Summary
We recently reported that in addition to its classical cytoplasmic location, the fast skeletal muscle Troponin T3 (TnT3) shuttles to the nucleus, where it appears to perform nonclassical transcription regulatory functions. Importantly, changes in the composition of the nucleus-localized pool of TnT3 and its fragments contribute to age-dependent muscle damage and wasting. Here, using ChIP-Seq, we demonstrate that TnT3 associates with DNA consensus sequences including the TGCCT motif, which is required for p53 binding to the promoter area of p53-related genes. Gene set enrichment analysis further demonstrated that the p53 pathway was the most significantly enriched pathway among genes annotated to the TnT3 ChIP-Seq peaks. We further demonstrated a strong correlation (r = 0.78, P = 1×10−4) between the expression levels of TNNT3 and TP53-inducible ribonucleotide reductase regulatory subunit M2B (RRM2B) in skeletal muscle tissue of 21 lean non-diabetic human subjects and a significant (P < 0.05) reduction in the levels of both gene transcripts in the third age-tertile group [42.3 - 70 years of age (yoa)] as compared to the second age-tertile (31.3 - 42.3 yoa). Of note, both TNNT3 and RRM2B expression levels negatively associated with total body fat mass (each with r = 0.49, P < 0.05), whereas RRM2B positively correlated with pancreatic β cell function (rRRM2B~HOMA-B= 0.47, P = 0.047). This work suggests that reduced TNNT3 gene expression is another mechanism leading to reduced TnT3 and excitation-contraction coupling with aging. Consequently, TnT3 appears to contribute to age-related sarcopenia and possibly other age-related deficiencies such as muscle insulin resistance and β cell dysfunction by interacting with TnT3-binding sequences in the promoter area of p53-related genes, among others, and consequently modulating the transcriptional regulation of these target genes.
Keywords: Aging, Troponin T3, skeletal muscle, gene transcription, calcium channel
Introduction
Tropomyosin (TM)-binding troponin (TnT) together with the calcium-binding troponin C (TnC) and the inhibitory subunit troponin I (TnI) form a complex that regulates muscle contraction. Specifically, the troponin complex interacts with actin and binds calcium to trigger production of muscle force (Gordon and others 2000). In addition to this classical cytoplasmic location and function, the fast skeletal muscle TnT3 shuttles to the nucleus (Zhang and others 2013c). As TnT3 exhibits a relaxed leucine zipper DNA-binding domain (Zhang and others 2013a), we hypothesized that TnT3 plays a nonclassical role in gene transcription regulation. (Latchman 2008)
TnI, TnT, Tm, and other cytoskeletal proteins shuttle to the nucleus in various cells. TnI has been reported in the nucleus of Drosophila nonmuscle cells (Sahota and others 2009) and TnI and TnT in rat and human cardiomyocytes (Asumda and Chase 2012; Bergmann and others 2009); however, their function in these locations is largely unknown. Consistently, we demonstrated that the nonmyofilament-associated fast skeletal muscle TnT3 enters the nucleus through a COOH-terminus nuclear localization sequence (NLS), KLKRQK (Zhang and others 2013a; Zhang and others 2013c). We reported that, by engaging a functional leucine zipper domain conserved among species (Zhang and others 2013b), TnT3 directly binds to characteristic sequence motifs in the nuclear DNA and may, therefore, regulates gene transcription (a nonclassical role for this protein) and/or function in nuclear signaling (Zhang and others 2016). In addition, studies of TnT modulation of gene expression in human iPSC-derived cardiomyocytes suggested that nucleus-translocated cardiac type TNNT2 may contribute to novel epigenetic mechanisms that underlie the pathogenesis of dilated cardiomyopathy (Wu and others 2015). Notably, TnI was also recently demonstrated to translocate to the nucleus and play a role in normal and tumor growth by transcriptionally upregulating the expression of several genes involved in cell proliferation (Casas-Tinto and others 2016). These reports therefore, support novel non-canonical nuclear roles for troponins in general.
Our previous work further revealed that nuclear TnT3 functions as a transcription factor, which may change over time since the nuclear TnT3 abundance and integrity changes with aging in a mouse model (Zhang and others 2013c). Whether TnT3 regulates genomic motifs for major transcription factors involved in cell senescence, cancer cell proliferation, and/or replication of mitochondrial DNA, is unknown. Using ChIP-sequencing (ChIP-Seq) in differentiated mouse muscle cells and quantitative expression profiling in biopsied human muscle tissue, the present study examines potential genes and pathway targets of nucleus-translocated TnT3.
Results
Genomic occupancy of TnT3
To understand how TnT3 regulate gene expression in muscle cells, we used ChIP-Seq to profile its genomic occupancy in C2C12 cells. Chromatin was immunoprecipitated from differentiated C2C12 myotubes using a TnT3-specific antibody and compared to chromatin immuno-precipitated by a control IgG antibody. A QC report for the raw reads is presented in Supplementary Figure SF1. TnT3 occupied 688 genomic sites with FDR≤0.1 (Figure 1A, Supplementary Table ST1), of which 111 were localized to the promoter region, 275 to introns, 32 to exons, 9 to the 3′-UTR, 2 to the 5′UTR, 5 to downstream regions up to 3kb distant from the end of the 3′-UTR, and 254 were localized to distal intergenic sequences (Figure 1B–C). The binding sites in the promoter area tend to locate in 1kb proximity of the transcription start sites (TSS) (Figure 1D).
Figure 1.
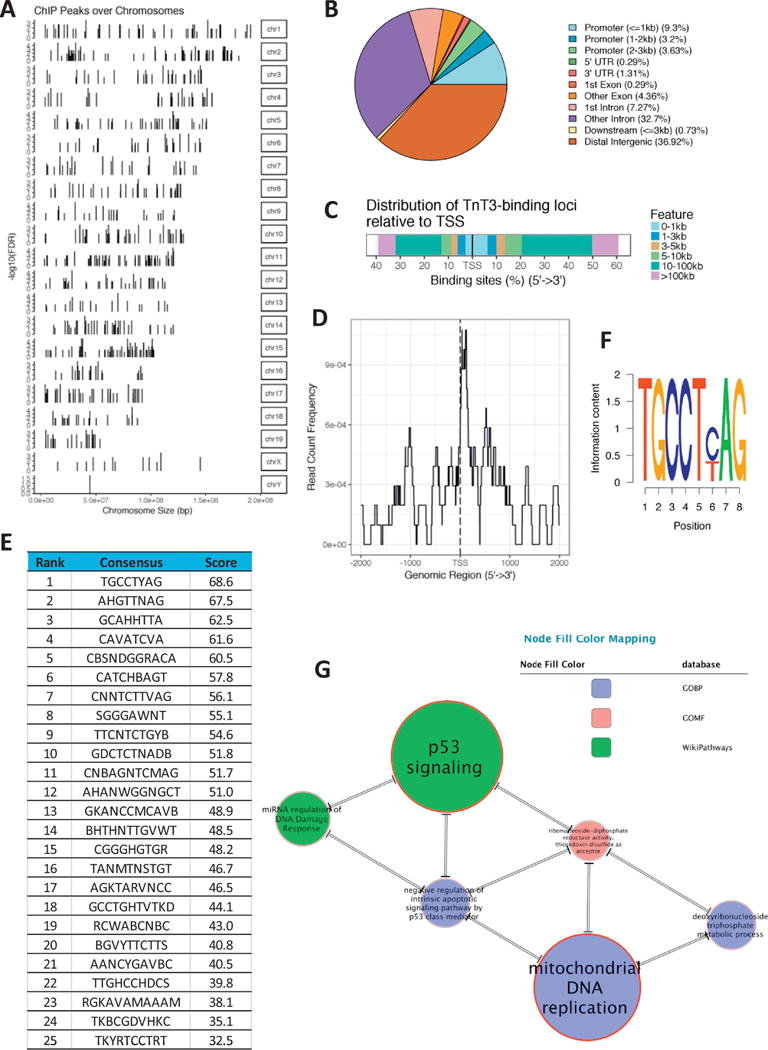
TnT3 ChIP-Seq Analysis. Localization of genomic sites occupied by TnT3 relative to chromosomal location (A); distribution of peak annotations in terms of genomic features (B); distribution of binding loci relative to the transcription start site (TSS) (C-D). List of the top 25 DNA binding motifs for TnT3 (E) and top sequence logo (F). Top SetRank Subnetwork displaying significant intersections among enriched categories. Double-line edges indicate that the significance of both gene sets was only in the intersection. The size of the nodes and node labels is proportional to the respective SetRank score (G).
De novo discovery of TnT3′s DNA binding motifs
Using the Bioconductor package BCRANK we conducted de novo discovery of DNA binding motifs in peak regions of the TnT3 ChIP-Seq experiment. The list of the top 25 motifs identified for TnT3 and the sequence logo for the top motif are presented in Figure 1E–F. Notably, the top motif sequence TGCCT(CT)AG includes the sequence TGCCT, which is required for p53 binding to the promoter area of p53 related genes (Funk and others 1992; Kern and others 1991).
Functional Enrichment Analysis
To find significant functional trends among the genes annotated in the ChIP peak regions identified by ChIP-Seq we used the R SetRank package. This package implements an advanced GSEA algorithm that eliminates many false positive hits, a common problem to functional enrichment tools based on gene sets compiled from gene and pathway annotation databases. Table 1 and Figure 1G show the main results from the functional enrichment analysis of the significant ChiP-Seq peaks identified in our study. Notably, two categories related to p53 signaling were among the top five enriched categories as defined by the SetRank score (a value that reflects the prominence of a gene set in a gene set network and is calculated using the PageRank algorithm (Brin and Page 1998)). The mouse gene ribonucleotide reductase M2 B (TP53 inducible) (Rrm2b), one of the top genes significantly enriched for TnT3 binding (Supplementary Table ST1) was also detected among the key network genes at the intersection of the top SetRank nodes: the p53 signaling and the mitochondrial DNA replication nodes, among others (Supplementary Table ST2).
Table 1.
Functional enrichment analysis of ChIP-Seq peaks using SetRank
| Category Name | Description | Database | Size | SetRank | P Value | Adjusted P Value |
|---|---|---|---|---|---|---|
| WP2902 | p53 signaling | WikiPathways | 67 | 0.097 | 0.0017 | 0.0259 |
| GO:0006264 | mitochondrial DNA replication | GOBP | 10 | 0.093 | 0.0017 | 0.0259 |
| WP2087 | miRNA regulation of DNA Damage Response | WikiPathways | 65 | 0.048 | 0.0017 | 0.0259 |
| GO:0009200 | deoxyribonucleoside triphosphate metabolic process | GOBP | 19 | 0.044 | 0.0017 | 0.0259 |
| GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | GOBP | 17 | 0.044 | 0.0017 | 0.0259 |
| GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor | GOMF | 3 | 0.036 | 0.0017 | 0.0259 |
| GO:0044291 | cell-cell contact zone | GOCC | 75 | 0.036 | 0.0020 | 0.0282 |
| GO:0019748 | secondary metabolic process | GOBP | 37 | 0.036 | 0.0023 | 0.0294 |
| GO:0031497 | chromatin assembly | GOBP | 116 | 0.036 | 0.0024 | 0.0294 |
| GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity | GOMF | 3 | 0.067 | 0.0034 | 0.0379 |
| GO:0002315 | marginal zone B cell differentiation | GOBP | 10 | 0.036 | 0.0034 | 0.0379 |
| GO:0043020 | NADPH oxidase complex | GOCC | 9 | 0.067 | 0.0035 | 0.0379 |
| R-MMU-5668599 | RHO GTPases Activate NADPH Oxidases | REACTOME | 13 | 0.036 | 0.0035 | 0.0379 |
| GO:0060828 | regulation of canonical Wnt signaling pathway | GOBP | 183 | 0.036 | 0.0063 | 0.0563 |
| GO:0005759 | mitochondrial matrix | GOCC | 243 | 0.036 | 0.0063 | 0.0563 |
| GO:0030178 | negative regulation of Wnt signaling pathway | GOBP | 137 | 0.036 | 0.0070 | 0.0563 |
| GO:0046661 | male sex differentiation | GOBP | 142 | 0.036 | 0.0071 | 0.0563 |
| GO:0016323 | basolateral plasma membrane | GOCC | 214 | 0.036 | 0.0072 | 0.0563 |
| GO:0003014 | renal system process | GOBP | 85 | 0.036 | 0.0086 | 0.0563 |
| GO:0050796 | regulation of insulin secretion | GOBP | 171 | 0.036 | 0.0091 | 0.0563 |
| GO:0005741 | mitochondrial outer membrane | GOCC | 147 | 0.036 | 0.0096 | 0.0563 |
| GO:0000323 | lytic vacuole | GOCC | 456 | 0.036 | 0.0099 | 0.0563 |
Table is sorted by Adjusted P value and SetRank score
TNNT3 and RRM2B expression in human skeletal muscle
To assess the translational value of our in vitro studies using the mouse muscle cells and ChIP-Seq, we additionally profiled (by quantitative TaqMan RT-PCR) the expression of the human TNNT3 and RRM2B genes in the vastus lateralis muscle of 21 lean non-diabetic human subjects (Figure 2A–G). The summary of clinical characteristics for this human cohort is presented in Table 2. Gender and BMI had been a priori designated as potential confounders and therefore included as covariates in the linear models and the calculation of partial correlations. We demonstrated a strong correlation (r = 0.78, P = 1×10−4, Figure 2D) between the expression levels of TNNT3 and RRM2B in the skeletal muscle tissue of the human subjects and a significant (P < 0.05) downregulation of both gene transcripts in the third age-tertile group [42.3 - 70 years of age (yoa)] as compared to the second age-tertile (31.3 - 42.3 yoa) (Figure 2A–C). A similar but statistically non-significant trend was observed when comparing the third and first (23 - 31.3 yoa) age-tertile groups. In addition, human muscle levels of both TNNT3 and RRM2B transcripts significantly and negatively correlated with total body fat mass determined by DXA (rTNNT3~Fat Mass = −0.49, rRRM2B~Fat Mass = −0.49, both P < 0.05, Figure 2E–F). Of note, RRM2B transcript levels also significantly correlated with pancreatic β cell function (rRRMB~HOMA-B = 0.47, P = 0.0473, Figure 2G), as estimated by the homeostatic model assessment (HOMA). The HOMA method is based on the physiological relationship between glucose and insulin in the basal (fasting) state, which reflects the balance between hepatic glucose output and insulin secretion that is maintained by feedback loops between the liver and the pancreas (Turner and others 1979; Wallace and others 2004). Since its inception in 1985 (Matthews and others 1985), the HOMA model has proven useful for assessing pancreatic β cell function (HOMA-B) and insulin resistance (HOMA-IR) and is widely used in clinical and epidemiological studies.
Figure 2.
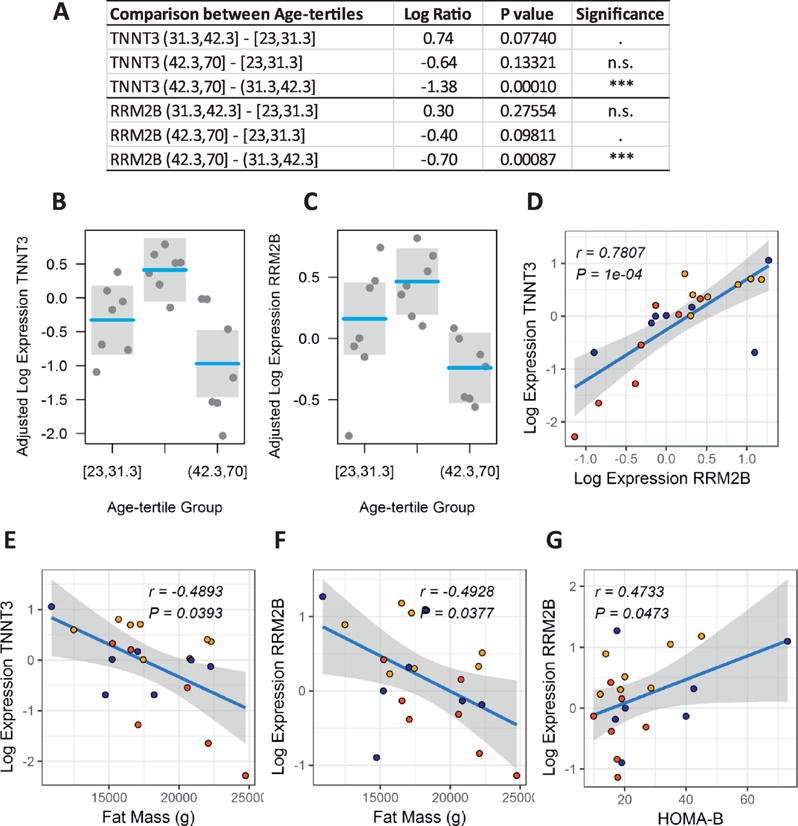
Expression profiles of TNNT3 and RRM2B in human vastus lateralis muscle. Differential expression analysis by age-tertile group after adjustment for confounding effects of gender and BMI (A-C). Correlation plots between muscle expression levels of TNNT3 and RRM2B (D), TNNT3 and whole body fat mass determined by DXA (E), RRM2B and whole body fat mass determined by DXA (E), and between RRM2B and pancreatic β cell function as determined by the homeostasis model assessment of β cell function (HOMA-B) (G). Reported correlation coefficients r are adjusted for gender and BMI. Symbol colors in correlation plots represent the first age-tertile (blue), second age-tertile (orange), and third age-tertile (red) groups.
Table 2.
Clinical characteristics of the human cohort
| Group (sample size) Age range (years) |
1st Age-Tertile (N=7) [23.0, 31.3] |
2nd Age-Tertile (N=7) (31.3, 42.3] |
3rd Age-Tertile (N=7) (42.3, 70] |
P |
|---|---|---|---|---|
| Gender (%) F | 5 (71.4) | 6 (85.7) | 5 (71.4) | 0.769 |
| M | 2 (28.6) | 1 (14.3) | 2 (28.6) | – |
| Age (years) | 25.0 [23.5, 26.0] | 35.0 [32.5, 38.5] | 49.0 [46.0, 55.5] | <0.001 |
| BMI (kg/m2) | 21.40 [20.75, 23.15] | 21.80 [20.80, 23.15] | 22.60 [20.50, 24.05] | 0.822 |
| Waist Circumference (cm) | 78.45 [74.80, 79.50] | 72.25 [69.70, 77.62] | 78.10 [73.28, 81.40] | 0.444 |
| Fat Percentage (%) | 31.90 [26.45, 32.60] | 32.00 [27.65, 33.15] | 31.40 [29.50, 36.25] | 0.919 |
| Fat Mass (kg) | 17.05 [15.00, 19.55] | 17.24 [16.11, 19.74] | 20.60 [16.82, 21.43] | 0.528 |
| Lean Mass (kg) | 38.74 [35.61, 50.12] | 36.23 [34.68, 43.43] | 40.38 [36.81, 44.94] | 0.713 |
| HDL (mg/dL) | 67.00 [57.50, 68.00] | 64.00 [61.50, 70.00] | 79.00 [63.00, 82.50] | 0.360 |
| LDL (mg/dL) | 95.00 [88.00, 104.00] | 94.00 [72.00, 121.00] | 136.00 [109.00, 137.00] | 0.256 |
| Triglycerides (mg/dL) | 96.00 [73.00, 111.00] | 61.00 [51.00, 72.00] | 99.00 [73.00, 108.50] | 0.152 |
| Fasting Glucose (mg/dL) | 86.53 [83.78, 91.42] | 84.93 [83.75, 89.47] | 91.90 [89.30, 94.12] | 0.251 |
| Fasting Insulin (μIU/mL) | 1.91 [1.28, 2.33] | 1.17 [1.04, 1.70] | 1.34 [1.24, 1.42] | 0.393 |
| HbA1C (%) | 5.20 [5.10, 5.30] | 5.30 [5.05, 5.45] | 5.80 [5.50, 5.85] | 0.020 |
| HOMA-IR | 0.46 [0.28, 0.48] | 0.26 [0.21, 0.37] | 0.32 [0.28, 0.34] | 0.462 |
| HOMA-B | 20.22 [18.28, 41.25] | 20.08 [16.28, 31.75] | 17.53 [15.59, 18.42] | 0.186 |
| QUICKI | 0.44 [0.44, 0.49] | 0.49 [0.46, 0.52] | 0.47 [0.47, 0.49] | 0.462 |
| MATSUDA | 11.31 [10.70, 18.11] | 21.78 [16.32, 24.63] | 19.60 [17.55, 23.72] | 0.175 |
Table constructed using the R package tableone. Data expressed as median and interquartile range inside square brackets. P values calculated using the non-parametric Kruskal-Wallis rank sum test. Square bracket symbols in age ranges indicate that the respective age limit is included; parenthesis indicates the limit is not included. Gender expressed as count and percentage inside parentheses. F: female, M: male.
Discussion
We have recently demonstrated that the fast skeletal muscle TnT3 isoform can translocate to the nuclei of mature myofibers and consequently induce muscle cell apoptosis in an age-dependent manner (Zhang and others 2013a; Zhang and others 2013c). We further demonstrated that TnT3 contains bona fide nuclear and nucleolar localization signals and mediates binding to the DNA through a leucine zipper (LZD) domain to function as a transcription factor (Zhang and others 2013a; Zhang and others 2016). These studies suggested a possible link between the deleterious effects of TnT3 overexpression and age-related sarcopenia.
In the current study, we identify genome-wide sequence motifs targeted by nuclear TnT3 in differentiated C2C12 muscle cells using ChIP-Seq analysis. Interestingly, the top motif identified [TGCCT(C/T)AG] includes the sequence TGCCT, which is required for p53 binding to the promoter area of p53 related genes (Funk and others 1992; Kern and others 1991). Supporting the potential binding of nuclear TnT3 to this particular ChIP-Seq motif, we previously clearly demonstrated that TnT3 directly binds a related sequence (i.e., ATCTGCC--AG, bold fonts denote the matching aligned consensus sequence) in the P5 promoter region of the calcium voltage-gated channel subunit alpha1 S (Cacna1s) gene (Zhang and others 2016). This previous work also showed that the effect of reduced/fragmented nuclear TnT3 leading to loss in muscle force, was rescued by in vivo treatment with an anti-calpain agent in old mice.
Further supporting our findings, functional enrichment analysis using an advanced GSEA algorithm highlights a p53 signaling-related subnetwork (Figure 1E) including the top SetRank categories enriched among the annotated TnT3 ChIP-Seq peaks. We reason that the overlapping of TnT3 and p53 DNA binding motifs might cause interference (e.g., competition) between the two transcription factors, therefore negatively affecting each other regulatory functions at specific DNA loci under specific conditions. Remarkably, a negative association between peak serum troponin T levels and circulating Bnip3L (the transcription of which is driven by p53) has been recently reported in acute myocardial infarction patients (Pereg and others 2015). We speculate that TnT3 might differentially bind to TGCCT-related sequences in the promoter area of p53-regulated genes and consequently interfere with their p53-induced transcriptional activation and respective novel roles in cardioprotection [suggested by the work of (Pereg and others 2015) and (Gogna and others 2013)]. Alternatively, and likely depending on genomic context, nucleus-translocated TnT3 might also contribute to activating the expression of specific p53-controlled genes in an p53-independent manner. Of note, the p53 pathway plays a crucial role at the intersection among longevity, cellular senescence, and tumor suppression.
The gene Rrm2b, encoding p53-controlled ribonucleotide reductase M2 B (also known as p53R2), identified in this study as a key TnT3-ChIP-Seq network gene at the intersection of the p53 signaling and the mitochondrial DNA replication nodes, has been reported to play a crucial role in dNTP supply for mitochondrial DNA (mtDNA) synthesis in both human and mouse. Mutations in this gene cause severe heritable mtDNA depletion in muscle of humans and similar effects are observed in the Rrm2b−/− mouse (Bourdon and others 2007). Through the production of dATP, the ribonucleotide reductase complex (of which the Rrm2b protein is a subunit) can also contribute to the enhancement of actomyosin function and muscle contractility (Regnier and others 2000). Consequently, genetic overexpression of the Rrm2b gene improved cardiac function in failing rodent hearts (Kolwicz and others 2016; Korte and others 2011; Nowakowski and others 2013). We reason that the TnT3-mediated transcriptional regulation of Rrm2b expression may therefore contribute, at least in part, to mtDNA maintenance defects and loss of muscle function in the elderly, with potential impact in the development of increased risk for cardiomyopathies.
Our validation experiments in a human cohort of lean people with age ranging from 23 to 70 yoa demonstrated a strong association between the expression levels of TNNT3 and RRM2B in the human skeletal muscle tissue and a significant reduction of both gene transcripts in the older age group. These results indirectly support the suggested transcriptional regulation of RRM2B by TnT3 in both mouse and humans, and underscore the role that these two molecules play during aging. Notably, the transcriptional regulation of RRM2B by TnT3 in skeletal muscle, their age-dependent expression levels, and the correlation between muscle RRM2B and pancreatic β cell function (HOMA-B) in humans, may provide mechanistic insight into the pathophysiology of insulin resistance of skeletal muscle and the progressive β cell dysfunction observed in mitochondrial diabetes (Lindroos and others 2009; Szendroedi and others 2009). This relationship may also play a role in age-related (late onset) diabetes, as sarcopenic muscle has been suggested to cause diabetes in aging populations (Cuthbertson and others 2016; Lee and others 2011; Scott and others 2016). The negative correlations between whole body fat mass and muscle TNNT3 and RRM2B (independent of gender and BMI) are also relevant and consistent with this hypotheses, as obesity and fat mass are associated with chronic inflammation and muscle/systemic insulin resistance (Kim and others 2000; Kim and Park 2018; Nicholas and others 2016; Virtanen and others 2005; Wellen and Hotamisligil 2005). By the same token, these associations may provide insights into pathophysiological mechanisms in sarcopenic obesity (Kim and others 2013; Levine and Crimmins 2012; Sakuma and Yamaguchi 2013).
In summary, we now identify the sequence motifs targeted by nuclear TnT3 in differentiated muscle C2C12 cells and uncover the transcriptional targeting of p53-related genes and pathways, including the negative regulation of apoptotic p53 signaling and the p53-controlled replication of mtDNA, as top candidate mechanism by which nuclear TnT3 might contribute to muscle dysfunction and sarcopenia. The validation, in a human cohort, of an age-dependent effect and a strong correlation between the expression levels of TNNT3 and its suggested transcriptional target p53-dependent RRM2B, as well as associations with total body fat mass and β cell function, enhances the translational value of our findings. However, our study has limitations that need to be addressed in future studies. Specifically, the direct binding of TnT3 isoforms to the consensus DNA motifs need to be demonstrated by gel shift assays, and the gene-specific transcriptional regulatory functions interrogated with luciferase reporter assays, among others. This study together with previously published work from our group set the stage for future work addressing the potential mechanistic involvement of TnT3 and RRM2B in the development of age-related sarcopenia, sarcopenic obesity, and the progressive β cell dysfunction observed in mitochondrial and late-onset diabetes.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants, R01AG057013 and R01AG15820 to Osvaldo Delbono, the Wake Forest Claude D. Pepper Older Americans Independence Center P30-AG21332.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests
The authors declare no competing financial interests.
References
- Asumda FZ, Chase PB. Nuclear cardiac troponin and tropomyosin are expressed early in cardiac differentiation of rat mesenchymal stem cells. Differentiation. 2012;83:106–115. doi: 10.1016/j.diff.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Brin S, Page L. The Anatomy of a Large-Scale Hypertextual Web Search Engine. Proceedings of the 7th World-Wide Web Conference; Brisbane. 1998. [Google Scholar]
- Casas-Tinto S, Maraver A, Serrano M, Ferrus A. Troponin-I enhances and is required for oncogenic overgrowth. Oncotarget. 2016;7:52631–52642. doi: 10.18632/oncotarget.10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson DJ, Bell JA, Ng SY, Kemp GJ, Kivimaki M, Hamer M. Dynapenic obesity and the risk of incident Type 2 diabetes: the English Longitudinal Study of Ageing. Diabetic medicine: a journal of the British Diabetic Association. 2016;33:1052–1059. doi: 10.1111/dme.12991. [DOI] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogna R, Madan E, Khan M, Pati U, Kuppusamy P. p53’s choice of myocardial death or survival: Oxygen protects infarct myocardium by recruiting p53 on NOS3 promoter through regulation of p53-Lys(118) acetylation. EMBO Molecular Medicine. 2013;5:1662–1683. doi: 10.1002/emmm.201202055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiological reviews. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Kern SE, Kinzler KW, Baker SJ, Nigro JM, Rotter V, Levine AJ, Friedman P, Prives C, Vogelstein B. Mutant p53 proteins bind DNA abnormally in vitro. Oncogene. 1991;6:131–136. [PubMed] [Google Scholar]
- Kim JY, Nolte LA, Hansen PA, Han DH, Ferguson K, Thompson PA, Holloszy JO. High-fat diet-induced muscle insulin resistance: relationship to visceral fat mass. American journal of physiology Regulatory, integrative and comparative physiology. 2000;279:R2057–2065. doi: 10.1152/ajpregu.2000.279.6.R2057. [DOI] [PubMed] [Google Scholar]
- Kim K, Park SM. Association of muscle mass and fat mass with insulin resistance and the prevalence of metabolic syndrome in Korean adults: a cross-sectional study. Scientific reports. 2018;8:2703. doi: 10.1038/s41598-018-21168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TN, Park MS, Lim KI, Choi HY, Yang SJ, Yoo HJ, Kang HJ, Song W, Choi H, Baik SH, Choi DS, Choi KM. Relationships between sarcopenic obesity and insulin resistance, inflammation, and vitamin D status: the Korean Sarcopenic Obesity Study. Clinical endocrinology. 2013;78:525–532. doi: 10.1111/j.1365-2265.2012.04433.x. [DOI] [PubMed] [Google Scholar]
- Kolwicz SC, Jr, Odom GL, Nowakowski SG, Moussavi-Harami F, Chen X, Reinecke H, Hauschka SD, Murry CE, Mahairas GG, Regnier M. AAV6-mediated Cardiac-specific Overexpression of Ribonucleotide Reductase Enhances Myocardial Contractility. Molecular therapy: the journal of the American Society of Gene Therapy. 2016;24:240–250. doi: 10.1038/mt.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte FS, Dai J, Buckley K, Feest ER, Adamek N, Geeves MA, Murry CE, Regnier M. Upregulation of cardiomyocyte ribonucleotide reductase increases intracellular 2 deoxy-ATP, contractility, and relaxation. Journal of molecular and cellular cardiology. 2011;51:894–901. doi: 10.1016/j.yjmcc.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman DS. Eukaryotic transcription factors. London: Academic Press; 2008. [Google Scholar]
- Lee CG, Boyko EJ, Strotmeyer ES, Lewis CE, Cawthon PM, Hoffman AR, Everson-Rose SA, Barrett-Connor E, Orwoll ES. Association between insulin resistance and lean mass loss and fat mass gain in older men without diabetes mellitus. Journal of the American Geriatrics Society. 2011;59:1217–1224. doi: 10.1111/j.1532-5415.2011.03472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Crimmins EM. Sarcopenic obesity and cognitive functioning: the mediating roles of insulin resistance and inflammation? Current gerontology and geriatrics research. 2012;2012:826398. doi: 10.1155/2012/826398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos MM, Majamaa K, Tura A, Mari A, Kalliokoski KK, Taittonen MT, Iozzo P, Nuutila P. m.3243A>G mutation in mitochondrial DNA leads to decreased insulin sensitivity in skeletal muscle and to progressive beta-cell dysfunction. Diabetes. 2009;58:543–549. doi: 10.2337/db08-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Nicholas LM, Morrison JL, Rattanatray L, Zhang S, Ozanne SE, McMillen IC. The early origins of obesity and insulin resistance: timing, programming and mechanisms. International journal of obesity. 2016;40:229–238. doi: 10.1038/ijo.2015.178. [DOI] [PubMed] [Google Scholar]
- Nowakowski SG, Kolwicz SC, Korte FS, Luo Z, Robinson-Hamm JN, Page JL, Brozovich F, Weiss RS, Tian R, Murry CE, Regnier M. Transgenic overexpression of ribonucleotide reductase improves cardiac performance. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6187–6192. doi: 10.1073/pnas.1220693110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg D, Cohen K, Mosseri M, Berlin TM, Steinberg D, Ellis M, Ashur-Fabian O. Incidence and Expression of Circulating Cell Free p53-Related Genes in Acute Myocardial Infarction Patients. Journal of Atherosclerosis and Thrombosis. 2015;22:981–998. doi: 10.5551/jat.29223. [DOI] [PubMed] [Google Scholar]
- Regnier M, Rivera AJ, Chen Y, Chase PB. 2-deoxy-ATP enhances contractility of rat cardiac muscle. Circulation research. 2000;86:1211–1217. doi: 10.1161/01.res.86.12.1211. [DOI] [PubMed] [Google Scholar]
- Sahota VK, Grau BF, Mansilla A, Ferrus A. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J Cell Sci. 2009;122:2623–2631. doi: 10.1242/jcs.050880. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Yamaguchi A. Sarcopenic obesity and endocrinal adaptation with age. International journal of endocrinology. 2013;2013:204164. doi: 10.1155/2013/204164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D, de Courten B, Ebeling PR. Sarcopenia: a potential cause and consequence of type 2 diabetes in Australia’s ageing population? The Medical journal of Australia. 2016;205:329–333. doi: 10.5694/mja16.00446. [DOI] [PubMed] [Google Scholar]
- Szendroedi J, Schmid AI, Meyerspeer M, Cervin C, Kacerovsky M, Smekal G, Graser-Lang S, Groop L, Roden M. Impaired mitochondrial function and insulin resistance of skeletal muscle in mitochondrial diabetes. Diabetes care. 2009;32:677–679. doi: 10.2337/dc08-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RC, Holman RR, Matthews D, Hockaday TD, Peto J. Insulin deficiency and insulin resistance interaction in diabetes: estimation of their relative contribution by feedback analysis from basal plasma insulin and glucose concentrations. Metabolism: clinical and experimental. 1979;28:1086–1096. doi: 10.1016/0026-0495(79)90146-x. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Iozzo P, Hallsten K, Huupponen R, Parkkola R, Janatuinen T, Lonnqvist F, Viljanen T, Ronnemaa T, Lonnroth P, Knuuti J, Ferrannini E, Nuutila P. Increased fat mass compensates for insulin resistance in abdominal obesity and type 2 diabetes: a positron-emitting tomography study. Diabetes. 2005;54:2720–2726. doi: 10.2337/diabetes.54.9.2720. [DOI] [PubMed] [Google Scholar]
- Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes care. 2004;27:1487–1495. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. The Journal of clinical investigation. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised beta-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell stem cell. 2015;17:89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Birbrair A, Delbono O. Nonmyofilament-associated troponin T3 nuclear and nucleolar localization sequence and leucine zipper domain mediate muscle cell apoptosis. Cytoskeleton. 2013a;70:134–147. doi: 10.1002/cm.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Birbrair A, Delbono O. Nonmyofilament-associated troponin T3 nuclear and nucleolar localization sequence and leucine zipper domain mediate muscle cell apoptosis. Cytoskeleton (Hoboken, NJ) 2013b;70:134–147. doi: 10.1002/cm.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Birbrair A, Wang ZM, Taylor J, Messi M, Delbono O. Troponin T nuclear localization and its role in aging skeletal muscle. AGE. 2013c;35:353–370. doi: 10.1007/s11357-011-9368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Pereyra AS, Wang ZM, Birbrair A, Reisz JA, Files DC, Purcell L, Feng X, Messi ML, Feng H, Chalovich J, Jin JP, Furdui C, Delbono O. Calpain inhibition rescues troponin T3 fragmentation, increases Cav1.1, and enhances skeletal muscle force in aging sedentary mice. Aging cell. 2016;15:488–498. doi: 10.1111/acel.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


