Abstract
A major part of the genome is known to be transcribed into non-protein coding RNAs (ncRNAs), such as microRNA and long non-coding RNA (lncRNA). The importance of ncRNAs is being increasingly recognized in physiological and pathological processes. lncRNAs are a novel class of ncRNAs that do not code for proteins and are important regulators of gene expression. In the past, these molecules were thought to be transcriptional “noise” with low levels of evolutionary conservation. However, recent studies provide strong evidence indicating that lncRNAs are (i) regulated during various cellular processes, (ii) exhibit cell type-specific expression, (iii) localize to specific organelles, and (iv) associated with human diseases. Emerging evidence indicates an aberrant expression of lncRNAs in diabetes and diabetes-related microvascular complications. In the present review, we discuss the current state of knowledge of lncRNAs, their genesis from genome, and the mechanism of action of individual lncRNAs in the pathogenesis of microvascular complications of diabetes and therapeutic approaches.
Keywords: long non-coding RNA, diabetes complications, diabetic retinopathy, diabetic nephropathy, diabetic cardiomyopathy, diabetic neuropathy
Introduction
Diabetes mellitus is a metabolic disorder resulting from a deficiency or failure to maintain normal glucose homeostasis (1). Diabetes is known to be associated with an increased risk of cardiovascular, renal, and cerebrovascular diseases resulting in high morbidity and mortality (2). The deleterious effects of diabetes include both microvascular (diabetes induced nephropathy, cardiomyopathy, neuropathy, and retinopathy) and macrovascular (CAD, peripheral arterial disease, and stroke) complications (1). Diabetes-induced microvascular complications are a consequence of deregulated expression of genes of several molecular pathways involved in their pathogenesis (3).
Long non-coding RNAs (lncRNAs), which are more than ≥200 nucleotides long, are a novel class of functional RNAs that do not code for any proteins. However, several ribosome-associated annotated lncRNAs contain a coding region that translates a peptide (4). lncRNAs regulate expression of several genes by binding to specific DNA/RNA or protein moieties (5). Unlike other small non-protein coding RNAs (ncRNAs), lncRNAs are not well conserved and function through diverse mechanisms (6). They may act (i) as scaffolds to bring protein complexes together, (ii) as sponges for microRNAs, (iii) as host genes for microRNAs, (iv) as controllers of mRNA decay, DNA sequestration of transcription factors, and epigenetic regulation of chromatin compaction, and (v) as stabilizers of mRNA through miRNA binding site masking (6–8).
Long non-coding RNAs have been implicated in a wide range of physiological processes and in the pathophysiology of several diseases, including diabetes and diabetes-induced microvascular complications (9–11). This review discusses the current state of knowledge of lncRNAs, their genesis from the genome, and the mechanism of action of individual lncRNAs in the pathogenesis of microvascular complications of diabetes and their potential as new therapeutics in diabetes-induced microvascular complications.
lncRNAs and Biogenesis
Nearly 2% of the genome is transcribed into protein coding RNAs and remaining 70–90% is transcribed into ncRNAs (12). Non-coding RNAs such as tRNAs, rRNAs, and spliceosomal RNA are critical components of many cellular machines (13). Apart from these classical ncRNAs, microRNAs and lncRNAs are being recognized as important regulators of gene expression. For a long time, lncRNAs were presumed to be transcriptional “noise” lacking protein coding potentials and with low levels of evolutionary conservation (14). However, in the past decade, strong evidence has emerged indicating that lncRNAs are (i) regulated during various cellular processes, (ii) exhibit cell type-specific expression, (iii) localize to specific organelles, and (iv) associated with human diseases (13). Current advanced transcriptomic technologies such as RNAseq, transcriptomics, and next-generation sequencing have led to the identification of several novel lncRNAs transcripts which seem to be evolved from intronic regions of genes (15). These non-protein coding gene loci transcribe upto 50,000 lncRNAs in humans and are poorly conserved in other species. There is uncertainty about how many of these lncRNAs are functional (16). Based on their genomic location, lncRNAs have been classified into five subclasses: intergenic lncRNAs, transcription start site-associated RNAs, intronic lncRNAs, natural antisense transcripts (NATs), and some transcribed pseudogenes (17) (Table 1).
Table 1.
Classification of long non-coding RNAs (lncRNAs) based on genomic location.
| lncRNA | Genomic context | Examples |
|---|---|---|
| Intergenic lncRNAs | Transcribed inter genetically from both strands by RNA Pol II | Xist, H19, HOTAIR, MALAT1 |
| Intronic lncRNAs | Originates from introns of protein coding genes | COLDAIR, PPP3CB, MAP3K14 and DAPK1 loci |
| Transcription start site-associated lncRNAs | Transcribed from promoter upstream region and enhancer region by RNA pol II | Associated with EXT1 and RBM39 genes, associated with RNAF12 and CCDC52 genes |
| Natural antisense transcripts | Transcribed from the opponent strand of either protein coding or non-protein coding genes | Xist/Tsix, Kcnq1/Kcnq1ot1, Igf2r/Air |
| Pseudogene lncRNAs | Residues of their parental genes that lost encoding function due to mutations | Xist evolved from protein-coding gene Lnx3 |
Intergenic lncRNAs
These are large intervening distinct ncRNAs or lincRNAs, which are found in sequence spaces and do not overlap protein-coding genes. Most of the intergenic lncRNAs, characterized till now, are found to be transcribed by RNA polymerase II and give rise to splice, poly A containing lncRNAs and are of average length of approximately 1,000 bp. Examples are Xist, H19, HOTAIR, and MALAT1 (15).
Transcription Start Site-Associated lncRNAs
The short-lived medium-length lncRNAs (200–2,000 nt) are transcribed from promoter upstream region and enhancer region by RNA pol II (18). These short transcripts (promoter-associated transcripts), i.e., PROMPTs are processed from the promoter upstream transcripts (15). These transcripts have protein coding like features such as 5′ capping and poly A tailing and occurs in low copy number and easily get degraded by exosomes (15). The function of promoter-associated transcripts is not known. It is not clear whether they have some regulatory function or these are simply a transcriptional by-product (18). The short transcripts (eRNAs) of average length less than 2 kb are the bidirectional transcripts. However, their processivity and biological function are also not known (15).
Intronic lncRNAs
These consist of small ncRNAs, known to originate from introns of protein-coding genes. Recent studies show that lncRNAs are also encoded within the introns of annotated genes (19). For example, lncRNAs COLDAIR has been shown to be involved in plant vernalization and located in the first intron of FLC; a flowering repressor locus (15, 20). These lncRNAs have been shown to have a role in various cellular and pathological processes. For example, many of these lncRNAs have been found to be differentially expressed in cancer (15).
Natural Antisense Transcripts
These lncRNAs get transcribed from the opponent strand of either protein coding or non-protein coding genes, in the genome and modulates the function of sense strand of the gene, and/or several other genes (21). These, too, have features like protein coding RNAs, such as splicing, 5′ capping, and polyadenylation and were originally discovered in bacteria (15). The well known examples of NATs are (i) Xist/Tsix which control X chromosome inactivation (22) and (ii) Kcnq1/Kcnq1ot1 and Igf2r/Air which are involved in imprinting (15, 23, 24).
Pseudogene lncRNAs
Pseudogenes are residues of their parental genes that have lost their encoding function due to nonsense, frameshift, and other mutations (25). Some pseudogenes are transcribed into lncRNAs of more than 200 nt in length and have high levels of sequence conservation (26). Two types of pseudogenes are known: (a) expressed pseudogenes which are intermediates on their way to complete pseudonization and (b) dead pseudogenes that have adopted new mutations (15). An example of pseudogene is the Xist lncRNAs which have evolved from the protein-coding gene Lnx3, by pseudogenization (27).
Diabetes-Related lncRNA
Diabetes mellitus includes (i) type 1 diabetes mellitus (T1DM), (ii) type 2 diabetes mellitus (T2DM), (iii) gestational diabetes mellitus, and (iv) other specific types of diabetes mellitus (28). Type I diabetes is also known as insulin-dependent diabetes and occurs commonly in childhood and early adulthood. Although the exact etiology of T1DM is not known, it is considered to be primarily due to genetic factors and involves immuno-destruction of insulin producing pancreatic B cells. T2DM is adult onset, non-insulin-dependent DM and is the most common type of diabetes, exceeding 90% of all cases of diabetes and is attributed to environmental and genetic factors (28). lncRNAs have recently gained widespread attention in a variety of human diseases including diabetes. Aberrant expression of several lncRNAs has been observed in both T1DM and T2DM. Some of the altered lncRNAs were found to be common in both. Table 2 shows the list of deregulated lncRNAs and their functions in different types of DM.
Table 2.
Long non-coding RNA (lncRNA) associated with various types of diabetes.
| lncRNAs | Findings | Diabetes type | Reference |
|---|---|---|---|
| ERBB3 locus-associated lncRNA (NONHSAG011351) | Prevent β-cell destruction | Type 1 diabetes | (69) |
| Plasmacytoma variant translocation 1 | Associated with end stage renal disease attributed to type 1 diabetes mellitus (T1DM) | Type 1 diabetes | (70) |
| Maternally expressed 3 gene | Alters susceptibility to T1DM | Type 1 diabetes | (71, 72) |
| Regulates β cell identity and function via insulin production and apoptosis in mouse MIN6 cells and isolated mouse islets | |||
| Associated with impaired glucose tolerance, and insulin synthesis and secretion | Type 2 diabetes | (73) | |
| HI-LNC25 | Positively regulates GLIS3 (which contains both T1D and T2D risk variants) in EndoC-βH1 human β cell line | Type 1 diabetes | (74) |
| β-cell long intergenic non-coding RNA (βlinc1) | Regulates β cell identity and function in mouse MIN6 cells and EndoC-βH1 human β cell line; also regulates its neighboring gene NKX2.2 (an islet transcription factor) | Type 1 diabetes | (75) |
| Associated with β-cell loss | Type 2 diabetes | (75) | |
| TUNAR (HI-LNC78) | Knockdown of TUNAR leads to impaired glucose-stimulated insulin secretion in human islets | Type 1 diabetes | (76) |
| PLUT (HI-LNC71) | Regulates transcription of PDX1, a key pancreatic β cell transcriptional regulator, in EndoC-βH1 cells, primary islet cells, mouse β cell line MIN6 | Type 1 diabetes | (76) |
| MALAT1 | Upregulation of MALAT1 is associated with microvascular dysfunction (diabetic retinopathy) in STZ-induced diabetic rats and db/db mice | Type 1 diabetes | (53) |
| TUG1 | Downregulation of lncRNA TUG1 expression increased apoptosis and reduced insulin secretion in mouse β cells | Type 1 diabetes | (77) |
| NONRATT021972 | Exacerbated neuropathic pain via TNF-α related pathways | Type 2 diabetes | (78) |
| ENST00000550337.1 | Biomarker | Pre-diabetes and Type 2 diabetes | (79) |
| lncRNA-ROR | Maintenance of human amniotic epithelial stem cell pluripotency and β islet-like cell differentiation | Type 2 diabetes | (80) |
| Antisense non-coding RNA (ANRIL) | Affect β-cell mass | Type 2 diabetes | (81) |
| E330013P06 (E33) | Promotes macrophage inflammation | Type 2 diabetes | (82) |
| Imprinted maternally expressed transcript (H19) | Associated with increased birth weight; higher expression in T2D patients | Type 2 diabetes | (83) |
| HI-LNC901 | Implicated in islet function | Type 2 diabetes | (84) |
| Nuclear paraspeckle assembly transcript 1 | Regulates mTOR signaling pathway | Type 2 diabetes | (85) |
| PDX1-associated lncRNA, upregulator of transcription (PLUTO) | Regulates PDX1 expression | Type 2 diabetes | (76) |
lncRNA and Microvascular Complications of Diabetes
Recent studies have shown dysregulation of several lncRNAs in diabetic nephropathy (DN), retinopathy, neuropathy, and cardiomyopathy suggesting their potential role in the pathophysiology and hence a potential therapeutic target of diabetes-induced microvascular complications (19).
lncRNAs and Diabetic Nephropathy
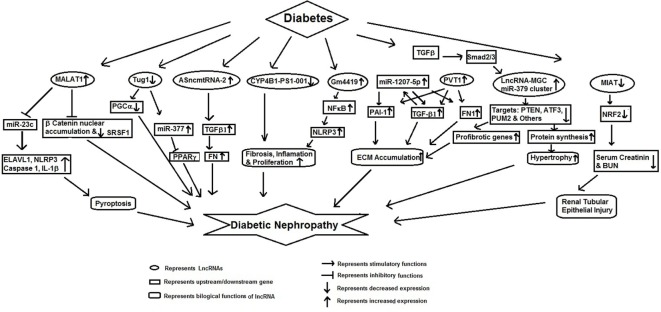
Several lncRNAs have been shown to regulate epigenetic changes and associated metabolic memory in DN (Figure 1). It has been suggested that DN may induce differential expression of lncRNAs in renal tissues leading to deregulation of multiple molecular pathways involved in its pathophysiology. For example, Chen et al. showed differential expression of 311 altered lncRNAs in a mouse model of DN as compared to the db/m control mouse (29). These lncRNAs were found to target several different pathways enriched in glutathione metabolism signaling, indicating lncRNAs may deregulate expression of several DN related pathways. In another study, Wang et al. have also found differential expression of 1,018 lncRNAs in the kidney tissues of db/db mice having DN. Among these differentially expressed lncRNAs, CYP4B1-PS1-001 was significantly downregulated. The overexpression of CYP4B1-PS1-001 suppressed proliferation and fibrosis of mesangial cells (MCs). All these findings suggest the pivotal role of CYP4B1-PS1-001 in the proliferation and fibrosis of mice MCs (30).
Figure 1.
Schematic model showing functional role of long non-coding RNAs in diabetic nephropathy.
Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is expressed in a variety of cells. An increased expression of MALAT1 has been reported in diabetic rat (31) and STZ-induced mouse model of DN (32). Li et al. observed an increased renal MALAT1 expression in STZ-induced diabetic rats and in hyperglycemia (HG) exposed HK-2 cells. The increased MALAT1 expression was associated with an increased pyroptosis of the renal epithelial cells and decreased expression of miR-23c. Further downregulation of MALAT1 resulted in the decreased expression of ELAVL1, NLRP3, Caspase-1 and the pro-inflammatory cytokine IL-1β, and upregulated miR-23c expression and inhibited pyroptosis. These results suggested that upregulation of MALAT1 was promoting renal cell apoptosis and DN by targeting miR-23c and its downstream target EVAL1. The role of MALAT1 in DN was further confirmed by Hu et al. who showed increased renal cortical MALAT1 levels in diabetic mice. Mouse podocytes showed an initial increase in MALAT1 followed by a decline. The decrease in MALAT1 was followed by translocation of β-catenin to the nucleus and increased expression of MALAT1 RNA-binding protein, serine/arginine splicing factor 1 (SRSF1) (32). Inhibition of MALAT1 corrected the podocyte damage via suppressing an MALAT1 lncRNA-binding protein (SRSF1). MALAT1 levels were found to be under the regulation of β-catenin and knockdown of β-catenin led to decreased MALAT1 levels (32). These results provide evidence for a potential role of MALAT1 in DN.
Antisense mitochondrial non-coding RNA-2 (ASncmtRNA-2) has been also shown to be upregulated in the animal model of DN and in high glucose treated MCs human renal mesangial cells. Gao et al. showed that the expression of ASncmtRNA-2 could be suppressed by nitric oxide synthase inhibitor, NG-nitro-l-arginine methylester (L-NAME). Furthermore, ASncmtRNA-2 upregulation was accompanied by increase in TGFβ1 and its downstream gene, fibronectin. These authors have proposed that ROS mediated upregulation of ASncmtRNA-2 promotes DN through increased transcription of pro-fibrotic factors (33).
Yi et al. reported differential expression of 14 lncRNAs in renal tissues from db/db diabetic mice with nephropathy and in MCs in response to HG (34). Among these, LincRNA-Gm4419 showed NF-κB as its target, suggesting its role in renal fibrosis. Knockdown of Gm4419 inhibited the expression of pro-inflammatory cytokines, biomarkers of renal fibrosis, and reduced cell proliferation in MCs, suggesting that lincRNA-Gm4419 promoted MC inflammation, fibrosis and proliferation hyperglycemic condition through NF-κB/NLRP3 inflammasome pathway (34).
Long non-coding RNA taurine-upregulated gene 1 (Tug1) regulates PPARγ coactivator α (PGC-1α, encoded by Ppargc1a) and was shown to be differentially expressed in glomeruli from diabetic milieu (35). Overexpression of this lncRNA was associated with marked improvement in biochemical and histological features of diabetic mice along with elevated expression of PGC-1α and its downstream target genes, suggesting that Tug1 acts by targeting mitochondrial bioenergetics in the podocyte cells of diabetic mice (35). Duan et al. have also recently shown that Tug1 ameliorated ECM accumulation through microRNA-377, targeting PPARγ in DN (36).
One of the major characteristic of DN is the excessive accumulation of ECM in the kidney glomeruli. Alvarez and DiStefano reported that lncRNA plasmacytoma variant translocation 1 (PVT1) was mediating ECM accumulation in glomeruli in DN (37). These authors showed that PVT1 regulated the expression of ECM-related proteins (TGF-β1, PAI-1, and FN1). Their findings suggest that PVT1 contributes to the progression of DN by regulating ECM expression (37). Alvarez et al. reported that miR-1207-5p is derived from PVT1 lncRNA and was upregulated by high glucose and TGFβ1 in kidney cells. Further they showed that like PVT1, miR-1207-5p also increased the expression of TGF-β1, PAI-1, and FN1 independently of PVT1. This indicates that glucose and TGFβ1 regulates miR-1207-5p expression but in an independent manner of its host gene PVT1. Taken together, these results show that miR-1207-5p and its host gene play an important role in the pathogenesis of DN (38).
Genotype loci rs13447075 and rs2648862 have been reported to strongly associated with diabetes-induced end stage renal disease (ESRD) (39). rs13447075 is situated in the coding portion of one of the transcript variants of PVT1, indicating an association of PVT1 in mediating susceptibility to ESRD attributable to diabetes (39).
Wang et al. observed decreased renal cortical expression of lncRNA ENSMUST00000147869 in db/db and db/m mice which was associated with Cyp4a12a levels (40). Overexpression of this lncRNA in mouse MCs rectified the fibroblast proliferation and fibrosis indices, suggesting a role of ENSMUST00000147869 in MC proliferation and fibrosis and as a potential biomarker of DN (40). Kato et al. have reported an increased expression of 40 microRNAs and their host lncRNA transcript (lnc-MGC) in glomeruli and in MC’s exposed to TGFβ1 or high glucose. Furthermore, a decreased expression of cluster microRNAs and lnc-MGC was seen in diabetic Chop−/− mice, which conferred protection from DN. Knockdown of lnc-MGC inhibited the expression of cluster microRNAs, and also decreases ECM and hypertrophy in diabetic mice (41). They proposed that lnc-MGC could be used as a therapeutic target for controlling the progression of DN. Zhou et al. have examined the role of lncRNA-myocardial infarction-associated transcript (MIAT) in diabetes-induced renal tubular injury. They found decreased expression of both MIAT and Nrf2 in diabetes-induced renal tubule, as well as in HG exposed HK-2 cells, which inversely correlated with serum creatinine and BUN. Overexpression of MIAT reversed inhibitory action of high glucose induced Nrf2 expression, indicates that MIAT/Nrf2 axis acts as an important signaling pathway for HG induced renal tubular epithelial injury (42).
lncRNAs and Diabetic Retinopathy (DR)
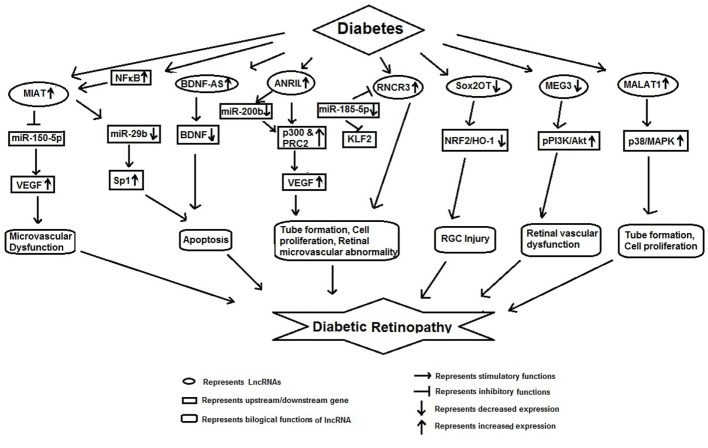
Diabetic retinopathy is a major complication of diabetes and leads to vision loss globally (43). Recent studies show that lncRNAs may be involved in its pathophysiology thus adding another dimension to its therapeutic targeting (Figure 2).
Figure 2.
Schematic model showing functional role of long non-coding RNAs in diabetic retinopathy.
Yan et al. demonstrated that high glucose augmented MIAT expression in diabetic retinas and endothelial cells. Silencing MIAT improved diabetes-induced retinal microvascular dysfunction in vivo. Similarly, it inhibited endothelial cell proliferation, migration, and tube formation in vitro. Further MIAT was found to be a competing endogenous RNA, which formed a feedback loop with VEGF and miR-150-5p to regulate endothelial cell function (44).
Apoptosis is a prominent feature of DR. Recently Zhang et al. showed that MIAT mediates the cellular apoptosis regulatory network. These authors observed an increased expression of MIAT and NF-κB (p-p65) in DR. They observed that NF-κB directly binds with MIAT, and treatment of rCM-1 cells with Bay11-7082 inhibited binding between NF-κB and MIAT, suggesting that Bay11-7082 acted as an inhibitor for NF-κB which suppressed the MIAT binding activity. Moreover, MIAT was shown to be regulated by miR-29b. Furthermore, MIAT inhibition significantly reversed the high glucose induced low expression of miR-29b, high expression of Sp1 (target of miR-29b) and apoptosis. These findings suggest that MIAT regulates cell apoptosis via its association with NF-κB (p-p65), and miR-29b (45).
Li et al. have recently reported that lncRNAs, brain-derived neurotrophic factor antisense (BDNF-AS) was upregulated whereas BDNF was markedly downregulated in human RPE cell lines in response to HG. Inhibition of BDNF-AS ameliorated d-glucose induced apoptosis and upregulated BDNF in ARPE-19 cells, suggesting that BDNF-AS, plays a critical role in the process of glucose induced apoptosis in DR through inverse regulation of BDNF (46).
Hyperglycemia increases the expression of lncRNA ANRIL in the retina and in retinal endothelial cells (47). This increased expression of ANRIL is responsible for altered expression and function of VEGF. In glucose-exposed ECs and in the retinal tissue of diabetic animals, ANRIL regulated glucose-mediated upregulation of VEGF through its interaction with p300 and PRC2 components. Inhibition of ANRIL in HRECs and in ANRILKO mice, significantly reduced VEGF EZH2 and p300 mRNA expression (47). These results suggest potentially new targeted method to prevent DR using an RNA-based approach in DR.
Like apoptosis, retinal microvascular abnormality is also an important pathological feature of DR (48). Shan et al. have shown HG-induced RNCR3 upregulation in vivo and in vitro. They further demonstrated that endothelial cell function was regulated through RNCR3/KLF2/miR-185-5p regulatory network. Knockdown of RNCR3 decreased cell proliferation, migration, and tube formation in vitro and was shown to alleviate vascular dysfunction in retina. Similarly, Liu et al. have also shown that knockdown of RNCR3 significantly inhibited retinal reactive gliosis. Collectively, these observations suggest that RNCR3 knockdown may be a potential therapeutic molecule for the prevention of diabetes-induced retinal microvascular abnormalities (48, 49).
Retinal ganglion cell (RGC) injury is also one of the distinguished pathological features of DR (50). Li et al. reported decreased expression lncRNA Sox2OT in the retinas of STZ-induced diabetic mice as well as RGCs in the presence of high glucose or oxidative stress. Inhibition of Sox2OT was found to protect high glucose-induced RGCs in vitro and appeared to play a neuroprotective role in diabetes-related retinal neurodegeneration in vivo. Sox2OT knockdown also regulated NRF2/HO-1 signaling activity, indicating that it also had anti-oxidant function. Thus, Sox2OT knockdown may be a potential treatment option for diabetes-induced retinal neurodegeneration (50).
Similarly, high glucose and oxidative stress were found to downregulate lncRNA-MEG3 in the retinas of STZ-induced diabetic mice and endothelial cells. Inhibition of MEG3 aggravated retinal vessel dysfunction in vivo, and also regulated retinal endothelial cell proliferation, migration, and tube formation in vitro (51). These studies suggest that overexpression of MEG3 may act as a therapeutic target for the treatment of DR.
Yan et al. have reported aberrant ocular expression of nearly 300 lncRNAs in diabetic mice. Among these 214 lncRNAs were downregulated and 89 lncRNAs upregulated. lncRNA MALAT1 was significantly increased in various models of diabetic mellitus, such as high glucose treated RF/6A cell, in the samples of aqueous humor, and in fibrovascular membranes of diabetic patients (52). lncRNA MALAT1 has been shown to play a pathogenic role in diabetes-induced endothelial cell dysfunction and its inhibition was found to inhibit retinal endothelial cell proliferation, migration, and tube formation and ameliorate DR (53). These authors suggested that crosstalk between the MALAT1 and p38 MAPK signaling pathway was involved in regulating retinal endothelial cell function and silencing MALAT1 had potential in anti-angiogenic therapy in DR (53).
lncRNAs and Diabetic Neuropathy (DNP)
Diabetic neuropathy is also a chronic complication of diabetes and has potential life-threatening consequences (54). Several reports have shown that lncRNAs act as important players in the pathophysiology of DNP (Figure 3). For example, Xu et al. reported increased expression of NONRATT021972 lncRNA in sympathetic neuronal-like PC12 cells in response to HG and high free fatty acid (HF) exposure. Inhibition of this lncRNA in PC12 cells significantly relieved HG-HF-induced tumor necrosis factor-α, interlukin-6, and recovered cell viability mediated by P2X7 receptor. They also observed increased expression of NONRATT021972 lncRNA in superior cervical ganglia of diabetic rats and found that treatment with NONRATT021972 siRNA led to decrease the expression of TNF-α, blocked serine phosphorylation of insulin receptor substrate (IRS) 1 and increased IRS1 expression in SCG of diabetic rats (55). These results suggest that inhibition of NONRATT021972 lncRNA could be targeted for prevention of DNP (54, 55).
Figure 3.
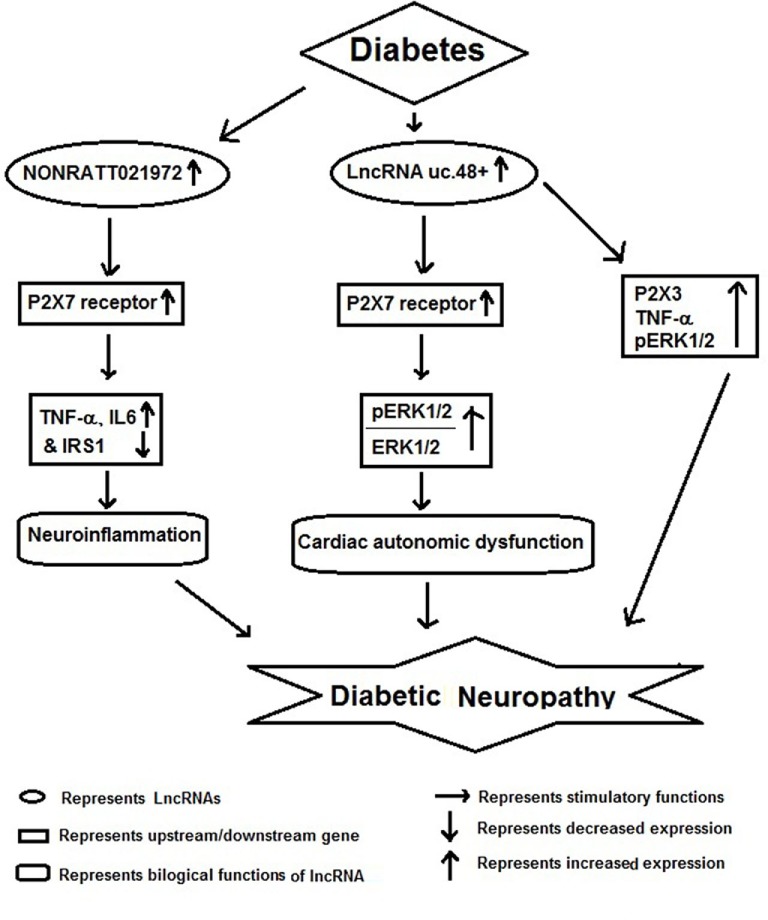
Schematic model showing functional role of long non-coding RNAs (lncRNAs) in diabetic neuropathy.
Recent studies show that DNP results in differential expression of several lncRNAs in SCG, which may result in deregulation of multiple molecular pathways involved in its pathophysiology. Li et al. found a differential expression of several lncRNAs regulating gene expression of pathways involved in immune response, cell migration, defense response, taxis, and chemotaxis. According to Kyoto Encyclopedia of Genes and Genomes, many of the target genes of differentially expressed lncRNAs were located in cytokine–cytokine receptor interactions, chemokine signaling pathway, and cell adhesion molecules. Furthermore, by gene co-expression network analysis, these authors showed 268 regulation edges among 105 lncRNAs and 11 mRNAs. These observations show that there is a co-expression of specific lncRNAs along with their target mRNAs in diabetic cardiac autonomic ganglia. In conclusion, authors have suggested a possible role for multiple lncRNAs as potential therapeutic targets or biomarkers for diabetic cardiac autonomic neuropathy (56).
A similar study by Wu et al. also found that several lncRNAs were differentially expressed in SCG of diabetic rats and among them uc.48+ lncRNA was significantly increased. The increased expression of uc.48+ lncRNA and P2X7 receptor was associated with the cardiac dysfunction. Silencing uc.48+ alleviated cardiac dysfunction, decreased its target gene P2X7 and the ratio of phosphorylated extracellular regulated protein kinases1/2 to extracellular regulated protein kinases1/2 in SCG of diabetic rats. Furthermore, they observed that silencing uc.48+ lncRNA improved diabetic sympathetic neuropathy through P2X7 and ERK signaling pathway (57). lncRNA uc.48+ inhibition has been also shown to decrease TNF-α and P2X3 expression and activation of ERK1/2 in the DRG of DM rats, indicating that uc.48+ inhibition can alleviate the DNP by inhibiting the excitatory transmission mediated by the P2X3 receptor in DRG (58).
lncRNAs and Diabetic Cardiomyopathy
Diabetic cardiomyopathy along with vascular inflammation is a characteristic feature of diabetes and the key cells involved in these pathological conditions are monocyte/macrophages and vascular smooth muscle cells (Figure 4). Recently, Kesherwani et al. have reported that several coding and non-coding RNAs (including lncRNA) were differentially expressed in insulin 2 mutant (Ins2±) Akita mouse model of diabetes (59). They performed global gene expression profiling and found that 488 transcripts were differentially expressed, including 12 non-coding RNAs. Among these, bone morphogenic protein-10 (BMP10) and vomeronasal-1 receptor-180 (VMN1R180) were highly upregulated genes; whereas hairy and enhancer of split-related (HELT) and WD repeat domain 83 opposite strand (WDR83OS) were the most downregulated genes. In addition, they have noticed that H19 lncRNA and miR-101c were found to be significantly upregulated; whereas Neat1 lncRNA was the most downregulated in Akita heart (59). In conclusion, they revealed that these results could be used as a platform to initiate focused future studies for the identification of novel therapeutic molecule in DbCM. HG has been shown to induce MIAT expression and apoptosis in vitro and in vivo model of diabetic cardiomyopathy. Furthermore, MIAT and DAPK2 were shown to be a target of miR-22-3p by luciferase assay. Overexpression of MIAT antagonized the inhibitory effect of miR-22-3p on DAPK2. Knockdown of MIAT led to the decrease of DAPK2 expression, apoptosis, and improves cardiac function in diabetic rats. Their finding suggests that MIAT functions as a competing endogenous RNA to upregulate DAPK2 expression by sponging miR-22-3p in DbCM (60).
Figure 4.
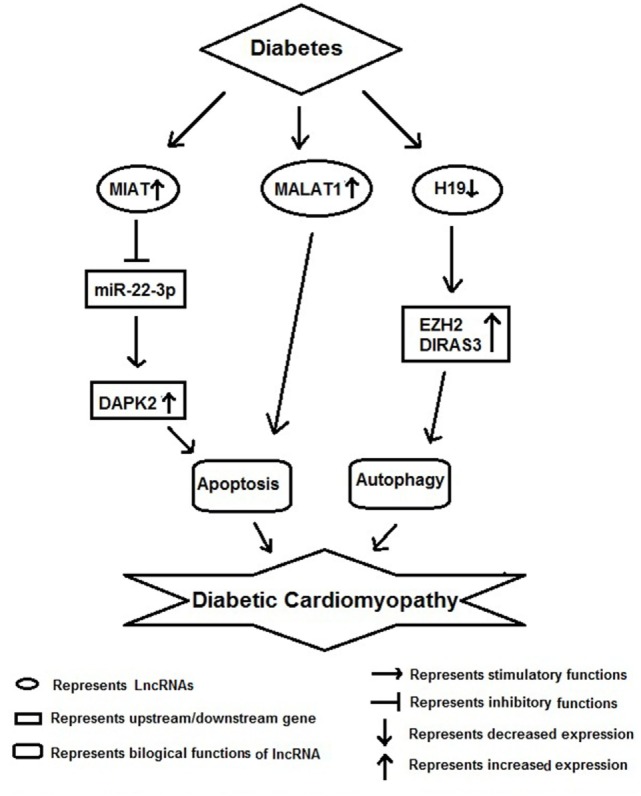
Schematic model showing functional role of long non-coding RNAs (lncRNAs) in diabetic cardiomyopathy.
In another study, Zhang et al. found an increased expression of MALAT1 in the heart tissue of diabetic rats. Knockdown of MALAT1 showed improvement in left ventricular function, partly through the attenuation of cardiomyocyte apoptosis (61). Zhuo et al. have shown decreased expression of H19 in hearts of diabetic rats, as well as in neonatal cardiomyocytes exposed to high glucose. As a result of it there was an increase in autophagy was noticed in vitro. Their RNA-binding protein immunoprecipitation studies showed direct binding of H19 with EZH2 in cardiomyocyte cells. Augmentation of H19 in cardiomyocytes in the presence of high glucose resulted in decrease DIRAS3 expression, suppress autophagy activation, and promote mTOR phosphorylation. In conclusion, they suggested that lncRNA H19 suppresses autophagy activation by epigenetically silencing DIRAS3 in DbCM (62).
Long non-coding RNAs have been suggested as circulating biomarker in DbCM (63). These authors reported that circulating lncRNAs such as long intergenic predicting cardiac remodeling (LIPCAR), MIAT, and smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA (SENCR) were found to be affiliated with grade I diastolic dysfunction. Further these lncRNAs were validated in a sample of 30 patients of diabetes and normal subjects. Indicating that these lncRNAs are independent biomarkers for diastolic function and remodeling in diabetic patients.
Angiotensin II is an important vasoconstrictor and pro-fibrotic and proliferative peptide. In vascular smooth muscle cells, several lncRNAs have shown to be deregulated by angiotensin II signaling pathway (64). For example, angiotensin II was shown to increase lnc-Ang362 expression in vascular smooth muscle cells. This induction of lnc-Ang362 increased expression of two miRNAs (mir-221 and mir-222) and cell proliferation. This suggests that lnc-Ang362 has an important role in vascular smooth muscle cell proliferation (65). In human vascular smooth muscle cells, lncRNA SENCR was involved in regulation of both contractile genes and genes involved in regulation of MYOCD (66, 67). Recently, Ballantyne et al. reported lncRNA SMILR in atherosclerotic plaques and its inhibition decreased vascular proliferation, suggesting this lncRNA promoted cellular proliferation (68).
Conclusion
Long non-coding RNA are important regulators of gene expression and control gene expression by binding to specific cellular moieties. The discovery of lncRNAs adds a new layer of complexity to the molecular etiology of microvascular complications of diabetes. Several lncRNAs have been identified and implicated in microvascular complications of diabetes by deregulating pathways, such as fibrosis, oxidative stress, endothelial dysfunction, etc. However, the mechanisms of many of deregulated lncRNAs are yet to be delineated. Thus, further research focusing on their mode of action in disease etiology and pathology is needed to understand their role in the pathogenesis of diabetes. A better understanding the mechanisms of newly identified lncRNAs can pave the way for early diagnosis and the design of better therapeutics.
Author Contributions
SR: data collection, writing, figure preparation, and references. MK: data collection, writing, editing, figure preparation, and references.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Chawla A, Chawla R, Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: distinct or continuum? Indian J Endocrinol Metab (2016) 20:546–51. 10.4103/2230-8210.183480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia (2015) 58:443–55. 10.1007/s00125-014-3462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract (2014) 2014:943162. 10.1155/2014/943162 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nam JW, Choi SW, You BH. Incredible RNA: dual functions of coding and noncoding. Mol Cells (2016) 39:367–74. 10.14348/molcells.2016.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moura J, Børsheim E, Carvalho E. The role of microRNAs in diabetic complications—special emphasis on wound healing. Genes (2014) 5:926–56. 10.3390/genes5040926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornfeld J-W, Brüning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet (2014) 5:57. 10.3389/fgene.2014.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morlando M, Fatica A. Alteration of epigenetic regulation by long noncoding RNAs in cancer. Int J Mol Sci (2018) 19:E570. 10.3390/ijms19020570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol (2010) 11:R56. 10.1186/gb-2010-11-5-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panchapakesan U, Pollock C. Long non-coding RNAs–towards precision medicine in diabetic kidney disease? Clin Sci (2016) 130:1599–602. 10.1042/CS20160261 [DOI] [PubMed] [Google Scholar]
- 10.Dechamethakun S, Muramatsu M. Long noncoding RNA variations in cardiometabolic diseases. J Hum Genet (2017) 62:97–104. 10.1038/jhg.2016.70 [DOI] [PubMed] [Google Scholar]
- 11.Li F, Wen X, Zhang H, Fan X. Novel insights into the role of long noncoding RNA in ocular diseases. Int J Mol Sci (2016) 17:478. 10.3390/ijms17040478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci (2015) 16:3251–66. 10.3390/ijms16023251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev (2009) 23:1494–504. 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics (2013) 193:651–69. 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry RB-T, Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development (2016) 143:3882–94. 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, Xu Z, Li Z, Sun L, Gong Z. An insight into the increasing role of lncRNAs in the pathogenesis of gliomas. Front Mol Neurosci (2017) 10:53. 10.3389/fnmol.2017.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Yang L, Chen L-L. The diversity of long noncoding RNAs and their generation. Trends Genet (2017) 33:540–52. 10.1016/j.tig.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 19.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res (2017) 77:3965–81. 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science (2011) 331:76–9. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Li D, Zhang R, Peng S, Zhang G, Yang T, et al. Strategies to identify natural antisense transcripts. Biochimie (2017) 132:131–51. 10.1016/j.biochi.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 22.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell (1999) 99:47–57. 10.1016/S0092-8674(00)80061-6 [DOI] [PubMed] [Google Scholar]
- 23.Kanduri C, Thakur N, Pandey RR. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J (2006) 25:2096–106. 10.1038/sj.emboj.7601090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyle R, Watanabe D, Te Vruchte D, Lerchner W, Smrzka OW, Wutz A, et al. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet (2000) 25:19–21. 10.1038/75546 [DOI] [PubMed] [Google Scholar]
- 25.Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, et al. Pseudogenes in the ENCODE regions: consensus annotation, analysis of transcription, and evolution. Genome Res (2007) 17:839–51. 10.1101/gr.5586307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumor Biol (2015) 36:503–13. 10.1007/s13277-015-3106-y [DOI] [PubMed] [Google Scholar]
- 27.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science (2006) 312:1653–5. 10.1126/science.1126316 [DOI] [PubMed] [Google Scholar]
- 28.He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. lncRNAs: key players and novel insights into diabetes mellitus. Oncotarget (2017) 8:71325–41. 10.18632/oncotarget.19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Dong C, Qian X, Huang S, Feng Y, Ye X, et al. Microarray analysis of long noncoding RNA expression patterns in diabetic nephropathy. J Diabetes Complications (2017) 31:569–76. 10.1016/j.jdiacomp.2016.11.017 [DOI] [PubMed] [Google Scholar]
- 30.Wang M, Wang S, Yao D, Yan Q, Lu W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol Cell Endocrinol (2016) 426:136–45. 10.1016/j.mce.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 31.Li X, Zeng L, Cao C, Lu C, Lian W, Han J, et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated mir-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res (2017) 350:327–35. 10.1016/j.yexcr.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 32.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, et al. lncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med (2017) 21:2732–47. 10.1111/jcmm.13189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Chen Z-Y, Wang Y, Liu Y, Ma J-X, Li Y-K. Long non-coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp Ther Med (2017) 13:581–7. 10.3892/etm.2017.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi H, Peng R, Zhang L, Sun Y, Peng H, Liu H, et al. lincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis (2017) 8:e2583. 10.1038/cddis.2016.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest (2016) 126:4205–18. 10.1172/JCI87927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan L-J, Ding M, Hou L-J, Cui Y-T, Li C-J, Yu D-M. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun (2017) 484:598–604. 10.1016/j.bbrc.2017.01.145 [DOI] [PubMed] [Google Scholar]
- 37.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One (2011) 6:e18671. 10.1371/journal.pone.0018671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez ML, Khosroheidari M, Eddy E, Kiefer J. Role of microRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS One (2013) 8:e77468. 10.1371/journal.pone.0077468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millis MP, Bowen D, Kingsley C, Watanabe RM, Wolford JK. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes (2007) 56:3027–32. 10.2337/db07-0675 [DOI] [PubMed] [Google Scholar]
- 40.Wang M, Yao D, Wang S, Yan Q, Lu W. Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine (2016) 54:81–92. 10.1007/s12020-016-0950-5 [DOI] [PubMed] [Google Scholar]
- 41.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun (2016) 7:12864. 10.1038/ncomms12864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Xu D, Sha W, Shen L, Lu G, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun (2015) 468:726–32. 10.1016/j.bbrc.2015.11.023 [DOI] [PubMed] [Google Scholar]
- 43.Zheng Y, He M, Congdon N. The worldwide epidemic of diabetic retinopathy. Indian J Ophthalmol (2012) 60:428–31. 10.4103/0301-4738.100542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan B, Liu J, Yao J, Li X, Wang X, Li Y, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res (2015) 116:1143–56. 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Chen M, Chen J, Lin S, Cai D, Chen C, et al. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci Rep (2017) 37. 10.1042/BSR20170036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Xu F, Xiao H, Han F. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J Cell Biochem (2018) 119(1):817–23. 10.1002/jcb.26245 [DOI] [PubMed] [Google Scholar]
- 47.Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy. Invest Ophthalmol Vis Sci (2017) 58:470–80. 10.1167/iovs.16-20569 [DOI] [PubMed] [Google Scholar]
- 48.Shan K, Li C-P, Liu C, Liu X, Yan B. RNCR3: a regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem Biophys Res Commun (2017) 482:777–83. 10.1016/j.bbrc.2016.11.110 [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Li C, Wang J-J, Shan K, Liu X, Yan B. RNCR3 knockdown inhibits diabetes mellitus-induced retinal reactive gliosis. Biochem Biophys Res Commun (2016) 479:198–203. 10.1016/j.bbrc.2016.09.032 [DOI] [PubMed] [Google Scholar]
- 50.Li C-P, Wang S-H, Wang W-Q, Song S-G, Liu X-M. Long noncoding RNA-SOX2OT knockdown alleviates diabetes mellitus-induced retinal ganglion cell (RGC) injury. Cell Mol Neurobiol (2017) 37:361–9. 10.1007/s10571-016-0380-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu G-Z, Tian W, Fu H-T, Li C-P, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun (2016) 471:135–41. 10.1016/j.bbrc.2016.01.164 [DOI] [PubMed] [Google Scholar]
- 52.Yan B, Tao Z-F, Li X-M, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci (2014) 55:941–51. 10.1167/iovs.13-13221 [DOI] [PubMed] [Google Scholar]
- 53.Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis (2014) 5:e1506. 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu H, He L, Liu C, Tang L, Xu Y, Xiong M, et al. lncRNA NONRATT021972 siRNA attenuates P2X7 receptor expression and inflammatory cytokine production induced by combined high glucose and free fatty acids in PC12 cells. Purinergic Signal (2016) 12:259–68. 10.1007/s11302-016-9500-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H, Liu C, Rao S, He L, Zhang T, Sun S, et al. lncRNA NONRATT021972 siRNA rescued decreased heart rate variability in diabetic rats in superior cervical ganglia. Auton Neurosci (2016) 201:1–7. 10.1016/j.autneu.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 56.Li G, Sheng X, Xu Y, Jiang H, Zheng C, Guo J, et al. Co-expression changes of lncRNAs and mRNAs in the cervical sympathetic ganglia in diabetic cardiac autonomic neuropathic rats. J Neurosci Res (2017) 95:1690–9. 10.1002/jnr.24000 [DOI] [PubMed] [Google Scholar]
- 57.Wu B, Zhang C, Zou L, Ma Y, Huang K, Lv Q, et al. lncRNA uc. 48+ siRNA improved diabetic sympathetic neuropathy in type 2 diabetic rats mediated by P2X 7 receptor in SCG. Auton Neurosci (2016) 197:14–8. 10.1016/j.autneu.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 58.Wang S, Xu H, Zou L, Xie J, Wu H, Wu B, et al. lncRNA uc. 48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal (2016) 12:139–48. 10.1007/s11302-015-9488-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kesherwani V, Shahshahan HR, Mishra PK. Cardiac transcriptome profiling of diabetic Akita mice using microarray and next generation sequencing. PLoS One (2017) 12:e0182828. 10.1371/journal.pone.0182828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis (2017) 8:e2929. 10.1038/cddis.2017.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Gu H, Chen J, Zhou X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int J Cardiol (2016) 202:753–5. 10.1016/j.ijcard.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 62.Zhuo C, Jiang R, Lin X, Shao M. lncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget (2017) 8:1429–37. 10.18632/oncotarget.13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, Van Der Meer RW, Rijzewijk LJ, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep (2016) 6:37354. 10.1038/srep37354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leung A, Natarajan R. Long noncoding RNAs in diabetes and diabetic complications. Antioxid Redox Signal (2017). 10.1089/ars.2017.7315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leung A, Trac C, Jin W, Lanting L, Akbany A, Sætrom P, et al. Novel long non-coding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res (2013) 113:266–78. 10.1161/CIRCRESAHA.112.300849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thum T, Kumarswamy R. The smooth long noncoding RNA SENCR. Arterioscler Thromb Vasc Biol (2014) 34:1124–5. 10.1161/ATVBAHA.114.303504 [DOI] [PubMed] [Google Scholar]
- 67.Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, et al. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther (2016) 24:978–90. 10.1038/mt.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, et al. Smooth muscle enriched long non-coding RNA (SMILR) regulates cell proliferation. Circulation (2016) 133:2050–65. 10.1161/CIRCULATIONAHA.115.021019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaur S, Mirza AH, Brorsson CA, Fløyel T, Størling J, Mortensen HB, et al. The genetic and regulatory architecture of ERBB3-type 1 diabetes susceptibility locus. Mol Cell Endocrinol (2016) 419:83–91. 10.1016/j.mce.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 70.Yamanaka H. TNF as a target of inflammation in rheumatoid arthritis. Endocr Metab Immune Disord Drug Targets (2015) 15:129–34. 10.2174/1871530315666150316121808 [DOI] [PubMed] [Google Scholar]
- 71.Stuhlmüller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol (2003) 163:901–11. 10.1016/S0002-9440(10)63450-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.You L, Wang N, Yin D, Wang L, Jin F, Zhu Y, et al. Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J Cell Physiol (2016) 231:852–62. 10.1002/jcp.25175 [DOI] [PubMed] [Google Scholar]
- 73.Zhu X, Wu Y-B, Zhou J, Kang D-M. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun (2016) 469:319–25. 10.1016/j.bbrc.2015.11.048 [DOI] [PubMed] [Google Scholar]
- 74.Morán I, Akerman İ, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab (2012) 16:435–48. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arnes L, Akerman I, Balderes DA, Ferrer J, Sussel L. βlinc1 encodes a long noncoding RNA that regulates islet β-cell formation and function. Genes Dev (2016) 30:502–7. 10.1101/gad.273821.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akerman I, Tu Z, Beucher A, Rolando DM, Sauty-Colace C, Benazra M, et al. Human pancreatic β cell lncRNAs control cell-specific regulatory networks. Cell Metab (2017) 25:400–11. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin D, Zhang E, You L, Wang N, Wang L, Jin F, et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell Physiol Biochem (2015) 35:1892–904. 10.1159/000373999 [DOI] [PubMed] [Google Scholar]
- 78.Yu W, Zhao G, Cao R, Zhu Z, Li K. lncRNA NONRATT021972 was associated with neuropathic pain scoring in patients with type 2 diabetes. Behav Neurol (2017) 2017:2941297. 10.1155/2017/2941297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Zhao Z, Gao C, Rao L, Hao P, Jian D, et al. The diagnostic value of whole blood lncRNA ENST00000550337. 1 for pre-diabetes and Type 2 diabetes mellitus. Exp Clin Endocrinol Diabetes (2017) 125:377–83. 10.1055/s-0043-100018 [DOI] [PubMed] [Google Scholar]
- 80.Zou G, Liu T, Guo L, Huang Y, Feng Y, Huang Q, et al. miR-145 modulates lncRNA-ROR and Sox2 expression to maintain human amniotic epithelial stem cell pluripotency and β islet-like cell differentiation efficiency. Gene (2016) 591:48–57. 10.1016/j.gene.2016.06.047 [DOI] [PubMed] [Google Scholar]
- 81.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15p15 INK4B tumor suppressor gene. Oncogene (2011) 30:1956. 10.1038/onc.2010.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reddy MA, Chen Z, Park JT, Wang M, Lanting L, Zhang Q, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long non-coding RNA. Diabetes (2014) 12:4249–61. 10.2337/db14-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer (2015) 14:184. 10.1186/s12943-015-0458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A (2014) 111:13924–9. 10.1073/pnas.1402665111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin Z, Li X, Zhan X, Sun L, Gao J, Cao Y, et al. Construction of competitive endogenous RNA network reveals regulatory role of long non-coding RNAs in type 2 diabetes mellitus. J Cell Mol Med (2017) 21:3204–13. 10.1111/jcmm.13224 [DOI] [PMC free article] [PubMed] [Google Scholar]