Abstract
Background
The contribution of phenotypic variation of peanut-specific T cells to clinical allergy or tolerance to peanut is not well understood.
Objectives
Our objective was to comprehensively phenotype peanut-specific T cells in the peripheral blood of individuals with and without peanut allergy (PA).
Methods
We obtained samples from PA individuals, including a cohort undergoing baseline peanut challenges for an immunotherapy trial (CoFAR6). Subjects were confirmed as PA, or if they passed a 1 g peanut challenge they were termed high-threshold (HT). Healthy controls (HC) were also recruited. Peanut-responsive T cells were identified by CD154 expression after 6–18h of stimulation with peanut extract. Cells were analyzed by flow cytometry and single cell RNA sequencing.
Results
PA individuals had tissue and follicle-homing peanut-responsive CD4+ T cells with a heterogeneous pattern of Th2 differentiation, while controls had undetectable T cell responses to peanut. The PA group had a delayed and IL-2-dependent upregulation of CD154 on cells expressing Treg markers, which was absent in HC or HT individuals. Depletion of Tregs in vitro enhanced cytokine production in HC and PA subjects, but cytokines associated with highly differentiated Th2 cells were more resistant to Treg suppression in PA subjects. Analysis of gene expression by single cell RNAseq identified T cells with highly correlated expression of IL4, IL5, IL9, IL13 and the IL-25 receptor IL17RB.
Conclusions
These results demonstrate the presence of highly differentiated Th2 cells producing Th2-associated cytokines with functions beyond IgE-class switch in peanut allergy. A multi-functional Th2 response was more evident than a Treg deficit among peanut-responsive T cells.
Keywords: Food allergy, peanut allergy, Th2, Treg, tolerance
Graphical Abstract
INTRODUCTION
Peanut allergy (PA) is believed to arise from defective oral tolerance pathways or a lack of dietary exposure in early life. In mouse models, oral tolerance is an active state of immune regulation mediated by peripherally-induced Foxp3+ Tregs that are educated by gastrointestinal DCs via TGF-beta and retinoic acid-dependent mechanisms 1, 2. Support for the clinical importance of oral tolerance comes from the LEAP study, which demonstrated that dietary exposure to peanut early in life (4–11 months) could suppress the development of PA in infants with elevated risk of PA 3. The failure or lack of oral tolerance is thought to be a requisite for the development of Th2-skewed immunity that underlies the pathologic immune responses in PA and other allergic diseases 4. The Th2 cytokine IL-4 is necessary for B cell class-switching to IgE, and a peanut-specific Th2-skewed CD4+ T cell profile has been found in subjects with PA using methods from proliferation-based assays to MHC II tetramers 5–8.
Th2-biased immunity can be a benign feature of the young immune system, particularly when partnered with IL-10 production 9. Features beyond production of IL-4 may therefore be necessary for pathogenicity, such as homing to B cell follicles or tissues. Other Th2-associated cytokines may also contribute to pathogenesis. IL-9 has begun to emerge as an important cytokine associated with food allergy in both mouse models and allergic subjects 10–12. Regulatory T cells have been reported as a source of IL-4 in mice and humans, and these “reprogrammed” Th2 cells may contribute to food allergy both through Th2 cytokine production as well as defective regulatory function 13. These studies highlight a growing appreciation of the complexity of the food-specific T cell response in food allergy.
We hypothesized that clinical PA results from a combination of disordered pro-inflammatory Th2 immunity and an antigen-specific defect in regulatory activity, and we tested this hypothesis in well-characterized patient cohorts and healthy controls, including 84 primarily pediatric subjects who underwent a double-blind placebo controlled food challenge to peanut. We performed single cell analysis by flow cytometry and RNA-seq to determine the phenotype of the T cell response underlying allergy or tolerance to peanut. Our data show that clinical tolerance to foods is associated with immunologic ignorance or anergy, while the defining feature of PA is the presence of antigen-specific cells including highly differentiated, tissue-homing, granulocyte growth factor-producing Th2 cells that may be less susceptible to regulation by Tregs. The clinical implication is that strategies to eliminate effector Th2 cells may be more effective for the treatment of PA than strategies aimed at expanding the antigen-specific regulatory response.
METHODS
Human subjects
Informed consent was obtained from all subjects or parents/guardians, and all procedures were approved by the Institutional Review Boards at each of the 5 clinical sites. Blood samples were obtained at baseline from a clinical trial conducted by the Consortium of Food Allergy Research (CoFAR) investigating epicutaneous immunotherapy for peanut allergy (CoFAR6, ClinicalTrials.gov identifier NCT01904604, 14). Subjects who responded to a double-blind placebo controlled food challenge with a cumulative dose up to 1 gram of peanut were categorized as peanut allergic (PA), while those who tolerated the challenge were termed high threshold (HT). Additional PA and healthy control (HC) subjects were recruited from Mount Sinai (MSSM Cohort) under other IRB-approved protocols. MSSM PA subjects were recruited based on a convincing clinical history, but did not undergo oral food challenge as part of this study. Subject demographics including peanut-specific IgE are detailed in Table 1. An experimental overview is shown in Figure E1.
TABLE 1.
DEMOGRAPHIC CHARACTERISTICS OF THE STUDY POPULATION
| Healthy Controls (HC) | Peanut Allergic (PA), CoFAR6 Cohort | High Threshold (HT), CoFAR6 Cohort | Peanut Allergic (PA), MSSM Cohort | |
|---|---|---|---|---|
| N* | 21 | 75 | 9 | 51 |
| Mean Age in | 30.5 | 9.5 | 9.7 | 10.5 |
| years (min, max) | (23, 44) | (4, 20) | (4, 16) | (3, 20) |
| Gender (M, F) | 13, 7 | 46, 29 | 7, 2 | 31, 19** |
| Allergy Assessment | Self-Report | Oral Food Challenge | Oral Food Challenge | Convincing Clinical History |
| Median Peanut sIgE in kUA/L (min, max) | 0 (0, 0.29) | 78.6 (0.41, >100***) | 5.51 (0.25, 13.3) | 46.2 (0, >100***) |
| Median Peanut sIgG4 in mgA/L, (min, max) | 0.13 (0.04, 2.58) | 0.64 (0.02, 7.01) | 0.51 (0.13, 2.24) | ND |
N is cumulative, the N for each experiment is given in the figure legends.
Gender and age unreported for one subject.
Values > 100 kUA/L were truncated
ND = not measured
Blood processing and peripheral blood mononuclear cell (PBMC) isolation
For COFAR6 samples, blood was shipped overnight from participating sites in temperature-controlled boxes (GreenBox, ThermoSafe, Arlington Heights, IL) with temperature monitors. PBMCs were isolated by density centrifugation with Ficoll-Paque Plus (GE Healthcare, Pittsburgh, PA) and cultured in AIM V® medium (Gibco, Grand Island, NY) with 2.5% autologous plasma.
PBMC stimulations
PBMCs were stimulated with 100 μg/mL crude peanut extract (CPE) or 5μL/mL anti-CD3/CD28 beads (Invitrogen, Carlsbad, CA) for 6 hours, 18 hours, 24 hours, or 5 days. Endotoxin was removed from peanut extract using Detoxi-Gel columns (Thermo Scientific, Waltham, MA). Residual endotoxin levels measured by LAL assay (Thermo Scientific) were 14 pg/ml. GolgiPlug (BD Biosciences, San Jose, CA) was added 4 hours prior to harvesting. For surface CD154 staining (sorting), blocking anti- CD40 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) was added to the culture to maintain CD154 at the surface. In some experiments, cells were stimulated with rhIL2 (StemCell, Vancouver, Canada) at 5 ng/ml, or neutralizing anti-IL2 (R&D Systems, Minneapolis, MN) was added at 500 ng/ml. PMA/ionomycin/brefeldin cocktail (BD Biosciences) was applied for 6h to stimulate cells in some experiments.
Flow Cytometry
Harvested PBMCs were stained for viability (Live/Dead Fixable stain, Invitrogen), washed and stained for surface markers, and washed for fixation and permeabilization. To detect intracellular CD154 and cytokines, cells were fixed in 4% paraformaldehyde (Electron Microscopy Services, Hatfield, PA) and treated with permeabilization buffer (eBioscience, San Diego, CA) before staining with labeled antibodies. For FoxP3 staining, cells were processed using the FoxP3/Transcription Factor Staining Buffer Set (eBioscience) before staining with antibodies. Antibody panels used for surface and cytoplasmic staining are shown in Tables E1–E4. Stained cells were subsequently analyzed on a LSR Fortessa (BD Biosciences). For Treg sorting or depletion, harvested PBMCs were stained with CD4 Alexa Fluor 488 (Clone OKT4, BioLegend, San Diego, CA), CD25 Alexa Fluor 700 (Clone BC96, BioLegend), and CD127 PE-Cy7 (Clone eBioRDR5, eBioscience). Cells were sorted on the FACS Aria IIu (BD Biosciences).
Peanut-specific IgE and IgG4
Frozen plasma samples were thawed and peanut-specific IgE and IgG4 levels were measured by ImmunoCAP (Thermo Fisher Scientific).
Secreted Cytokine Measurement
Cell culture supernatants were centrifuged and stored at −80°C. Supernatant cytokines were measured using the ProcartaPlex Human Cytokine Panel 1B (eBioscience) and the Luminex® 200™ System (Luminex Corporation, Austin, TX) in Mount Sinai’s Human Immune Monitoring Core Facility. In other experiments, cytokines were measured with the LEGENDplexTM Human Th Cytokine Panel according to manufacturer’s instructions (BioLegend) and acquired on the BD LSRFortessa (BD Biosciences)15. Sensitivity of the two cytokine measurement kits is shown in Tables E5–6.
Enrichment and FACS sorting of CD154+ T cells
PBMCs stimulated for 18h were harvested and enriched for CD154+ cells using the CD154 MicroBead Kit (Miltenyi Biotec). Enriched cells were stained with CD3 APC-Cy7 (Clone SK7, eBioscience), CD4 Alexa Fluor 488 (Clone OKT4, BioLegend) and CD154 streptavidin PE (Clone 24–31, eBioscience). CD3+CD4+CD154+ T cells were sorted on the BD FACSAria IIu (BD Biosciences) in Mount Sinai’s Flow Cytometry Core Facility, and resuspended according to manufacturer protocols (Fluidigm, South San Francisco, CA).
CD154+ T cells were captured after 18h of stimulation and sort-purified to obtain CD154+ cells at >99% purity. We used the Fluidigm C1 pipeline to obtain single cell cDNA, prepared barcoded cDNA libraries, and performed 100 NT paired end read sequencing on multiplexed cells (48 cells/lane) using Illumina HiSeq (Illumina, San Diego, CA).
Fluidigm C1 processing and cDNA library construction of single-cell RNA-sequencing
Sorted Tregs and CD154+ T cells were resuspended and loaded onto a Fluidigm C1TM Single-Cell Auto Prep IFC for Fluidigm C1TM processing as per manufacturer protocols (Fluidigm). Single-cells were visualized by microscopy and wells containing >1 cell were noted and excluded from further steps. cDNA was generated according to Fluidigm C1™ protocols with SMARTer® Ultra™ Low RNA Kit for the Fluidigm C1™ System reagents (Clontech Laboratories, Mountain View, CA). Resultant cDNA from Fluidigm C1™ processing was subsequently assessed for quantity and quality using a Qubit Fluorometer 3.0 (Thermo Fisher Scientific) and 2100 Bioanalyzer or 2200 TapeStation (Agilent Technologies, Santa Clara, CA). cDNA which passed QC were subsequently processed for library construction using the Nextera XT DNA Library Prep and Nextera XT Index Kits (all from Illumina). Amplified single-cell libraries were then pooled and submitted to Mount Sinai’s Genomics Core Facility for sequencing on the Illumina HiSeq 2500 (100bp paired-end reads).
Single-cell analysis
RNAseq raw fastq were aligned to a hg38 reference genome (UCSC) using STAR (2.4.2a). Aligned reads were mapped to hg38 genes using featureCounts from the subread package (1.4.4) 16. Cells with reads < 5 x 105 were removed. Negative Binomial (NB) model based method edgeR (3.10.0) 15 was used for single cell differential expression analysis. Genes expressed in at least one cell with at least one read per million mapped were kept. The Relative Log Expression (RLE) method was used to calculate normalization factors between samples. We included plates as covariates to account for any confounding effect of individuals in the generalized linear model (GLM) and performed quasi-likelihood F-tests for hypothesis testing. Genes with FDR < 0.05 were defined as differentially expressed genes (DEG). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus 17 and are accessible through GEO Series accession number GSE98852 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE98852).
Molecular states clustering
Monocle (1.2.0) toolkit was used to find differentially expressed genes between cell types/states and to cluster CD154+ cells subgroups 18. Genes expressed at low levels (less than 1 read per million mapped in 1 individual) were filtered out and the read counts converted into reads per kilo base per million mapped reads (RPKM). Independent Component Analysis (ICA) was carried out for dimension reduction of gene expression. DEG generated using edgeR-QL were fed as guide genes to order the cells.
TCR clonal inference
TCR clonal analysis was based on TraCeR (0.1) 19. T cell receptor sequences were reconstructed by alignment of reads to synthetic human TCR genomes (all possible V-J combinations provided by TraCeR) using bowtie2 (2.1.0) 20, and assembled reads into TCR contigs using trinity (2.2.0) 21. The assembled contigs were then aligned against the IMGT human database using igblast (1.5.0) 22 and quantified as transcript per million (tpm) using kallisto (0.43.0) 23. Only the two most highly expressed TCR from each locus were retained. We searched for cells sharing exact TCR sequences for each cell type.
RESULTS
Peanut-specific T cell responses and clinical phenotype
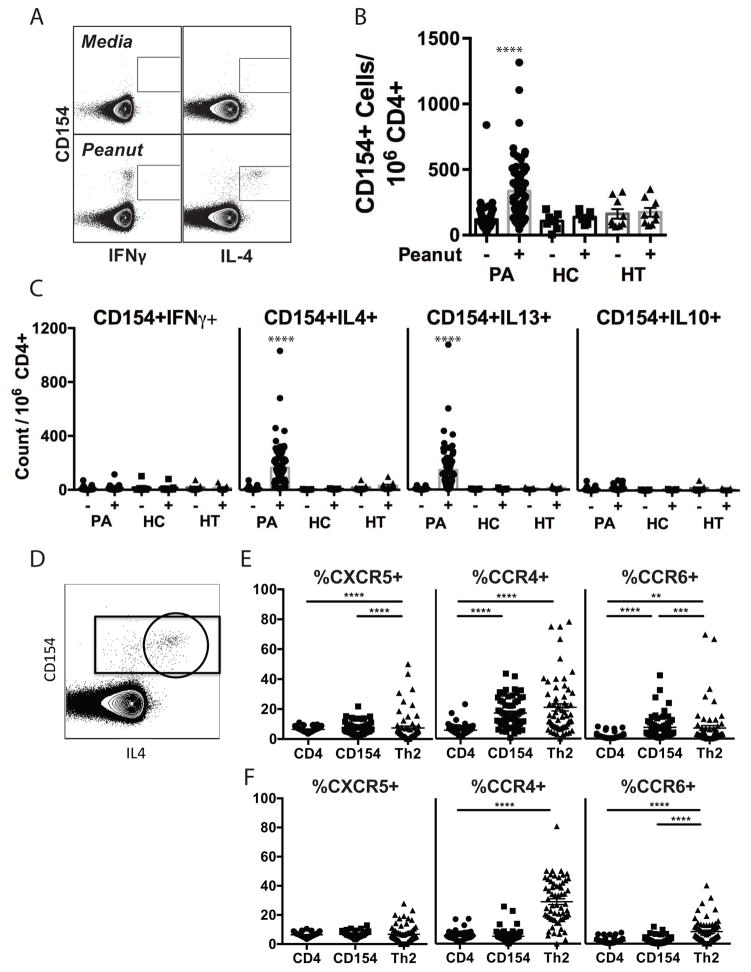
Peripheral blood was obtained from a cohort of primarily pediatric subjects with PA who reacted to an oral challenge dose of less than 1 g peanut (CoFAR Cohort); subjects with a history of PA and detectable peanut-specific IgE who tolerated a cumulative dose of 1 g peanut (High Threshold, or HT); and healthy adult controls who self-reported tolerance to peanut (HC). PA and HT groups were obtained from subjects undergoing baseline peanut challenges as part of enrollment screening for CoFAR6, a trial of epicutaneous peanut immunotherapy 14. Demographic information is shown in Table 1. Freshly isolated peripheral blood mononuclear cells (PBMCs) were cultured with peanut extract for 6h, harvested and stained for surface markers and intracellular cytokines. Peanut-responsive T cells were identified by expression of CD154, also known as CD40L, which is upregulated on T cells in response to TCR engagement and is essential for providing T cell help to B cells. Co-expression of cytokines IL-4, IL-13, IFNγ, and IL-10 was determined (representative plots of CD154 vs. IL-4 and IFN-γ are shown in Fig 1A). The 6h stimulation time-point was chosen as optimal for intracellular cytokine detection (Fig E2). We observed a significant increase in CD154+ CD4+ T cells after peanut stimulation in PA, but not HT or HC subjects (Fig 1B). Stimulation with egg white protein induced CD154 expression in PA subjects who were egg allergic but not in those who were egg tolerant (data not shown), demonstrating specificity for clinical reactivity. Cytokine expression by CD154+ T cells in PA subjects was dominated by IL-4 and IL-13, with a low but statistically significant increase in IL-10 and no significant IFNγ response (Fig 1C). No increase in CD154+ cells of any cytokine phenotype was elicited from HT or HC CD4+ T cells. The frequency of peanut-responsive CD154+IL-4+ or CD154+IL-13+ CD4+ T cells significantly correlated with the level of peanut-specific IgE in PA subjects (Spearman r=0.68 for both IL-4 and IL-13, p < 0.0001). There was no correlation of peanut-responsive Th2 cells with age (Spearman r=0.093, p=0.46; and r=0.089, p=0.48 for IL-13 and IL-4, respectively, Fig E3).
Figure 1. Immunologic responses to peanut stimulation in allergic and control subjects.
A. Representative dot plot of CD154 response to peanut in CD3+CD4+ cells. B. Quantification of CD154+ T cells in peanut allergic (PA, n=69, CoFAR cohort), healthy control (HC, n=7), and high threshold (HT, n=9) subjects after culture with peanut (+) or media (−). C. Quantification of total cytokine+CD154+ T cells in PA, HC, and HT subjects for IFNγ, IL-10, IL-4, and IL-13 in response to peanut stimulation. Statistics calculated by Kruskal-Wallis with Dunn’s multiple comparison test. As illustrated in D, total CD3+CD4+ T cells, CD154+ T cells (gated rectangle), and Th2 cells (IL-4+CD154+ T cells, gated circle) were evaluated for CXCR5 (n=60), CCR4 (n=61), and CCR6 (n=61) expression (calculated as % positive within indicated populations) after stimulation for 6h with peanut extract (E) or anti-CD3/CD28 (F). Statistics calculated by Friedman’s test with Dunn’s post-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
We examined chemokine receptor expression on peanut-responsive T cells from PA subjects to assess homing capacity (Fig 1D–F). We examined the marker CCR4 that supports homing to skin and lung, the mucosal homing marker CCR6, and the B cell follicle homing marker CXCR5. A significantly greater frequency of peanut-responsive CD4+ T cells expressed CCR4 and CCR6 compared to the total CD4+ T cell population (Fig 1E). This was not due to activation, since polyclonally activated CD4+ T cells did not show a similar enrichment in chemokine receptor frequency (Fig 1F). CCR4 and CCR6 were both enriched on activated Th2 cells (defined as CD154+IL4+ cells, circled in Fig 1D). CXCR5 was significantly enriched on peanut-responsive Th2 cells, but not polyclonally activated Th2 cells. These data reveal that peanut-responsive Th2 cells from peanut allergic individuals include skin, lung, and intestinal homing populations, as well as follicle-homing T cells with the potential to regulate IgE production.
Phenotypic Heterogeneity of Peanut-Responsive Th2 Cells in Peanut Allergy
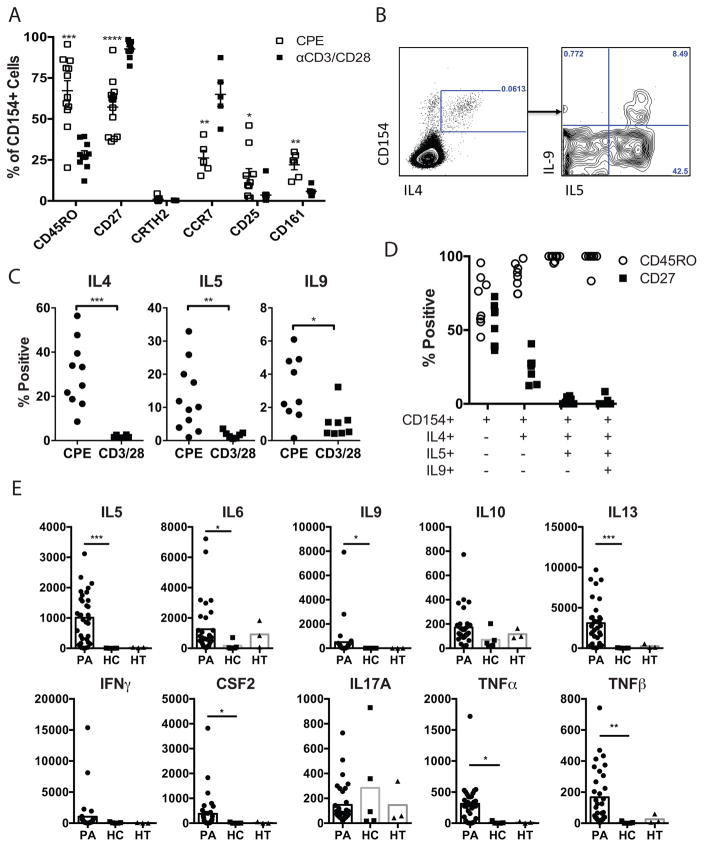
We recruited additional PA subjects to perform phenotypic analysis of peanut-responsive T cells (MSSM cohort, demographic information in Table 1). Memory and differentiation markers including CD45RO, CD27, CCR7, CD25, CD161, and CRTH2 were examined. CD4+ T cells upregulating CD154 after peanut exposure were compared to CD4+ T cells upregulating CD154 after polyclonal stimulation, to control for effects of activation (Fig 2A). Peanut-activated cells were enriched for the memory marker CD45RO, and expressed less CD27 and CCR7 compared to polyclonally-activated T cells. Expression of CD25 and CD161 were also enriched on peanut-activated T cells, although these markers were expressed by a minority of peanut-responsive T cells. The marker CRTH2, which is constitutively expressed on basophils, eosinophils, and a small subset of CD4+ T cells (Fig E4, and 24, 25), was present on a population of T cells distinct from peanut-responsive T cells (Fig 2A, Fig E4). Peanut-responsive CD154+ T cells are therefore selectively enriched for effector memory T cells in PA subjects, but are heterogeneous and lack unified surface markers.
Figure 2. Phenotypic heterogeneity of peanut-responsive Th2 cells.
A. Frequency of expression of memory and differentiation markers on CD154+ cells after stimulation with peanut (CPE) or anti-CD3/CD28. Each symbol represents one subject (n=6–13, MSSM PA cohort). B. Representative flow cytometry plot showing co-expression of IL-5 and IL-9 in peanut-responsive Th2 (CD154+IL-4+) cells. C. Percent frequency of CD154+ T cells co-expressing indicated cytokines after stimulation with peanut (CPE) or anti-CD3/CD28. Each symbol represents an individual subject (n=6–11, MSSM PA Cohort). D. Memory marker expression on each cell subset. Each bar represents the mean and SEM of 8 subjects (MSSM PA cohort). E. Quantification of secreted cytokines after peanut stimulation of PBMCs from PA (n=33), HC (n=5), and HT (n=3) subjects for 5 days. Statistics calculated by Mann-Whitney U test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
We examined co-expression of IL-5 and IL-9 with IL-4 in peanut-responsive T cells (Fig 2B–D). After 6h of stimulation with peanut extract, we observed a subset of CD154+IL-4+ cells co-expressing IL-5, and within that subset, a smaller subset of cells co-expressing IL-9 (Fig 2B). We did not observe IL-5 or IL-9 expression in CD154+ cells that did not express IL-4, suggesting that these cytokines were derived from Th2 cells rather than Th9 subsets. As cells acquired co-expression of IL-5 and IL-9, they became uniformly positive for the memory marker CD45RO, and negative for CD27, indicative of terminal differentiation (Fig 2D). We will refer to these Th2 cells co-expressing IL-5 and/or IL-9 as Th2+ cells.
For a more comprehensive profile of the effector phenotype of the peanut-specific immune response, we measured cytokines secreted into the culture supernatant by multiplex analysis 5 days after peanut stimulation. A time course of cytokine response demonstrated that Th2 cytokines were first detectable in supernatants after 4 days of stimulation, and were maximal at 5 days (data not shown). PBMCs from PA subjects secreted an array of Th2-associated (IL-5, IL-9, IL-13) as well as pro-inflammatory (IL-6, GM-CSF, TNFα, TNFβ) cytokines (Fig 2E). IL-4 was below the level of detection in most samples. Like IL-2, IL-4 consumption in ex vivo cultures has been described 26 and may explain the lack of detectable IL-4. IFN-γ, IL-10, and IL-17 were detectable but not significantly increased in response to peanut stimulation, even in HT or HC subjects.
Identification of peanut-responsive CD4+ T cells bearing regulatory markers
CD154 expression has been reported to be expressed on regulatory T cells with slower kinetics than effector cells 27. We did not observe expression of CD154 on CD4+CD25hiCD127low cells at 6h of stimulation, but at 18h of peanut or polyclonal stimulation we observed upregulation of CD154 on these cells (Fig E5). Importantly, depletion of CD25+ cells prior to stimulation abolished the population of CD154+CD3+CD4+CD25hiCD127lowFoxp3+ cells, indicating that CD25 was present on the cells prior to stimulation (Fig E5).
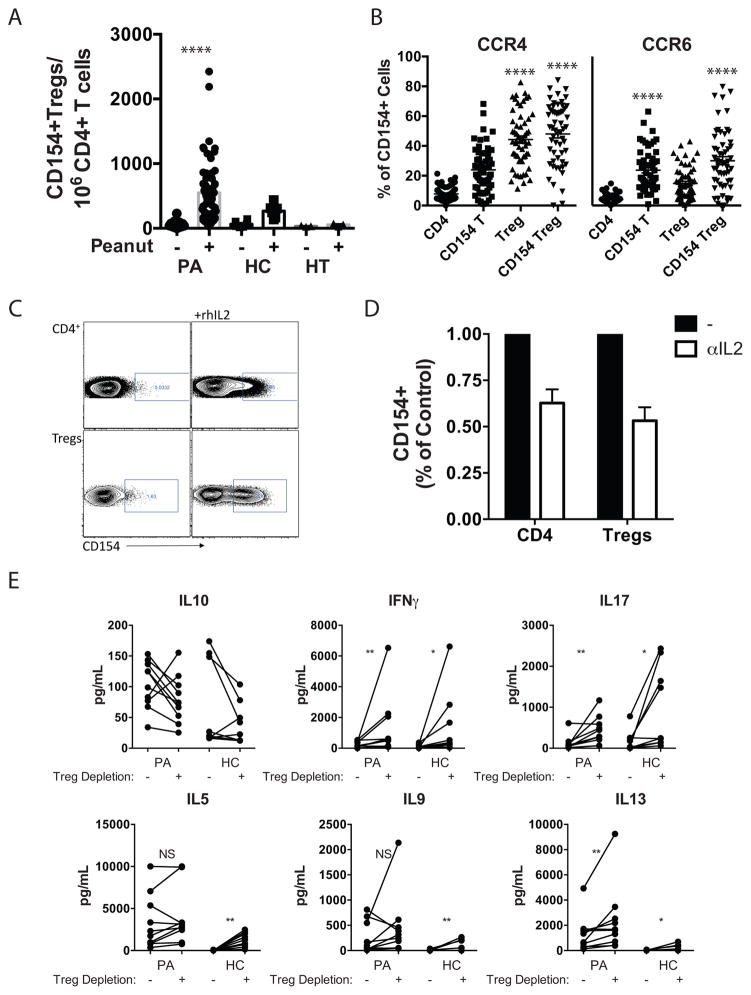
We examined the frequency of peanut-responsive cells with Treg markers in PA, HT, and HC subjects. We observed a significant increase in CD154 expression after 18h of peanut stimulation on CD3+CD4+CD25hiCD127lowFoxp3+ cells from PA subjects, which was lower or absent in HC and HT subjects (Fig 3A). Assessment of chemokine receptor expression on CD154+ cells with regulatory markers showed high expression of CCR4, similar to the total population of Tregs, and levels of CCR6 that were enriched compared to either total CD4+ T cells or total Tregs (Fig 3B). Peanut-responsive cells with Treg markers expressed high levels of the memory marker CD45RO, intermediate levels of CD27, and low levels of CCR7, consistent with a tissue-homing memory T cell phenotype (Fig E6). Similar to self-reactive Tregs identified using tetramers 28, these peanut-responsive Tregs expressed neither IL-10 nor IFN-γ (data not shown).
Figure 3. Identification and phenotypic analysis of peanut-responsive Tregs.
A. Quantification of CD154+FoxP3+CD25+CD127lowCD4+ T cells after stimulation with peanut (+) for 18 h in PA (n=62, CoFAR cohort), HC (n=6), and HT (n=3) subjects. B. Expression of CCR4 and CCR6 (n=57, CoFAR PA cohort) on CD4+ T cells, CD154+CD4+ T cells (CD154 T), FoxP3+CD25+CD127low Tregs (Treg), and CD154+FoxP3+CD25+CD127− cells (CD154 Treg) after peanut stimulation. C. Representative dot plots showing the impact of rhIL2 on CD154 expression in CD4+ T cells or Tregs after 18h. D. Impact of IL-2 neutralization on CD154 expression on CD4+ T cells or Tregs after 18h of peanut stimulation (n = 4 PA subjects). E. Impact of Treg depletion (removal of CD3+CD4+CD25highCD127low by FACS) on peanut-induced cytokine secretion. Individual values are shown for PA (MSSM cohort, n=10) or HC (n=9) subjects. *p<0.05, **p<0.01 ***p<0.001 **** p<0.0001. Statistics calculated with Wilcoxon matched pairs signed rank test (A,E) or Friedman’s test with Dunn’s post-test correction (B).
It has been reported that CD154 can be regulated by IL-2 29. Because of the slow kinetics of the Treg response to peanut, relatively high frequency of cells as a percentage of total Tregs, and activation of Tregs only in PA subjects, we investigated the link between IL-2 and CD154 expression on Tregs. Treatment of PBMCs with rhIL-2 for 18h increased CD154 expression on CD4+ T cells and more strikingly on Tregs (Fig 3C). Neutralization of IL-2 suppressed CD154 expression on CD4+ T cells after anti-CD3/CD28 stimulation at 18h but not 6h (data not shown), and reduced by approximately 50% the frequency of peanut-responsive Tregs identified after 18h of stimulation with peanut extract (Fig 3D). These results indicate that Tregs upregulate CD154 as a secondary response to release of IL-2, likely from effector cells observed to be activated at 6h.
The low frequency of antigen-responsive cells (500–600 cells per million CD4+ T cells) and limited blood volumes available precluded the performance of suppression assays with purified Tregs responding to peanut stimulation. As an alternative approach to assess Treg function, we depleted PA and HC PBMCs of CD4+CD25hiCD127low cells by FACS sorting prior to stimulation with peanut extract and determined the effect on cytokine secretion (Fig 3E). Peanut-induced IFN-γ and IL-17 production was significantly increased in cultures of Treg-depleted PA and HC PBMCs whereas IL-10 production was slightly but not significantly decreased. IL-13, which was produced at higher levels in PA subjects, was also significantly increased by Treg depletion in both PA and HC subjects. In contrast, secretion of IL-5 and IL-9 were significantly enhanced by Treg removal in HC, but not in PA subjects.
Single Cell Transcriptional Profile of Peanut-Responsive CD4+ T cells
We observed substantial heterogeneity in the phenotype of peanut-responsive cells as identified by CD154 expression after stimulation with peanut. To exhaustively phenotype these cells at molecular resolution, we performed single cell RNA-seq on CD154+ T cells captured after 18h of stimulation. After applying quality control measures, single cell sequencing data were available for 212 peanut-responsive CD154+ cells from 5 PA subjects and 122 αCD3/CD28-activated CD154+ cells from 3 PA subjects. As an additional reference population, we sorted CD4+CD25hiCD127low Tregs from freshly isolated PBMCs from 4 PA or 4 HC subjects (97 resting Tregs), for a total of 431 cells (sequencing information in Table E7).
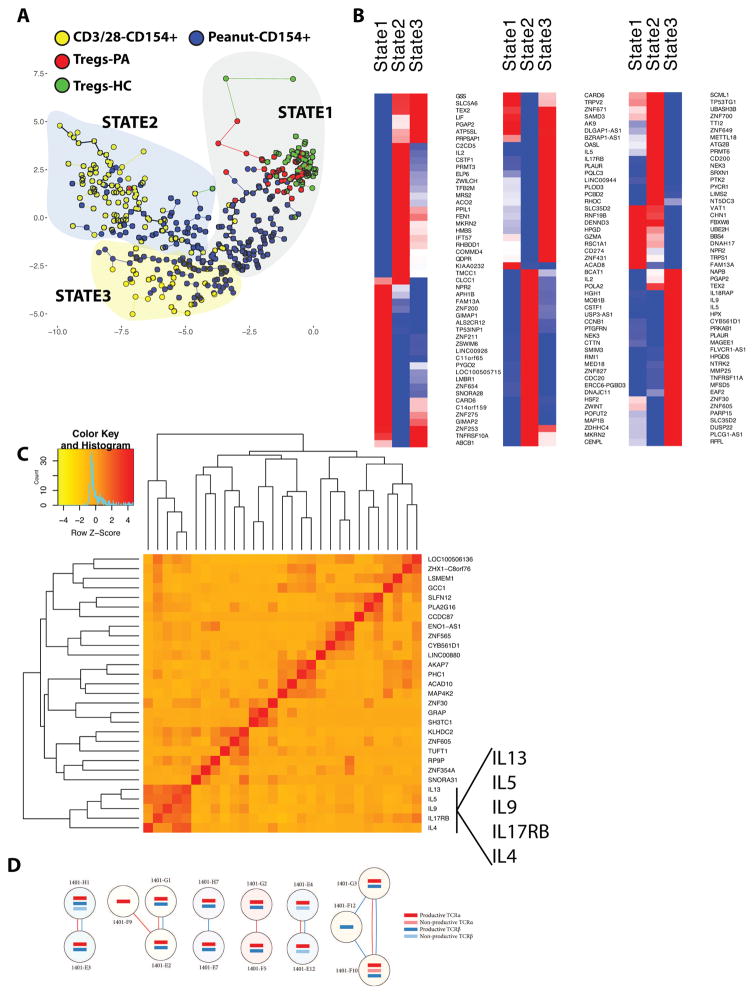
Next, using Monocle 18 a notion of expression-state clustering and evolution was generated using guide genes identified as differentially expressed between resting HC Tregs and polyclonally activated T cells from PA subjects (Fig 4A, Fig E7A). This comparison was chosen to examine how peanut-responsive T cells would cluster between two biologically distinct cell populations, and indeed three states or clusters of cells based on this notion of transcriptional similarity were identified. Cells from different subjects were distributed across all three states (Fig E7B). State 1 contained 98% of the resting Treg populations (Fig 4A), only 4% of the polyclonally activated CD154+ cells, and 45% of the peanut-activated CD154+ cells. State 2 contained 2% of the resting Treg population, 75% of the polyclonally activated CD154+ cells, and 25% of the peanut-activated CD154+ cells. State 3 contained no resting Tregs, 25% of the polyclonally activated CD154+ cells, and 30% of the peanut-activated CD154+ cells. Therefore, Tregs were primarily associated with State 1, most of the polyclonally activated cells were associated with State 2, while peanut-activated cells were distributed across the three states. Other analysis methods such as Seurat 30 primarily identified the three input cell types but were not able to effectively sub-cluster peanut-responsive T cells (data not shown).
Figure 4. Single-Cell Sequencing Analysis of CD154+ T cells and Tregs.
A. Minimum spanning tree showing clustering and ordering of peanut-activated T cells, polyclonally-activated T cells, and Tregs from PA or HC into 3 states, and the distribution of cells by phenotype within those states. B. Heat maps showing genes differentially expressed between peanut-activated T cells in the 3 states. C. Pearson correlation of genes significantly upregulated in State 3. The x and y axes depict a hierarchy of genes that exhibit similar correlation values and are thus co-expressed. Only genes with a correlation >0.5 with another significantly upregulated gene are shown. D. TCR analysis of peanut-activated cells from a peanut allergic donor (Subject ID 1401) indicating shared TCR α and/or β chains. Each circle indicates one cell, with red line indicating a shared α chain and blue line indicating shared β chain.
We selected only the peanut-activated T cells to identify differentially expressed genes between states (Figure 4B, complete DEG list provided in SuppExcelFile1. A DEG list comparing all peanut-activated T cells to all polyclonally-activated T cells is provided in SuppExcelFile2). State 3, which was comprised mostly but not exclusively of peanut-activated T cells, was a clearly pro-inflammatory Th2-associated state. Differentially expressed genes (FDR < 5%) included IL5, IL9, IL13, IL4, CSF2, and IL3, as well as the IL-25 receptor, IL17RB. Enrichment analysis identified pathways including “regulation of immunoglobulin production” and “regulation of JAK-STAT” cascade (Table E8). Fig E8 shows the gene expression of IL2, IL5, and IL9 by individual cells across states. The correlation of gene expression in peanut-activated T cells in State 3 is shown in Fig 4C. A cluster of 5 genes including IL4, IL5, IL9, IL13, and IL17RB comprised the dominant cluster of correlated genes.
Pathway analysis of differentially expressed genes in State 2 identified cell cycle or cell division (Table E9), and differentially expressed genes included IL-2 and chemokines and cytokines such as CCL22 and IL17F. Peanut-activated T cells in State 1, which clustered together with resting Tregs, were not defined by a regulatory gene signature. The top pathways associated with differentially expressed genes in peanut-activated cells in State 1 were mitochondrion organization, metabolic process, and DNA strand elongation (Table E10), suggesting an early activation state. Identification of 5 clusters rather than 3 did not separate the peanut-responsive T cells from Tregs in State 1 (data not shown). Genes differentially expressed between peanut-activated T cells and Tregs within State 1 are provided in SuppExcelFile3. Differentially expressed genes upregulated in peanut-responsive cells in State 1 compared to peanut-responsive cells in the other two states include several surface receptors or channels (NPR2, GIMAP2, CLCC1). Although genes upregulated in the 3 different states suggest association with cell cycle, a formal analysis of cell cycle genes did not indicate significant contribution to variance. Application of a machine learning classifier for cell cycle allocation to predict cell cycle 31 demonstrated that cells were predominantly in G1, and that clustering was independent of cell cycle (Fig E9).
We reconstructed the full complementarity determining region 3 (CDR3) directly from the sc-RNA-seq data to characterize the TCR sequence of alpha and beta chains (Table E11, Fig E10). Using TraCeR 19 (https://www.github.com/teichlab/tracer), we identified 4 clonal expansions each comprised of a pair of cells with identical α and β TCR sequences from a single PA individual (Fig 4D, Fig E11). Clones were found in all 3 states, and interestingly pairs of clones were found in different states (Fig E11). Clonality was not uniquely associated with Th2 polarized cells, and instead the most highly upregulated gene was PLA2G15 (Fig E11), a phospholipase that results in a lymphoproliferative and autoimmune disorder when deleted 32.
DISCUSSION
We studied the T cell response to peanut at the cellular and molecular level in a total of 156 human subjects, including 84 who underwent a double-blind placebo controlled food challenge to determine their threshold of clinical reactivity to peanut, and 21 healthy controls. In peanut allergic individuals, we identified a peanut-responsive population of highly differentiated Th2 cells, and a delayed IL-2-dependent activation of cells expressing regulatory markers, while non-allergic controls had a marked absence of peanut-reactive immune response. A summary model that places our findings in the pathogenesis of peanut allergy is shown in Figure 5.
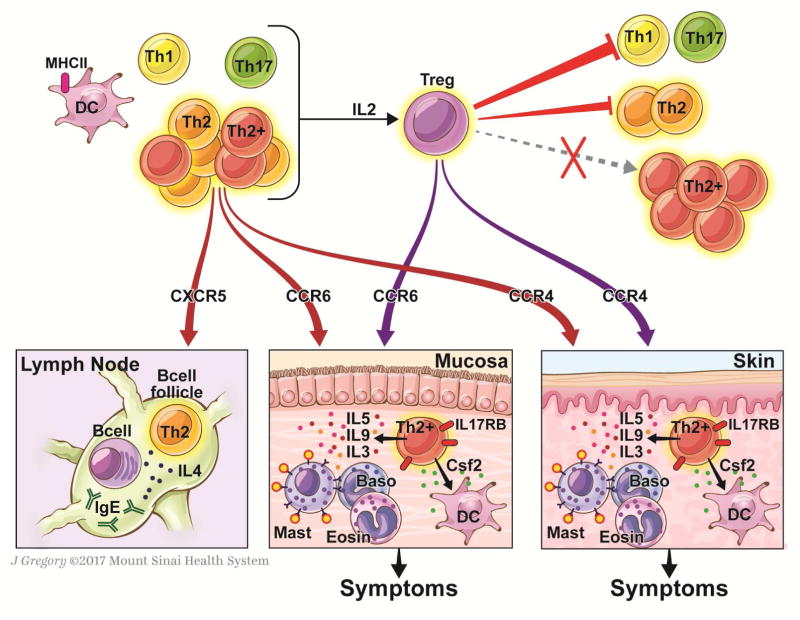
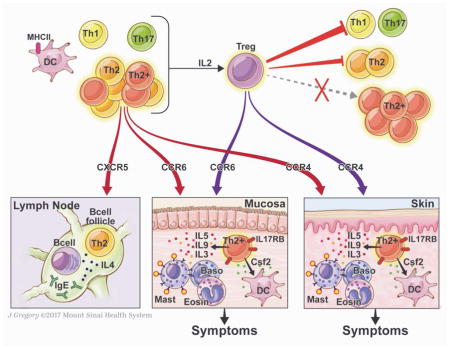
Figure 5. Proposed model of the contribution of peanut-responsive T cells to peanut allergy.
Stimulation of PBMCs with peanut leads to early (6h) activation of memory Th2 cells expressing IL-4 and IL-13, and memory Th2+ cells co-expressing IL-4 and IL-13 with IL-5 +/− IL-9. Th1 and Th17 cells remain quiescent. Th2 cells express molecules to facilitate homing to skin, lung, intestine, or B cell follicles, where we hypothesize that they may promote allergic effector cell expansion (skin, mucosa) or IgE-class switch (follicle). The heterogeneous T cell effector T cell response to peanut also includes IL-3 and Csf2 that may also contribute to tissue inflammation through actions on allergic effector cells and DCs, respectively. These distal effects on effector cells and IgE production would be expected to contribute to symptoms on peanut re-exposure. Release of IL-2 promotes the subsequent activation of Tregs. Tregs completely suppress (red arrows) Th1 and Th17 responses, partially suppress Th2 responses, but do not significantly suppress Th2+ responses. We speculate that targeting the heterogeneous Th2 response, including the Th2+ cells through molecules such as IL17RB, may be effective in the treatment of peanut allergy.
Using a multifaceted approach to immune profiling the effector response to peanut including detection of intracellular and secreted cytokines as well as single cell profiling, we identified a highly heterogeneous Th2 response to peanut in PA subjects including cells co-expressing IL-5 +/− IL-9, that we refer to as Th2+ cells (Fig 5). This is to our knowledge the first report of single cell RNA sequencing of food allergen-responsive T cells. Previously, transcriptional profiling of peanut-responsive CD4+ T cells was done on the bulk level using microarray analysis 12, and on the single cell level using a panel of allergy and immune tolerance focused genes 11. The identification of peanut-responsive T cell clones in this exploratory analysis is remarkable as we sequenced a relatively small number of cells (40–50 per patient), cells were activated with a crude peanut extract containing many peanut allergens, and stimulation time did not allow for expansion of allergen-responsive T cells. Although preliminary, the positive identification of peanut-responsive clones prompts additional investigation of more patients with higher throughput single cell capture approaches, such as DropSeq 30 to comprehensively study association of phenotype with TCR specificity.
We found that peanut-responsive cells clustered into three states, one of which was enriched for highly differentiated Th2 cells (State 3). Correlation analysis showed that IL4, IL5, IL9, IL13, and IL17RB were highly co-expressed in peanut-responsive T cells. IL-3 and Csf2 (GM-CSF) were also differentially expressed in this Th2 state, but did not correlate with the cluster of other Th2-related cytokines and may be distinct effector cell phenotypes. Using bulk analysis methods, we recently identified a highly differentiated Th2 (IL-5, IL-9) and pro-inflammatory (TNFα, Csf2) immune profile in cohorts of patients with egg allergy10, 33. IL-9 is differentially expressed by peanut-responsive T cells from PA subjects as identified through bulk analysis by microarray 34. Wisniewski et al also identified a multi-functional Th2 response to peanut in peanut-allergic subjects using a proliferation-based flow cytometry approach 6. The Th2-associated cytokines we identified as upregulated by sc-RNA-seq (IL3, IL5, IL9, Csf2) are critical for the differentiation of granulocytes including mast cells, eosinophils, and basophils, and we hypothesize that these T cell-derived granulocyte growth factors control the allergic milieu of the skin or mucosa (Fig 5). IL-9 drives intestinal mastocytosis, is elevated in the intestine of patients with food allergy, and is critical for food allergic symptoms 10, 33. OX40L-expressing DCs promote the production of IL-3 from naive CD4+ T cells, which then recruit basophils and promote Th2 priming of CD4+ T cells 35. The role of T cell-derived IL-5, IL-3, Csf2, and TNFα in food allergy remains to be identified. The unique association of the IL-25 receptor IL17RB on these highly differentiated Th2 cells suggests a role for tissue-derived IL-25 in peanut allergy pathogenesis, which is supported by work in animal models of food allergy 36, 37. IL17RB was also previously reported as highly differentially expressed in peanut-responsive T cells analyzed by bulk microarray 11.
The peanut-responsive Th2 cells described here share several features of cells described as “Th2A” and “pathogenic effector Th2 or peTh2” 7, 38–40. Common features include multi-cytokine potential including IL-5 and IL-9, lack of expression of CD27, and expression of hPGDS and IL17RB. Both Th2A and peTH2 cells are CRTH2+CD161high 38, 40, which is distinct from the peanut-responsive Th2+ cells described here. peTH2 cells correlate with eosinophilia, and were proposed as a Th2 phenotype that distinguished Th2 diseases characterized by allergic inflammation of tissues (eosinophilic gastroenteritis or atopic dermatitis) from an IgE-mediated Th2 disease (peanut allergy) 7, 40. TH2A cells have been described across distinct allergies, including aeroallergens and peanut 38. The reason for this discrepancy in expression of CD161 and CRTH2 between peTH2, TH2A, and the Th2+ cells we describe here is not clear. We used an activation-based detection approach, while Th2A cells were identified with a tetramer-based approach, and it is possible that activation downregulates CRTH2 expression. However, we did observe CRTH2 expression on PMA/ionomycin-stimulated CD154+ cells, and peTH2 cells have been described to emerge in vitro through multiple rounds of stimulation 40. Despite the discrepancy in expression of CRTH2 and CD161 which may be related to method of antigen-specific cell detection or culture conditions, expression of CD45RO+/CD45RA−, CD27−, hPGDS+ and IL17RB+ are consistent markers of these allergen-specific Th2 cells with multi-cytokine production across multiple studies (including peTH2, TH2A, Th2+).
Homing plays a key role in T cell function. We found that peanut-responsive Th2 cells were enriched for the homing markers CCR4, CCR6, and CXCR5. Although CCR6 is often used as a surface marker of Th17 cells, we previously showed that CCR6 is required for homing of pathogenic Th2 cells to the small intestine and development of food allergic symptoms in mice 41. CCR4 is expressed on both Th2 cells and Tregs, and facilitates homing to the skin 42 and lungs 43. The skin, lungs, and gastrointestinal tract are common sites of manifestations of food allergy, and we speculate that the homing of T cells producing an array of Th2-related cytokines contributes to allergic inflammation in the tissues that contributes to acute responses to food allergen (Fig 5). Provision of B cell help by T cells is dependent on homing to B cell follicles in a CXCR5-dependent manner. IL-4 production by Th2 cells contributes to allergy in large part through support of IgE class switching, and T cells require CXCR5 expression to interact with B cells in the follicle of the lymph node 42. We observed significantly enriched expression of CXCR5 on peanut-responsive Th2 cells in the blood of peanut allergic subjects, although the frequency of these cells was low compared to the total population of peanut-responsive cells. Lymph nodes such as tonsil would likely be more informative than blood on the frequency and phenotype of peanut-specific Tfh cells. It would be of interest to compare this CXCR5+Th2 population in those with IgE-mediated versus non-IgE-mediated allergic diseases to foods (i.e. milk allergy with immediate hypersensitivity reactions versus milk allergy contributing to eosinophilic esophagitis). We would hypothesize that this CXCR5+Th2 population would be uniquely associated with IgE-mediated disease.
Tregs are thought to be deficient or “reprogrammed” in food allergy 13. We identified a population of cells bearing regulatory markers that upregulated CD154 in a delayed manner compared to Th2 cells (detectable at 18h vs 6 h). Deletion of CD25+ cells prior to stimulation completely abolished this response, indicating that they expressed Treg markers prior to stimulation. These cells expressed markers consistent with peripherally induced Tregs. The delayed kinetics of activation, frequency (~5% of the Treg population), and IL-2-dependence suggest that these cells are the result of bystander activation rather than antigen-specific Tregs.
There is conflicting evidence on the antigen specificity of Tregs. Antigen-specific Tregs (identified as Tregs based on regulatory marker expression and lack of cytokine production upon stimulation) were identified in humans using an array of MHC II tetramers specific for self-antigens or microbial antigens 28. Tetramer-positive Tregs were identifiable, but were very rare cells (<1% of tetramer-positive cells for any given antigen). Bacher et al used CD137 (4–1BB) as an activation marker of antigen-specific Tregs, and examined the frequency of Tregs specific for a variety of antigens and allergens. Tregs specific for aeroallergens or microbial antigens could be readily found in peripheral blood of healthy subjects, but food-specific Tregs were markedly low to absent 44. In contrast to these findings in healthy controls, food allergen-responsive cells with a Treg phenotype and regulatory function in suppression assays have been identified by proliferation assays (CFSE dilution) in allergic individuals 45, 46. In addition, single cell transcriptional profiling using tetramer-selected cells and a curated panel of genes identified a population of peanut-specific cells with expression of regulatory genes (Foxp3, CD25, IL-10, as well as IFN-γ) in peanut allergic subjects 47. While our data suggest that Tregs may be bystander activated, Tregs as a population are functional and dampen the immune response to peanut. The potential suppressive role of CD154 on Tregs remains to be clarified. The fact that cytokines commonly produced by highly differentiated Th2 cells (IL-5 and IL-9) were less affected by removal of Tregs in PA individuals suggests that Th2+ cells may be more resistant to regulation by Tregs than other T cell subsets. Memory T cells have been described to be less susceptible to suppression than naïve T cells 48, and we speculate that as Th2 cells differentiate to become Th2+ cells, they become less susceptible to the regulatory effects of Tregs. Bacher et al have proposed, based on their findings in the context of birch allergy, that Th2 cells escape Treg control due to differing antigen specificities 44. There is little information available on the immune basis of tolerance to foods under homeostatic conditions in healthy human subjects, although a lack of food-specific Tregs has been documented in healthy subjects 44. In contrast to PA subjects, we observed a marked absence of T cell reactivity to peanut in healthy controls (HC) as well as those we termed “high threshold” (HT) who were sensitized to peanut, had a clinical history of peanut allergy, but passed a food challenge with a cumulative dose of 1 g of peanut protein. The HT group was not re-challenged with a higher dose of peanut protein, so we do not know what proportion of this group would react at a higher dose of peanut versus being fully tolerant. Importantly, the HT cohort was consistent in age and diet to the PA cohort, while the HC cohort was adult and not avoiding peanut in the diet. Despite the diversity of these two control groups in age and diet, they exhibited a consistent absence of T cell response as measured by CD154-based detection or cytokine production. Although the HC group was not matched for age, we did not observe any trend of an association between peanut-specific T cell response and age within the PA group. Furthermore, the age range of the PA group was 4–20, with 14 individuals above the age of 13. Therefore, it is unlikely that the absence of peanut-responsive T cells in the HC group, also observed in the HT group, was due to age. This lack of T cell reactivity was accompanied by relatively low (HT) or no (HC) peanut-specific IgE, and a low level of peanut-specific IgG4. The relative absence of allergen-specific T cell responses in HC subjects is consistent with two previous reports that used MHC II tetramers or CD154 expression to detect peanut-reactive T cells in healthy controls 7, 8. Our data suggest that naturally occurring clinical tolerance, whether complete tolerance in adult HC individuals not restricting peanut in the diet or a higher threshold of reactivity to peanut in HT subjects, is characterized by immunologic ignorance or anergy rather than an antigen-specific counter-regulatory Th1 response or Treg response. This does not rule out a role for Tregs in the healthy response to foods, as our data show that removal of the entire Treg compartment enhances the immune response to peanut in HC subjects. Indeed, patients with a variant of IPEX syndrome, who lack functional Tregs, have been reported to develop allergies to multiple foods, clearly showing a role for Tregs in immune tolerance to foods 49. However, our data suggest that this regulatory activity may not be limited to antigen-specific Tregs.
In summary, we have identified a heterogeneous population of Th2, Th2+, and other growth-factor producing peanut-responsive cells with capacity for tissue and lymph node homing that we postulate play a central role in PA pathogenesis. We did not find evidence for a lack of peanut-responsive Tregs or disrupted Treg response in PA subjects. This is consistent with the findings of Bacher et al, who identified the presence of Th2 effector cells with different antigen specificity than Tregs in birch-allergic individuals, with no deficiency in number or suppressive capacity of Tregs 44. These results have several implications for therapy. The relative resistance of IL-5 and IL-9 to regulation by Tregs in PA subjects, as well as the lack of detectable antigen-responsive Tregs in HC and HT subjects, suggests that eliminating or targeting peanut-specific Th2 cells would be more effective for the treatment of PA than boosting the pool of peanut-specific Tregs. Tight correlation between the IL-25 receptor and cytokine expression in highly differentiated Th2 cells, as well as the common finding of IL-25 receptor expression by allergen-specific T cells across multiple studies, suggests IL17RB as a target for modifying the phenotype or eliminating these cells.
Supplementary Material
Key Messages.
Peanut allergy was associated with a multi-functional T cell response including highly differentiated Th2 cells producing IL-5 and IL-9, cells expressing CSF2 and IL3, as well as a delayed IL-2-dependent activation of Tregs
Individuals with a higher threshold of reactivity to peanut as well as healthy controls exhibited a lack of peanut-specific Th2 or Treg response, suggesting anergy or deletion as a basis of tolerance
The contribution of highly differentiated multi-cytokine producing Th2 cells to the pathogenesis of peanut allergy needs further investigation
Acknowledgments
We thank Michelle Mishoe, Zara Atal, Sandra Hatem for assistance with the MSSM cohort; Jordi Ochando, Christopher Bare, Venu Pothula of the Flow Cytometry Core for assistance with sorting; Jing-Jing Qi, Manishkumar Patel, and Seunghee Kim-Schulze of the Human Immune Monitoring Center for flow cytometry analysis, and the Genomics Core for RNA sequencing. Many thanks to Calman Prussin for helpful discussions and advice on CD154-based detection of cells. The study was supported by U24AI118644 (to MCB and MM), and the David and Julia Koch Research Program in Food Allergy Therapeutics (HAS). Facilities were supported by a NCI P30 Cancer Center Support Grant (P30 CA196521). MSSM PA cohort samples were kindly supplied by the Food Allergy Resource Initiative, which is supported by funding from Food Allergy Research and Education and maintained at the Elliot and Roslyn Jaffe Food Allergy Institute, Icahn School of Medicine at Mount Sinai. The clinical study (CoFAR6) providing samples for this study is registered with ClinicalTrials.gov with ID NCT01904604 and is supported by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) grant U19AI066738 and U01AI066560. The project was also supported by NIH/NCATS grant numbers UL1 TR0001082 (Colorado), UL1 TR000067 (Mount Sinai), UL1 TR000039 (Arkansas), UL1 TR000083 (North Carolina), and UL1 TR000424 (Johns Hopkins) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Support for this trial was also provided by DBV Technologies (Montrouge, France) through funds provided to the Consortium of Food Allergy Research (CoFAR).
Abbreviations
- CoFAR
Consortium of Food Allergy Research
- CPE
Crude peanut extract
- HC
Healthy control group
- HT
High threshold group
- MSSM
Mount Sinai School of Medicine
- PA
Peanut allergy
- RNA-seq
RNA sequencing
- sc-RNA-seq
single cell RNA sequencing
Footnotes
AUTHOR CONTRIBUTIONS
DC designed experiments, performed all experimental procedures with assistance from CA, analyzed data, and co-wrote the manuscript. XC and BL helped design RNAseq experiments, analyzed the single cell RNAseq data, and co-wrote the manuscript. RS and LN provided guidance and assistance with single cell isolation and library preparation. SMJ, RAW, SJS, AWB, and DYML were site principal investigators, and HAS was overall principal investigator of CoFAR6, the clinical trial from which samples were obtained. AG coordinated samples from the CoFAR central laboratory and prepared research reagents for the study. PD curated datasets related to the CoFAR study. WFD and MM provided advice on experiment design. MCB conceived the project, co-designed experiments, supervised analysis, and co-wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Esterhazy D, Loschko J, London M, Jove V, Oliveira TY, Mucida D. Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat Immunol. 2016;17:545–55. doi: 10.1038/ni.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372:803–13. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noval Rivas M, Chatila TA. Regulatory T cells in allergic diseases. J Allergy Clin Immunol. 2016;138:639–52. doi: 10.1016/j.jaci.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turcanu V, Maleki SJ, Lack G. Characterization of lymphocyte responses to peanuts in normal children, peanut-allergic children, and allergic children who acquired tolerance to peanuts. J Clin Invest. 2003;111:1065–72. doi: 10.1172/JCI16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wisniewski JA, Commins SP, Agrawal R, Hulse KE, Yu MD, Cronin J, et al. Analysis of cytokine production by peanut-reactive T cells identifies residual Th2 effectors in highly allergic children who received peanut oral immunotherapy. Clin Exp Allergy. 2015;45:1201–13. doi: 10.1111/cea.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prussin C, Lee J, Foster B. Eosinophilic gastrointestinal disease and peanut allergy are alternatively associated with IL-5+ and IL-5(−) T(H)2 responses. J Allergy Clin Immunol. 2009;124:1326–32. e6. doi: 10.1016/j.jaci.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. Ara h 1-reactive T cells in individuals with peanut allergy. J Allergy Clin Immunol. 2011;127:1211–8. e3. doi: 10.1016/j.jaci.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt PG, Strickland D, Bosco A, Belgrave D, Hales B, Simpson A, et al. Distinguishing benign from pathologic TH2 immunity in atopic children. J Allergy Clin Immunol. 2016;137:379–87. doi: 10.1016/j.jaci.2015.08.044. [DOI] [PubMed] [Google Scholar]
- 10.Chen CY, Lee JB, Liu B, Ohta S, Wang PY, Kartashov AV, et al. Induction of Interleukin-9-Producing Mucosal Mast Cells Promotes Susceptibility to IgE-Mediated Experimental Food Allergy. Immunity. 2015;43:788–802. doi: 10.1016/j.immuni.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brough HA, Cousins DJ, Munteanu A, Wong YF, Sudra A, Makinson K, et al. IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol. 2014;134:1329–38. e10. doi: 10.1016/j.jaci.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 12.Kosoy R, Agashe C, Grishin A, Leung DY, Wood RA, Sicherer SH, et al. Transcriptional Profiling of Egg Allergy and Relationship to Disease Phenotype. PLoS One. 2016;11:e0163831. doi: 10.1371/journal.pone.0163831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noval Rivas M, Burton OT, Wise P, Charbonnier LM, Georgiev P, Oettgen HC, et al. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity. 2015;42:512–23. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones SM, Sicherer SH, Burks AW, Leung DY, Lindblad RW, Dawson P, et al. Epicutaneous immunotherapy for the treatment of peanut allergy in children and young adults. J Allergy Clin Immunol. 2017;139:1242–52. e9. doi: 10.1016/j.jaci.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stubbington MJ, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–32. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 24.Rahman AH, Tordesillas L, Berin MC. Heparin reduces nonspecific eosinophil staining artifacts in mass cytometry experiments. Cytometry A. 2016;89:601–7. doi: 10.1002/cyto.a.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tordesillas L, Rahman AH, Hartmann BM, Sampson HA, Berin MC. Mass cytometry profiling the response of basophils and the complete peripheral blood compartment to peanut. J Allergy Clin Immunol. 2016;138:1741–4. e9. doi: 10.1016/j.jaci.2016.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewen C, Baca-Estrada ME. Evaluation of interleukin-4 concentration by ELISA is influenced by the consumption of IL-4 by cultured cells. J Interferon Cytokine Res. 2001;21:39–43. doi: 10.1089/107999001459141. [DOI] [PubMed] [Google Scholar]
- 27.Litjens NH, Boer K, Betjes MG. Identification of circulating human antigen-reactive CD4+ FOXP3+ natural regulatory T cells. J Immunol. 2012;188:1083–90. doi: 10.4049/jimmunol.1101974. [DOI] [PubMed] [Google Scholar]
- 28.Su LF, Del Alcazar D, Stelekati E, Wherry EJ, Davis MM. Antigen exposure shapes the ratio between antigen-specific Tregs and conventional T cells in human peripheral blood. Proc Natl Acad Sci U S A. 2016;113:E6192–E8. doi: 10.1073/pnas.1611723113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fayen JD. Multiple cytokines sharing the common receptor gamma chain can induce CD154/CD40 ligand expression by human CD4+ T lymphocytes via a cyclosporin A-resistant pathway. Immunology. 2001;104:299–306. doi: 10.1046/j.1365-2567.2001.01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scialdone A, Natarajan KN, Saraiva LR, Proserpio V, Teichmann SA, Stegle O, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Shayman JA, Kelly R, Kollmeyer J, He Y, Abe A. Group XV phospholipase A(2), a lysosomal phospholipase A(2) Prog Lipid Res. 2011;50:1–13. doi: 10.1016/j.plipres.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterfeld H, Ahrens R, Strait R, Finkelman FD, Renauld JC, Hogan SP. Differential roles for the IL-9/IL-9 receptor alpha-chain pathway in systemic and oral antigen-induced anaphylaxis. J Allergy Clin Immunol. 2010;125:469–76. e2. doi: 10.1016/j.jaci.2009.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otani IM, Anilkumar AA, Newbury RO, Bhagat M, Beppu LY, Dohil R, et al. Anti-IL-5 therapy reduces mast cell and IL-9 cell numbers in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2013;131:1576–82. doi: 10.1016/j.jaci.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leyva-Castillo JM, Hener P, Michea P, Karasuyama H, Chan S, Soumelis V, et al. Skin thymic stromal lymphopoietin initiates Th2 responses through an orchestrated immune cascade. Nat Commun. 2013;4:2847. doi: 10.1038/ncomms3847. [DOI] [PubMed] [Google Scholar]
- 36.Lee JB, Chen CY, Liu B, Mugge L, Angkasekwinai P, Facchinetti V, et al. IL-25 and CD4(+) TH2 cells enhance type 2 innate lymphoid cell-derived IL-13 production, which promotes IgE-mediated experimental food allergy. J Allergy Clin Immunol. 2016;137:1216–25. e1–5. doi: 10.1016/j.jaci.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han H, Thelen TD, Comeau MR, Ziegler SF. Thymic stromal lymphopoietin-mediated epicutaneous inflammation promotes acute diarrhea and anaphylaxis. J Clin Invest. 2014;124:5442–52. doi: 10.1172/JCI77798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wambre E, Bajzik V, DeLong JH, O'Brien K, Nguyen QA, Speake C, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med. 2017:9. doi: 10.1126/scitranslmed.aam9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wambre E, DeLong JH, James EA, LaFond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J Allergy Clin Immunol. 2012;129:544–51. 51 e1–7. doi: 10.1016/j.jaci.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, et al. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human T(H)2 cell subpopulation with enhanced function. J Allergy Clin Immunol. 2016;137:907–18. e9. doi: 10.1016/j.jaci.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Blazquez AB, Knight AK, Getachew H, Bromberg JS, Lira SA, Mayer L, et al. A functional role for CCR6 on proallergic T cells in the gastrointestinal tract. Gastroenterology. 2010;138:275–84. e1–4. doi: 10.1053/j.gastro.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–21. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikhak Z, Strassner JP, Luster AD. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J Exp Med. 2013;210:1855–69. doi: 10.1084/jem.20130091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, et al. Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell. 2016;167:1067–78. e16. doi: 10.1016/j.cell.2016.09.050. [DOI] [PubMed] [Google Scholar]
- 45.Shreffler WG, Wanich N, Moloney M, Nowak-Wegrzyn A, Sampson HA. Association of allergen-specific regulatory T cells with the onset of clinical tolerance to milk protein. J Allergy Clin Immunol. 2009;123:43–52. e7. doi: 10.1016/j.jaci.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 46.Syed A, Garcia MA, Lyu SC, Bucayu R, Kohli A, Ishida S, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3) J Allergy Clin Immunol. 2014;133:500–10. e11. doi: 10.1016/j.jaci.2013.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan JF, Hovde R, Glanville J, Lyu SC, Ji X, Gupta S, et al. Successful immunotherapy induces previously unidentified allergen-specific CD4+ T-cell subsets. Proc Natl Acad Sci U S A. 2016;113:E1286–95. doi: 10.1073/pnas.1520180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Brook MO, Carvalho-Gaspar M, Zhang J, Ramon HE, Sayegh MH, et al. Allograft rejection mediated by memory T cells is resistant to regulation. Proc Natl Acad Sci U S A. 2007;104:19954–9. doi: 10.1073/pnas.0704397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torgerson TR, Linane A, Moes N, Anover S, Mateo V, Rieux-Laucat F, et al. Severe food allergy as a variant of IPEX syndrome caused by a deletion in a noncoding region of the FOXP3 gene. Gastroenterology. 2007;132:1705–17. doi: 10.1053/j.gastro.2007.02.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.