling activity. Biochem Biophys Res Commun. 2015;464:208–13. doi: 10.1016/j.bbrc.2015.06.118. [DOI] [PubMed] [Google Scholar]
- 27.Wollenberg A, Zoch C, Wetzel S, Plewig G, Przybilla B. Predisposing factors and clinical features of eczema herpeticum: a retrospective analysis of 100 cases. J Am Acad Dermatol. 2003;49:198–205. doi: 10.1067/s0190-9622(03)00896-x. [DOI] [PubMed] [Google Scholar]
- 28.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–8. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Howell MD, Kim BE, Gao P, Grant AV, Boguniewicz M, Debenedetto A, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2007;120:150–5. doi: 10.1016/j.jaci.2007.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung DY, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in IFN-gamma response. J Allergy Clin Immunol. 2011;127:965–73. e1–5. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bang ML, Gu Y, Dalton ND, Peterson KL, Chien KR, Chen J. The muscle ankyrin repeat proteins CARP, Ankrd2, and DARP are not essential for normal cardiac development and function at basal conditions and in response to pressure overload. PLoS One. 2014;9:e93638. doi: 10.1371/journal.pone.0093638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aihara Y, Kurabayashi M, Saito Y, Ohyama Y, Tanaka T, Takeda S, et al. Cardiac ankyrin repeat protein is a novel marker of cardiac hypertrophy: role of M-CAT element within the promoter. Hypertension. 2000;36:48–53. doi: 10.1161/01.hyp.36.1.48. [DOI] [PubMed] [Google Scholar]
- 33.Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, et al. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–62. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- 34.Zolk O, Marx M, Jackel E, El-Armouche A, Eschenhagen T. Beta-adrenergic stimulation induces cardiac ankyrin repeat protein expression: involvement of protein kinase A and calmodulin-dependent kinase. Cardiovasc Res. 2003;59:563–72. doi: 10.1016/s0008-6363(03)00476-0. [DOI] [PubMed] [Google Scholar]
- 35.Samaras SE, Almodovar-Garcia K, Wu N, Yu F, Davidson JM. Global deletion of Ankrd1 results in a wound-healing phenotype associated with dermal fibroblast dysfunction. Am J Pathol. 2015;185:96–109. doi: 10.1016/j.ajpath.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almodovar-Garcia K, Kwon M, Samaras SE, Davidson JM. ANKRD1 acts as a transcriptional repressor of MMP13 via the AP-1 site. Mol Cell Biol. 2014;34:1500–11. doi: 10.1128/MCB.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei Y, Henderson BR, Emmanuel C, Harnett PR, deFazio A. Inhibition of ANKRD1 sensitizes human ovarian cancer cells to endoplasmic reticulum stress-induced apoptosis. Oncogene. 2015;34:485–95. doi: 10.1038/onc.2013.566. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Ruggiero CL, Bauman WA, Cardozo C. Ankrd1 is a transcriptional repressor for the androgen receptor that is downregulated by testosterone. Biochem Biophys Res Commun. 2013;437:355–60. doi: 10.1016/j.bbrc.2013.06.079. [DOI] [PubMed] [Google Scholar]
- 39.Kojic S, Nestorovic A, Rakicevic L, Belgrano A, Stankovic M, Divac A, et al. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch Biochem Biophys. 2010;502:60–7. doi: 10.1016/j.abb.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Zhang SY, Boisson-Dupuis S, Chapgier A, Yang K, Bustamante J, Puel A, et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 41.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–5. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 42.Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J Immunol. 2005;175:4189–93. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen SB, Sorensen LN, Malmgaard L, Ank N, Baines JD, Chen ZJ, et al. Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J Virol. 2007;81:13315–24. doi: 10.1128/JVI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y, et al. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe. 2014;16:450–61. doi: 10.1016/j.chom.2014.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.