Abstract
HIV-1 subtype B virus is the most prevalent subtype in Puerto Rico (PR), accounting for about 90% of infection in the island. Recently, other subtypes and circulating recombinant forms (CRFs), including F(12_BF), A (01_BF), and CRF-39 BF-like, have been identified. The purpose of this study is to assess the distribution of drug resistance mutations and subtypes in PR. A total of 846 nucleotide sequences from the period comprising 2013 through 2017 were obtained from our “HIV Genotyping” test file. Phylogenetic and molecular epidemiology analyses were performed to evaluate the evolutionary dynamics and prevalence of drug resistance mutations. According to our results, we detected a decrease in the prevalence of protease inhibitor, nucleoside reverse transcriptase inhibitor (NRTI), and non-NRTI (NNRTI) resistance mutations over time. In addition, we also detected recombinant forms and, for the first time, identified subtypes C, D, and CRF-24BG in PR. Recent studies suggest that non-subtypes B are associated with a high risk of treatment failure and disease progression. The constant monitoring of viral evolution and drug resistance mutation dynamics is important to establish appropriate efforts for controlling viral expansion.
Keywords: : HIV-1, drug resistance mutations, HIV-1 subtypes
Introduction
HIV-1 is a highly variable virus and drug-resistant mutants readily become predominant under antiretroviral therapies.1,2 HIV heterogeneity is a result of the virus rapid rate of evolution, short generation time, and low fidelity of its reverse transcriptase.3–5 HIV-1 genetic variability is used to classify the virus into four distinct groups: M, N, O and P; group M (for major) has nine subtypes (A1, A2, A3, A4, A5, A6, B, C, D, F1, F2, G, H, J, and K).6–14 Nevertheless, recombination is the major determinant of viral genetic diversity, which occurs during reverse transcription of co-packaged viral RNA.15,16 A possible source of recombination is the co-infection between two different strains.17–19 In addition, current molecular data suggest that recombination can introduce or eliminate drug resistance mutations, which could jeopardize antiretroviral treatment (ART).20 The remarkable genetic variability of HIV-1 influences its infectivity, progression, transmissibility and patient's response to antiretroviral treatment.21,22
HIV-1 epidemics in the Caribbean region seem to be mediated by clade B.23,24 Puerto Rico (PR), which is a commonwealth of the United States, has one of the highest prevalence rates of HIV-1 in the United States and the Caribbean.25 In 2014, the prevalence of infection rate in PR was twice that of the average rate of the combined states and territories of the United States, ranking number 6 (567.3 per 100,000 population).26 In the case of males, the most prevalent route of transmission is through heterosexual and male-to-male sexual contact. Among females, heterosexual contact is the most frequent mode of HIV transmission.27 According to Los Alamos database, HIV-1 B is the most common variant and represents 99.7% of the virus subtypes in the infected population.28 Recently, other subtypes and circulating recombinant forms (CRFs), including F(12-BF), A (01-BF), and CRF-39 BF-like, have been identified in the island.29 Because of the high levels of migration to and immigration from the continental United States, it is important for the citizens of both areas to understand how the virus evolves. The purpose of this study was to assess the distribution of drug resistance mutations and subtypes in PR. Surveillance of HIV-1 genetic diversity is necessary to initiate public health efforts.30
Materials and Methods
Ethics statement
The protocol was approved by the Institutional Review Board of Ponce Medical School Foundation.
Data source and variables
We analyzed 846 nucleotide sequences obtained from our secured “HIV Genotyping” test database, which contains de-identified sequence data from the years 2013–2017. The sequences were obtained from samples collected in PR between 2013–2017 and were amplified and analyzed as described in the subsequent sections. Descriptive statistics were used for demographic parameters.
Viral RNA extraction, reverse transcription polymerase chain reaction and sequencing
HIV-1 viral RNA was extracted using the QIAamp viral RNA kit (QIAGEN), following the manufacturer's instructions. Purified RNA was amplified by reverse transcription polymerase chain reaction (RT-PCR) using Titan OneStep RT-PCR kit (Roche). Briefly, first-round RT-PCR conditions were as follows: reverse transcription at 50°C for 10 min, inactivation at 94°C for 2 min, 35 cycles of 94°C for 10 s, 53°C for 30 s, and 68°C for 1 min; after the first 10 cycles, 5 s were added to the elongation step of each cycle, with a final extension at 68°C for 7 min. The first-round amplicon (2 μL) was reamplified using the Roche Fast Start PCR Mastermix. Second-round PCR conditions were as follows: 95°C for 15 min followed by 35 cycles of (94°C for 30 s, 53°C for 30 s, and 72°C for 2 min) and a final elongation at 72°C for 10 min. The sequences of the primers used for amplification and sequencing were as described in the World Health Organization (WHO) manual for HIV drug resistance testing using dried blood specimens. First-round primers: forward protease: 5′-TGAARGAITGYACTGARAGRCAGGCTAAT-3′; reverse protease: 5′-AYCTIATYCCTGGTGTYTCATTRTT-3′; forward RT: 5′-TTTYAGRGARCTYAATAARAGAACTCA-3′; reverse RT: 5′-CCTCITTYTTGCATAYTTYCCTGTT-3′. Second-round primers: forward protease: 5′-YTCAGRCAGRCCRGARCCAACAGC-3′; reverse protease: 5′-CTGGTGTYTCATTRTTKRTACTAGGT-3′; forward RT: 5′-TTYTGGGARGTYCARYTAGGRATACC-3′; reverse RT: 5′-GGYTCTTGRTAAATTTGRTATGTCCA-3′. Confirmed amplicons were directly sequenced using our WHO-accredited HIV genotyping protocols. Sequences were determined by using ABI 3730 XL. Sequences generated were uploaded onto our secured “HIV Genotyping” test database.
Analysis of drug resistance mutations and viral subtypes
Drug resistance mutations associated with protease inhibitors (PIs), nucleoside reverse transcriptase inhibitors (NRTIs), and non-NRTIs (NNRTIs) were determined by using Calibrated Population Resistance Tool (CPR) available in Stanford HIV Database program (http://HIVDB.stanford.edu).31 The sequences that covered part of the pol gene were aligned using BioEdit (v.7.0.9). Viral subtypes were evaluated using bootscanning methods available in REGA v3.0, Context-based Modeling for Expeditious Typing (COMET) HIV-1, and by the jumping profile hidden Markov model (jpHMM) available at the jpHMM web server at GOBICS.32,33
Phylogenetic analysis
The phylogenetic tree was inferred using the maximum likelihood (ML) method implemented by MEGA software v6.34 We used bootstrap ML percentages to assess the robustness of each branch. Branches with bootstrap values above .70 were considered robustly supported. A reference set of 87 sequences was downloaded from REGA v3 database. The sequences were 1,030 bp long (nucleotides 2253–3452 relative to the reference sequence HXB2) and covered pol gene.
Results
During the period of 2013 through 2017, data were collected on 846 subjects. Demographic data show that the sample was predominantly male (74%) and the mean ages for males and females were 41 and 44 years, respectively (Table 1).
Table 1.
Demographic Characteristics
| Parameter | Sequences |
|---|---|
| Gender, n (%) | |
| Male | 627 (74.1) |
| Female | 209 (24.7) |
| N/A | 10 (1.2) |
| Age, mean | |
| Male | 41 (17–92)a |
| Female | 44 (14–77)a |
Age data available in 94% (males) and 89% (females) of the sequences, respectively.
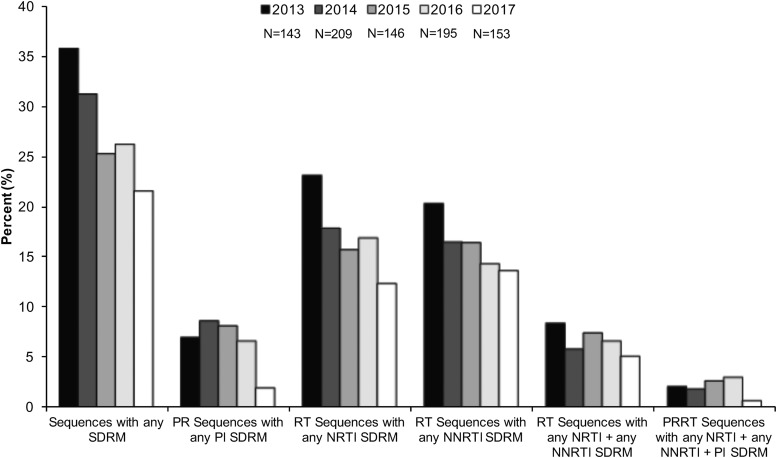
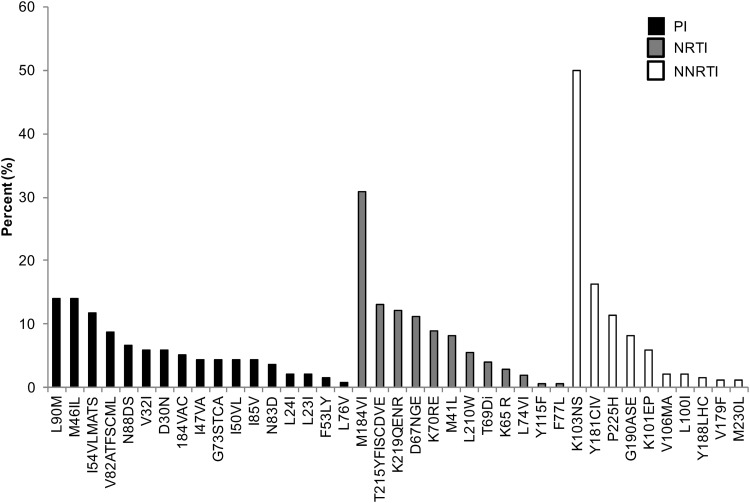
CPR available from the Stanford HIV Database program was used to evaluate the percentage of expression of HIV-1 NRTI, NNRTI, and PI drug resistance mutations.31 We detected a decrease in the prevalence of drug resistance mutations over time. In a period of 5 years, from 2013 to 2017, sequences with any PI, NRTI, and NNRTI SDRMs decreased 5%, 10.7%, and 6.6%, respectively (Fig. 1). However, each year, more than 21% of sequences had one or more drug resistance mutations (Fig. 1). Cross-class resistance, which involves two or three classes of resistance mutations, was observed in less than 10% of the samples. The drug resistance mutations associated with NRTI and NNRTI were detected more commonly than PI resistance mutations (Fig. 1). The most frequently observed PI drug resistance mutations during this period were L90 M and M46IL (∼14% each), whereas the most common NRTI and NNRTI drug resistance mutations were M184VI (30%) and K103NS (50%), respectively (Fig. 2).
FIG. 1.
Prevalence of PI, NRTI, and NNRTI mutations over time. The prevalence of SDRM was evaluated using Calibrate Population Resistance Tool (CRP), Stanford HIV Database program. NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; SDRM, surveillance drug resistance mutations.
FIG. 2.
Frequency of specific HIV-1 drug resistance mutations among sequences analyzed during the period of 2013 through 2017. PI, NRTI, and NNRTI drug resistance mutations were evaluated using Calibrate Population Resistance Tool (CRP), Stanford HIV Database program.
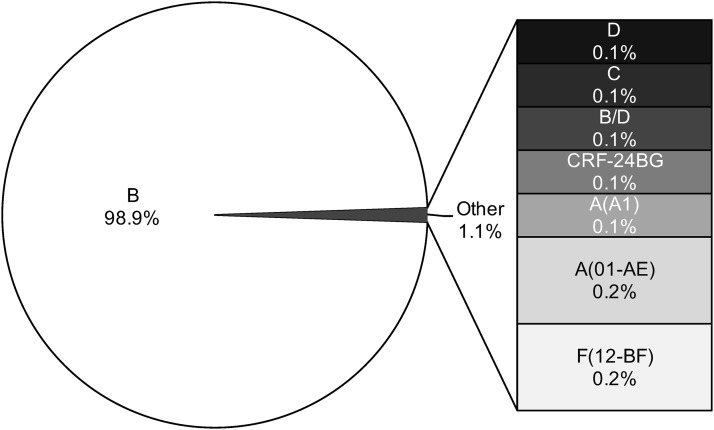
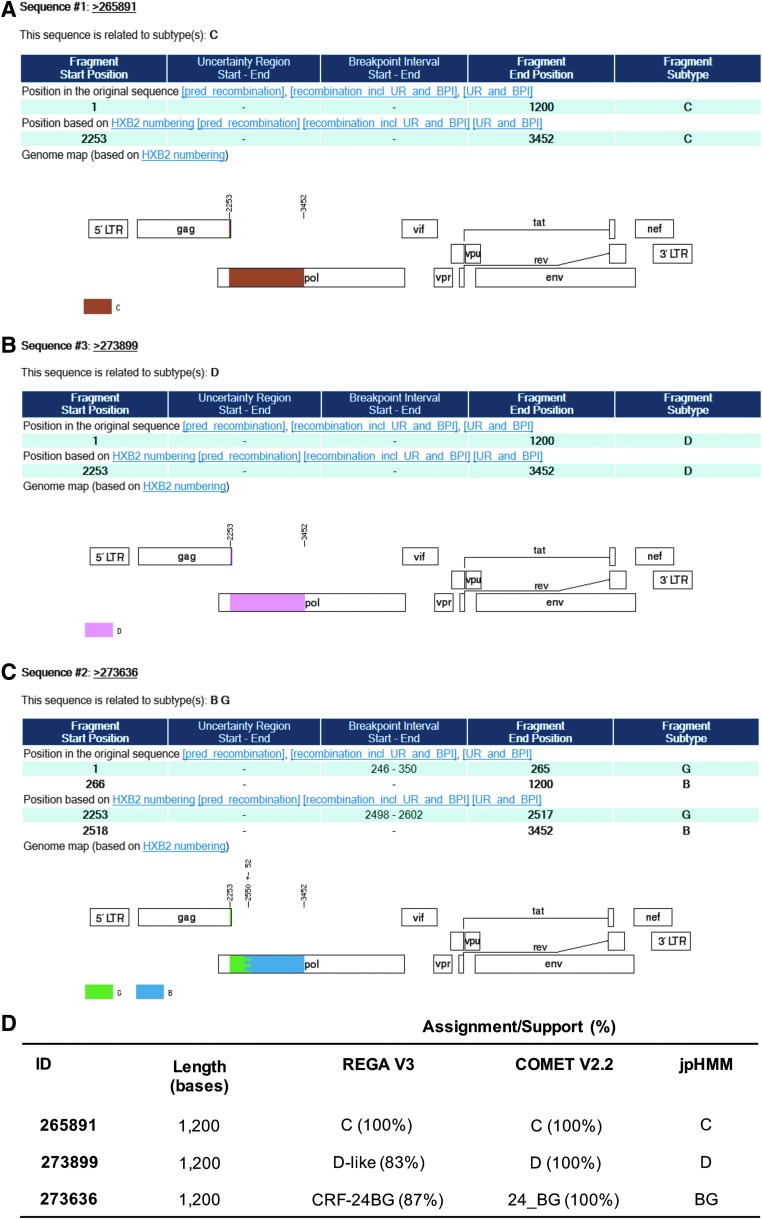
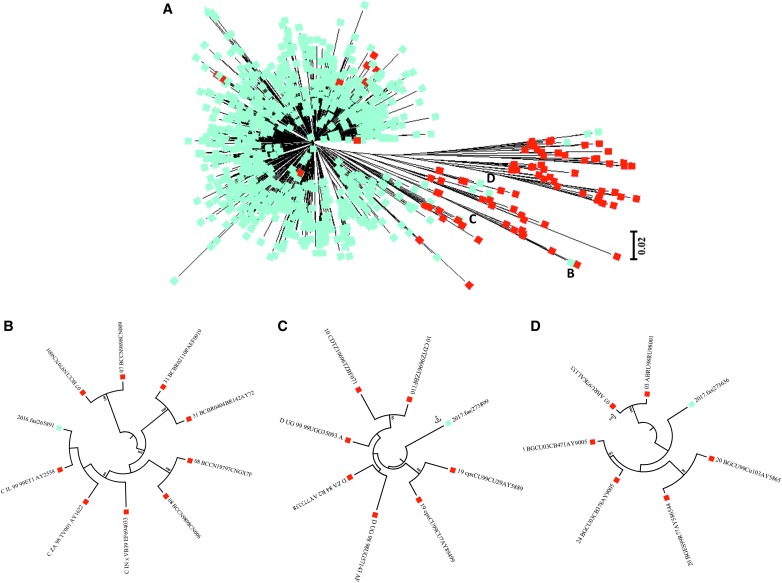
Viral subtypes were assessed using the bootscanning method available in REGA v3 and confirmed using the COMET HIV-1 v2.2 and by jpHMM platforms.32 The majority of HIV infections are associated with subtype B (n = 837; 98.9%). However, we detected other subtypes and recombinant forms in the island as follows: recombinant B/D (n = 1; 0.118%), F(12-BF) (n = 2; 0.236%), A(01-AE) (n = 2; 0.236%), A(A1) (n = 1; 0.118%), C (n = 1; 0.118%), D (n = 1; 0.118%), and CRF-24BG (n = 1; 0.118%) (Fig. 3). For the cases in which we obtained discordant results between platforms, we relied on the results obtained with the COMET platform for subtype assignment. Recent analyses suggest that COMET HIV-1 v2.2 has shown superior specificity in short sequences.32,35 Despite the fact that subtype and inter-subtypes had been reported in PR [B/F, B/D, A1/B, A(A2), A(01-AE), F(12-BF), and CRF39-BF] previously, this is the first report of subtypes C, D, and CRF-24BG (Fig. 3).29,36 The three cases were evaluated by using REGA v3, COMET HIV-1 v2.2, and by jpHMM available in jpHMM web server at GOBICS, to confirm the results33,37 (Fig. 4). The breakpoint interval in CRF-24BG obtained by using jpHMM is located between the nucleotide positions 2498–2602 (based on HXB2 numbering). Subtype determination based on the pol sequences was visualized by using ML tree, generated using MEGA v .6, under a General Time-Reversible nucleotide substitution model (suggested by ModelTest) with a gamma-distributed rate variation for each of these tree alignments and a resampling process (100 bootstraps).38,39 We compared samples processed in our site (n = 846) and reference sequences (n = 87, REGA v3 references samples). The three new cases reported in the island clustered with related sequences from around the world (Fig. 5).
FIG. 3.
HIV-1 subtype assignment was performed using the bootscanning method within REGA v3 and confirmed using COMET HIV-1 v2.2 and jpHMM. Results indicate the percentage of a particular subtype represented in the sample (n = 846 sequences). COMET, Context-based Modeling for Expeditious Typing; jpHMM, jumping profile hidden Markov model.
FIG. 4.
Detection of novel HIV-1 subtypes in Puerto Rico. (A) HIV-1 subtype C. (B) HIV-1 subtype D. (C) HIV-1 CRF-24BG. (D) The three cases were evaluated by using REGA v3, COMET HIV-1 v2.2, and by jpHMM available in jpHMM web server at GOBICS. The visualizations are the examples of the assignment by jpHMM. CRF, circulating recombinant form.
FIG. 5.
Maximum likelihood phylogenetic tree of HIV-1 pol gene (protease/reverse transcriptase) performed using MEGA software (v 6). (A) Sequences analyzed during the period of 2013 through 2017 (blue, n = 846) and reference sequences (red, n = 87). The reference set of 87 sequences was downloaded from REGA v3 database. (B) Subtype C. (C) Subtype D. (D) CRF-24BG.
Discussion
After a positive diagnosis of HIV-1 infection, three different classes of drugs (NRTI, NNRTI, and PI) are used in different combination to treat patients. This treatment markedly inhibited replication and reduced transmission risk.40,41 However, high viral turnover, error rate of the reverse transcriptase enzyme, poor adherence to medications, and selective pressure exerted by medications lead to genetic variants with drug resistance mutations.42 The routine surveillance of the genetic diversity of HIV-1 virus is necessary to initiate public health efforts. The decline in PI, NRTI, and NNRTI mutations in PR (2013–2017) is consistent with recent studies that support that HIV-1 major drug resistance mutations are declining in resource-rich settings.43,44 This finding would reflect the improvement of treatment regimens, leading to increases in treatment adherence.44–47 In addition, prevention programs that provide health insurance, care, and support to the HIV-1 infected patients have been established effectively by local health departments and nongovernmental organizations.
Our finding that M184VI and K103NS were the most frequent reverse transcriptase mutations is in agreement with results obtained by Sepulveda-Torres et al., who performed an analysis on PR samples during the period of 2002–2011 with data generated using TRUGENE HIV-1 Genotyping Kit and OpenGene DNA sequencing system.48 In their study, M184VI and K103NS were also identified as the predominant drug resistance mutations. The M184VI resistance mutation is an NRTI-resistance mutation associated with high level of resistance to lamivudine and emtricitabine and cause low level of resistance to abacavir and didanosine.49–51 However, treatment with lamivudine and emtricitabine increases susceptibility to zidovudine, tenofovir, and stavudine and is associated with reduced HIV-1 replication.52–58 The K103N is an NNRTI resistance mutation associated with reduced susceptibility to nevirapine and efavirenz; however, K103S causes high-level resistance to nevirapine and intermediate resistance to efavirenz.57,59–62 In addition, K103N shows a viral fitness, which is the ability of the virus to adapt and reproduce in the host, very close to levels comparable to that of the wild-type virus.63 The highest PI mutations were L90M and M46IL, which are associated with significant reductions in fitness.63–66 The L90M is associated with reduction of susceptibility of PI medications, except tipranavir and darunavir.67,68 The M46IL mutations occur alone or in combination with others, which are associated with reduced susceptibility to atazanavir, fosamprenavir, indinavir, lopinavir, and nelfinavir.69,70 The high prevalence of L90M and M46IL mutations in our study is similar with the results obtained in the INSIGHT Strategic Timing of ART trial.71 The finding that reverse transcriptase drug resistance mutations were more commonly expressed than resistance mutations to PI has been observed in other studies. The increase in reverse transcriptase drug resistance mutations may be related to the elevated use of NNRTs/NNRTIs, and in the case of NNRTI resistance mutations, to the increased transmission in newly diagnosed patients, or to the interruption of a suppressive NNRTI-based regimen.44,71–73 A limitation in this study is unavailability of sequence data to assess integrase strand inhibitor resistance mutations for all sequences under the study period (2013–2017). Not all sequences analyzed in this study included integrase gene sequences. Thus, additional studies are needed to evaluate the molecular evolution of HIV-1 integrase at the interpatient level to assess the ongoing adaptation of the enzyme.
PR has one of the highest prevalence rates of HIV-1 in United States, with male-to-male and heterosexual transmissions as the major routes of the infection. While the HIV-1 epidemic affecting the Caribbean is largely mediated by B-clade virus,74–76 only a small number of non-B-clade HIV-1 infections have thus far been identified in the region.29,36,76 Although a previous study demonstrated that HIV-1 isolates from PR were closely associated with U.S. B-clade HIV-1 strains, the Caribbean B-clade HIV isolates are distinct from the U.S. B-clade isolates.23,77 Cuba is an exception, with high genetic diversity and circulation of several viral variants of non B-clade.78 To gain information that may be useful for developing strategies to prevent further spreading of HIV-1 epidemic in PR, it is important to focus our efforts on identifying the viruses circulating through the island. Understanding of how the virus evolves is crucial due to high levels of migration to and immigration from the continental United States. In the Caribbean region, subtype C has been previously reported in Cuba and Saint Lucia, while subtype D and CRF-24BG have been reported only in Cuba.28,76,79,80 Our current report of the first cases of subtype C, D, and CRF-24BG in PR highlights the importance of molecular monitoring of new subtypes spreading in the island.
The most relevant biological properties among each HIV subtypes are their rate of adaptive evolution, neutral mutations, tropism, and acquisition of antiretroviral resistance.6,81–83 In addition, transmissibility of the virus and patient's response to antiretroviral treatment have been implicated with the HIV-1 genetic subtype classification. Subtype C, which is the most prevalent subtype in the world (nearly 50%), is very common in Southern Africa and India.7 This subtype has been associated with increased infectivity during heterosexual intercourse and poorer ART outcomes, and develops resistance to antiretroviral therapy faster than other subtypes.84–87 Recent studies suggest that subtype D may be associated with a faster virological rebound, CD4 decline, prevalence of CXCR4-using virus, and high rates of disease progressions.88–91 The CRFs are increasing in the past decade in various parts of the world.92,93 The CRF-24BG was previously reported in Cuba and other recombinant BG strains in Portugal, Spain, and Germany, which were associated with intravenous drug users.94–96
The possible role of tourism in the spread of non B-clade strains needs to be addressed. Recent studies have emphasized the important role of tourism in spreading the HIV-1 epidemic to and throughout the Caribbean.48,97–99 In addition, the migration of the HIV-1 at-risk population among the different countries is undoubtedly related with the flow of the epidemic.100–102 The establishment of local and regional (Caribbean) programs to monitor how the virus evolves in our countries can help us for developing strategies to prevent the HIV-1 epidemic from spreading in the Caribbean region. An example of unusual mutations in the Caribbean region is the signature of threonine deletion (env gene) in Trinidad and Tobago, which is predominantly associated with heterosexual transmission.103 The circulation of HIV-1 non-B clade in the island, which favors recombination, may have repercussions in diagnostic and clinical management of HIV-infected individuals.104–106 While epidemiological and clinical information about patients infected with these novel subtypes were not available for this study, our current analysis provides new data that will be useful for understanding how the HIV-1 virus evolves in PR. This information can help us develop strategies to prevent the impact of HIV-1 epidemic spread in PR into a more complex epidemiological landscape.
Acknowledgments
The study was supported by the AIDS Research Infrastructure Program (RCMI Program; NIMHD-G12-MD007579).
Results included in this article were presented at the Research Center in Minority Institutions Translational Science Conference, October 28–November 1, 2017.
Sequence Data
The sequences are available at GenBank with accession numbers MF960930-MF961775.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Stebbing J, Patterson S, Gotch F: New insights into the immunology and evolution of HIV. Cell Res 2003;13:1–7 [DOI] [PubMed] [Google Scholar]
- 2.Ho DD, Huang Y: The HIV-1 vaccine race. Cell 2002;110:135–138 [DOI] [PubMed] [Google Scholar]
- 3.Seillier-Moiseiwitsch F, Margolin BH, Swanstrom R: Genetic variability of the human immunodeficiency virus: Statistical and biological issues. Annu Rev Genet 1994;28:559–596 [DOI] [PubMed] [Google Scholar]
- 4.Peeters M, Sharp PM: Genetic diversity of HIV-1: The moving target. AIDS 2000;14 Suppl 3:S129–S140 [PubMed] [Google Scholar]
- 5.Coffin JM: Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol 1992;176:143–164 [DOI] [PubMed] [Google Scholar]
- 6.Leal E, Villanova FE: Diversity of HIV-1 subtype B: Implications to the origin of BF recombinants. PLoS One 2010;5:e11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buonaguro L, Tornesello ML, Buonaguro FM: Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: Pathogenetic and therapeutic implications. J Virol 2007;81:10209–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plantier JC, Leoz M, Dickerson JE, et al. : A new human immunodeficiency virus derived from gorillas. Nat Med 2009;15:871–872 [DOI] [PubMed] [Google Scholar]
- 9.Vidal N, Mulanga C, Bazepeo SE, Lepira F, Delaporte E, Peeters M: Identification and molecular characterization of subsubtype A4 in central Africa. AIDS Res Hum Retroviruses 2006;22:182–187 [DOI] [PubMed] [Google Scholar]
- 10.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM: The challenge of HIV-1 subtype diversity. N Engl J Med 2008;358:1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaacs T, Abera AB, Muloiwa R, Katz AA, Todd G: Genetic diversity of HHV8 subtypes in South Africa: A5 subtype is associated with extensive disease in AIDS-KS. J Med Virol 2016;88:292–303 [DOI] [PubMed] [Google Scholar]
- 12.Bartolo I, Rocha C, Bartolomeu J, et al. : Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: New insights into the origins of the AIDS pandemic. Infect Genet Evol 2009;9:672–682 [DOI] [PubMed] [Google Scholar]
- 13.Kazennova E, Laga V, Gromov K, et al. : Genetic variants of HIV type 1 in men who have sex with men in Russia. AIDS Res Hum Retroviruses 2017;33:1061–1064 [DOI] [PubMed] [Google Scholar]
- 14.Lapovok I, Laga V, Kazennova E, Bobkova M: HIV type 1 integrase natural polymorphisms in viral variants circulating in FSU countries. Curr HIV Res 2017;15:318–326 [DOI] [PubMed] [Google Scholar]
- 15.Preston BD: Reverse transcriptase fidelity and HIV-1 variation. Science 1997;275:228–229; author reply 230–221 [DOI] [PubMed] [Google Scholar]
- 16.Schlub TE, Smyth RP, Grimm AJ, Mak J, Davenport MP: Accurately measuring recombination between closely related HIV-1 genomes. PLoS Comput Biol 2010;6:e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragupathy V, Zhao J, Wood O, et al. : Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virol J 2011;8:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang G, Weiser B, Kuiken C, et al. : Recombination following superinfection by HIV-1. AIDS 2004;18:153–159 [DOI] [PubMed] [Google Scholar]
- 19.van der Kuyl AC, Cornelissen M: Identifying HIV-1 dual infections. Retrovirology 2007;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor BS, Hammer SM: The challenge of HIV-1 subtype diversity. N Engl J Med 2008;359:1965–1966 [DOI] [PubMed] [Google Scholar]
- 21.Bretscher MT, Althaus CL, Muller V, Bonhoeffer S: Recombination in HIV and the evolution of drug resistance: For better or for worse? Bioessays 2004;26:180–188 [DOI] [PubMed] [Google Scholar]
- 22.Cortez KJ, Maldarelli F: Clinical management of HIV drug resistance. Viruses 2011;3:347–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadai Y, Eyzaguirre LM, Sill A, et al. : HIV-1 epidemic in the Caribbean is dominated by subtype B. PLoS One 2009;4:e4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magiorkinis G, Angelis K, Mamais I, et al. : The global spread of HIV-1 subtype B epidemic. Infect Genet Evol 2016;46:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Diaz CE, Andrinopoulos K: HIV and incarceration in the Caribbean: The experiences of Puerto Rico and Jamaica. P R Health Sci J 2012;31:161–169 [PubMed] [Google Scholar]
- 26.CDC: Available at https://gis.cdc.gov/grasp/nchhstpatlas/charts.html accessed April2017
- 27.Rico DdSdP: Perfil Epidemiológico Integrado para la Prevención del VIH en Puerto Rico, 2008–2014. Available at www.salud.gov.pr/Estadisticas-Registros-y-Publicaciones/Estadisticas VIH/Perfil Epidemiol%C3%B3gico/Perfil Epidemiol%C3%B3gico Integrado para la Prevenci%C3%B3n del VIH en Puerto Rico.pdf accessed April2017
- 28.database H: Geography Search Interface. www.hiv.lanl.gov/ accessed April2017
- 29.Lopez P, Rivera-Amill V, Rodriguez N, Vargas F, Yamamura Y: The genetic diversity and evolution of HIV-1 subtype B epidemic in Puerto Rico. Int J Environ Res Public Health 2015;13:ijerph13010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao CF, Chang SY, Hsia KT, et al. : Surveillance of HIV type 1 recent infection and molecular epidemiology among different risk behaviors between 2007 and 2009 after the HIV type 1 CRF07_BC outbreak in Taiwan. AIDS Res Hum Retroviruses 2011;27:745–749 [DOI] [PubMed] [Google Scholar]
- 31.Gifford RJ, de Oliveira T, Rambaut A, et al. : Phylogenetic surveillance of viral genetic diversity and the evolving molecular epidemiology of human immunodeficiency virus type 1. J Virol 2007;81:13050–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pineda-Pena AC, Faria NR, Imbrechts S, et al. : Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: Performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013;19:337–348 [DOI] [PubMed] [Google Scholar]
- 33.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP: COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014;42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sallam M, Sahin GO, Ingman M, Widell A, Esbjornsson J, Medstrand P: Genetic characterization of human immunodeficiency virus type 1 transmission in the Middle East and North Africa. Heliyon 2017;3:e00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flores I, Pieniazek D, Moran N, et al. : HIV-1 subtype F in single and dual infections in Puerto Rico: A potential sentinel site for monitoring novel genetic HIV variants in North America. Emerg Infect Dis 1999;5:481–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Schultz AK, Calef C, et al. : jpHMM at GOBICS: A web server to detect genomic recombinations in HIV-1. Nucleic Acids Res 2006;34(Web Server issue):W463–W465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posada D, Crandall KA: MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998;14:817–818 [DOI] [PubMed] [Google Scholar]
- 39.Salemi M, de Oliveira T, Ciccozzi M, Rezza G, Goodenow MM: High-resolution molecular epidemiology and evolutionary history of HIV-1 subtypes in Albania. PLoS One 2008;3:e1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergne L, Peeters M, Mpoudi-Ngole E, et al. : Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: Evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol 2000;38:3919–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albert J, Berglund T, Gisslen M, et al. : Risk of HIV transmission from patients on antiretroviral therapy: A position statement from the Public Health Agency of Sweden and the Swedish Reference Group for Antiviral Therapy. Scand J Infect Dis 2014;46:673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammer SM: HIV drug resistance: Implications for management. Top HIV Med 2002;10:10–15 [PubMed] [Google Scholar]
- 43.Montagna C, Mazzuti L, Falasca F, et al. : Trends in drug resistance-associated mutations in a real-life cohort of Italian patients infected with HIV-1. J Glob Antimicrob Resist 2015;3:267–272 [DOI] [PubMed] [Google Scholar]
- 44.Frentz D, Boucher CA, van de Vijver DA: Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS Rev 2012;14:17–27 [PubMed] [Google Scholar]
- 45.Maggiolo F, Ripamonti D, Arici C, et al. : Simpler regimens may enhance adherence to antiretrovirals in HIV-infected patients. HIV Clin Trials 2002;3:371–378 [DOI] [PubMed] [Google Scholar]
- 46.Julg B, Bogner JR: Atriplatrade mark—HIV therapy in one pill. Ther Clin Risk Manag 2008;4:573–577 [PMC free article] [PubMed] [Google Scholar]
- 47.Calva JJ, Larrea S, Tapia-Maltos MA, et al. : The decline in HIV-1 drug resistance in heavily antiretroviral-experienced patients is associated with optimized prescriptions in a treatment roll-out program in Mexico. AIDS Res Hum Retroviruses 2017;33:675–680 [DOI] [PubMed] [Google Scholar]
- 48.Sepulveda-Torres LDC, Rishishwar L, Rogers ML, et al. : A decade of viral mutations and associated drug resistance in a population of HIV-1+Puerto Ricans: 2002–2011. PLoS One 2017;12:e0177452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller V, Ait-Khaled M, Stone C, et al. : HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 2000;14:163–171 [DOI] [PubMed] [Google Scholar]
- 50.Molina JM, Marcelin AG, Pavie J, et al. : Didanosine in HIV-1-infected patients experiencing failure of antiretroviral therapy: A randomized placebo-controlled trial. J Infect Dis 2005;191:840–847 [DOI] [PubMed] [Google Scholar]
- 51.Eron JJ, Jr, Bosch RJ, Bettendorf D, et al. : The effect of lamivudine therapy and M184 V on the antiretroviral activity of didanosine. J Acquir Immune Defic Syndr 2007;45:249–251 [DOI] [PubMed] [Google Scholar]
- 52.Larder BA, Kemp SD, Harrigan PR: Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 1995;269:696–699 [DOI] [PubMed] [Google Scholar]
- 53.Gotte M, Arion D, Parniak MA, Wainberg MA: The M184 V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J Virol 2000;74:3579–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frost SD, Nijhuis M, Schuurman R, Boucher CA, Brown AJ: Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: The relative roles of drift and selection. J Virol 2000;74:6262–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diallo K, Gotte M, Wainberg MA: Molecular impact of the M184 V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 2003;47:3377–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Tamura K, Nei M: MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci 1994;10:189–191 [DOI] [PubMed] [Google Scholar]
- 57.Database SUHDR: Available at https://hivdb.stanford.edu/dr-summary/resistance-notes/NRTI/ accessed April2017
- 58.Harrigan PR, Stone C, Griffin P, et al. : Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592 U89) after monotherapy and combination therapy. CNA2001 Investigative Group. J Infect Dis 2000;181:912–920 [DOI] [PubMed] [Google Scholar]
- 59.Rhee SY, Liu T, Ravela J, Gonzales MJ, Shafer RW: Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob Agents Chemother 2004;48:3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eshleman SH, Jones D, Galovich J, et al. : Phenotypic drug resistance patterns in subtype A HIV-1 clones with nonnucleoside reverse transcriptase resistance mutations. AIDS Res Hum Retroviruses 2006;22:289–293 [DOI] [PubMed] [Google Scholar]
- 61.Harrigan PR, Mo T, Wynhoven B, et al. : Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2005;19:549–554 [DOI] [PubMed] [Google Scholar]
- 62.Melikian GL, Rhee SY, Varghese V, et al. : Non-nucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance: Implications for preclinical evaluation of novel NNRTIs and clinical genotypic resistance testing. J Antimicrob Chemother 2014;69:12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Luca A: The impact of resistance on viral fitness and its clinical implications. In: Antiretroviral Resistance in Clinical Practice (Geretti AM, ed.) London, Mediscript, 2006 [PubMed] [Google Scholar]
- 64.Martinez-Picado J, Savara AV, Sutton L, D'Aquila RT: Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol 1999;73:3744–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menzo S, Monachetti A, Balotta C, et al. : Processivity and drug-dependence of HIV-1 protease: Determinants of viral fitness in variants resistant to protease inhibitors. AIDS 2003;17:663–671 [DOI] [PubMed] [Google Scholar]
- 66.Devereux HL, Emery VC, Johnson MA, Loveday C: Replicative fitness in vivo of HIV-1 variants with multiple drug resistance-associated mutations. J Med Virol 2001;65:218–224 [DOI] [PubMed] [Google Scholar]
- 67.Craig C, Race E, Sheldon J, et al. : HIV protease genotype and viral sensitivity to HIV protease inhibitors following saquinavir therapy. AIDS 1998;12:1611–1618 [DOI] [PubMed] [Google Scholar]
- 68.Rhee SY, Taylor J, Fessel WJ, et al. : HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother 2010;54:4253–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vermeiren H, Van Craenenbroeck E, Alen P, et al. : Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J Virol Methods 2007;145:47–55 [DOI] [PubMed] [Google Scholar]
- 70.Dandache S, Sevigny G, Yelle J, et al. : In vitro antiviral activity and cross-resistance profile of PL-100, a novel protease inhibitor of human immunodeficiency virus type 1. Antimicrob Agents Chemother 2007;51:4036–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baxter JD, Dunn D, White E, et al. : Global HIV-1 transmitted drug resistance in the INSIGHT Strategic Timing of AntiRetroviral Treatment (START) trial. HIV Med 2015;16 Suppl 1:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fox Z, Phillips A, Cohen C, et al. : Viral resuppression and detection of drug resistance following interruption of a suppressive non-nucleoside reverse transcriptase inhibitor-based regimen. AIDS 2008;22:2279–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wheeler WH, Ziebell RA, Zabina H, et al. : Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses U.S.-2006. AIDS 2010;24:1203–1212 [DOI] [PubMed] [Google Scholar]
- 74.Wheeler VW, Radcliffe KW: HIV infection in the Caribbean. Int J STD AIDS 1994;5:79–89 [DOI] [PubMed] [Google Scholar]
- 75.Heslop OD, Smikle MF, Deer D, et al. : Human immunodeficiency virus type-1 (HIV-1) subtypes in Jamaica. West Indian Med J 2005;54:279–282 [DOI] [PubMed] [Google Scholar]
- 76.Vaughan HE, Cane P, Pillay D, Tedder RS: Characterization of HIV type 1 clades in the Caribbean using pol gene sequences. AIDS Res Hum Retroviruses 2003;19:929–932 [DOI] [PubMed] [Google Scholar]
- 77.Noel RJ, Jr, Chaudhary S, Rodriguez N, Kumar A, Yamamura Y: Phylogenetic relationships between Puerto Rico and continental USA HIV-1 pol sequences: A shared HIV-1 infection. Cell Mol Biol (Noisy-le-grand) 2003;49:1193–1198 [PubMed] [Google Scholar]
- 78.Blanco M, Machado LY, Diaz H, Ruiz N, Romay D, Silva E: HIV-1 genetic variability in cuba and implications for transmission and clinical progression. MEDICC Rev 2015;17:25–31 [DOI] [PubMed] [Google Scholar]
- 79.Machado LY, Blanco M, Dubed M, et al. : HIV type 1 genetic diversity in newly diagnosed Cuban patients. AIDS Res Hum Retroviruses 2012;28:956–960 [DOI] [PubMed] [Google Scholar]
- 80.Cuevas MT, Ruibal I, Villahermosa ML, et al. : High HIV-1 genetic diversity in Cuba. AIDS 2002;16:1643–1653 [DOI] [PubMed] [Google Scholar]
- 81.Walker PR, Pybus OG, Rambaut A, Holmes EC: Comparative population dynamics of HIV-1 subtypes B and C: Subtype-specific differences in patterns of epidemic growth. Infect Genet Evol 2005;5:199–208 [DOI] [PubMed] [Google Scholar]
- 82.Martinez-Cajas JL, Pant-Pai N, Klein MB, Wainberg MA: Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: A systematic review of virologic and biochemical evidence. AIDS Rev 2008;10:212–223 [PubMed] [Google Scholar]
- 83.Riemenschneider M, Cashin KY, Budeus B, et al. : Genotypic prediction of co-receptor tropism of HIV-1 subtypes A and C. Sci Rep 2016;6:24883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walter BL, Armitage AE, Graham SC, et al. : Functional characteristics of HIV-1 subtype C compatible with increased heterosexual transmissibility. AIDS 2009;23:1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Montano SM, Sanchez JL, Laguna-Torres A, et al. : Prevalences, genotypes, and risk factors for HIV transmission in South America. J Acquir Immune Defic Syndr 2005;40:57–64 [DOI] [PubMed] [Google Scholar]
- 86.Haggblom A, Svedhem V, Singh K, Sonnerborg A, Neogi U: Virological failure in patients with HIV-1 subtype C receiving antiretroviral therapy: An analysis of a prospective national cohort in Sweden. Lancet HIV 2016;3:e166–e174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sutherland KA, Ghosn J, Gregson J, et al. : HIV-1 subtype influences susceptibility and response to monotherapy with the protease inhibitor lopinavir/ritonavir. J Antimicrob Chemother 2015;70:243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Easterbrook PJ, Smith M, Mullen J, et al. : Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc 2010;13:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bousheri S, Burke C, Ssewanyana I, et al. : Infection with different hiv subtypes is associated with CD4 activation-associated dysfunction and apoptosis. J Acquir Immune Defic Syndr 2009;52:548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vasan A, Renjifo B, Hertzmark E, et al. : Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis 2006;42:843–852 [DOI] [PubMed] [Google Scholar]
- 91.Raymond S, Delobel P, Chaix ML, et al. : Genotypic prediction of HIV-1 subtype D tropism. Retrovirology 2011;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Requejo HI: Worldwide molecular epidemiology of HIV. Rev Saude Publica 2006;40:331–345 [DOI] [PubMed] [Google Scholar]
- 93.Lau KA, Wong JJ: Current trends of HIV recombination worldwide. Infect Dis Rep 2013;5(Suppl 1):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Delgado E, Thomson MM, Villahermosa ML, et al. : Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J Acquir Immune Defic Syndr 2002;29:536–543 [DOI] [PubMed] [Google Scholar]
- 95.Harris B, von Truchsess I, Schatzl HM, Devare SG, Hackett J, Jr: Genomic characterization of a novel HIV type 1 B/G intersubtype recombinant strain from an injecting drug user in Germany. AIDS Res Hum Retroviruses 2005;21:654–660 [DOI] [PubMed] [Google Scholar]
- 96.Esteves A, Parreira R, Piedade J, et al. : Spreading of HIV-1 subtype G and envB/gagG recombinant strains among injecting drug users in Lisbon, Portugal. AIDS Res Hum Retroviruses 2003;19:511–517 [DOI] [PubMed] [Google Scholar]
- 97.Orisatoki RO, Oguntibeju OO, Truter EJ: The contributing role of tourism in the HIV/AIDS epidemic in the Caribbean. Niger J Med 2009;18:143–148 [DOI] [PubMed] [Google Scholar]
- 98.Padilla MB, Guilamo-Ramos V, Bouris A, Reyes AM: HIV/AIDS and tourism in the Caribbean: An ecological systems perspective. Am J Public Health 2010;100:70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guilamo-Ramos V, Jaccard J, McCarthy K, et al. : Taxonomy of Caribbean tourism alcohol venues: Implications for HIV transmission. Drug Alcohol Depend 2013;132:238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Surratt H: Sex work in the Caribbean Basin: Patterns of substance use and HIV risk among migrant sex workers in the US Virgin Islands. AIDS Care 2007;19:1274–1282 [DOI] [PubMed] [Google Scholar]
- 101.Deren S, Kang SY, Colon HM, Robles RR: The Puerto Rico-New York airbridge for drug users: Description and relationship to HIV risk behaviors. J Urban Health 2007;84:243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wolfers ME, van den Hoek C, Brug J, de Zwart O: Using Intervention Mapping to develop a programme to prevent sexually transmittable infections, including HIV, among heterosexual migrant men. BMC Public Health 2007;7:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cleghorn FR, Jack N, Carr JK, et al. : A distinctive clade B HIV type 1 is heterosexually transmitted in Trinidad and Tobago. Proc Natl Acad Sci U S A 2000;97:10532–10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yebra G, de Mulder M, Martin L, et al. : Most HIV type 1 non-B infections in the Spanish cohort of antiretroviral treatment-naive HIV-infected patients (CoRIS) are due to recombinant viruses. J Clin Microbiol 2012;50:407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Candotti D, Adu-Sarkodie Y, Davies F, et al. : AIDS in an HIV-seronegative Ghanaian woman with intersubtype A/G recombinant HIV-1 infection. J Med Virol 2000;62:1–8 [DOI] [PubMed] [Google Scholar]
- 106.Zhang M, Foley B, Schultz AK, et al. : The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 2010;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]