Abstract
The majority of high-content imaging (HCI) assays have been performed on two-dimensional (2D) cell monolayers for its convenience and throughput. However, 2D-cultured cell models often do not represent the in vivo characteristics accurately and therefore reduce the predictability of drug toxicity/efficacy in vivo. Recently, three-dimensional (3D) cell-based HCI assays have been demonstrated to improve predictability, but its use is limited due to difficulty in maneuverability and low throughput in cell imaging. To alleviate these issues, we have developed miniaturized 3D cell culture on a micropillar/microwell chip and demonstrated high-throughput HCI assays for mechanistic toxicity. Briefly, Hep3B human hepatoma cell line was encapsulated in a mixture of alginate and fibrin gel on the micropillar chip, cultured in 3D, and exposed to six model compounds in the microwell chip for rapidly assessing mechanistic hepatotoxicity. Several toxicity parameters, including DNA damage, mitochondrial impairment, intracellular glutathione level, and cell membrane integrity were measured on the chip, and the IC50 values of the compounds at different readouts were determined to investigate the mechanism of toxicity. Overall, the Z’ factors were between 0.6 – 0.8 for the HCI assays, and the coefficient of variation (CV) were below 20%. These results indicate high robustness and reproducibility of the HCI assays established on the miniaturized 3D cell culture chip. In addition, it was possible to determine the predominant mechanism of toxicity using the 3D HCI assays. Therefore, our miniaturized 3D cell culture coupled with HCI assays has great potential for high-throughput screening (HTS) of compounds and mechanistic toxicity profiling.
Keywords: High-content imaging, high-throughput screening, mechanistic toxicity, 3D cell culture, miniaturization, microarray chip platform
1. Introduction
Current drug development has become an expensive business with costs around $2.6 billion per FDA-approved drug and development time nearly 10–12 years. The cost of drug development has increased tremendously over the past 20 years with the approval rate of new drugs rapidly declining (Paul et al., 2010). With the current attrition rate standing at 90% for new molecular entities, decreasing the high attrition rate is a major challenge for pharmaceutical industries. Toxicity is one of the leading causes for attrition of lead candidates in drug discovery processes. Lack of highly predictive in vitro toxicity assay platforms in an early preclinical drug discovery stage has been attributed to the high failure of lead compounds in animals and humans (Astashkina et al., 2012). To overcome this issue, multi-parametric mechanistic toxicity assays, also known as high-content imaging (HCI) assays, have been implemented in vitro to weed out potentially toxic compounds in vivo. Its capability to analyze multiple endpoints such as target specific signals (e.g., mitochondrial impairment, DNA damage, glutathione level, oxidative stress, calcium homeostasis, apoptosis/necrosis, etc.) as well as morphological changes in nuclear structure and organelle structure along with various reporter signal has enhanced our understanding of the mechanism of action of drug candidates. Thus, HCI assays have become an important tool in drug discovery process in pharmaceutical industry (Vliet et al., 2014). Nonetheless, the majority of HCI assays have been still carried out on two-dimensional (2D) cell monolayer culture for their low cost, high throughput, and convenience, despite of the enormous reports on the lack of morphological, physiological, protein/gene expression, and metabolic properties along with limited cell-cell and cell-extracellular matrix (ECM) interactions as compared to their 3D counterparts (Alépée et al., 2014; Breslin and O’Driscoll, 2013; Haycock, 2011; Justice et al., 2009; Lee et al., 2013; Page et al., 2013; Pampaloni et al., 2007).
Due to the limitations of 2D cell culture in terms of mimicking in vivo phenotypic and genotypic characteristics, the HCI assays in 2D cell models do not provide the information of complex biological mechanism inside the human body and limit the predictability of drug toxicity/efficacy in vivo (Page et al., 2013). As an alternative approach, 3D cell cultures including spheroid cultures in hanging droplet plates and non-adherent well plates have been demonstrated to maintain physiological relevance in terms of cell growth, proliferation, migration, and differentiation along with biological cues from ECMs in response to external stimuli (Astashkina and Grainger, 2014; Booij et al., 2016; Page et al., 2013). For example, various literatures have reported the maintenance of long-term liver-specific function and high predictivity towards drug-induced hepatotoxicity with 3D cell models (Gunness et al., 2013; Mueller et al., 2014; Takayama et al., 2013). Therefore, performing HCI assays on 3D cell cultures (3D HCI) help to analyze the morphological and functional features of human tissues and enable the understanding of mechanisms of potential toxicity of drug candidates and adverse drug reactions in vivo (Justice et al., 2009). Although 3D HCI is a highly useful tool for identifying and evaluating mechanistic drug toxicity and safety in humans, only limited HCI assays have been implemented in 3D cells due to difficulty in cell culture maneuverability and low throughput in cell imaging. Recently, 3D cell culture models in conjunction with HCI assays have been used for evaluating the efficacy of anticancer drugs and observing morphological changes in tumor spheroids. The examples of 3D cell models include liquid overlay in 96-well (Celli et al., 2014; Reid et al., 2014) and 384-well plates (Wenzel et al., 2014), hanging droplet plate culture (Cavnar et al., 2014; Horman et al., 2013; Hsiao et al., 2012), and cell encapsulation in hydrogels (Di et al., 2014; Sirenko et al., 2016).
High throughput in 3D cell culture and imaging is of paramount importance when it comes to implementing 3D HCI in large-scale compound screening. Conventional 3D cell culture platforms face several technical challenges due to low throughput in imaging 3D cells in XYZ directions and difficulty in dispensing relatively large volumes of cells in viscous hydrogel solutions and changing growth media periodically without disturbing spheroids. In particular, acquisition of images from 3D cells on hydrogel scaffold poses a big challenge as the cells are not grown in a single focal plane. Although confocal microscopy is widely used in imaging 3D cells and tissues due to its superior ability to acquire high resolution images in different optical sections (Lang et al., 2006), its 3D HCI application for large-scale compound screening is still limited due to low throughput by slow point scanning, potential photobleaching, and phototoxicity (Jahr et al., 2015; Scherf and Huisken, 2015). Light-sheet microscopy has recently been reported in HCI as a promising imaging technology capable of imaging 3D samples in high throughput without damaging the cell samples. In spite of its high performance, implementing this technology requires complete changes in experimental methods being used, and the commercial systems are still not fully accessible (Reynaud et al., 2015). In addition to the throughput and imaging issues, relatively large assay volumes required in conventional 3D cell culture systems and the cost of expensive reagents limit the widespread use of 3D HCI (Montanez-Sauri et al., 2015). To address these issues, we have developed miniaturized 3D cell cultures on a micropillar/microwell chip platform and demonstrated HCI capability for mechanistic toxicity studies in 3D-cultured hepatic cells in the present study. The miniaturization of 3D cell culture allowed the whole sample depth to fit within the focus depth of a normal objective due to its small dimension (e.g., typical cell spots are 700 μm in diameter and 100 μm in height) and thus, allowed the use of an automated wide-field fluorescent microscope. In addition, the miniaturization of 3D cell culture allowed for high control of microenvironmental cues, enabling more reproducible outcomes (Håkanson et al., 2014; Montanez-Sauri et al., 2015). Furthermore, it reduced reagent consumption, easily facilitated combinatorial approaches, and minimized the use of valuable materials, such as patient-derived cells.
2. Materials & Methods
2.1. Materials
Hep3B human hepatoma cell line was obtained from ATCC (Manassas, VA). RPMI-1640 and model compounds, including acetaminophen, lovastatin, rotenone, tamoxifen, menadione, and sodium citrate, were purchased from Sigma Aldrich (St. Louis, MO). Coating materials including poly(maleic anhydride alt-1-octadecene) (PMA-OD), 0.01% poly-L-lysine (PLL), and barium chloride (BaCl2) were also purchased from Sigma Aldrich. Fluorescent probes, including calcein AM, tetramethyl rhodamine methyl ester (TMRM), Hoechst 33342, and monochlorobimane (mBCl), were purchased from Thermo Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) was purchased from Corning (Tewksbury, MA).
2.2. Hep3B cell culture and preparation of cell suspension
Hep3B cells between passage number 10 - 15 were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 0.1% gentamicin in T-75 cell culture flasks at 37°C in a 5% CO2 incubator (Thermo Fisher Scientific). Once the cells reached 80 – 90% confluency, they were detached using 1 mL of 0.05% trypsin with 0.53 mM EDTA and suspended in 5 mL of RPMI with 10% FBS. The cell suspension was centrifuged at 1200 rpm (200 g) for 4 min, and the supernatant was removed. The cell pellets were then re-suspended in RPMI with 10% FBS to a final concentration of 6 x 106 cells/mL. After preparing the cell suspension, 1 million cells were seeded in 15 mL of the growth medium in a new T-75 flask.
2.3. Selection of model compounds
To demonstrate mechanistic hepatotoxicity by compounds on 3D-cultured Hep3B cells on a micropillar/microwell chip platform, six model compounds that have a wide range of toxicity mechanisms were selected. Acetaminophen, lovastatin, rotenone, tamoxifen, menadione, and sodium citrate were used, among which sodium citrate was used as a negative control (Table 1).
Table 1.
Six model compounds used in the study and their mechanism of toxicity.
| Compound | Classification | Mechanism of toxicity | Test concentration (μM) | LD50 (rat oral) | References |
|---|---|---|---|---|---|
| Acetaminophen | Analgesic and antipyretic drug | AP, BA, OS | 8 – 2,124 | 1,944 | (Park et al., 2005; Wang et al., 2002) |
| Lovastatin | Hypolipidemic drug | AP, DD, MI | 2 – 472 | 5,000 | (Kallas-Kivi et al., 2016; Niknejad et al., 2014; Walther et al., 2016; Wei et al., 2007; Zhao et al., 2015) |
| Rotenone | Pesticide, insecticide, and piscicide | AP, MI, OS | 0.12 – 30 | 30 | (Cabezas et al., 2012; Isenberg and Klaunig, 2000; Moon et al., 2005) |
| Tamoxifen | Hormonal drug for breast cancer | AP, MI, OS | 0.48 – 118 | 4,100 | (Lee et al., 2000; Tabassum et al., 2006; Tolosa et al., 2015, 2012) |
| Menadione | Vitamin K precursor | MI, OS | 0.7 – 117 | 500 | (Akiyoshi et al., 2009; De Assis et al., 2015; Tolosa et al., 2012; Woods et al., 1997) |
| Sodium Citrate | Intermediate in the Krebs cycle | Nontoxic control | 5.6 – 1,417 | 6,730 | (Tolosa et al., 2012) |
Apoptosis (AP), bioactivation (BA), DNA damage (DD), mitochondrial impairment (MI), and oxidative stress (OS).
2.4. Three-dimensional (3D) bioprinting with a microarray spotter
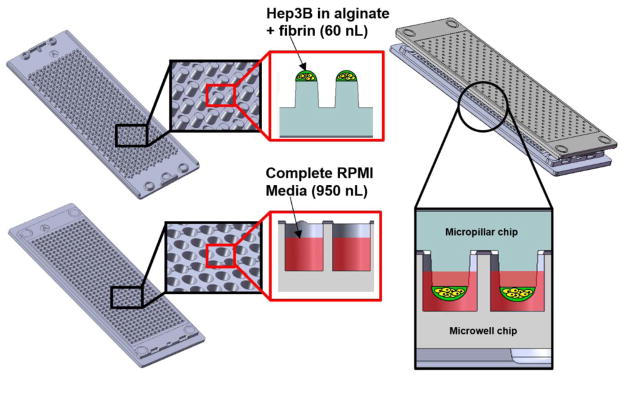
For robust cell spot attachment and miniaturized 3D cell culture on the microarray chip platform, micropillar chips were coated with 0.01% PMA-OD followed by spotting 60 nL volume of a mixture of 0.003% PLL and 16.66 mM BaCl2 onto the micropillar chips using a microarray spotter (S+ microarrayer from Samsung Electro-Mechanic Co. or SEMCO). Hep3B cell suspension was mixed with low-viscosity 3% (w/v) alginate and 100 mg/mL fibrinogen to get a final cell concentration of 2 x 106 cells/mL in 0.75% (w/v) alginate and 25 mg/mL fibrinogen. Hep3B cells suspended in the mixture of alginate and fibrinogen was then printed on top of dried PLL-BaCl2 spots for cell encapsulation and spot attachment while maintaining the slide deck at 7°C for minimizing water evaporation. After leaving the chips on the slide deck for 2 min for gelation, the micropillar chips with cell spots were sandwiched (or stamped) with the microwell chips containing 950 nL of RPMI-1640 supplemented with 10% FBS, 1% penicillin/streptomycin, 0.1% gentamicin, and 10 NIH units/mL thrombin (i.e., complete RPMI media with thrombin) (Fig. 1). The stamped micropillar/microwell chips were stored in a humidified petri dish and incubated at 37°C in a 5% CO2 incubator for two different time periods (i.e., 24 and 72 h) before drug treatment. The two-different pre-incubation time points were used to determine the effect of spheroid formation/size on compound toxicity.
Fig. 1.
Schematics of a micropillar chip with cell spots and a microwell chip containing growth media.
2.5. Treatment of 3D-cultured Hep3B cells with model compounds
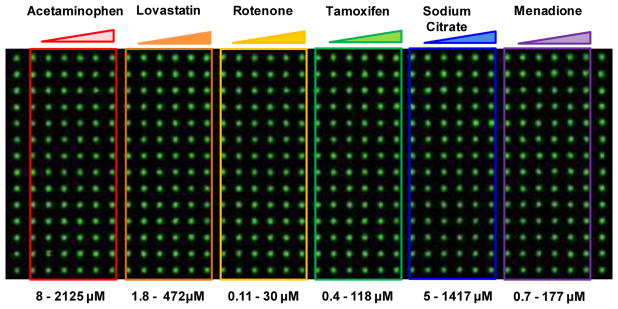
After 24 and 72 h pre-incubation of Hep3B cells, the cells were treated with the six model compounds for 48 h prior to cell staining for HCI assays. A powder form of the compounds was dissolved in DMSO except for sodium citrate dissolved in water to prepare compound stock solutions. Five concentrations and one solvent-alone control (DMSO or water) were prepared to obtain dose response curves and calculate IC50 values. Briefly, each compound stock solution was 4-fold serially diluted in DMSO (or water for sodium citrate) in a 96-well plate to prepare five dosages. Solvent-alone control was prepared by adding 100% DMSO (or water for sodium citrate). The working concentrations were prepared by mixing 1.5 μL of compounds in DMSO with 298.5 μL of complete RPMI media in a 96-well plate to achieve the final concentration of DMSO less than 0.5% (v/v). The final concentration ranges of the compounds used were as follows: acetaminophen (8.3 – 2,125 μM), lovastatin (1.8 – 472 μM), rotenone (0.12 – 30 μM), tamoxifen (0.46 – 118 μM), sodium citrate (5.5 – 1,417 μM), and menadione (0.7 – 177 μM). The dosage range of these compounds were selected based on their known IC50 values obtained from literatures. After preparing the compound plate in complete RPMI media, 950 nL of these compounds at five dosages and one control were printed in the microwell chips in a layout shown in Figure 2. For each dosage, 14 replicates of the compounds were printed. To assess compound toxicity, the micropillar chips pre-incubated for 24 and 72 h were sandwiched with the microwell chips containing compounds and then incubated for 48 h at 37°C in a 5% CO2 incubator. For the micropillar chips pre-incubated for 24 and 72 h, the same compound treatment and cell staining with fluorescent dyes were performed for direct comparison.
Fig. 2.
The scanned image of spheroids on the micropillar chip and the layout of the compounds in the microwell chip.
2.6. High-content imaging (HCI) assays on 3D-cultured Hep3B cells exposed to the compounds
Mechanisms of compound-induced toxicity were determined by evaluating multiple parameters, including mitochondrial impairment, DNA damage, intracellular glutathione level, and cell membrane integrity. Briefly, Hep3B cells treated with the compounds were stained with TMRM for the assessment of mitochondrial impairment as a function of mitochondrial membrane potential, Hoechst 33342 for DNA damage, mBCl for intracellular glutathione level, and calcein AM for cell membrane integrity. Staining solutions were prepared by adding the stock solution of the fluorescent dyes in a saline solution containing 140 mM sodium chloride (NaCl) and 20 mM calcium chloride (CaCl2) and obtaining the desired concentrations of 0.25 μM TMRM, 40 μM Hoechst 33342, 100 μM mBCl, and 1 μM calcein AM. The micropillar chips were rinsed twice for 10 min each with 5 mL of the saline solution followed by staining with 1.5 mL of the fluorescent dye solutions for 45–60 min. The stained micropillar chips were rinsed twice for 15 min each after staining to remove excess dyes from the cells and dried in the dark for at least 2 h before image acquisition.
2.7. Immunofluorescence assay for measuring the expression of extracellular matrices (ECMs)
The expression of ECMs by 3D-cultured Hep3B cells was determined by immunofluorescence assays. Briefly, Hep3B cells were encapsulated in the mixture of 0.75% (w/v) alginate and 25 mg/mL fibrinogen as mentioned above and seeded in a 6-well plate. Complete RPMI media was added, and the cells were grown for 72 h to form spheroids. After 72 h incubation, Hep3B cells were fixed with 4% paraformaldehyde (PFA) for 10 min and a mixture of methanol and acetone (1:1, v/v) for 10 min. After fixation, the samples were rinsed and blocked with 3% bovine serum albumin in 1× tris-buffered saline (TBS) for 1 h at room temperature. The samples were then incubated overnight with anti-collagen I antibody, anti-fibronectin antibody for collagen 1 and fibronectin, which were diluted in the blocking solution at 1:200 ratio. After overnight incubation with the primary antibodies, Hep3B cells were rinsed with 1× TBS containing 0.1% Tween 20 (T-TBS) and incubated with goat anti-mouse IgG (H+L) superclonal secondary antibody Alexa fluor conjugate for 2 h at room temperature. The cells were then stained with 10 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) for 10 min and rinsed with T-TBS. Cell images were obtained immediately after rinsing with Zeiss A1 epi-fluorescence microscope (Carl Zeiss).
2.8. High-throughput image acquisition with a chip scanner
Images of individual cell spots on the micropillar chips were acquired rapidly using a chip scanner (S+ scanner from SEMCO, South Korea), which is an automated fluorescence microscope specifically designed for the micropillar/microwell chip for image acquisition. Fluorescent dyes excited were monitored at excitation and emission wavelengths with appropriate filter settings. The S+ scanner contains four filter channels for detecting multicolor, blue, green, and red fluorescent dyes, individually or simultaneously. A multiband filter set (DA/FI/TR/Cy5-A-000 from Semrock) for measuring blue, green, orange, and red fluorescent dyes simultaneously, a red filter (TxRed-4040C-000 from Semrock) for measuring deep red fluorophores, a blue filter (DAPI-5060C-000 from Semrock) for measuring deep blue fluorophores, a green filter (XF404 from Omega) for green fluorophores were used to obtain fluorescent images from the cells stained with the fluorescent dyes. A 4x objective lens (UPLFLN 4X, Olympus, Japan) was used to obtain the image of the entire spots from the micropillar chips. Exposure times for the filter channels (80 ms for blue filter, 50 ms for red filter, and 80 ms for green filter) were adjusted based on histogram to obtain optimum fluorescence intensity and prevent photobleaching of fluorescence. The fluorescent dyes were uniformly distributed in 3D-cultured cell spots (i.e., 60 nL) without diffusion issues due to small dimensions.
2.9. Image processing and data analysis
Fluorescence intensity of the images obtained from the micropillar chips were quantified using ImageJ software (NIH), and standard dose-response curves for each compound per readout were plotted using image analysis software (S+ chip analysis from SEMCO, South Korea). A batch processing plugin was developed and used to obtain fluorescence intensity from the micropillar chips stained for determining various mechanisms of toxicity (Yu et al., 2016). Briefly, the images were first corrected to eliminate background fluorescence, converted to a gray scale and applied a threshold to limit the intensity extraction from fluorescently labelled cellular regions using the ImageJ plugin. After extraction of fluorescence intensity from the images, S+ chip analysis was used to plot dose response curves and calculate IC50 values (concentration of compound that inhibits 50% of cellular response). Changes in fluorescence intensity from TMRM, Hoechst 33342, mBCl, and calcein AM were evaluated to determine mitochondrial impairment, DNA damage, intracellular glutathione level, and cell membrane integrity, respectively. We have focused on extracting various mechanistic toxicity information from cell spheroids stained with multiple fluorescent dyes on the chip platform unlike our precedent work on colony morphology analysis (Lee et al., 2014b).
2.10. Statistical analysis
Data obtained from S+ chip analysis including IC50 values and standard deviations were compared to understand the effect of spheroid formation/size and the mechanisms of compound toxicity. Statistical difference in IC50 values was calculated with one-way ANOVA using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA), and p values were obtained for comparison. The difference was considered significant only in case of p<0.05 with a confidence interval of 95%. Robustness and reproducibility of the assays on the chip platform were determined by calculating the Z’ factor and the coefficient of variation (CV).
3. Results
3.1. Spheroid formation over time on the chip platform
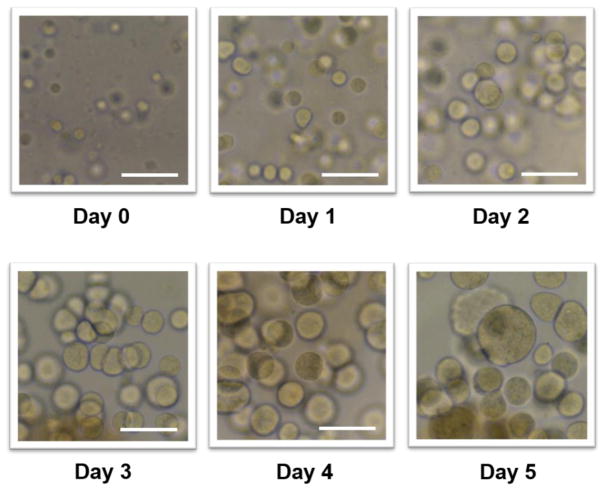
Hep3B cells were encapsulated in the mixture of 0.75% (w/v) alginate and 25 mg/mL fibrinogen and cultured on the microarray chip platform for miniaturized 3D cell culture. Fibrinogen was supplemented in alginate to provide better cellular microenvironments for spheroid formation. Fibrinogen transformed into fibrin by the addition of thrombin and formed a gel. Bright field images were taken every day for 5 days to monitor cell growth and spheroid formation (Fig. 3). The cells encapsulated in alginate-fibrin gel started to form compact aggregates from day 2 and continued to grow and form larger spheroids over 5 days. As the doubling times of Hep3B cell monolayers and 3D-cultured Hep3B cells on the micropillar/microwell chip are ~24 h (Sagawa et al., 2008) and 28 h (Lee et al., 2016) respectively, we decided to use 5-day culture period in this study. Over-growth of Hep3B cells on the chip led to spot detachment perhaps due to the activity of matrix metalloproteinases (Ha et al., 2004; Mason and Joyce, 2011; Weng et al., 2012). Within 5 days of normal Hep3B cell culture on the chip, there was no spot detachment observed. This result indicates that our PMA-OD and PLL surface chemistry is robust for cell spot attachment, 3D cell culture, and cell staining with fluorescent dyes.
Fig. 3.
Bright field microscopic images representing cell growth and spheroid formation over the period of 5 days on the micropillar chip. Hep3B cells on the micropillar chip were encapsulated in the mixture of 0.75 % (w/v) alginate and 25 mg/mL fibrinogen. The scale bar is 250 μm.
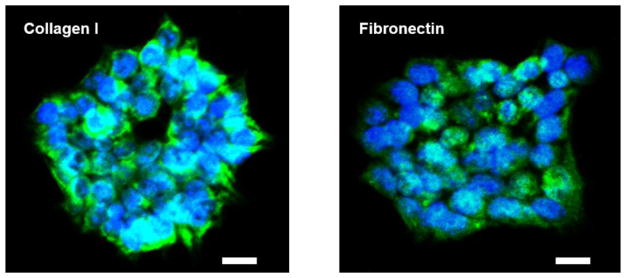
The expression level of ECM components from 3D-cultured Hep3B cells were determined by the immunofluorescence assays in the 6-well plate. Various literatures have reported the presence of collagen and fibronectin as major ECM components in liver tissues (Gessner et al., 2013; Martinez-Hernandez and Amenta, 1993; Wells, 2008). From the immunofluorescence staining we found the expression of collagen I and fibronectin in 3D-cultured Hep3B cells (Fig. 4). Unfortunately, it was difficult to measure ECM expression from the micropillar/microwell chip directly due to excessive spot detachment in the presence of surfactants such as Tween 20 used in the immunofluorescence staining. Thus, we decided to perform the immunofluorescence assays in the 6-well plate instead of the micropillar/microwell chip. Since the morphology of Hep3B cell spheroids on the micropillar/microwell chip and in the 6-well plate was similar, we hypothesized that 3D-cultured Hep3B cells on the chip also express the hepatic ECMs.
Fig. 4.
Immunofluorescence staining to determine the expression of major ECM components including collagen I and fibronectin expressed by 3D-cultured Hep3B cells in a 6-well plate. Green colors represent collagen I and fibronectin expression, and blue dots indicate nucleus. The scale bar is 20 μm.
3.2. Robustness of the high-content imaging (HCI) assays
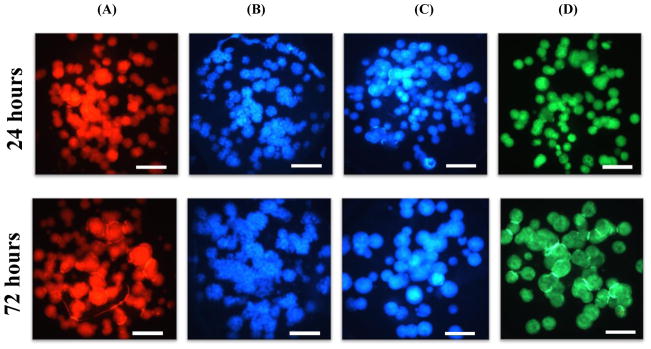
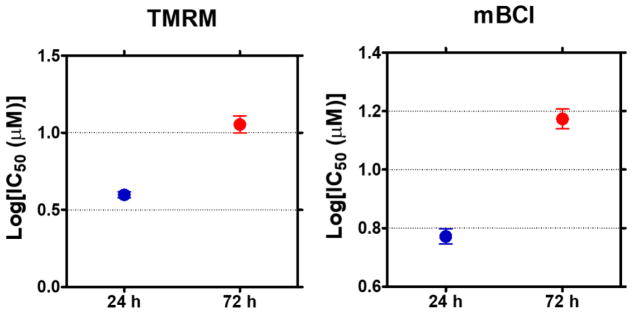
Prior to testing the mechanism of action of the model compounds using the four fluorescent dyes, we evaluated the robustness of the HCI assays and their reproducibility by calculating Z’ factors and the coefficient of variation (CV). Briefly, Hep3B cells printed on the micropillar chips were incubated with complete RPMI media for 24 h (Condition I) and 72 h (Condition II) for evaluating the effects of spheroid sizes on compound toxicity. The six model compounds were exposed to 24 and 72 h-incubated micropillar chips with 3D-cultured Hep3B cells for 48 h, which was followed by the HCI assays with the four fluorescent dyes. There was distinctive difference in terms of spheroid sizes and their fluorescent signal intensities at different incubation times (Fig. 5).
Fig. 5.
The micropillar chips with Hep3B cells cultured for 24 h and 72 h and stained with fluorescent dyes: (A) TMRM, (B) Hoechst 33342, (C) mBCl, and (D) calcein AM. The scale bar is 200 μm.
The robustness of the HCI assays on the micropillar/microwell chip was measured by calculating Z’ factors calculated as follows:
where Avg.Max is an average of maximum fluorescence intensity from fully viable Hep3B cells on the chip, SDMax is a standard deviation of maximum fluorescence intensity, Avg.Min is an average of minimum fluorescence intensity from the dead cells affected by the highest dose of menadione, SDMin is a standard deviation of minimum fluorescence intensity. The calculated Z’ factors from menadione were between 0.61 – 0.80 (Table 2). The Z’ factor between 0.5 – 1 is considered highly robust for an assay. Thus, it indicates that the HCI assays on the chip platform is robust and suitable for accurately identifying the mechanisms of compound toxicity.
Table 2.
The Z’ factors and the CV values of the HCI assays performed on the microarray chip platform.
| HCI assays | Z’ factor | CV value (%) |
|---|---|---|
| TMRM | 0.64 | 11.4 |
| Hoechst 33342 | 0.61 | 9.7 |
| mBCl | 0.80 | 5.6 |
| Calcein AM | 0.63 | 10.9 |
The CV is defined as the ratio of the standard deviation (SD) to the average (Avg.).
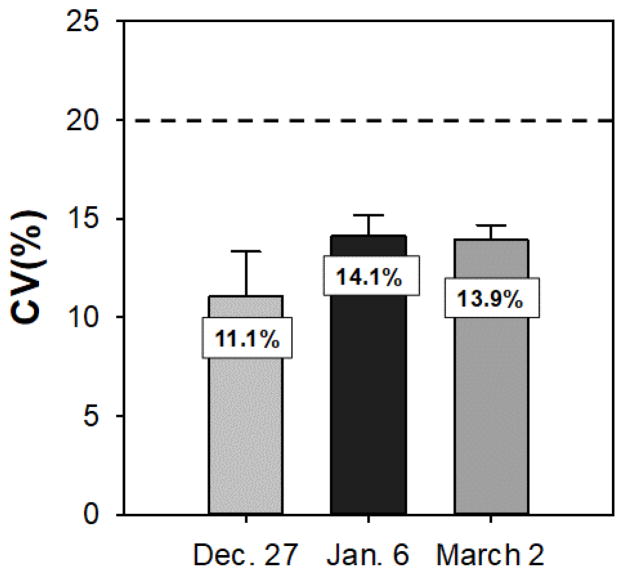
The HCI assay data obtained from 3D-cultured Hep3B cells on the chip platform exposed to no compound was used to calculate CV values of cell printing and assess day-to-day variability (Table 2 and Fig. 6). The result indicates that the experimental errors are below 20%, thus excellent and acceptable. The overall CV values obtained from different days for the calcein AM assay was 13.03%, which indicates high reproducibility of the HCI assay performed on chip platform.
Fig. 6.
Day-to-day reproducibility of CV values determined by the cell membrane integrity assay with calcein AM. Multiple micropillar chips were stained and analyzed on December 27, January 6, and March 2, 2017. The CV values obtained from different days were 11.1, 14.1, and 13.9 respectively. In addition, the overall CV value was 13.03% which shows high reproducibility of the assay performed on the chip platform.
With 24 h and 72 h-incubated micropillar chips, dose response experiments were performed with the model compounds and IC50 values were calculated from the dose-response curves obtained (Figs. 7, 8). Briefly, fluorescence intensity from cell images was quantified using the ImageJ plugin, and then the intensity vs. compound concentrations graphs (dose-response curves) were plotted to determine IC50 values (Fig. 7). The response of single concentration of a compound was calculated from 14 replicates on one chip, and three replicates of the micropillar chips were used to demonstrate the robustness and reproducibility of each HCI assay.
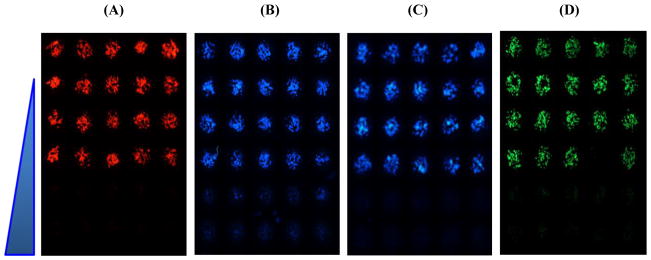
Fig. 7.
Representative images from 72 h-incubated micropillar chips with 3D-cultured Hep3B cells, exposed to menadione (0.7 – 177 μM) for 48 h, and stained with the four fluorescent dyes: (A) TMRM for mitochondrial impairment, (B) Hoechst 33342 for DNA damage, (C) mBCl for changes in intracellular glutathione level, and (D) calcein AM for cell membrane integrity. The dose-dependent effect of menadione for all four assays was clearly observed from the microarray images obtained.
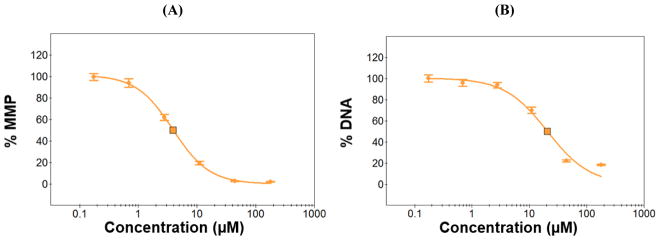
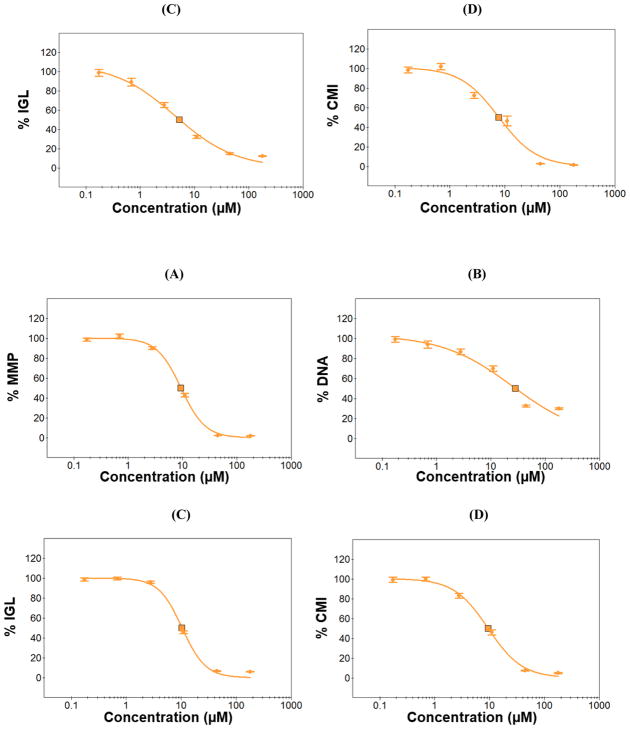
Fig. 8.
Dose-response curves of menadione obtained from the four HCI assays: (A) TMRM for mitochondrial impairment as a function of mitochondrial membrane potential (MMP), (B) Hoechst 33342 for DNA damage, (C) mBCl for changes in intracellular glutathione level (IGL), and (D) calcein AM for cell membrane integrity (CMI). The 24 h and 72 h-incubated micropillar chips with 3D-cultured Hep3B cells were exposed to menadione (0.7 – 177 μM) for 48 h and then stained with the four fluorescent dyes to calculate IC50 values.
3.3. Mechanisms of compound toxicity and the effect of spheroid sizes
The IC50 values of all six compounds obtained from the four HCI assays were compared at 24 h (I) and 72 h (II) pre-incubation conditions to evaluate the effect of spheroid sizes on the mechanisms of action of the model compounds (Table 3). The numbers in the table represent IC50 values in μM along with standard deviation across three micropillar chips. The variance of IC50 values with changes in time of pre-incubation and mechanism of toxicity were observed, and statistical analysis were performed to compare the significance (Figs. 9, 10). These values were further compared with rat oral LD50 values, IC50 data from 2D-cultured Hep3B cell monolayers in 96-well plate, and literature review to analyze cellular responses of the model compounds in vitro and in vivo. For example, lovastatin showed slight increase in IC50 values with increase in spheroid sizes resulting from 72 h pre-incubation as compared to those from 24 h pre-incubation for the four HCI assays. However, the difference in IC50 was insignificant (p > 0.5) among the assays evaluated. Rotenone showed statistically significant increase in IC50 only for DNA impairment (p < 0.5) whereas there was no difference observed among the other three assays for the two pre-incubation conditions. Menadione showed significant increase in IC50 values with increase in spheroid sizes for both mitochondrial impairment with TMRM staining and intracellular glutathione level with mBCl staining whereas there was no difference in IC50 for the other two assays (Fig. 9). Tamoxifen showed no significant difference in IC50 for intracellular glutathione level and DNA damage, while showing decrease in IC50 for cell membrane integrity with calcein AM and mitochondrial impairment with TMRM. Acetaminophen was nontoxic for all conditions tested due to lack of drug metabolizing enzymes (DMEs) in Hep3B cells. In addition, sodium citrate, the negative control compound, was also nontoxic as expected.
Table 3.
A summary of IC50 values (μM) obtained from the chip platform with the four HCI assays at 24 h (I) and 72 h (II) pre-incubation. The control data was obtained from 2D Hep3B cell monolayers with a presto blue assay.
| Compound | TMRM | Hoechst 33342 | mBCl | Calcein AM | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| I (24 h) | II (72 h) | I (24 h) | II (72 h) | I (24 h) | II (72 h) | I (24 h) | II (72 h) | 2D (prest o blue) | 2D (literature review) | LD50 mg/kg (rat oral) | |
| Acetaminophen | >2100 | >2100 | >2100 | >2100 | >2100 | >2100 | >2100 | >2100 | >2100 | 29,775±1775 | 1944 |
| Lovastatin | 141± 21.2 | 152± 6.7 | 146± 0.0 | 188± 17.7 | 149± 11.0 | 195± 5.0 | 123± 16.4 | 150± 0.0 | 133± 5.7 | 20 ± 3.7 | 5000 |
| Rotenone | 0.9± 0.2 | 0.6± 0.3 | 1.3± 1.2 | 3.0± 0.0 | 1.4± 0.16 | 1.7± 0.7 | 4.3± 2.1 | 8.0± 6.0 | 1.2± 0.02 | 1.7± 1.2 | 30 |
| Tamoxifen | 66± 2.0 | 50± 0.5 | 110± 15 | 102± 6.2 | 92± 20 | 83± 18.5 | 67± 8.0 | 57± 4.3 | 72± 2.0 | 45 | 4100 |
| Menadione | 4.0± 0.3 | 11.5± 2.5 | 22± 2.3 | 20± 1.0 | 5.3± 1.2 | 15± 2.3 | 8.7± 4.7 | 11± 3.8 | 5.5± 2.8 | 13.5± 3.6 | 500 (mice oral) |
| Sodium citrate | >1400 | >1400 | >1400 | >1400 | >1400 | >1400 | >1400 | >1400 | >1400 | >1400 | 6730 |
Fig. 9.
One-way ANOVA analysis of IC50 values obtained from 3D-cultured Hep3B cells pre-incubated for 24 h and 72 h, exposed to menadione for 48 h, and stained with TMRM and mBCl for mitochondrial impairment and glutathione level (n= 3, ** p<0.01). Statistically significant difference in IC50 values of menadione was observed at the two pre-incubation conditions, indicating that increase in spheroid sizes reduce mitochondrial impairment and glutathione level-induced toxicity of menadione.
Fig. 10.
One-way ANOVA analysis of IC50 values obtained from 3D-cultured Hep3B cells pre-incubated for 72 h, exposed to rotenone and tamoxifen for 48 h, and stained with calcein AM, TMRM, Hoechst 33342, and mBCl (n= 3, *p<0.05, ** p<0.01). Statistically significant difference in IC50 values of rotenone was observed from TMRM staining, indicating that the main mechanism of rotenone toxicity is mitochondrial impairment. Statistically significant difference in IC50 values of tamoxifen was observed from calcein AM and TMRM staining, indicating that the main mechanism of tamoxifen toxicity is cell membrane integrity and mitochondrial impairment.
3.4. Determination of the main mechanism of compound toxicity
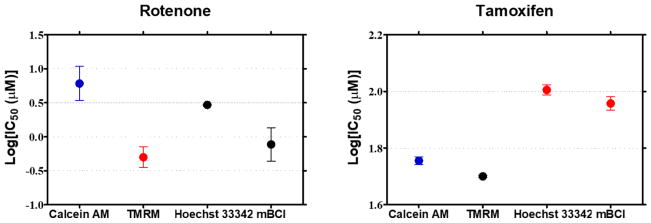
Among the four mechanisms of compound toxicity, we determined the main mechanisms at the two pre-incubation time periods by comparing the IC50 values statistically. One-way ANOVA analysis was performed to evaluate statistically significant difference in IC50 values obtained from the four HCI assays. The IC50 values obtained from calcein AM staining (i.e., cell membrane integrity assay) were used as a control, and the other IC50 values from the three remaining assays were compared. Statistically significant difference in IC50 values were observed from rotenone and tamoxifen whereas IC50 values from lovastatin and menadione showed insignificant difference when compared among the four assays. For rotenone, mitochondrial impairment was shown to be the main mechanism of toxicity with p < 0.05 whereas mitochondrial impairment and cell membrane integrity were the main mechanisms of tamoxifen toxicity with p<0.01 (Fig. 10).
4. Discussion
The main goal of the present work was to establish HCI assays on 3D-cultured human cells on the micropillar/microwell chip platform for high-throughput compound screening and predictive assessment of toxicity and efficacy. There is an urgent demand in pharmaceutical industries to develop a highly predictive in vitro cell-based assay platform for early stage detection of mechanistic toxicity of lead candidates. The miniaturized 3D cell culture chip platform developed in this study was robust and high throughput for mechanistic compound toxicity screening. On the micropillar/microwell chip platform, 532 individual spheroid spots can be cultured rapidly, exposed to multiple compounds at several dosages, and then stained with fluorescent dyes to investigate mechanistic toxicity of compounds. Due to its small dimension, the miniaturized 3D cell cultures on the chip platform allow the whole sample depth to fit within the focus depth of a normal objective as compared to conventional 3D cell culture platforms such as 96-well plates. In addition, accurate control over cellular microenvironments can be achieved on the chip, which enabled highly reproducible outcomes. The micropillar/microwell chip platform has been implemented for several applications with different cell lines, including metabolism-induced compound toxicity screening with THLE-2 human liver cell lines (Kwon et al., 2014), high-throughput screening of anticancer drug efficacy with U251 brain cancer cell lines (Lee et al., 2014a), evaluation of ajoene toxicity with Hep3B human hepatoma cell lines (Lee et al., 2014c), and screening of neurotoxic compounds with ReNcell VM human neural progenitor cell lines (Nierode et al., 2016). Nonetheless, this is the first demonstration of high-throughput, HCI capability on the micropillar/microwell chip platform with 3D-cultured cells for mechanistic study of compound-induced toxicity. Compound-induced toxicity may occur via several mechanisms, including but not limited to mitochondrial impairment, DNA damage, apoptosis/necrosis, and oxidative stress. These mechanisms may be interrelated so that induction of one mechanism can trigger induction of other mechanisms. For example, the formation of reactive oxygen species (ROS) due to drug metabolism in hepatic cells can lead to decrease in antioxidant levels, lowering the glutathione concentration within the cells (Park et al., 2005). Depletion of the glutathione level further increases the accumulation of ROS, which can damage lipid bilayers, proteins, DNAs, and mitochondrial membranes, and can alter calcium homeostasis, ultimately resulting in necrosis/apoptosis (Tolosa et al., 2012). In this study, TMRM, Hoechst 33342, mBCl, and calcein AM have been selected to assess mitochondrial impairment via measuring changes in mitochondrial membrane potential, DNA damage, decrease in intracellular glutathione levels, and cell membrane integrity which will be lost with the onset of necrosis.
The importance of 3D cell culture models in the assessment of mechanisms underlying diseases and mechanisms of action of drugs has been highlighted by several literatures (Booij et al., 2016; Godoy et al., 2013; McCracken et al., 2014; Mueller et al., 2014; Schwartz et al., 2015; Shelper et al., 2016) The ability of 3D cell models to mimic cancer tissue microenvironments, improved cell-to-cell and cell-to ECM interactions are unmatched by conventional 2D culture models with their spatial limitations (Baker and Chen, 2012; Booij et al., 2016; Justice et al., 2009; Nyga et al., 2011; Shelper et al., 2016). It is important to understand the properties and functions of ECM components for better 3D cell culture. In response to the previous discovery, we decided to use a combination of alginate and fibrin for cell encapsulation. Alginate is biologically inert and provide structural rigidity for cell encapsulation but does not interact with actin from the cells while fibrin provides necessary ECM properties to support cell growth and spheroid formation.
The study by Kobayashi et al. demonstrated the effect of spheroid sizes on the actions of drugs (Kobayashi et al., 1992). Mueller et al. compared 2D and 3D culture of HepG2 human liver cancer cells and fully differentiated HepaRG liver cells and demonstrated the effect of spheroid formation on drug toxicity (Mueller et al., 2014). In general, bigger spheroids provided protective environments to the cells, thereby reducing the toxic effect of drugs. We observed a similar trend on our chip platform. We studied the effect of spheroid sizes on compound-induced toxicity using the two pre-incubation conditions such that Hep3B cells could grow an extended period of time for bigger spheroid formation. Overall, IC50 values obtained from 72 h pre-incubation were higher than those obtained from 24 h pre-incubation, indicating that bigger spheroids are more protective against the model compounds tested. In particular, statistically significant difference in IC50 values (p<0.05) was observed for menadione and rotenone by spheroid sizes. It has been known that multicellular spheroids express cell adhesion molecules like e-cadherin and CD44 which play important role in cell survival (LI et al., 2008). Moreover, cells in 3D culture has been shown to express various ECM proteins which are known for providing environment-mediated drug resistance (EMDR) in tumors (Meads et al., 2009; Kang et al., 2016).
Surprisingly, difference in IC50 values from the other compounds were minimal. This could be presumably because the formation of bigger spheroid size in the small cell spots may not contribute to the diffusion issues and drug binding to ECM components, apart from drug resistance issues from transporter overexpression (Kovalev et al., 2013; Xia and Smith, 2012; Xiong et al., 2015).
As a control for the four HCI assays performed on 3D-cultured Hep3B cells, a PrestoBlue® cell viability assay was used for 2D-cultured Hep3B cells in a 96-well plate. The PrestoBlue® assay is an absorbance or fluorescence assay measuring biotransformation of cell permeable resazurin to resorufin by reducing agents (e.g., NADH) in living cells. The PrestoBlue® assay allowed to avoid growth media removal and cell detachment and obtain accurate dose-response data from metabolically active cells (Xu et al., 2015). Given the different mechanism of readouts and difference in cell culture, the values from the PrestoBlue® assay and the four HCI assays were significantly IC50 different, particularly for rotenone, menadione, and tamoxifen. The IC50 values from 3D-cultured Hep3B cells were several times higher in some cases, indicating again the important role of protective environments from spheroids, which might be a better mimic of liver tumors. In addition to the 2D control, rat oral LD50 values were used as another control for in vivo data. Interestingly, the toxicity profiles from the animal and 3D-cultured human cells were significantly different, particularly for lovastatin, tamoxifen, and menadione, which may represent the discrepancy between animals and humans in terms of toxicity and efficacy assessment. Moreover, the IC50 values of the model compounds obtained from the four assays were in line with the mechanism of toxicity mentioned in literature (Table 1).
The six model compounds were selected based on their wide range of mechanism of action reported in literature. Acetaminophen is a widely used medication for pain and fever and has been extensively studied due to its hepatotoxic side effects in the presence of DMEs (Chen et al., 1998). It is relatively nontoxic at low dosages but causes hepatic centrilobular necrosis at high dosages, resulting in liver failure and death (Hinson and Al, 2003; Botting, 2000). It transformed into a reactive metabolite, N-acetyl-p-benzoquinone imine (NAPQI) in the presence of cytochrome P450 (CYP450) isoforms and NADPH (Botting, 2000). If not neutralized by cellular glutathione, NAPQI can target mitochondrial proteins, which can cause loss of energy and result in cell death. Oxidative stress by the formation of superoxide and hydrogen peroxide is another mechanism of acetaminophen-medicated toxicity (James et al., 2003), leading to cell death via apoptosis in the liver (Jaeschke et al., 2002). Acetaminophen may undergo glucuronidation and sulfation via phase II DMEs including UDP-glucuronosyltransferases (UGTs) and sulfotransferases (SULTs), which can eliminate it through conjugate reactions (Hinson and Al, 2003; Chen et al., 1998). In this study, we could not observe any dose-dependent effect from acetaminophen, presumably because of lack of metabolism competency in Hep3B cells. Hep3B cells are known to express no major CYP450 isoforms (Guo et al., 2011).
Lovastatin is a lipid-lowering therapeutic drug used in the treatment of hyperlipidemia and is known to have anticancer properties. Multiple in vitro studies have shown that statins regulate cell growth, proliferation, and apoptosis via different mechanisms (Kallas-Kivi et al., 2016; Niknejad et al., 2014; Walther et al., 2016; Wei et al., 2007). Lovastatin is relatively nontoxic, but several literature has reported dose-dependent adverse effects and varying sensitivity of this compound based on cell types and culture conditions (Kallas-Kivi et al., 2016; Niknejad et al., 2014). Lovastatin is known to lower the growth rate, reduce intracellular calcium concentrations, and decrease mitochondrial membrane potential and affect cell viability (Wei et al., 2007; Walther et al., 2016). Lovastatin was significantly less toxic in our study as compared to other in vitro studies from literature, but it was significantly more toxic than the results obtained with in vivo animal studies (Table 3). This result might be due to lack of metabolic capability of Hep3B cells as CYP450 enzymes have been well known for metabolizing lovastatin (Gazzerro et al., 2012; Paoletti et al., 2002).
Rotenone is used as an insecticide and pesticide. The main mechanisms of rotenone toxicity observed in this study was correlated with those from the literatures, which are mitochondrial impairment, oxidative stress, and apoptosis (Isenberg and Klaunig, 2000; Li et al., 2003). The IC50 values of rotenone from the mitochondrial impairment assay and the intracellular glutathione level assay were particularly lower than those from the other two assays performed on 3D-cultured Hep3B cells on the chip. Thus, mitochondrial impairment and associated oxidative stress could be the main mechanisms of rotenone toxicity. Interestingly, mitochondrial impairment was shown to be the most sensitive assay among the other assays tested for rotenone, tamoxifen, and menadione. Rotenone is widely studied for its neurotoxic effect via mitochondrial damage and apoptosis induction (Ahmadi et al., 2003; Jin et al., 2007; Johnson and Bobrovskaya, 2015; Sherer et al., 2003). For example, Jin et. al. reported rotenone-induced mitochondrial damage in dopaminergic cells via Caspase-3 activation (Jin et al., 2007). Rotenone is also known to be a mitochondrial complex I inhibitor, which affects the electron transport chain in mitochondria and causes ROS generation. Accumulation of ROS inside cells decreases the glutathione level resulting in oxidative stress and causes mitochondrial impairment thereby releasing cytochrome-c and inducing apoptotic cell death (Cabezas et al., 2012; Jin et al., 2007; Li et al., 2003; Moon et al., 2005).
Tamoxifen is an anti-estrogen drug used in the treatment of breast cancer. Hepatotoxicity is a commonly reported side effect of tamoxifen (Ford et al., 2016; Henderson et al., 2016; Khuroo et al., 2014; Lin et al., 2014; Tabassum et al., 2006; Villegas et al., 2016). In our study, mitochondrial impairment and disruption of cell membrane integrity were the main mechanisms of tamoxifen-induced toxicity. Tamoxifen was less sensitive to the DNA damage assay and the intracellular glutathione level assay. According to literature, mitochondrial impairment is the main mechanism of tamoxifen-induced toxicity (Lee et al., 2000; Tabassum et al., 2006; Tolosa et al., 2015, 2012). For example, Tolosa et al. used tamoxifen as a positive control compound for mitochondrial impairment (Tolosa et al., 2015).
Similarly, mitochondrial impairment and glutathione depletion by oxidative stress were the main mechanisms of menadione toxicity, which was in line with literature. Depletion of intracellular glutathione (Cho et al., 1997) and mitochondrial impairment (Tolosa et al., 2012) by menadione have been reported to be the major mechanisms of menadione-induced toxicity in vitro (Niemczyk et al., 2004). Menadione is a vitamin K3 supplement investigated widely as an oxidative stress inducer (De Assis et al., 2015) and for its efficacy in the treatment of cancers (Niemczyk et al., 2004; Delwar et al., 2012; Akiyoshi et al., 2009). Its cytotoxic effect via generation of ROS has been reported in several studies (Castro et al., 2008; Ip et al., 2004). For example, oxidation of menadione by molecular oxygen has been shown to produce superoxide anions, leading to depletion of intracellular glutathione and then DNA damage (Fischer-Nielsen et al., 1995; Morgan et al., 1992). In addition, Morgan et al. and Akiyoshi et al. demonstrated menadione-induced toxicity in rat hepatocytes and human immortalized leukemia cell line via DNA damage, specifically single-stranded breaks mediated by hydroxyl radicals (Morgan et al., 1992; Akiyoshi et al., 2009). However, in our study DNA damage was shown to be the least sensitive mechanism of toxicity among the four mechanisms for menadione-treated Hep3B cells as seen from the Hoechst 33342 staining.
Overall, our high-throughput 3D cell culture platform was suitable for HCI assays to generate highly predictive, mechanistic toxicity of the model compounds. However, it was unable to detect the toxicity of metabolism-sensitive compounds such as acetaminophen. It is mainly because of lack of metabolism competence from Hep3B cells which do not express major DMEs necessary for bioactivation of the compounds (Guo et al., 2011; Tolosa et al., 2012). The Hep3B cell line was selected in this study as a model cell line because of its human liver origin, availability, long life span, and easy maintenance. However, inability to express major DMEs in the hepatoma cell line compared to primary hepatocytes can influence the predictivity of hepatotoxicity in vivo from our chip platform (Rodríguez-Antona et al., 2002). The results from tamoxifen, lovastatin, and menadione, all of which are mildly toxic or nontoxic in vivo, but highly toxic on our chip platform, may be attributed to lack of metabolism competence in Hep3B cells. This issue can be avoided by incorporating DME microarrays on the chip platform (Yu et al., 2017). Since the outcomes from this study were based on merely six compounds, thus a larger set of model compounds would be required to determine sensitivity and selectivity of the HCI assays tested.
Apart from the distinctive benefits offered by the microarray chip platforms for 3D cell culture and high-throughput, high-content screening, there are some limitations which needs to be considered when implementing the chip platform for specific applications. One of major challenges lies in maintaining robust spot attachment for long-term cell culture. Hydrogel spots tend to degrade and detach when cultured for more than two weeks, posing limitation in performing long-term 3D cell culture. Degradable hydrogels such as Matrigel and Collagen can be easily degraded by matrix metalloproteinases produced by hepatoma cell lines in culture, which further facilitate spot detachment. In addition, growth media in the microwell chip may evaporate during incubation due to small volume (typically 950 nL), thus requiring incubation in a high humidity chamber and replacing growth media every 2–3 days. Finally, it is unavoidable to dry cell spots after staining for image acquisition due to small cell spot volume (typically 60 nL).
5. Conclusions
We demonstrated high-throughput, HCI capability for the first time on the micropillar/microwell chip platform for the assessment of mechanistic, compound-induced toxicity. With uniform and healthy, 3D-cultured Hep3B cells on the micropillar chip and the four HCI assays, including TMRM, Hoechst 33342, mBCl, and calcein AM, we successfully assessed mechanistic toxicity such as mitochondrial impairment, DNA damage, intracellular glutathione level, and cell membrane integrity, and generated reproducible and consistent IC50 values from the chip platform. The in vitro data obtained from the chip platform was well correlated with mechanistic toxicity from other studies. There would be a larger number of model compounds necessary to further validate HCI capability on the chip and accurately calculate predictivity of mechanistic toxicity for use in pharmaceutical industries. In addition, other hepatic cell types with metabolism competence such as primary hepatocytes and HepaRG cells should be used on the chip platform to improve predictivity of metabolism-induced compound toxicity as Hep3B cells lack major DME expression. With additional HCI assays on the chip, such as genotypic and phenotypic assays, we envision that our miniaturized 3D cell culture platform can be implemented in an early stage of drug screening processes to weed out lead candidates that could be potentially toxic to humans.
Highlights.
Miniaturized cell culture in three dimension (3D) on a chip has been demonstrated
High-throughput high-content imaging assays are established for mechanistic toxicity
Robust and reproducible, 3D cell-based toxicity assays have been demonstrated
Acknowledgments
This study was supported by a R01 grant from National Institutes of Health (NIEHS R01ES025779) and institutional grants from Cleveland State University (Faculty Research Development and Faculty Innovation Fund).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadi FA, Linseman DA, Grammatopoulos TN, Jones SM, Bouchard RJ, Freed CR, Heidenreich KA, Zawada WM. The pesticide rotenone induces caspase-3-mediated apoptosis in ventral mesencephalic dopaminergic neurons. J Neurochem. 2003;87:914–921. doi: 10.1046/j.1471-4159.2003.02068.x. [DOI] [PubMed] [Google Scholar]
- Akiyoshi T, Matzno S, Sakai M, Okamura N, Matsuyama K. The potential of vitamin K3 as an anticancer agent against breast cancer that acts via the mitochondria-related apoptotic pathway. Cancer Chemother Pharmacol. 2009;65:143–150. doi: 10.1007/s00280-009-1016-7. [DOI] [PubMed] [Google Scholar]
- Alépée N, Bahinski A, Daneshian M, De Wever B, Fritsche E, Goldberg A, Hansmann J, Hartung T, Haycock J, Hogberg HT, Hoelting L, Kelm JM, Kadereit S, Mcvey E, Landsiedel R, Leist M, Lübberstedt M, Noor F, Pellevoisin C, Petersohn D, Zurich MG. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX. 2014;31:441–477. doi: 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astashkina A, Grainger DW. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv Drug Deliv Rev. 2014;69–70:1–18. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Astashkina A, Mann B, Grainger DW. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol Ther. 2012;134:82–106. doi: 10.1016/j.pharmthera.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij TH, Klop MJD, Yan K, Szántai-Kis C, Szokol B, Orfi L, Van De Water B, Keri G, Price LS. Development of a 3D tissue culture-based high-content screening platform that uses phenotypic profiling to discriminate selective inhibitors of receptor tyrosine kinases. J Biomol Screen. 2016;21:912–922. doi: 10.1177/1087057116657269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botting RM. Mechanism of action of acetaminophen : Is there a cyclooxygenase 3 ? Clin Infect Dis. 2000;31:202–210. doi: 10.1086/317520. [DOI] [PubMed] [Google Scholar]
- Breslin S, O’Driscoll L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov Today. 2013;18:240–249. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Cabezas R, El-Bachá RS, González J, Barreto GE. Mitochondrial functions in astrocytes: Neuroprotective implications from oxidative damage by rotenone. Neurosci Res. 2012;74:80–90. doi: 10.1016/j.neures.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Castro FAV, Mariani D, Panek AD, Eleutherio ECA, Pereira MD. Cytotoxicity mechanism of two naphthoquinones (menadione and plumbagin) in saccharomyces cerevisiae. PLoS One. 2008;3:1–6. doi: 10.1371/journal.pone.0003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavnar SP, Salomonsson E, Luker KE, Luker GD, Takayama S. Transfer, Imaging, and Analysis Plate for Facile Handling of 384 Hanging Drop 3D Tissue Spheroids. J Lab Autom. 2014;19:208–14. doi: 10.1177/2211068213504296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli JP, Rizvi I, Blanden AR, Massodi I, Glidden MD, Pogue BW, Hasan T. An imaging-based platform for high-content, quantitative evaluation of therapeutic response in 3D tumour models. Sci Rep. 2014;4:3751. doi: 10.1038/srep03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Koenigs LL, Thompson SJ, Peter RM, Rettie AE, Trager WF, Nelson SD. Oxidation of acetaminophen to its toxic quinone imine and nontoxic catechol metabolites by baculovirus-expressed and purified human cytochromes P450 2E1 and 2A6. Chem Res Toxicol. 1998;11:295–301. doi: 10.1021/tx9701687. [DOI] [PubMed] [Google Scholar]
- Cho Y, Kim M, Lee J, Chung J. The Role of Thiols in Protecting against Simultaneous Toxicity of Menadione to Platelet Plasma and Intracellular Membranes1. J Pharmacol Exp Ther. 1997;280:1335–1340. [PubMed] [Google Scholar]
- De Assis PM, Castro LS, Siqueira AFP, De Carvalho Delgado J, Dos Santos Hamilton TR, Goissis MD, Mendes CM, Nichi M, Visintin JA, Assumpção ME. System for evaluation of oxidative stress on in-vitro-produced bovine embryos. Reprod Biomed Online. 2015;31:577–580. doi: 10.1016/j.rbmo.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Delwar ZM, Avramidis D, Follin E, Hua Y, Siden Å, Cruz M, Paulsson KM, Yakisich JS. Cytotoxic effect of menadione and sodium orthovanadate in combination on human glioma cells. Invest New Drugs. 2012;30:1302–1310. doi: 10.1007/s10637-011-9680-y. [DOI] [PubMed] [Google Scholar]
- Di Z, Klop MJD, Rogkoti V, Le Devedec SE, van de Water B, Verbeek FJ, Price LS, Meerman JHN. Ultra High Content Image Analysis and Phenotype Profiling of 3D Cultured Micro-Tissues. PLoS One. 2014;9:1–10. doi: 10.1371/journal.pone.0109688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Nielsen A, Corcoran GB, Poulsen E, Kamendulist LM, Loft S. Menadione-Induced Dna Fragmentation Without 8-0X0-2’-Deoxyguanosine Formation in Isolated Rat Hepatocytes. Biochem Pharmacol. 1995;49:1469–1474. doi: 10.1016/0006-2952(94)00525-q. [DOI] [PubMed] [Google Scholar]
- Ford BM, Franks LN, Radominska-Pandya A, Prather PL. Tamoxifen Isomers and Metabolites Exhibit Distinct Affinity and Activity at Cannabinoid Receptors: Potential Scaffold for Drug Development. PLoS One. 2016;11:1–23. doi: 10.1371/journal.pone.0167240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzerro P, Proto MC, Gangemi G, Malfitano AM, Ciaglia E, Pisanti S, Santoro A, Laezza C, Bifulco M. Pharmacological Actions of Statins: A Critical Appraisal in the Management of Cancer. Pharmacol Rev. 2012;64:102–146. doi: 10.1124/pr.111.004994. [DOI] [PubMed] [Google Scholar]
- Gessner RC, Hanson AD, Feingold S, Cashion AT, Corcimaru A, Wu BT, Mullins CR, Aylward SR, Reid LM, Dayton PA. Functional Ultrasound Imaging for Assessment of Extracellular Matrix Scaffolds Used for Liver Organoid Formation. Biomaterials. 2013;34:9341–9351. doi: 10.1126/scisignal.274pe36.Insulin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J, Braeuning A, Budinsky Ra, Burkhardt B, Cameron NR, Camussi G, Cho CS, Choi YJ, Craig Rowlands J, Dahmen U, Damm G, Dirsch O, Donato MT, Dong J, Dooley S, Drasdo D, Eakins R, Ferreira KS, Fonsato V, Fraczek J, Gebhardt R, Gibson A, Glanemann M, Goldring CEP, Gómez-Lechón MJ, Groothuis GMM, Gustavsson L, Guyot C, Hallifax D, Hammad S, Hayward A, Häussinger D, Hellerbrand C, Hewitt P, Hoehme S, Holzhütter HG, Houston JB, Hrach J, Ito K, Jaeschke H, Keitel V, Kelm JM, Kevin Park B, Kordes C, Kullak-Ublick Ga, Lecluyse EL, Lu P, Luebke-Wheeler J, Lutz A, Maltman DJ, Matz-Soja M, McMullen P, Merfort I, Messner S, Meyer C, Mwinyi J, Naisbitt DJ, Nussler AK, Olinga P, Pampaloni F, Pi J, Pluta L, Przyborski Sa, Ramachandran A, Rogiers V, Rowe C, Schelcher C, Schmich K, Schwarz M, Singh B, Stelzer EHK, Stieger B, Stöber R, Sugiyama Y, Tetta C, Thasler WE, Vanhaecke T, Vinken M, Weiss TS, Widera A, Woods CG, Xu JJ, Yarborough KM, Hengstler JG. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunness P, Mueller D, Shevchenko V, Heinzle E, Ingelman-Sundberg M, Noor F. 3D organotypic cultures of human heparg cells: A tool for in vitro toxicity studies. Toxicol Sci. 2013;133:67–78. doi: 10.1093/toxsci/kft021. [DOI] [PubMed] [Google Scholar]
- Guo L, Dial S, Shi L, Branham W, Liu J, Fang JL, Green B, Deng H, Kaput J, Ning B. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab Dispos. 2011;39:528–538. doi: 10.1124/dmd.110.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KT, Kim JK, Lee YC, Kim CH. Inhibitory effect of Daesungki-Tang on the invasiveness potential of hepatocellular carcinoma through inhibition of matrix metalloproteinase-2 and -9 activities. Toxicol Appl Pharmacol. 2004;200:1–6. doi: 10.1016/j.taap.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Håkanson M, Cukierman E, Charnley M. Miniaturized pre-clinical cancer models as research and diagnostic tools. Adv Drug Deliv Rev. 2014;69–70:52–66. doi: 10.1016/j.addr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock J. 3D Cell Culture: A Review of Current Approaches and Techniques. Methods in Molecular Biology. 2011:243–259. doi: 10.1007/978-1-60761-984-0. [DOI] [PubMed] [Google Scholar]
- Henderson SL, Wendy TA, Kim RB. Profound reduction in tamoxifen active metabolite endoxifen in a breast cancer patient treated with rifampin prior to initiation of an anti-TNFalpha biologic for ulcerative colitis: a case report. BMC Cancer. 2016;16:1–6. doi: 10.1186/s12885-016-2342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman SR, To J, Orth AP, Cuddihy MJ, Caracino D. High-content analysis of three-dimensional tumor spheroids : investigating signaling pathways using small hairpin RNA. Nat Methods. 2013;10:v–vi. doi: 10.1038/nmeth.f.370. [DOI] [Google Scholar]
- Hsiao AY, Tung YC, Qu X, Patel LR, Pienta KJ, Takayama S. 384 hanging drop arrays give excellent Z-factors and allow versatile formation of co-culture spheroids. Biotechnol Bioeng. 2012;109:1293–304. doi: 10.1002/bit.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SP, Woo KY, Lau OW, Che CT. Hepatoprotective Effect of Sabina przewalskii against Menadione-induced Toxicity. Phyther Res. 2004;18:329–331. doi: 10.1002/ptr.1326. [DOI] [PubMed] [Google Scholar]
- Isenberg JS, Klaunig JE. Role of the mitochondrial membrane permeability transition (MPT) in rotenone-induced apoptosis in liver cells. Toxicol Sci. 2000;53:340–351. doi: 10.1093/toxsci/53.2.340. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, Gores GJ, Cederbaum AI, Hinson Ja, Pessayre D, Lemasters JJ. Mechanisms of hepatotoxicity. Toxicol Sci. 2002;65:166–176. doi: 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- Jahr W, Schmid B, Schmied C, Fahrbach FO, Huisken J. Hyperspectral light sheet microscopy. Nat Commun. 2015;6:1–7. doi: 10.1038/ncomms8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LP, Mayeux PR, Hinson JA. ACETAMINOPHEN-INDUCED HEPATOTOXICITY. Drug Metab Dispos. 2003;31:1499–1506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- Jin J, Davis J, Zhu D, Kashima DT, Leroueil M, Pan C, Montine KS, Zhang J. Identification of novel proteins affected by rotenone in mitochondria of dopaminergic cells. BMC Neurosci. 2007;8:67. doi: 10.1186/1471-2202-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ME, Bobrovskaya L. An update on the rotenone models of Parkinson’s disease: Their ability to reproduce the features of clinical disease and model gene-environment interactions. Neurotoxicology. 2015;46:101–116. doi: 10.1016/j.neuro.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Justice BA, Badr NA, Felder RA. 3D cell culture opens new dimensions in cell-based assays. Drug Discov Today. 2009;14:102–7. doi: 10.1016/j.drudis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Kallas-Kivi A, Trei A, Maimets T. Lovastatin Decreases the Expression of CD133 and Influences the Differentiation Potential of Human Embryonic Stem Cells. Stem Cells Int. 2016 doi: 10.1155/2016/1580701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lee DW, Hwang HJ, Yeon SE, Lee MY, Kuh HJ. Mini-pillar array for hydrogel-supported 3D culture and high-content histologic analysis of human tumor spheroids. Lab Chip. 2016;16:2265–2276. doi: 10.1039/C6LC00526H. [DOI] [PubMed] [Google Scholar]
- Khuroo T, Verma D, Talegaonkar S, Padhi S, Panda AK, Iqbal Z. Topotecan-tamoxifen duple PLGA polymeric nanoparticles: Investigation of in vitro, in vivo and cellular uptake potential. Int J Pharm. 2014;473:384–394. doi: 10.1016/j.ijpharm.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Takemura Y, Ohnuma T. Relationship between tumor cell density and drug concentration and the cytotoxic effects of doxorubicin or vincristine: mechanism of inoculum effects. Cancer Chemother Pharmacol. 1992;31:6–10. doi: 10.1007/BF00695987. [DOI] [PubMed] [Google Scholar]
- Kovalev AA, Tsvetaeva DA, Grudinskaja TV. Role of ABC-cassette transporters (MDR1, MRP1, BCRP) in the development of primary and acquired multiple drug resistance in patients with early and metastatic breast cancer. Exp Oncol. 2013;35:287–290. [PubMed] [Google Scholar]
- Kwon SJ, Lee DW, Shah DA, Ku B, Jeon SY, Solanki K, Ryan JD, Clark DS, Dordick JS, Lee MY. High-Throughput and Combinatorial Gene Expression on a Chip for Metabolism-Induced Toxicology Screening. Nat Commun. 2014;5:1–16. doi: 10.1037/a0013262.Open. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1053/j.gastro.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Lee DW, Choi YS, Seo YJ, Lee MY, Jeon SY, Ku B, Kim S, Yi SH, Nam DH. High-throughput screening (HTS) of anticancer drug efficacy on a micropillar/microwell chip platform. Anal Chem. 2014a doi: 10.1021/ac402546b. [DOI] [PubMed] [Google Scholar]
- Lee DW, Choi YS, Seo YJ, Lee MY, Jeon SY, Ku B, Nam DH. High-throughput, miniaturized clonogenic analysis of a limiting dilution assay on a micropillar/microwell chip with brain tumor cells. Small. 2014b;10:5098–5105. doi: 10.1002/smll.201401074. [DOI] [PubMed] [Google Scholar]
- Lee DW, Lee MY, Ku B, Yi SH, Ryu JH, Jeon R, Yang M. Application of the DataChip/MetaChip technology for the evaluation of ajoene toxicity in vitro. Arch Toxicol. 2014c;88:283–290. doi: 10.1007/s00204-013-1102-9. [DOI] [PubMed] [Google Scholar]
- Lee DW, Oh WY, Yi SH, Ku B, Lee MY, Cho YH, Yang M. Estimation of bisphenol A—Human toxicity by 3D cell culture arrays, high throughput alternatives to animal tests. Toxicol Lett. 2016;259:87–94. doi: 10.1016/j.toxlet.2016.07.711. [DOI] [PubMed] [Google Scholar]
- Lee DW, Yi SH, Jeong SH, Ku B, Kim J, Lee MY. Plastic pillar inserts for three-dimensional (3D) cell cultures in 96-well plates. Sensors Actuators B Chem. 2013;177:78–85. doi: 10.1016/j.snb.2012.10.129. [DOI] [Google Scholar]
- Lee YS, Kang YS, Lee SH, Kim JA. Role of NAD(P)H oxidase in the tamoxifen-induced generation of reactive oxygen species and apoptosis in HepG2 human hepatoblastoma cells. Cell Death Differ. 2000;7:925–932. doi: 10.1038/sj.cdd.4400717. [DOI] [PubMed] [Google Scholar]
- LI CL, TIAN T, NAN KJ, ZHAO N, GUO YH, CUI J, WANG J, ZHANG WG. Survival advantages of multicellular spheroids vs. monolayers of HepG2 cells in vitro. Oncol Rep. 2008;20:1465–1471. doi: 10.3892/or. [DOI] [PubMed] [Google Scholar]
- Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- Lin Y, Liu J, Zhang X, Li L, Hu R, Liu J, Deng Y, Chen D, Zhao Y, Sun S, Ma R, Zhao Y, Liu J, Zhang Y, Wang X, Li Y, He P, Li E, Xu Z, Wu Y, Tong Z, Wang X, Huang T, Liang Z, Wang S, Su F, Lu Y, Zhang H, Feng G, Wang S. A prospective, randomized study on hepatotoxicity of anastrozole compared with tamoxifen in women with breast cancer. Cancer Sci. 2014;105:1182–1188. doi: 10.1111/cas.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Amenta PS. The hepatic extracellular matrix. I Components and distribution in normal liver. Virchows Arch A Pathol Anat. 1993;423:1–11. doi: 10.1007/BF01606425. [DOI] [PubMed] [Google Scholar]
- Mason SD, Joyce Ja. Proteolytic Networks in Cancer. Trends Cell Biol. 2011;21:1–18. doi: 10.1016/j.tcb.2010.12.002.Proteolytic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665–674. doi: 10.1038/leusup.2012.5. [DOI] [PubMed] [Google Scholar]
- Montanez-Sauri SI, Beebe DJ, Sung KE. Microscale screening systems for 3D cellular microenvironments: platforms, advances, and challenges. Cell Mol Life Sci. 2015;72:237–249. doi: 10.1007/s00018-014-1738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y, Lee KH, Park JH, Geum D, Kim K. Mitochondrial membrane depolarization and the selective death of dopaminergic neurons by rotenone: Protective effect of coenzyme Q10. J Neurochem. 2005;93:1199–1208. doi: 10.1111/j.1471-4159.2005.03112.x. [DOI] [PubMed] [Google Scholar]
- Morgan Wa, Hartley Ja, Cohen GM. Quinone-induced DNA single strand breaks in rat hepatocytes and human chronic myelogenous leukaemic K562 cells. Biochem Pharmacol. 1992;44:215–221. doi: 10.1016/0006-2952(92)90003-2. doi: http://dx.doi.org/10.1016/0006-2952(92)90003-2. [DOI] [PubMed] [Google Scholar]
- Mueller D, Krämer L, Hoffmann E, Klein S, Noor F. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol Vitr. 2014;28:104–112. doi: 10.1016/j.tiv.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Niemczyk E, Majczak A, Hallmann A, Kedzior J, Wozniak M, Wakabayashi T. A possible involvement of plasma membrane NAD(P)H oxidase in the switch mechanism of the cell death mode from apoptosis to necrosis in menadione-induced cell injury. Acta Biochim Pol. 2004;51:1015–1022. [PubMed] [Google Scholar]
- Nierode GJ, Perea BC, McFarland SK, Pascoal JF, Clark DS, Schaffer DV, Dordick JS. High-Throughput Toxicity and Phenotypic Screening of 3D Human Neural Progenitor Cell Cultures on a Microarray Chip Platform. Stem Cell Reports. 2016;7:970–982. doi: 10.1016/j.stemcr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknejad N, Gorn-Hondermann I, Ma L, Zahr S, Johnson-Obeseki S, Corsten M, Dimitroulakos J. Lovastatin-induced apoptosis is mediated by activating transcription factor 3 and enhanced in combination with salubrinal. Int J Cancer. 2014;134:268–279. doi: 10.1002/ijc.28369. [DOI] [PubMed] [Google Scholar]
- Nyga A, Cheema U, Loizidou M. 3D tumour models: Novel in vitro approaches to cancer studies. J Cell Commun Signal. 2011;5:239–248. doi: 10.1007/s12079-011-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page H, Flood P, Reynaud EG. Three-dimensional tissue cultures: current trends and beyond. Cell Tissue Res. 2013;352:123–31. doi: 10.1007/s00441-012-1441-5. [DOI] [PubMed] [Google Scholar]
- Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Paoletti R, Corsini A, Bellosta S. Pharmacological interactions of statins. Atheroscler Suppl. 2002;3:35–40. doi: 10.1016/S1567-5688(02)00002-8. [DOI] [PubMed] [Google Scholar]
- Park K, Williams DP, Naisbitt DJ, Kitteringham NR, Pirmohamed M. Investigation of toxic metabolites during drug development. Toxicol Appl Pharmacol. 2005;207:S425–434. doi: 10.1016/j.taap.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9:203–14. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- Reid BG, Jerjian T, Patel P, Zhou Q, Yoo BH, Kabos P, Sartorius Ca, Labarbera DV. Live multicellular tumor spheroid models for high-content imaging and screening in cancer drug discovery. Curr Chem genomics Transl Med. 2014;8:27–35. doi: 10.2174/2213988501408010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Peychl J, Huisken J, Tomancak P. Guide to light-sheet microscopy for adventurous biologists. Nat Methods. 2015;12:30–34. doi: 10.1038/nmeth.3222. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, Gómez-Lechón MJ. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- Sagawa T, Yamada Y, Takahashi M, Sato Y, Kobune M, Takimoto R, Fukaura J, Iyama S, Sato T, Miyanishi K, Matsunaga T, Takayama T, Kato J, Sasaki K, Hamada H, Niitsu Y. Treatment of hepatocellular carcinoma by AdAFPep/rep, AdAFPep/p53, and 5-fluorouracil in mice. Hepatology. 2008;48:828–840. doi: 10.1002/hep.22420. [DOI] [PubMed] [Google Scholar]
- Scherf N, Huisken J. The smart and gentle microscope. Nat Biotechnol. 2015;33:815–818. doi: 10.1038/nbt.3310. [DOI] [PubMed] [Google Scholar]
- Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Costa VS, Jiang P, Nguyen BK, Bolin JM, Daly W, Wang Y, Stewart R, Page CD, Murphy WL, Thomson JA. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci U S A. 2015;112:12516–21. doi: 10.1073/pnas.1516645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelper TB, Lovitt CJ, Avery VM. Assessing Drug Efficacy in a Miniaturized Pancreatic Cancer In Vitro 3D Cell Culture Model. Assay Drug Dev Technol. 2016;14:367–80. doi: 10.1089/adt.2016.737. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, Miller GW, Yagi T, Matsuno-Yagi A, Greenamyre JT. Mechanism of toxicity in rotenone models of Parkinson’s disease. J Neurosci. 2003;23:10756–10764. doi: 10.1523/JNEUROSCI.23-34-10756.2003. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O, Hancock MK, Hesley J, Hong D, Cohen A, Gentry J, Carlson CB, Mann DA. Phenotypic Characterization of Toxic Compound Effects on Liver Spheroids Derived from iPSC Using Confocal Imaging and Three-Dimensional Image Analysis. Assay Drug Dev Technol. 2016;14:1–14. doi: 10.1089/adt.2016.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum H, Rehman H, Banerjee BD, Raisuddin S, Parvez S. Attenuation of tamoxifen-induced hepatotoxicity by taurine in mice. Clin Chim Acta. 2006;370:129–136. doi: 10.1016/j.cca.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–9. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Tolosa L, Carmona A, Castell JV, Gómez-Lechón MJ, Donato MT. High-content screening of drug-induced mitochondrial impairment in hepatic cells: effects of statins. Arch Toxicol. 2015;89:1847–1860. doi: 10.1007/s00204-014-1334-3. [DOI] [PubMed] [Google Scholar]
- Tolosa L, Pinto S, Donato MT, Lahoz A, Castell JV, O’Connor JE, Gomez-Lechón MJ, Castell V, Connor JEO, Jose M. Development of a Multiparametric Cell-based Protocol to Screen and Classify the Hepatotoxicity Potential of Drugs. Toxicol Sci. 2012;127:187–198. doi: 10.1093/toxsci/kfs083. [DOI] [PubMed] [Google Scholar]
- Villegas VE, Rondón-Lagos M, Annaratone L, Castellano I, Grismaldo A, Sapino A, Zaphiropoulos PG. Tamoxifen treatment of breast cancer cells: Impact on Hedgehog/GLI1 signaling. Int J Mol Sci. 2016;17:1–12. doi: 10.3390/ijms17030308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet E, Danesian M, Beilmann M, Davies A, Fava E, Fleck R, Julé Y, Kansy M, Kustermann S, Macko P, Roth A, Sachse C, Shah I, Uteng M, Van De B. Current Approaches and Future Role of High Content Imaging in Safety Sciences and Drug Discovery. ALTEX. 2014;31:479–493. doi: 10.14573/altex.1405271. [DOI] [PubMed] [Google Scholar]
- Walther U, Emmrich K, Ramer R, Mittag N, Hinz B. Lovastatin lactone elicits human lung cancer cell apoptosis via a COX-2/PPARgamma-dependent pathway. Oncotarget. 2016;7:10345–10362. doi: 10.18632/oncotarget.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Shindoh H, Inoue T, Horii I. Advantages of in vitro cytotoxicity testing by using primary rat hepatocytes in comparison with established cell lines. J Toxicol Sci. 2002;27:229–237. doi: 10.2131/jts.27.229. [DOI] [PubMed] [Google Scholar]
- Wei N, Mi MT, Zhou Y. Influences of lovastatin on membrane ion flow and intracellular signaling in breast cancer cells. Cell Mol Biol Lett. 2007;12:1–15. doi: 10.2478/s11658-006-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RG. Cellular Sources of Extracellular Matrix in Hepatic Fibrosis. Clin Liver Dis. 2008;12:1–18. doi: 10.1016/j.cld.2008.07.008.Cellular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng CJ, Chou CP, Ho CT, Yen GC. Molecular mechanism inhibiting human hepatocarcinoma cell invasion by 6-shogaol and 6-gingerol. Mol Nutr Food Res. 2012;56:1304–1314. doi: 10.1002/mnfr.201200173. [DOI] [PubMed] [Google Scholar]
- Wenzel C, Riefke B, Gründemann S, Krebs A, Christian S, Prinz F, Osterland M, Golfier S, Räse S, Ansari N, Esner M, Bickle M, Pampaloni F, Mattheyer C, Stelzer EH, Parczyk K, Prechtl S, Steigemann P. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323:131–143. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Woods JA, Young AJ, Gilmore IT, Morris A, Bilton RF. Measurement of menadione-mediated DNA damage in human lymphocytes using the comet assay. Free Radic Res. 1997;26:113–124. doi: 10.3109/10715769709097790. [DOI] [PubMed] [Google Scholar]
- Xia CQ, Smith PG. Drug efflux transporters and multidrug resistance in acute leukemia: therapeutic impact and novel approaches to mediation. Mol Pharmacol. 2012;82:1008–21. doi: 10.1124/mol.112.079129. [DOI] [PubMed] [Google Scholar]
- Xiong J, Mao D, Liu L. Research Progress on the Role of ABC Transporters in the Drug Resistance Mechanism of Intractable Epilepsy. Biomed Res Int. 2015;2015:1–10. doi: 10.1155/2015/194541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, McCanna DJ, Sivak JG. Use of the viability reagent PrestoBlue in comparison with alamarBlue and MTT to assess the viability of human corneal epithelial cells. J Pharmacol Toxicol Methods. 2015;71:1–7. doi: 10.1016/j.vascn.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Yu KN, Nadanaciva S, Rana P, Lee DW, Ku B, Roth AD, Dordick JS, Will Y, Lee MY. Prediction of metabolism-induced hepatotoxicity on three-dimensional hepatic cell culture and enzyme microarrays. Arch Toxicol. 2017:1–16. doi: 10.1007/s00204-017-2126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Joshi P, Lee DW, Lee M-Y. High-Content Image Analysis. In: Lee M-Y, editor. Microarray Bioprinting Technology: Fundamentals and Practises. Springer International Publishing; 2016. pp. 143–160. [DOI] [Google Scholar]
- Zhao N, Dong Q, Qian C, Li S, Wu QF, Ding D, Li J, Wang BB, Guo KF, Xie JJ, Cheng X, Liao YH, Du YM. Lovastatin blocks Kv1.3 channel in human T cells: a new mechanism to explain its immunomodulatory properties. Sci Rep. 2015;5:1–14. doi: 10.1038/srep17381. [DOI] [PMC free article] [PubMed] [Google Scholar]