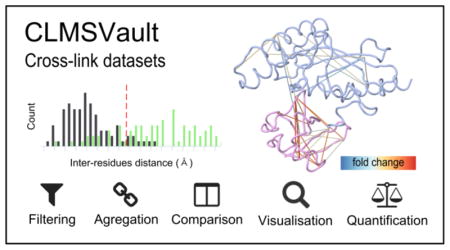
Abstract
Protein cross-linking mass spectrometry (CL-MS) enables the sensitive detection of protein interactions and the inference of protein complex topology. The detection of chemical cross-links between protein residues can identify intra- and inter-protein contact sites and/or provide physical constraints for molecular modeling of protein structure. Recent innovations in cross-linker design, sample preparation, mass spectrometry and software tools have significantly improved CL-MS approaches. Although a number of algorithms now exist for the identification of cross-linked peptides from mass spectral data, a dearth of user-friendly analysis tools represent a practical bottleneck to the broad adoption of the approach. To facilitate the analysis of CL-MS data, we developed CLMSVault, a software suite designed to leverage existing CL-MS algorithms and provide intuitive and flexible tools for cross-platform data interpretation. CLMSVault stores and combines complementary information obtained from different cross-linkers and search algorithms. CLMSVault provides filtering, comparison, and visualization tools to support CL-MS analyses, and includes a workflow for label-free quantification of cross-linked peptides. An embedded 3D viewer enables the visualization of quantitative data and the mapping of cross-linked sites onto PDB structural models. We demonstrate the application of CLMSVault for analysis of a non-covalent Cdc34-ubiquitin protein complex cross-linked under different conditions. CLMSVault is open source software (available at https://gitlab.com/courcelm/clmsvault.git) and a live demo is available at http://democlmsvault.tyerslab.com/.
Keywords: bioinformatics, mass spectrometry, cross-linker, quantification, protein interaction
Graphical Abstract
INTRODUCTION
Protein interactions orchestrate virtually all biological processes as required for cells to grow, divide, differentiate and respond to extracellular signals 1. Protein interactions may be extremely stable, as in the case of subunits of protein machines such as RNA polymerases or the ribosome. Regulatory interactions are typically much weaker, readily reversible and are typically controlled by post-translational modifications such as phosphorylation or ubiquitination. In order to understand the basis for cellular regulation, it is essential to detect protein interactions and to determine the structural contacts between proteins in both stable complexes and more transient regulatory contexts.
While many different experimental approaches are available to identify and characterize protein interactions, – including the yeast two-hybrid system, affinity purification mass spectrometry (MS), fluorescence-based methods, X-ray crystallography, NMR spectroscopy, and electron microscopy – the detection of weak interactions remains an important challenge 2, 3. Among various MS-based methods, cross-linking mass spectrometry (CL-MS) holds promise for the capture and detection of weak protein interactions and/or to reveal topological information on intra- and inter-protein contact sites. In this approach, purified protein complexes are treated with reactive bi-functional chemical cross-linkers to form covalent linkages between spatially proximate amino acid residues, followed by digestion with different proteases and mass spectrometric detection of cross-linked peptides. Protein cross-links provide distance constraints for the modeling of protein structures and have been used to infer the structure and dynamics of ribosome assembly 4, RNA polymerase II architecture 5, the INO80 chromatin remodeling complex 6, and protein phosphatase 2A complexes 7. CL-MS holds the prospect of determining protein interactions on a proteome-wide scale in complex cell lysates, and proof-of-concept results have been obtained in bacteria, nematodes, and humans 8, 9.
A variety of cross-linkers with different reactivities and lengths have been used to obtain complementary structural information 10. More sophisticated cross-linkers have been devised to facilitate the acquisition and interpretation of cross-linked peptide MS/MS spectra, including linkers that incorporate an isotopic label, cleavable linkage, biotin tag, photo-reactive group, or reporter ions. This diversity of cross-linker reagents has led to the development of several specialized algorithms that recognize specific properties to identify cross-linked peptides from MS/MS spectra 11, 12. In the simple case of non-cleavable cross-linkers, algorithms consider fragments from two peptides linked by the residual mass of one cross-linker molecule. The theoretical mass search space is the square of the total number of possible peptides, which becomes computationally limiting for systematic pairwise searches of large numbers of peptides. One solution to this problem is to match patterns corresponding to one linear peptide followed by identification of the second peptide such that all possible pairs need to be considered for identification of the cross-linked pair. This feature was implemented in Kojak 13, pLink 8 and Xi 5, 14 algorithms. In general, none of these strategies allow for multiple cross-links per peptide pair such that potentially useful data is discarded from the search process. For isotope-labeled cross-linkers, the doublet peak signals of the light and heavy cross-linkers in the MS spectrum is used by the xQuest algorithm 15 to narrow the search inter-peptide cross-links. Algorithms designed for cleavable cross-linkers, such as X-links 16 and Blinks 17 search for two modified linear peptides and a precursor mass that matches the sum of peptide masses plus the cross-linker mass, thereby circumventing the complex spectrum of the cross-linked peptide itself. False discovery rate (FDR) estimation based on the target/decoy database strategy was adopted for cross-linked peptides. FDR computations are either built in with the search engine in some case 8, 18 or provided as a separate tool (xProphet15, xiFDR19, Percolator13, 20).
Currently, there are only a few software tools available to pursue the analysis of identified cross-links to study protein interaction or structure. xiNET 21, XlinkDB 22, and ProXL DB 23 tool can generate a cross-link map/protein network. The XlinkDB 22 software can also query reference database for reported interaction. Tridimensional visualization on protein model is possible using Hekate 24, Xlink Analyzer 25, XlinkDB 22, and ProXL DB 23 each of which calculates the Euclidian distance between cross-linked residues to indicate whether a cross-link violates the maximum expected distance for the cross-linker used. Xwalk 26 uniquely calculates a solvent accessible surface distance. XlinkDB 2 27 now integrates a modeling workflow to propose protein complex structure based on cross-link length constraints. XlinkDB 22 and ProXL 23 are two integrated analysis platform for cross-links analyses.
Another issue, related to CL-MS analyses, is the complexity associated with data processing for quantification. Non-standardized file formats provided by identification software are not directly compatible with existing quantitative proteomics software tools, and only a limited number of data processing workflows have been created for this purpose. Recently, Maxquant decoupled protein identification and quantification steps to allow quantification of cross-links 28. Previous reports used manual peak area integration for label-free quantification 29 or MassChroQ for isotope-labeled cross-link quantification 30. The XiQ 31 and xTract 32 algorithms are the only tools specifically developed for the quantification of isotope-labeled cross-linkers. xTract also has the capability for label-free quantification.
To address the need for integrative analysis of CL-MS data, we developed a new software tool suite called CLMSVault that combines important functionalities for analysis of cross-linked peptides. CLMSVault serves as a local repository for CL-MS data and is able to parse and compare search results from five different search algorithms. CLMSVault adds three new unique features to complement current CL-MS analyses. First, it aggregates cross-link identifications from different cross-linkers and/or search algorithms in order to provide additional restraints for protein complex models. Second, CLMSVault provides a label-free quantification workflow for cross-linked peptides across each supported search algorithms and allows the visualization of cross-link abundance between experimental conditions. These differences can be either visualized as a simple list, as a cross-link map with xiNET 21 or in a new 3D cross-link viewer based on PDB structural models. Third, CLMSVault supports data dissemination of interaction data in the open PSI-MI TAB and XML file formats. We demonstrate the capabilities of CLMSVault on new cross-link data for a non-covalent complex between the Cdc34 ubiquitin-conjugating enzyme and ubiquitin. CLMSVault thus enables the analysis and interpretation of cross-linked mass spectrometry datasets.
SOFTWARE DESIGN AND FUNCTIONALITIES
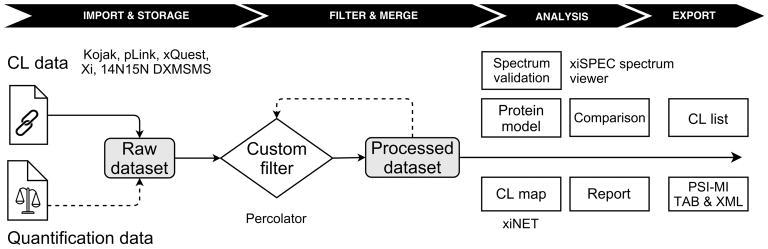
The CLMSVault data processing workflow and functionalities are summarized in Figure 1. An overview of the graphical user interface is provided in Figures S-1 and S-2. CLMSVault is implemented using the Python web framework Django (Django Software Foundation). CLMSVault can be installed on a local workstation or a remote server and accessed via common web browsers (tested on Chrome, Firefox, and Internet Explorer). Local installation is a lightweight bundle for quick deployment and portability that is based on SQLite for database storage. For multiple users and large datasets, CLMSVault should be installed on a dedicated server for best performance. Server deployment requires a pre-installed web server (e.g. Apache) and a relational database (e.g. MySQL, PostgresSQL). CLMSVault has been successfully tested on a Windows 7 workstation and Linux server (Ubuntu 14, Intel Xeon 2 GHz, 1 GB RAM). The application is user/password protected for one group of users. CLMSVault is fully open source and available to download at https://gitlab.com/courcelm/clmsvault.git. A manual was written to provide guidance how to use CLMSVault and to present the user interface for major functionalities (see the supplemental material or see the code source for the latest version).
Figure 1.
Data processing workflow for CL-MS analysis in CLMSVault
Datasets import, export, and processing
CLMSVault currently supports dataset import from five search algorithms. Custom result parsers were written for Kojak 13, pLink 8 and Xi 5, 14 for label-free cross-linkers and xQuest 15 for isotopically labeled cross-linkers. To detect inter-protein cross-links from protein homodimers, 14N15N DXMSMS match software 33 search results can be imported for cross-linked 14N15N labeled proteins. The CLMSVault data model supports storage of intra-, inter-protein cross-links and dead-end peptides. Users may specify any custom protein sequence database for data analysis in CLMSVault. Protein sequence databases used for MS/MS interpretation should be supplied as FASTA files to allow visualization of cross-links at a later stage.
Abundance fold changes or log fold changes for cross-linked peptides can be imported into CLMSVault with a simple CSV file for visualization and filtering. Results can thus be imported either from any quantification software tools with minimal reformatting or directly from the xTract label-free quantification workflow. CLMSVault automatically formats cross-link identifications as a compatible input format for xTract, and imports abundance change outputs.
Once imported by the user, the raw dataset can be either browsed or processed. Browsing offers a table view of cross-linked peptides with several actions, filters, search and sorting functionalities (see Figure S-1). Permanent filters can be applied to peptide lists through the creation of processed datasets generated either by a semi-supervised learning approach implemented in Percolator as recently proposed for cross-linked peptides 13 or by user-specified filtering criteria to produce a cross-linked peptides sub-dataset. Processed datasets can be merged to combine complementary results from different experiments, cross-linkers or search algorithms. It is recommended to filter each dataset before merging if their FDR level could be different and if search algorithms scoring schemes are compatible for filtering.
For dissemination of protein interaction data across public databases, such as BioGRID 34 and IMEx 35, users can export data as either PSI-MI TAB or XML 2.5 files.
Statistics, visualization, and comparison tools
Different analysis tools were integrated into CLMSVault to assess dataset quality after filtering and to provide an overview of identifications for each analysis or an entire dataset. A simple report with cross-links count (intra, inter, dead-end) and FDR level can be generated. For example, this can be used to find the optimal conditions (e.g. cross-linking reaction, digestion, chromatography, MS, data processing) that yield the highest number of high confidence cross-links. MS/MS spectrum of each identified cross-link can be manually validated using the integrated xiSPEC spectrum viewer (Rappsilber laboratory, http://spectrumviewer.org).
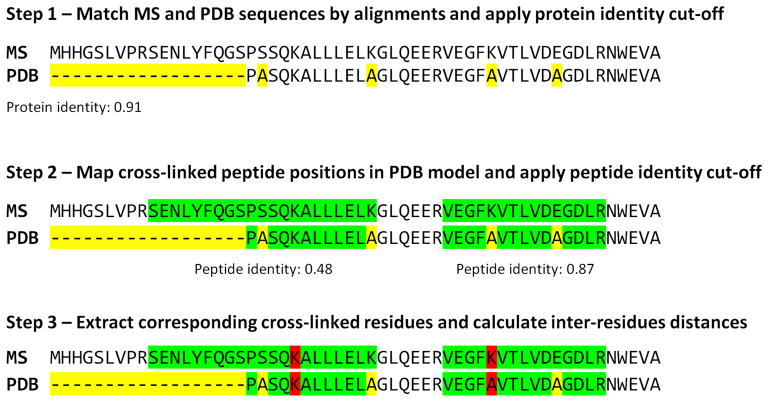
To visualize protein complex organization, the xiNET software 21 was integrated to CLMSVault for access without any file manipulation. xiNET draws interactive protein interaction networks and cross-link maps in two dimensions. It was modified to visualize cross-links with a color gradient based on cross-linked peptide score from a single search tool dataset (Figure 4A). For 3D visualization of cross-links on protein structural models, we developed an interactive viewer based on JSmol 36. A PDB model can be manually uploaded by the user or automatically retrieved from the RCSB protein data bank 37. Identified cross-linked peptides are mapped to the protein model using a three-step algorithm (Figure 2): 1) match corresponding protein sequences between MS and PDB by sequence identity, 2) map identified cross-linked peptides positions on PDB model, and 3) locate cross-linked residues positions. This algorithm is tolerant to positional shifts between protein models and MS/MS search protein sequences, missing parts, and residue substitutions. Homologous protein models can be used if protein sequence similarity is sufficient for accurate sequence alignment. The cross-link 3D viewer user interface provides the protein model overlaid with identified cross-links and a complete list of cross-linked residues (Figure S-2). Each cross-linked residue can be highlighted to reveal their position. This feature is particularly useful to locate a cross-linked residue where only a single residue of the cross-linked pair is mapped to the model (e.g. a protein missing from the model). It is also possible to highlight dead-end link to reveal information about solvent accessibility. The viewer also displays a histogram of the distributions of cross-link Cα inter-residues Euclidian distances accompanied by a Mann-Whitney rank test to assess whether cross-link distances are shorter than expected by chance (Figure S-2C). The inter-residue distance histogram reports four different distributions: 1) all, 2) minimum, 3) selected and 4) random. Selecting “All” displays all possible distances, including multiple mapping of the same cross-linked peptide at different positions or proteins. Multiple mapping of a single cross-link in a homo-dimer protein complex model has four possible distances (2 intra-proteins and 2 inter-proteins) since the model is not symmetric. In the case of an intra-protein cross-link, distances may be within the maximum expected distance for intra-protein mapping but can be exceeded when considering inter-protein mapping. This situation affects the shape of the distribution and wrongly suggests that many cross-links are not consistent with the model. To avoid this situation, the “minimum” distance distribution (e.g. minimum distance per cross-link retained) can be displayed. Given that detected cross-links can be interactively selected for display on the model by the user, their “selected” distance distribution is simultaneously updated. The last distribution plotted is a “random” distance distribution for comparison against the detected cross-links. The user can select the appropriate cross-linkable residues based on the cross-linker and search engine parameters for sampling the random distribution (e.g. DSS: K or KSTY). The minimum distances distribution is compared a thousand times with a random selection of equal cross-links using the Mann-Whitney rank test to determine if it is significantly different. This test verifies if the two samples means are equals or not. It was selected because the observed and random cross-links distance distributions are not normal. With the integrated xiNET application and our new 3D cross-links viewer, CLMSVault provides two complementary visualization tools to display cross-link locations in both 2D and 3D.
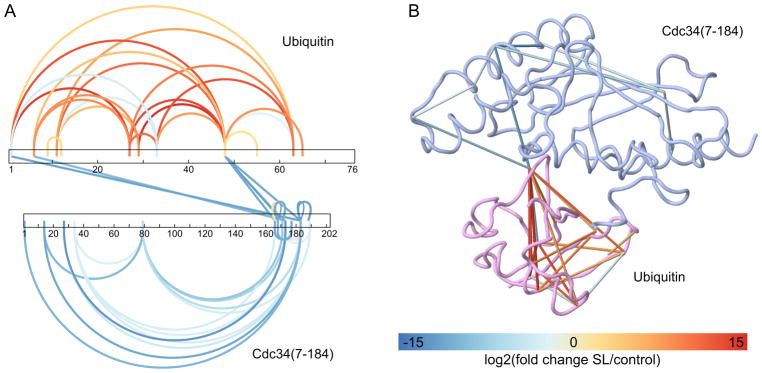
Figure 4. Visualization of cross-link abundance fold changes obtained by quantitative proteomics.
Label-free quantification of cross-linked peptides with xTract was done to evaluate the effect of 1% sodium laurate (SL) detergent solution on Cdc34/ubiquitin complex structure. Variation in cross-link abundance can be visualized in CLMSVault with either the modified xiNET (A) or our 3D viewer (B) with the fold change color scheme. Cross-links not quantified were omitted for clarity. Cross-links with a t-test significance ≤ 0.05 are displayed.
Figure 2. Algorithm for automatic mapping of cross-linked residues on PDB model.
This algorithm first finds corresponding sequences between the MS reference database and the PDB model using sequence alignment to avoid protein identifier translation issues (step 1). A user-defined protein identity cut-off is then applied and matching proteins kept. Protein sequence alignments are used as a guide to map cross-linked peptides on the PDB model to avoid issues with positional shifts, point mutations or model truncation (step 2). This algorithm allows for the use of any homologous protein models. Cross-linked residue positions are finally extracted and their Cα distance is calculated (step 3). Yellow corresponds to sequence mismatches, green to peptide sequences and red to cross-linked residues.
For quantitative comparisons of cross-linked peptide abundances, the fold change of cross-linked peptides between different experimental conditions can be imported and filtered within CLMSVault. These quantitative comparisons may also be visualized with the modified xiNET or on the 3D viewer, which display each cross-link with a color gradient based on fold change in abundance. The gradient is scaled automatically with minimum or maximum values or can be manually defined in the user interface.
EXPERIMENTAL METHODS
Cdc34-ubiquitin cross-linking analysis
Cdc34ACAT (residues 7–184) was produced in bacteria as previously described 38 and His-tagged ubiquitin was obtained from Boston Biochem. Disuccinimidyl suberate (DSS, Sigma-Aldrich) cross-linking reactions were performed by incubating 25 μM of Cdc34A, 25 μM of His-tagged ubiquitin and 1 mM cross-linker in reaction buffer (20 mM HEPES, pH 7.5) for 30 min at 22°C. For the quantification example under denaturing condition, sodium laurate detergent was added to a concentration of 1% before initiating the cross-linking reaction. Three cross-linking reactions were done for both the control and detergent condition on the same batch of proteins. Reactions were stopped by the addition of ammonium bicarbonate solution to a final concentration of 50 mM. Samples were reduced with TCEP (Thermo Scientific) and alkylated with 2-chloroacetamide (Sigma-Aldrich) in two steps, both at 5 mM for 20 min at 37°C with shaking. Free cross-linker, reducing and alkylating agents were removed using a Nanosep 3K spin device (Pall) and replaced with a fresh solution of 50 mM ammonium bicarbonate. Samples were digested at a 1:100 w/w ratio with sequencing grade modified trypsin (Promega), acidified with TFA (Sigma-Aldrich) to stop the digestion, and dried in a SpeedVac. Dried samples were resuspended in 0.2 % formic acid (EMD) before LC-MS/MS analysis on a Q-Exactive Plus mass spectrometer coupled to an Ultimate 3000 binary RSLCnano UHPLC instrument (Thermo Scientific). Peptides were loaded on an EXP Stem Trap column (0.36 mm × 13.5 mm, HALO ES-C18 2.7 μm, Optimize Technologies) and separated using an in-house packed analytical column (15 cm × 150 μm, Jupiter 3μm, C18, 300 Å, Phenomenex) and a linear gradient of 5–40 % aqueous acetonitrile (0.2 % formic acid) in 45 min. MS instrument parameters were the following: electrospray ionization voltage 3.6 kV, MS1 resolution (70,000), MS1 AGC (1e6), MS1 maximum injection time (100 ms), MS1 scan range (300–1500 m/z), MS2 resolution (35,000), MS2 AGC (5e5), MS2 maximum injection time (120 ms), loop count (12), isolation width (1 m/z), NCE (27), underfill ratio (2.5 %), charge exclusion (<= 2 and >= 8) and dynamic exclusion (10 s).
Raw MS/MS data were pre-processed with MaxQuant 1.3.0.5 to create APL peak list files. Precursor masses were recalibrated, the top 20 peaks per 100 Da were retained, and de-isotoping was disabled for MS/MS processing. APL files were then submitted to Xi for cross-linked peptide identification. A custom protein sequence database containing Cdc34ACAT (residues 7–184) and ubiquitin was used for Xi searches. Decoy entries were generated automatically by Xi. Mass tolerance was set to 6 ppm for both precursor and fragment masses. Carbamidomethyl (C) and oxidation (M) were added as variable modifications. DSS mass shift was set to 138.0681 Dalton, and reactive residues were restricted to [K, S, T, Y]. Xi results were exported in CSV file format and uploaded to CLMSVault for analysis (available on the demo site). The final list of cross-linked peptides was obtained by applying Percolator with a q-value filter <= 0.001.
Cross-link identifications were first formatted with CLMSVault to the CSV format prior to label-free quantification using xTract (v1.0.1). Identifications and MS raw files from the three cross-linking replicates for each experimental condition were used for quantification. Default xTract parameters were used for quantification. Cross-links with absolute fold change ≤ 2 and with t-test significance ≤ 0.05 were imported into CLMSVault for visualization.
RESULTS AND DISCUSSION
CLMSVault workflow and functionality
The CLMSVault data processing workflow is divided into four steps: 1) import and storage, 2) filter and merge, 3) data analysis and 4) data export (Figure 1). The first step extracts cross-linked peptide identifications from search outputs and stores these in a relational database for subsequent analyses. To accommodate various cross-linker features, CLMSVault integrates search outputs from different search engines that use label-free (Kojak, pLink, and Xi), stable isotope (xQuest) or 14N15N (14N15N DXMSMS match) labeled proteins. In the second step, different filtering criteria are applied to retrieve confident identifications and remove data redundancy for subsequent analyses. Report with cross-linked peptide count and FDR level is generated to assess dataset quality and report global changes in detected cross-links. Visualization tools such as xiNET and the CLMSVault cross-link 3D viewer can be used to observe cross-linked positions within a structural model. For comparative studies, identified cross-link lists can be inspected side-by-side to view multiple datasets or visualized quantitatively in xiNET or the 3D viewer, as illustrated below for the Cdc34-ubiquitin case example. These comparative tools can thus indicate regions that undergo conformational changes upon binding of another protein or small molecule. Finally, the last step in the workflow is the export of cross-link data for further analysis. Protein interactions can be exported to PSI-MI TAB or XML format for deposition in public databases such as BioGRID 34 or IMEx 35. Overall, the CLMSVault workflow provides versatile input, processing, and visualization features to analyze various CL-MS experiments.
Feature comparison against other processing pipelines
A few research groups have developed and published in-house platform for cross-links analysis. Since each one has been tailored for specific internal needs, they include different sets of tools. We have thus compared here CLMSVault features against two others platforms: XlinkDB, and ProXL DB (Table S-1). Common to all is a web user interface that allows cross-platform and remote access. CLMSVault and ProXL DB can be both installed on premise while the latest version of XlinkDB is only available remotely. Dataset import is performed with a simple custom tab-delimited text file in XlinkDB while CLMSVault and ProXL DB directly use the output files of multiple search algorithms. The latter two platforms thus store more details of CL-MS data set. ProXL DB also uses its own XML format for import/export. For dissemination of protein interactions to public databases, CLMSVault is the only tool that allows export to PSI-MI TAB and XML formats. CLMSVault and ProXL DB allow filtering of the cross-linked peptide to control false positive identification and to select a subset of data for downstream analysis. All platforms provide 2D and 3D visualization tools. Features unique to XlinkDB are the possibility to report known protein-protein interactions from IntAct 39/EciD 40 databases and to generate protein complex modeling. ProXL DB can report the type of secondary structure and if cross-links are located in disordered regions. This platform and CLMSVault integrate a spectrum viewer. The CLMSVault 3D viewer also has distinctive features. Our mapping algorithm has the advantage of tolerating mutations and is thus able to use models from homologous proteins. A histogram also shows the distribution of distances between cross-linked residues to verify consistency of cross-links with the model. Finally, CLMSVault provides a unique visualization and comparison tools for quantitative proteomics that facilitates the correlation of structural features between different conditions.
CL-MS analysis of the Cdc34-ubiquitin interaction
To demonstrate the data processing and visualization features of CLMSVault, we performed CL-MS experiments on a low-affinity non-covalent complex between the E2 ubiquitin-conjugating enzyme Cdc34 (a.k.a., Ube2R1) and ubiquitin. Cdc34 is the dedicated E2 enzyme for the cullin-RING (CRL) class of E3 ubiquitin ligases that target many hundreds of proteins for rapid degradation by the proteasome 41. Ubiquitin is transferred to Cdc34 as an activated thioester linkage and then conjugated to substrates in a fashion that depends on a non-covalent interaction between a conserved hydrophobic patch on ubiquitin and the ubiquitin donor site on the surface of Cdc34 38. This interaction between the catalytic domain of Cdc34 and ubiquitin can occur in the absence of the thioester linkage but cannot be detected by sensitive methods such as NMR 38. However, a recently described inhibitor of Cdc34 acts by stabilization of the Cdc34 donor site-ubiquitin interaction, and has allowed the X-ray structure of the complex to be determined 42. The Cdc34-ubiquitin interaction thus serves as a test case for the sensitivity of cross-linking methods to capture weak protein interactions and to map interactions onto a structural model.
Purified recombinant Cdc34cat and ubiquitin were cross-linked using the amine reactive cross-linker DSS and analyzed on a Q-Exactive Plus mass spectrometer. MS/MS spectra raw files were pre-processed using MaxQuant and then submitted to the Xi search engine to identify cross-linked peptides. Xi results were imported into CLMSVault for post-processing and data analysis. Selected filtering criteria were applied based on the protein database size, cross-linker residue specificity and the search engine employed. We used Percolator to separate true and false positive identifications, and only cross-linked peptides with a q-value ≤ 0.001 were retained. This stringent value was selected to avoid fortuitous matches in non-cross-linked control samples. We noted for non-cross-linked samples that the 1,036 false positive hits have been nearly eliminated by the Percolator filter. Redundant cross-link identifications were removed before the analysis step (for each unique cross-linked residues pairs, the best scoring entry was kept). These selection filters narrowed down the prospective hit list to 23 and 96 inter- and intra-protein cross-links respectively. All unfiltered cross-link data and unique filtered cross-link data are provided in Table S-2.
We subsequently verified that observed cross-links are consistent with the protein complex model obtained by X-ray crystallography with the CLMSVault viewer. Out of the 119 cross-links identified from our Cdc34/ubiquitin complex, we successfully mapped 89 (75%) to the 4MDK model (Figure 3A). Cross-links not mapped are explained by missing N-terminal and internal regions in the Cdc34 model. The CLMSVault cross-links 3D viewer was designed to handle these missing regions and residues substitutions that are frequent in the protein model. It can also deal with homologous proteins if no model is available for the studied species.
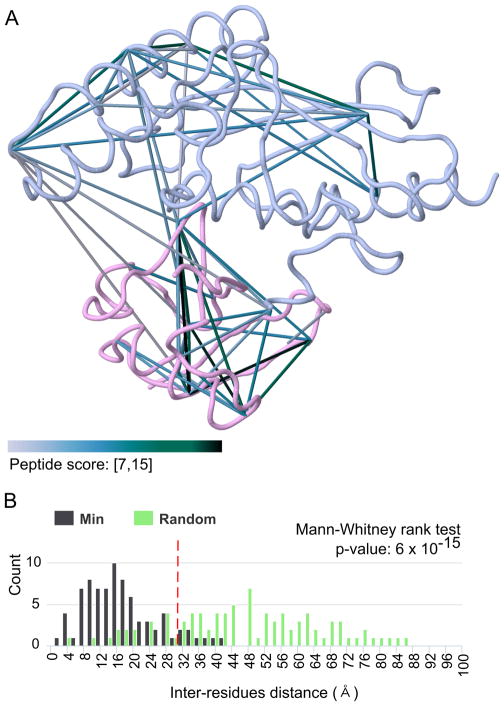
Figure 3. Visualization of identified cross-links on 3D protein complex model.
A) Overlay of identified cross-links on the PDB model 4MDK: Cdc34 (blue) and ubiquitin (pink). Cross-links are colored by their peptide score to indicate confidence in the cross-linked peptide-spectrum match. Automatic labelling of cross-link inter-residue Cα distance was removed for clarity. B) Euclidian distance distribution between cross-linked residues. This distribution is used to verify that most cross-link distances are consistent with the maximum expected length for the selected cross-linker (red dashed line for DSS). It is also compared to a random distance distribution with a Mann-Whitney rank test to evaluate if it is significantly different and to confirm that cross-links are not false-positive hits. These 2 panels are actual CLMSVault screenshots with minor modifications for enhanced print reading.
To verify that our cross-links results are in agreement with the model, we verified cross-links inter-residue Euclidian distances distribution. It is expected that most inter-residue distances should be consistent with the extended length of the cross-linker plus an extra length to account for protein flexibility. For the DSS cross-linker used in this experiment, the maximum Cα–Cα distance is 30 Å 43. In our experiment, 81 of the 89 mapped cross-links (91%) are within this limit and are consistent with the protein complex model 4MDK. The eight remaining cross-links could be explained by protein flexibility, where lysine residues can extend as far as 40 Å, consistent with recent molecular dynamics study 43. The observed distances distribution also differs significantly from the random distribution (Mann-Whitney rank test, p-value ≤ 6 × 10−15) (Figure 3B). Overall, CLMSVault helped us conclude that our CL-MS results are in agreement with the X-ray crystallography protein complex model.
To demonstrate how CLMSVault can be used to visualize the dynamics of protein structures and protein-protein interactions, Cdc34cat and ubiquitin were cross-linked in triplicates under denaturing conditions (1% sodium laurate detergent). Cross-links abundance variations measured by label-free quantification were obtained using xTract. Abundance changes of cross-link peptides can be easily monitored in CLMSVault from either xiNET or our 3D viewer (Figure 4). In the case of the Cdc34/ubiquitin protein complex, the addition of detergent concentration led to a reduced number of inter-protein cross-links with concurrent decrease and increase in intra-protein cross-links for Cdc34 and ubiquitin respectively (Figure 4). In short, CLMSVault proposes a new, unique and easy way to visualize quantitative changes of cross-link datasets to track structural changes.
CONCLUSIONS
CLMSVault is a novel platform that greatly facilitates the reporting and viewing of protein cross-linking mass spectrometry datasets. It can use data from 5 different MS/MS search engines. This provides a unique opportunity to aggregate results from different algorithms and cross-linkers to extend constraints for modeling. CLMSVault provides integrated storage, filtering, comparative and visualization tools for analyses of CL-MS datasets in a single software. We developed a new interactive 3D viewer for cross-links with the unique capability to highlight abundance changes between two different conditions. We also integrated a processing workflow for label-free quantification of cross-linked peptides to enable conditional and dynamic exploration of protein structures and interactions. CLMSVault will facilitate dissemination of new protein-protein interactions discovered by CL-MS to public databases using the PSI-MI standardized file format. Finally, CLMSVault is an enabling tool to manage and analyze protein-protein datasets obtained from large-scale cross-linking studies.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by Canadian Institutes of Health Research grant MOP-126129 (M.T.) and the Canadian Cancer Society Research Institute (to M.T.), by an award from the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec (M.T), by a Genome Canada Genomics Innovation Node award to the Centre for Advanced Proteomic Analyses (MT. and P.T.), by the Lea Reichmann Chair in Cancer Proteomics (A.C.G.), and by Canada Research Chairs in Proteomics and Bioanalytical Spectrometry (P.T.), Functional Proteomics (A.C.G), and Systems and Synthetic Biology (M.T.).
We thank members of the Rappsilber lab for providing access and assistance to Xi and xiNET software tools, Éric Bonneil for his assistance with LC-MS instrumentation and Rino Stocco for protein purification of Cdc34
ABBREVIATION
- ACN
acetonitrile
- Cdc34
ubiquitin-conjugating enzyme E2 R1
- CL-MS
cross-linking mass spectrometry
- CL PSM
cross-linked peptide spectrum match
- CSV
comma- separated values
- DSS
disuccinimidyl suberate
- FDR
false discovery rate
- PDB
protein data bank
- SL
sodium laurate
Footnotes
Author Contributions
MC developed CLMSVault software, did the CL-MS experiments and wrote the manuscript. JCH provided suggestions for the software. EC provided suggestions for CL-MS experiments. JCH, EC, ACG, PT and MT reviewed and edited the manuscript. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
The following files are available free of charge at ACS website http://pubs.acs.org:
Figure S-1–2 with screenshots of CLMSVault user interface and Table S-1 - Feature comparison between Xlink-DB and CLMSVault software tools (DOC).
Table S-2 - Cross-linked peptide identification (XLSX).
CLMSVault user manual (DOC)
References
- 1.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326(5957):1220–4. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehne B, Schlitt T. Protein-protein interaction databases: keeping up with growing interactomes. Hum Genomics. 2009;3(3):291–7. doi: 10.1186/1479-7364-3-3-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walzthoeni T, Leitner A, Stengel F, Aebersold R. Mass spectrometry supported determination of protein complex structure. Curr Opin Struct Biol. 2013;23(2):252–60. doi: 10.1016/j.sbi.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Lauber MA, Rappsilber J, Reilly JP. Dynamics of ribosomal protein S1 on a bacterial ribosome with cross-linking and mass spectrometry. Mol Cell Proteomics. 2012;11(12):1965–76. doi: 10.1074/mcp.M112.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29(4):717–26. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner KP. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154(6):1207–19. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, Malmstrom L, Aebersold R. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337(6100):1348–52. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9(9):904–6. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 9.Chavez JD, Weisbrod CR, Zheng C, Eng JK, Bruce JE. Protein interactions, post-translational modifications and topologies in human cells. Mol Cell Proteomics. 2013;12(5):1451–67. doi: 10.1074/mcp.M112.024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29(6):862–76. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 11.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9(8):1634–49. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barysz HM, Malmstroem J. Development of large-scale cross-linking mass spectrometry. Mol Cell Proteomics. 2017 doi: 10.1074/mcp.R116.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoopmann MR, Zelter A, Johnson RS, Riffle M, MacCoss MJ, Davis TN, Moritz RL. Kojak: efficient analysis of chemically cross-linked protein complexes. J Proteome Res. 2015;14(5):2190–8. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giese SH, Fischer L, Rappsilber J. A Study into the Collision-induced Dissociation (CID) Behavior of Cross-Linked Peptides. Mol Cell Proteomics. 2016;15(3):1094–104. doi: 10.1074/mcp.M115.049296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9(9):901–3. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
- 16.Anderson GA, Tolic N, Tang X, Zheng C, Bruce JE. Informatics strategies for large-scale novel cross-linking analysis. J Proteome Res. 2007;6(9):3412–21. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoopmann MR, Weisbrod CR, Bruce JE. Improved strategies for rapid identification of chemically cross-linked peptides using protein interaction reporter technology. J Proteome Res. 2010;9(12):6323–33. doi: 10.1021/pr100572u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, Sinz A. StavroX--a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23(1):76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 19.Fischer L, Rappsilber J. Quirks of Error Estimation in Cross-Linking/Mass Spectrometry. Anal Chem. 2017;89(7):3829–3833. doi: 10.1021/acs.analchem.6b03745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The M, MacCoss MJ, Noble WS, Kall L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3. 0. J Am Soc Mass Spectrom. 2016;27(11):1719–1727. doi: 10.1007/s13361-016-1460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Combe CW, Fischer L, Rappsilber J. xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics. 2015;14(4):1137–47. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng C, Weisbrod CR, Chavez JD, Eng JK, Sharma V, Wu X, Bruce JE. XLink-DB: database and software tools for storing and visualizing protein interaction topology data. J Proteome Res. 2013;12(4):1989–95. doi: 10.1021/pr301162j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riffle M, Jaschob D, Zelter A, Davis TN. ProXL (Protein Cross-Linking Database): A Platform for Analysis, Visualization, and Sharing of Protein Cross-Linking Mass Spectrometry Data. J Proteome Res. 2016;15(8):2863–70. doi: 10.1021/acs.jproteome.6b00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holding AN, Lamers MH, Stephens E, Skehel JM. Hekate: software suite for the mass spectrometric analysis and three-dimensional visualization of cross-linked protein samples. J Proteome Res. 2013;12(12):5923–33. doi: 10.1021/pr4003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosinski J, von Appen A, Ori A, Karius K, Muller CW, Beck M. Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J Struct Biol. 2015;189(3):177–83. doi: 10.1016/j.jsb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahraman A, Malmstrom L, Aebersold R. Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics. 2011;27(15):2163–4. doi: 10.1093/bioinformatics/btr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweppe DK, Zheng C, Chavez JD, Navare AT, Wu X, Eng JK, Bruce JE. XLinkDB 2. 0: integrated, large-scale structural analysis of protein crosslinking data. Bioinformatics. 2016;32(17):2716–8. doi: 10.1093/bioinformatics/btw232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZA, Fischer L, Cox J, Rappsilber J. Quantitative Cross-linking/Mass Spectrometry Using Isotope-labeled Cross-linkers and MaxQuant. Mol Cell Proteomics. 2016;15(8):2769–78. doi: 10.1074/mcp.M115.056481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chavez JD, Liu NL, Bruce JE. Quantification of protein-protein interactions with chemical cross-linking and mass spectrometry. J Proteome Res. 2011;10(4):1528–37. doi: 10.1021/pr100898e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valot B, Langella O, Nano E, Zivy M. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics. 2011;11(17):3572–7. doi: 10.1002/pmic.201100120. [DOI] [PubMed] [Google Scholar]
- 31.Fischer L, Chen ZA, Rappsilber J. Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers. J Proteomics. 2013;88:120–8. doi: 10.1016/j.jprot.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walzthoeni T, Joachimiak LA, Rosenberger G, Rost HL, Malmstrom L, Leitner A, Frydman J, Aebersold R. xTract: software for characterizing conformational changes of protein complexes by quantitative cross-linking mass spectrometry. Nat Methods. 2015;12(12):1185–90. doi: 10.1038/nmeth.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrotchenko EV, Serpa JJ, Makepeace KA, Brodie NI, Borchers CH. NN DXMSMS Match program for the automated analysis of LC/ESI-MS/MS crosslinking data from experiments using N metabolically labeled proteins. J Proteomics. 2014;109C:104–110. doi: 10.1016/j.jprot.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, O’Donnell L, Oster S, Theesfeld C, Sellam A, Stark C, Breitkreutz BJ, Dolinski K, Tyers M. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, Chatr-aryamontri A, Chautard E, Chen C, Dumousseau M, Goll J, Hancock RE, Hannick LI, Jurisica I, Khadake J, Lynn DJ, Mahadevan U, Perfetto L, Raghunath A, Ricard-Blum S, Roechert B, Salwinski L, Stumpflen V, Tyers M, Uetz P, Xenarios I, Hermjakob H. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9(4):345–50. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JmolCommunity JSmol: an open-source HTML5 viewer for chemical structures in 3D. http://wiki.jmol.org/index.php/JSmol#JSmol.
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Ceccarelli DF, Orlicky S, St-Cyr DJ, Ziemba A, Garg P, Plamondon S, Auer M, Sidhu S, Marinier A, Kleiger G, Tyers M, Sicheri F. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat Chem Biol. 2014;10(2):156–63. doi: 10.1038/nchembio.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(Database issue):D358–63. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andres Leon E, Ezkurdia I, Garcia B, Valencia A, Juan D. EcID. A database for the inference of functional interactions in E. coli. Nucleic Acids Res. 2009;37(Database issue):D629–35. doi: 10.1093/nar/gkn853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig KL, Tyers M. The F-box: a new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72(3):299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- 42.Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145(7):1075–87. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 43.Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23(6):747–59. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.