Abstract
Understanding the cellular uptake and intracellular trafficking of oligonucleotides provides an important basic underpinning for the developing field of oligonucleotide-based therapeutics. Whether delivered as “free” oligonucleotides, as ligand–oligonucleotide conjugates, or in association with various nanocarriers, all forms of oligonucleotide enter cells by endocytosis and are initially ensconced within membrane-limited vesicles. Accordingly, the locus and extent of release to the cytosol and nucleus are key determinants of the pharmacological actions of oligonucleotides. A number of recent studies have explored the intracellular trafficking of various forms of oligonucleotides and their release from endomembrane compartments. These studies reveal a surprising convergence on an early-intermediate compartment in the trafficking pathway as the key locus of release for oligonucleotides administered in “free” form as well as those delivered with lipid complexes. Thus, oligonucleotide release from multivesicular bodies or from late endosomes seems to be the crucial endogenous process for attaining pharmacological effects. This intrinsic process of oligonucleotide release may be amplified by delivery agents such as lipid complexes or small molecule enhancers.
Keywords: : antisense, trafficking, endosomal release
Introduction
It is apparent that antisense (ASO), splice switching (SSO), and siRNA oligonucleotides have the potential to address a variety of therapeutic targets that would be difficult to approach using small molecule drugs [1–4]. Currently, momentum in the oligonucleotide therapeutics field has increased with the FDA approval of several oligonucleotide-based drugs [5]. However, challenges remain to the widespread use of oligonucleotides in the clinic. One important challenge concerns attaining effective delivery of these, rather large, highly polar molecules to their therapeutic targets within cells [6,7].
A variety of delivery strategies for oligonucleotides have been developed, including lipid or polymer nanoparticles and receptor-targeted ligand–oligonucleotide conjugates [8–10]. In addition, new chemical modifications have enhanced the stability and deliverability of “free” oligonucleotides [11,12]. Nonetheless, all forms of oligonucleotides, whether “free” or associated with a delivery moiety, share the same mode of entry into cells, namely uptake by endocytosis [13,14]. This places the oligonucleotide within endomembrane vesicles that are part of a complex intracellular trafficking system; thus, the oligonucleotide is initially separated from its intended target in the cytosol or nucleus by a lipid bilayer membrane. However, it is clear that some escape takes place, thus allowing the observed pharmacological effects of oligonucleotides. How oligonucleotides exit endomembrane vesicles and the extent to which this happens are still poorly understood, but there has been some recent progress in this area. This article will review current understanding of the intracellular trafficking of oligonucleotides and their escape from endomembrane compartments. It will also offer a hypothesis concerning the underlying mechanisms. Several recent reviews have also discussed aspects of oligonucleotide trafficking and provide additional detail [7,15,16].
Overview of Intracellular Trafficking
Basic aspects of endocytosis and trafficking
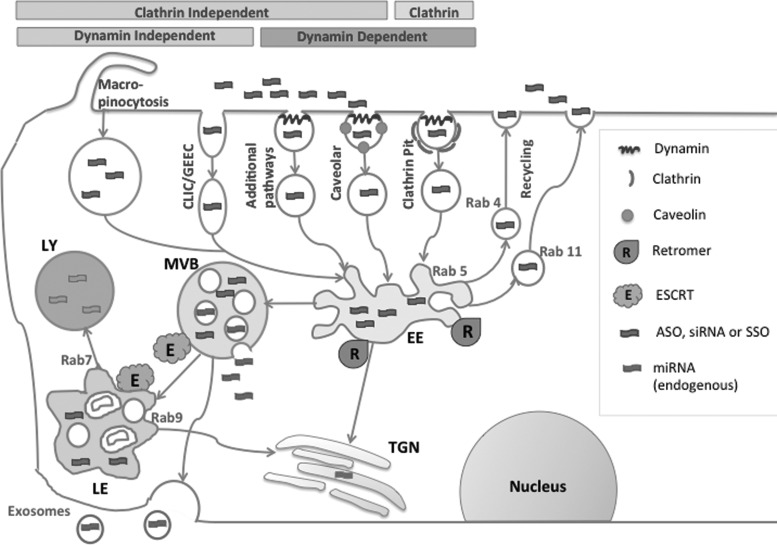
Most cells have several distinct pathways of endocytosis, including the well-known clathrin-coated pit and caveolar pathways as well as additional ones such as macropinocytosis and the CLIC/GEEC pathway that are important in fluid uptake [17]. Various forms of oligonucleotides use these pathways to different degrees.
All endocytotic pathways ultimately converge on early/recycling endosomes (EEs) that play a key role in the sorting of internalized materials [18]. EEs have a peripheral location, a highly tubulated structure, and an internal pH of 6.0–6.5. Materials destined for return to the cell surface enter the tubulations and are pinched off into small vesicles that will ultimately fuse with the plasma membrane. Materials in the lumen of the EE will be directed to downstream endomembrane compartments [19]. A third pathway from the EE is the retrograde route that carries materials to the trans-Golgi [20].
EEs primarily transition to multivesicular bodies (MVBs) that are nontubulated structures with a pH of about 5.5 and that contain multiple intraluminal vesicles (ILVs) that are formed by inward pinching off of the MVB membrane [21]. MVBs transition to late endosomes (LEs) that are pleomorphic structures having a perinuclear location and a pH of about 5 [22]. LEs can convey materials to the trans-Golgi, but the primary pathway involves fusion with lysosomes (LYs), the low pH (4.5), hydrolase-rich structures that are the cell's primary digestive apparatus [23]. Thus, the EE to MVB to LE to LY route is the primary pathway of intracellular trafficking [24]. However, as mentioned, there are branches from this main pathway.
The molecular machinery of trafficking
The endomembrane trafficking pathways comprise a very complex, highly dynamic aspect of cellular function. These pathways are mediated by a plethora of proteins and lipids that control the formation, coalescence, and scission of the various membrane compartments as well as the directed flow of internalized material through those compartments. Fortunately the molecular machinery underlying these events is becoming increasingly well understood.
The endomembrane trafficking pathway involves four basic steps: (i) pinching off (scission) of a membrane bound vesicle from a donor compartment; (ii) movement of the vesicle through the cytosol; (iii) recognition between the vesicle and a target compartment; and (iv) fusion of the vesicle and the target compartment. The first step is mediated by adaptor and coat proteins that recruit constituents of the donor membrane into a bud, followed by pinching off of the bud [25]. The coated pit process on the plasma membrane [26], which utilizes AP adaptors, clathrin coat proteins, and the dynamin GTPase to provide the drive for scission, is a familiar example, but many other types of coats function in vesicle budding in other compartments.
Movement of vesicles through the cytosol involves directed transport on microtubules or actin networks driven by kinesin, dynein, or myosin motor proteins [27]. Recognition between vesicle and target compartments is mediated by a functional class of proteins termed tethers [28]. The best understood examples are the golgins, tethers that function within the Golgi apparatus to direct vesicles to distinct Golgi subcompartments [29]. These extended proteins provide direct physical connection between the vesicle and the target compartment. Fusion between vesicle and target membranes is mediated by SNARE proteins, with v-SNARES on the vesicle interacting with t-SNARES on the target membrane to form a four–helix bundle. The bundle complex undergoes a conformational change that results in close apposition and eventual fusion of the two membranes [30].
These complex endomembrane trafficking events are guided by small GTPases, primarily those of the 80-member Rab family [31]. Various Rabs are involved in vesicle uncoating, interactions with the tubulin and actin networks, and the recognition and fusion events involving tether proteins and SNARES. Rabs also provide markers for individual endomembrane compartments [22]. For example Rab 5 and its activators and effectors are critically involved in the formation and maintenance of the early/recycling endosome compartment. As progression toward MVBs and LEs takes place, several protein complexes are recruited that displace Rab 5 and allow its replacement by Rab 7, which then serves as a marker and key functional molecule for LEs. Other Rab proteins also make significant contributions to trafficking. For example, Rab 4 and Rab 11, respectively, control fast and slow vesicle recycling pathways that carry material from the sorting endosome back to the plasma membrane, while Rab 9 controls a pathway that shuttles vesicles from LEs to the trans-Golgi.
Two other protein complexes deserve mention as they also play vital roles in trafficking. The ESCRT (endosomal sorting complex required for transport) machinery is composed of five multiprotein entities that have the capability of altering membrane topology [32]. The ESCRT machinery contributes to a number of cellular processes, including cytokinesis and formation of exosomes, but of interest here is its role in generating the ILVs that populate the lumens of both MVBs and LEs. Ubiquitination is a key signal for determining the molecules that are sorted to the ILVs. The Retromer is another multiprotein complex that can cause membrane deformation. Its role is to generate vesicles that shuttle from the early/recycling endosome to the trans-Golgi apparatus or to the cell surface [20].
In addition to proteins, various lipids play a role in trafficking. There is a clear gradient of lipid composition from EEs to LYs [33]. Variations in the amount of several phosphatidyl inositides seem to regulate the binding of key proteins to the membranes of particular compartments. In addition, variation in membrane deformability regulated by cholesterol and sphingomyelin may play a role in vesicle recognition and fusion. Certain lipids are predominately found in particular endomembrane compartments, with a good example being lysobisphosphatidic acid (LBPA) that is localized in LEs. An overview of the trafficking process is provided in Fig. 1.
FIG. 1.
Overview of the Endocytosis and Intracellular Trafficking of Oligonucleotides. Key elements of trafficking are depicted as described in the main text. The diagram does not show proposed sites of oligonucleotide release. EE, early endosome; MVB, multivesicular body; LE, late endosome, LY, lysosome; TGN, trans-Golgi network.
Trafficking of Oligonucleotides
Over the years since the advent of antisense technology, there have been a number of studies of the uptake, release, and subcellular distribution of oligonucleotides. Recently, these issues have been addressed with improved technologies, including advanced confocal microscopy as well as more selective chemical and molecular probes. Thus, this article will primarily deal with studies that were published during the last few years that have benefited from these newer technologies.
In considering the overall picture of oligonucleotide trafficking, some important distinctions must be made. First, there is a major difference in typical delivery approaches for siRNA versus single-stranded ASOs and SSOs. For siRNA, delivery usually involves a nanoscale carrier such as cationic lipid or polymer nanoparticles. In contrast, ASOs and SSOs, which usually have phosphorothioate (PS) backbones, can be taken up by cells by “gymnosis”, that is without assistance of a carrier. Clearly, there may be substantial differences in the trafficking of such different entities. However, as will be discussed below, recent evidence also suggests some commonalities in the mechanism of intracellular delivery. A second distinction needs to be made between trafficking within the endomembrane system and events that occur after the oligonucleotide leaves membrane-bound compartments. At that point, interactions with a variety of intracellular proteins will affect the pattern of oligonucleotide distribution between the cytosol and the nucleus.
Trafficking of oligonucleotides delivered with lipid complexes
Several excellent recent studies have used various advanced confocal fluorescence microscopy or electron microscopy techniques to explore the intracellular fate of siRNA delivered via small lipid nanoparticles (LNPs) or via larger lipoplexes (LPs). A 2013 study by Gilleron et al. used siRNA labeled either with a fluor or with gold nanoparticles and delivered into cells using ∼100 nm LNPs containing an ionizable lipid [34]. Using both chemical and siRNA inhibitors of components of the endocytosis machinery, the investigators determined that there was an early phase of uptake involving clathrin-coated pits and the LDL receptor followed by more extensive uptake involving macropinocytosis. Subsequent to initial uptake, siRNA was found first in EEs followed by progression to LEs and LYs. By examining the colocalization of fluor-tagged siRNA with marker proteins for specific endomembrane compartments, it was found that the LNPs induced formation of a hybrid EE/LE compartment. Escape from endomembrane compartments to the cytosol was quantitated and amounted to less than 2% of the siRNA that accumulated in cells. Mathematical modeling suggested that oligonucleotides were escaping from a specific intracellular compartment rather than from multiple compartments. By using inhibitors to block the progression of trafficking, it was determined that escape of siRNA took place from a relatively early compartment before transport to LEs or LYs.
In a somewhat similar 2013 study, Sahay et al. used advanced confocal microscopy to examine trafficking of fluorescent siRNA delivered with cationic LNPs [35]. Perturbation of the endocytotic machinery indicated that the LNPs were primarily taken up by macropinocytosis thus bypassing the coated pit machinery. This study suggested that most of the siRNA in the LNPs was routed to LEs and LYs. A striking discovery in this study was that the cholesterol transport protein NPC1 played an important role in the recycling and export of the LNP-siRNA from LEs. Thus, cells that were null for NPC1 expression accumulated increased amounts of fluor-tagged siRNA and were more susceptible to the “knock down” effects of a siRNA targeting EGFP. Further studies implicated a role for Rab 8a- and Rab 27b-regulated recycling pathways in controlling the level of intracellular siRNA. Thus, this study emphasized the role of recycling processes, as well as the initial uptake, in influencing subcellular levels of siRNA, particularly within LEs. A 2016 study from another group confirmed a key role for NPC1 in cationic LNP recycling. Thus, the small molecule NP3.47, an inhibitor of NPC1, caused in increased accumulation of labeled LNPs in LEs and LYs, increased effectiveness of LNP-siRNA, and reduced recycling of siRNA out of the cells [36]. Interestingly, in control cells, as much as 80% of the siRNA initially taken up by the cells was reexported to the medium over a 24 h period, thus confirming the importance of the recycling process.
In a 2015 publication, Wittrup et al. analyzed the uptake and trafficking of fluor-tagged siRNA delivered in relatively large cationic LPs [37]. The large amount of siRNA associated with each lipoplex permitted these investigators to follow the fate of siRNA in individual endocytotic vesicles. The siRNA was released as a burst followed by rapid diffusion throughout the cytosol. Only partial release was observed, and after one burst, there was no further release. Interestingly ‘knock down’ by the released siRNA was an all or nothing phenomenon as observed using cells containing a EGFP reporter. By expressing GFP-chimeras of proteins that are markers for individual membrane compartments, the investigators were able to identify the stage of trafficking, at which siRNA escape from individual vesicles took place. Thus, maximal association of the EE markers EEA1 and Rab 5 with the lipoplex-containing vesicle took place before bursting, while the LE marker Rab 7 was present during the burst period. The burst took place before any significant association of the lysosomal marker LAMP1 with the vesicle. Thus, as in the report by Gilleron et al., siRNA release was associated with a relatively early compartment in the trafficking pathway. The Wittrup article also examined the role of autophagy in siRNA trafficking. They found that autophagy markers such as LC3 were recruited to vesicles damaged by bursting, followed by formation of a typical autophagosome. As several galectin proteins bind to damaged membranes, the investigators used binding of YFP-galectins as a surrogate marker to trace the bursting process. This allowed them to evaluate trafficking using much smaller LNPs where the amount of fluorescent siRNA released is too small to be detected by microscopy. Overall, the process of uptake, bursting, and release was similar for LNPs and LPs. A rapid bursting event for siRNA release was also identified in another study using confocal FRET techniques to study the process [38].
Quite a different picture emerged from a study of mRNA delivery using LNPs [39]. The authors cleverly used haploid cells that were then gene-edited using CRISPR to delete expression of key trafficking modulators, including Rabs 4, 5, and 7. They found that deletion of Rab 7, but not Rabs 4 and 5, had a substantial effect on expression of the luciferase mRNA utilized in the study. The Rab 7 depleted cells also showed enlarged LEs and reduced LE to LY conversion. This led to impairment of LY-based signaling processes, particularly by the mTOR complex, which are important for mRNA translation. Thus, the limitation here is not so much delivery to the cytosol but rather the formation of a key endomembrane structure required for signaling.
Trafficking of oligonucleotides associated with other nanoparticles
Oligonucleotides can also be delivered using other types of nanoparticles, including cationic polymeric particles (polyplexes), gold nanospheres, and DNA nanostructures. There has been less mechanistic work regarding intracellular trafficking using these delivery methods than in the case of LNPs, but nonetheless some interesting reports have appeared. A recent review provides a good summary of the characteristics of polyplexes formed with polyethylenimine or other cationic polymers [40]. Positively charged cell penetrating peptides (CPPs) have also been used for oligonucleotide delivery both when conjugated to the nucleic acid and also in the form of oligo-CPP nanocomplexes, as recently reviewed [41].
Initial uptake of both polyplexes and oligo-CPP complexes involves scavenger receptors followed by endocytosis [42]. It is interesting to compare the uptake and trafficking of polyplexes versus LPs, a topic that was reviewed some time ago [43]. More recent work has examined some of the mechanistic differences between lipoplex and polyplex delivery. Thus, one study found that polyplexes enter cells by a pathway that is independent of both clathrin and caveolin [44]. However, another study using CPP-oligo complexes found a role for clathrin-mediated uptake [45]. Another study of polyplexes used live cell fluorescence microscopy to visualize the trafficking pathway of both the antisense oligonucleotide and the positive polymer [46]. Following initial uptake, the endosomal compartments containing the polyplexes undergo acidification. Release of oligonucleotide to the cytosol takes place via a rapid bursting mechanism with subsequent diffusion and accumulation in the nucleus. Interestingly, such bursting events are rare and only one of several polyplex-containing endosomes within a cell contribute to cytosolic and nuclear fluorescence. In contrast to the oligonucleotide, the polymer portion of the polyplex remains in the endomembrane compartment. Thus, the intracellular behavior of the antisense oligonucleotide polyplexes in this study is reminiscent of the behavior observed for siRNA in large LPs [37]. Trafficking of polyplexes to acidic compartments, including LEs and lysosomes, was also observed in another study [47].
A recent report used both fluorescent microscopy and a novel organelle tagging technique to follow polyplexes through the uptake and trafficking process [48]. This study showed rapid passage through EE and LE compartments and accumulation in lysosomes. Recycling and exocytosis have also been reported to play an important role in trafficking of polyplexes, with the intact polyplex being released from the cell [49]. In a surprising contrast to LNPs, the NPC1 transporter that is involved in lipid recycling enhances rather than inhibits the delivery capabilities of polyplexes [50]. An interesting emerging issue for both LNP and polyplex delivery is the potential role of autophagy. Thus, reports have indicated that damage to endosomes caused by delivery agents can trigger the autophagic process both for LPs and for polyplexes [37,51,52]. However, it is not clear to what degree autophagosome formation affects the functionality of the delivered oligonucleotide, with some reports indicating a significant effect via modulation of autophagy [51] and others indicating the opposite [37]. In summary, there seem to be some significant differences in the uptake and trafficking behavior of polyplexes versus small LNPs, with perhaps more similarity to the behavior of larger LPs. However, for polyplexes, there remain many unanswered questions concerning the precise locus of oligonucleotide release as well as the mechanism of release.
Another interesting type of oligonucleotide nanocarrier is the “spherical nucleic acid” (SNA) nanoparticle comprised siRNA or ASOs tightly, but noncovalently bound to gold nanospheres [53]. These have been tested extensively in animal models of cancer and other diseases and have some desirable characteristics, including a high payload of siRNA, and relatively small size [54]. In addition to gold, SNAs can be formed from other nanostructures. In one case, ASO SNAs based on gold nanoparticles and on quantum dots were carefully examined in terms of cellular uptake and trafficking [55]. The SNAs were found to traffick to LEs but did not progress to LYs. A tiny fraction of intact SNA escaped to the cytosol; the authors suggest that it is this fraction that is responsible for the pharmacological effects of the SNAs. The bulk of the material was degraded in LEs, and the oligonucleotide fragments were released from the cells while the nanoparticles were retained in endosomes. Obviously, the behavior of these solid nonbiodegradable nanoparticles is quite different from that of biodegradable LNPs or polyplexes.
DNA or RNA nanostructures offer another interesting alternative for oligonucleotide delivery [56]. Similar to SNAs, the nanostructures are considerably smaller than typical lipid or polymer nanoparticles, and this may offer some advantages for in vivo distribution. Also, nanostructures can directly incorporate the ASO or siRNA as part of the structure and thus may have a higher ratio of active agent to carrier than is the case for conventional nanoparticles. Thus far, there has been only very limited mechanistic work concerning oligonucleotide delivery using nanostructures. A very interesting report using tetrahedral nanostructures decorated with targeting ligands showed that the placement of the ligand had an important influence on the effectiveness of the siRNA component, although the reason for this was not clear [57]. Another report used single-particle tracking to investigate the fate of DNA nanostructures in cells, but this was not coupled to functional effects [58].
To summarize, while there has been great interest in using various types of polymeric or solid nanoparticles for oligonucleotide delivery, there has only been limited exploration of the underlying mechanisms of cellular uptake and trafficking.
Trafficking of free oligonucleotides
Cellular uptake of “free” siRNA is very poor since its phosphodiester backbone fails to bind significantly to cell surfaces. The same is true of single-stranded oligonucleotides having morpholino or peptide nucleic acid backbones. These types of molecules require either incorporation into a carrier complex or conjugation to a targeting ligand to permit efficient uptake. In contrast, free PS-based ASOs and SSOs are efficiently taken up by many types of cells by a process that is sometimes termed “gymnosis” [59]. Initial binding of PS oligos to the cell surface may involve a variety of receptors and may differ in various cell types [6–8]. The intracellular trafficking processes of PS oligos have recently been studied in some detail by groups at the City of Hope medical center and at Ionis Pharmaceuticals. There are both convergent and divergent aspects of the interpretations reached by the two groups.
An early study from the Ionis group delineated the existence of productive and nonproductive pathways for uptake and trafficking of PS-ASOs [60]. Cellular uptake involved an endocytotic process that was not dependent on clathrin or caveolin, and most of the ASO accumulated in membrane bound vesicles. The nonproductive uptake was thought to involve ASO accumulation in LYs. A subsequent study delineated a role for annexin 2, a membrane associated protein, in the trafficking of ASOs from early to LEs and suggested that release of ASO to the cytosol took place at the LE stage [61]. Further studies from this group examined the relationship between PS-ASO trafficking and the LE lipid LBPA [62]. Colocalization of this lipid and the ASO was observed in the ILVs that are found within LEs. In addition, treatments that reduced or inhibited LBPA interfered with the antisense effect. The authors suggest that back fusion of LBPA-rich ILVs with the LE membrane may provide a mechanism of release of the ASO to the cytosol. However, in this study, the authors found no role for the ESCRT machinery in the delivery of the ASOs. This is surprising in view of the importance of this complex in ILV generation and in view of work from another group that used a siRNA library to identify TSG101, an ESCRT component, as important in oligonucleotide delivery [63]. As an aside, it is interesting to note that LBPA has recently been identified as having a key role in the escape of a CPP from LEs [64], thus suggesting a commonality in oligonucleotide and peptide trafficking mechanisms. In summary, work from the Ionis group emphasizes the role of components of the endomembrane trafficking machinery and suggests that ASO access to the cytosol and nucleus takes place at the LE stage with the involvement of ILVs.
A somewhat different picture emerges from the work of the group at City of Hope. In their original publication, they emphasized that oligonucleotides taken up by gymnosis primarily colocalized with cytoplasmic bodies, initially identified as P-bodies, which are organelles associated with RNA processing [59]. In a subsequent report, the investigators observed uptake of ASOs into endomembrane compartments, particularly Rab 7 positive LEs, but also substantial colocalization with GW bodies, another RNA processing site [65]. They also found an association of the ASO with Ago2 and that reduction in Ago2 levels led to impaired antisense effects. Based on this and on other observations, the authors suggest a cytoplasmic Ago2-dependent mechanism for ASO-mediated message degradation; however, this mechanism is distinct from RNA cleavage via the RISC complex.
Another study from this group focused on trafficking of ASOs in the endomembrane system as regulated by protein kinase C alpha (PKC-α) [66]. It had been known previously that PKC isoforms play a role in regulating sorting in the intracellular trafficking pathway. In this study, using both chemical inhibitors and siRNA, levels of PKC-α activity were reduced, leading to inhibition of ASO activity. By contrast, increasing PKC-α activity improved ASO effects. These observations were ascribed to the role of PKC-α in promoting maturation of EEs to LEs. In summary, work from this group agrees with that of the Ionis group in emphasizing ASO trafficking to LEs as a critical step in delivery. However, it diverges in its emphasis on a key role for a cytoplasmic mechanism of mRNA degradation that involves Ago2 and in suggesting that high levels of ASO are associated with GW- or P-bodies. It should be noted that other groups investigating uptake by gymnosis have observed oligonucleotides primarily associated with endomembrane compartments rather than other cytoplasmic structures [67]. Another point to mention is that it is becoming clear that RNaseH-mediated RNA degradation can take place in the cytosol as well as the nucleus [68]; thus, it may be possible to reconcile divergent views on the importance of nuclear versus cytosolic ASO effects.
Trafficking of ligand-conjugated oligonucleotides
Conjugation of single- or double-stranded oligonucleotides with a ligand that displays high affinity for a particular receptor is potentially a powerful approach for increasing the extent and selectivity of delivery. In this study, we will focus on conjugates using relatively small peptide or carbohydrate ligands rather than antibodies, aptamers, or other macromolecular ligands.
The most advanced molecules to date are the siRNA glycoconjugates developed by Alnylam Pharmaceuticals. Conjugated siRNAs using triantennary N-acetyl galactosamine (GalNac) ligands were taken up by hepatic asialoglycoprotein receptors with great efficiency and attained effective target knockdown both in cultured hepatocytes and in mouse liver [69]. Although the first clinical trial with GalNac siRNA was terminated because of unanticipated mortality in the treated group, this exciting approach continues to generate a great deal of interest with additional preclinical work and clinical trials continuing [70]. In addition to siRNA conjugates, work is also proceeding with GalNac ASO conjugates [71] as well as with highly chemically modified charge neutralized versions of siRNA [72]. Surprisingly, given the great potential of this approach, there has been little detailed work on the intracellular trafficking of the siRNA glycoconjugates. One important question is whether the substantial pharmacological effectiveness of the GalNac oligonucleotide conjugates depends on some unique aspect of the hepatic endomembrane trafficking system that is not found in other cell types.
Small peptides that bind to specific receptors constitute another important class of ligands for oligonucleotide conjugates. Early studies utilized SSOs conjugated with bivalent RGD peptides that interact with certain integrins, as well as bombesin peptides that interact with a G-protein coupled receptor [73,74]. These studies demonstrated receptor specific uptake by clathrin or caveolar mechanisms followed by trafficking of the conjugates to late endosomal compartments. Also observed was a relatively slow onset of pharmacological effect compared to delivery via lipid transfection, being more similar to the case of gymnosis of unmodified ASOs [59]. Continued studies of this type revealed that the uptake pathway could significantly affect the downstream pharmacological activity. Thus, by varying the concentration of oligonucleotide, equal uptake was attained for RGD-conjugated versus unconjugated SSOs; however, the conjugate always produced a more robust effect [75]. A similar observation was made using RGD-conjugated siRNAs of varying valency. Thus, while bi-, tri-, and tetravalent conjugates accumulated in cells to an equal degree, only the tri- and tetravalent ones effectively ‘knocked down’ the target message [76]. Interestingly, such highly ligand-dependent pharmacological effects of oligonucleotides have been seen in other, very different, delivery systems, including DNA nanostructures [57] and CpG oligonucleotide conjugates [77].
There has been substantial interest in conjugation of siRNA with lipophilic ligands such as cholesterol. This favors association of the siRNA with lipoproteins in the blood and subsequent uptake by hepatic lipoprotein receptors [1]. However, until recently, the cellular trafficking mechanisms of cholesterol-siRNA had not been well explored. An interesting report has now shed some light on this topic by following the cellular fate of a fluor-tagged, chemically modified, cholesterol-conjugated siRNA [78]. Interestingly, the Chol-siRNA trafficked in a pathway similar to EGF peptide that involved EEs marked by the tethering protein EEA1. In contrast, the Chol-siRNA did not enter a pathway used by transferrin that involves EEs marked by Rabenosyn-5, a Rab 5 effector protein. EEA1 endosomes usually traffick toward LEs and LYs, while Rabenosyn-5 endosomes tend to recycle to the plasma membrane. Thus, this report also suggests the importance of the trafficking pathway and of delivery to LEs for robust siRNA action.
Conjugates of oligonucleotides with CPPs are also an interesting approach for delivery [79,80]. In particular, investigators have extensively studied conjugates of CPPs with uncharged morpholino or PNA oligonucleotides, especially in the context of therapy of Duchenne muscular dystrophy and of other neuromuscular disease [81]. Early studies confirmed that CPP-oligo conjugates were taken up by endocytosis and displayed limited release from endomembrane vesicles [82]. A more recent study compared uptake and trafficking of CPP-morpholino oligo conjugates in skeletal muscle cells versus cardiomyocytes and found differences in uptake pathway (caveolin vs clathrin) and in distribution to the nucleus and subsequent pharmacological effect [83]. Thus, despite their potential importance for in vivo delivery of uncharged oligonucleotides, there is only a limited amount of detailed information available concerning intracellular trafficking of CPP-oligonucleotide conjugates.
Small molecules that affect oligonucleotide trafficking
The complex protein and lipid machinery of intracellular trafficking affords a number of targets for small molecules that might be used to manipulate trafficking processes. Thus, during the last few years, several groups have pursued this approach in the context of oligonucleotide delivery. An early report from my group found that a compound termed Retro-1, that is known to block the retrograde trafficking pathway, could enhance the effectiveness of ASOs and SSOs in cell culture, as well as that of SSOs in a mouse model [84]. Retro-1 caused increased redistribution of oligonucleotides from endosomes to the nucleus, but did not strongly affect lysosomes.
Encouraged by these results, we undertook high-throughput screening (HTS) and discovered two novel families of small molecules that strongly enhanced the pharmacological effects of oligonucleotides by affecting their intracellular trafficking [85,86]; we term such molecules OECs (oligonucleotide-enhancing compounds). Other groups have also used HTS or directed discovery to find small molecules that enhance oligonucleotide effects by a variety of means [39,87–89]. In a recent report, we investigated the mechanism of action of OECs in some detail [90]. We determined that low but effective concentrations of OECs released oligonucleotides from an early compartment in the trafficking pathway, before entry into low pH compartments. However, EEs themselves were refractory to the effects of OECs. Thus, the most likely site of oligonucleotide release seems to be an intermediate compartment such as MVBs or a mixed EE/LE population. At higher concentrations, OECs could cause loss of protons from lysosomes, but the mechanism of toxicity was most closely associated with an increase in plasma membrane permeability.
Interactions of oligonucleotides with cytosolic or nuclear proteins
Following release from membrane-limited compartments, oligonucleotides can interact with multiple proteins in the cytosol or nucleus [6]. These interactions might affect the intracellular distribution and pharmacological capabilities of the oligonucleotide. Several interesting recent reports have explored this theme. One example of this is the report that Ago2 binds and affects the actions of ASOs [65]. However, this topic has been most extensively studied by Crooke and colleagues who have examined the association of ASOs with numerous proteins, including chaperonins, Hsp90, and RNA-binding proteins [91–93], and have shown effects on oligonucleotide distribution and on pharmacological actions.
Mechanistic Aspects of Oligonucleotide Trafficking and Release from Endosomes
As outlined above, the intracellular fate of oligonucleotides has been examined in most detail in three very distinct delivery contexts; first, siRNA associated with LNPs or LPs, second, “free” ASOs, and third, delivery using small molecules. Remarkably, there is a clear convergence on an early stage of the trafficking pathway as the key site of oligonucleotide release to the cytosol. This, despite the fact that both the delivery methodologies and the cell types used were quite varied. Thus, with lipid particles, oligonucleotide release has been described as being in a mixed EE/LE population [34], or after Rab 5, coincident with Rab 7, and before LAMP1 association with vesicles [37]. In the case of free ASOs, oligonucleotide release from LEs by means of back fusion of ILVs has been proposed [62]. For oligonucleotide enhancing, small molecules release was described as being triggered after the EE stage, but before entry into highly acidic downstream compartments [90]. Interestingly, despite the fact that a substantial amount of oligonucleotide accumulates in LYs release from this compartment does not seem to make a strong contribution to the pharmacological effects of the oligonucleotide, at least in the case of ASOs or SSOs taken up by gymnosis [60,86,90]. These varied observations strongly indicate that there is an early-intermediate locus in the trafficking pathway that is uniquely predisposed to release oligonucleotides from endomembrane vesicles. A question then is what factors determine the site of oligonucleotide release to the cytosol?
While the entire intracellular trafficking pathway is dynamic, there are some stages that are particularly active in terms of small vesicles budding from or fusing to larger membrane bound compartments. This includes the trafficking of vesicles to the plasma membrane from recycling endosomes, the retrograde diversion of materials from EEs to the Golgi, and the action of the ESCRT complex and of LBPA in the formation of ILVs within MVBs [20,32,94]. It seems likely that oligonucleotide release might take place at these highly dynamic loci. During the processes of vesicle scission and fusion, defects in the lipid bilayer can occur that may permit release of vesicle contents [95]. Thus, the “stalk” that develops between membrane compartments during scission or fusion [96] is known to contain nonbilayer regions [97,98], and such regions are known to have increased permeability [99,100]. In the case of delivery via LNPs or LPs, the cationic lipids used can promote formation of nonbilayer regions in the membrane [101], thus enhancing the endogenous process. Interestingly, both early/recycling endosomes and MVBs/LEs have multiprotein complexes that actively generate membrane buds. Yet, evidence to date suggests that EEs do not readily release their contents while MVBs or LEs do so. Is this difference based on differences in the trafficking machinery in these compartments?
The heterotrimeric Retromer complex has been known to play a key role in formation and delivery of membranous vesicles from EEs to the trans-Golgi, although recent evidence suggests that it also contributes to Rab 4- or Rab 11-mediated recycling from EEs to the plasma membrane [102]. The core complex is associated with several SNX proteins that contain BAR domains capable of membrane sensing and stabilizing membrane curvature, while PX domains allow recognition of the phosphoinositides typical of EEs. These characteristics support the ability of the Retromer to alter membrane topology and generate vesicles from EEs that then traffic to other sites.
The five multiprotein complexes of the ESCRT system (ESCRTs -0, -I, -II, -III, and the Vps4 complex) drive the formation of ILVs as well as cytokinesis and the budding of membrane vesicles or virus particles at the cell surface. Importantly, the topology of ESCRT-mediated budding is the converse of that which occurs during all other aspects of endocytosis and trafficking [103]. The ESCRT III complex is most directly involved in the membrane sculpting involved in ILV generation. Under the influence of the earlier ESCRT components, the multiple protein subunits of ESCRT III interact with acidic lipids on the endosome cytoplasmic surface and undergo oligomerization. This results in the formation of coiled protein filaments that drive invagination of a membrane bud into the endosome interior and ultimately scission of the bud to form an ILV. It should be noted that several alternative mechanisms of ILV formation have been described involving various components of the ESCRT system as well as accessory proteins [104].
An important aspect of membrane dynamics in trafficking concerns the role of lipids. For example, in studies of SNARE-mediated membrane fusion, it has become clear that the recruitment of fusogenic lipids to the fusion site plays a role in the process [98]. As mentioned previously, there are gradients of lipid composition in the endomembrane trafficking pathway [33]. Of particular interest is the unsaturated anionic lipid LBPA that has been implicated in the trafficking of both oligonucleotides [62] and peptides [64]. LBPA is located on the inner face of MVB/LE membranes and in ILVs [105]. LBPA cooperates with the ESCRT-associated protein Alix to regulate aspects of vesicle biogenesis and the sorting of endosomal constituents [106]. LBPA and Alix seem to be particularly important in the process of back fusion, whereby ILVs fuse with the limiting membrane of the MVB or LE compartments. The back fusion process has been suggested as a key site of release of “free” ASOs [62]. This would entail transfer of oligonucleotides across a lipid bilayer membrane into the lumen of the ILV. At first, this seems a daunting prospect considering the polar nature of oligonucleotides. However, it should be noted that ILVs are quite small (∼50 nanometers) and thus have a very high membrane curvature [107]. This produces strain on the bilayer [100] and, in conjugation with membrane perturbing lipids such as LBPA, may induce transient formation of leaky nonbilayer sites in the ILV membrane, thus permitting oligonucleotide escape. However, questions remain as to the precise role of LBPA in ASO release and whether LPBA/Alix also plays a role in release of oligonucleotides delivered by other means such as LNPs.
The main point of this short summary is to point out that the membrane budding processes catalyzed by the Retromer and those catalyzed by the ESCRT complex are very different both topologically and mechanistically. It seems possible that ESCRT-mediated budding creates membrane defects that are ‘leakier’ than those created by the Retromer complex, and that this is the basis for oligonucleotide escape from intermediate compartments rather than EEs.
It is worthwhile to consider how some of these questions about the site and mechanism of oligonucleotide release might be answered. One approach could be the use of model systems. Our understanding of the basic biology of intracellular trafficking owes a great deal to experiments where proteins or lipids thought to be involved in trafficking were reconstituted into model membranes (liposomes) [98]. An interesting relevant example is a study that showed that LPBA itself could cause invagination of liposome membranes to form ILV-like structures [108]. Thus, reconstitution of Retromer or ESCRT-III constituents, as well as LBPA, into oligonucleotide-loaded liposomes may provide some very interesting insights that could precisely identify the key molecules involved in oligonucleotide release from endomembrane compartments.
Another set of related questions concerns whether oligonucleotides are released as a burst or gradually, whether release from an endosome is complete or not, whether release alters the morphology of the endomembrane compartment, and in the case of delivery with a carrier, whether the carrier is also released. The existing literature is somewhat confused regarding these issues. Thus, one study with lipid complexes suggested a bursting mechanism [37], while another indicated gradual release [34]. Studies with polyplexes have suggested that the carrier moiety largely remains within endosomes [46], while studies with lipid carriers suggest extensive recycling of siRNA to the exterior of the cell via lipid transport mechanisms that presumably also transport the carrier [36]. Some reports have indicated that generation of leaky endosomes is followed by autophagy (and thus eventual digestion of the endosome)[37,51]. However, we have not observed any substantial changes in the extent or morphology of endomembrane structures during the oligonucleotide release process mediated by small molecules [90]. Yet, another issue is whether there are size limitations on escape of oligonucleotides from endosomes. Clearly cationic lipid carriers can deliver very large cargoes such as mRNA or plasmids. However, what about the endogenous mechanism that leads to escape of ASOs taken up by gymnosis or the escape mediated by small molecule enhancers? Many of these contradictions and questions should be addressable using current sophisticated quantitative microscopic techniques along with appropriate fluorescent probes. Thus, dual tagging of the oligonucleotide and its carrier, as well as simultaneous expression of a compatible fluorescent compartment marker protein, could yield insight into multiple unresolved questions about the site and extent of oligonucleotide release. Likewise conjugation of oligonucleotides to dextrans of different average molecular weight could help probe the size restrictions of the escape pathway.
As discussed above, recent studies have tracked the site of oligonucleotide release to an early-intermediate locus in the trafficking pathway, perhaps MVBs or an EE/LE hybrid. However, the broad definitions of compartments may need further refinement as it is becoming clear that each major endomembrane compartment likely has subcompartments. A very relevant example of this is the study that showed that cholesterol-conjugated oligonucleotides were associated with EEs marked with EEA1 but not those marked with Rabenosyn-5 [78]. Going further afield, within the Golgi system, it is clear that there are subpopulations of vesicles that traffick between different compartments and that are marked by different tethering proteins and SNARES [29]. As another example, in the trafficking of alpha-adrenorecptors, there are distinct pools of endosomes that signal differently [109]. Thus, a key question is whether oligonucleotide release takes place from an entire major compartment or whether there is a specific subcompartment that is responsible.
Another potential concern is whether attempts to enhance the delivery of oligonucleotides will disrupt endogenous trafficking pathways and the signaling processes that accompany them. An interesting, although rather extreme, example of this is the well-known study showing that very high levels of expression of shRNA can impair nuclear–cytosol trafficking of endogenous miRNA [110]. Thus, studies where oligonucleotide trafficking, with or without use of delivery agents, is examined in parallel with studies of trafficking of well-understood markers such as transferrin or LDL may help to identify potential deleterious effects on endogenous mechanisms.
Finally, playing the role of devil's advocate, one could ask whether it is really important to identify compartments associated with oligonucleotide release, and whether trying to enhance release is a good idea or not. The growing evidence that an intermediate compartment is involved may simply mean that there is more oligonucleotide in that compartment than anywhere else rather than that there is a selective release. Quantitative fluorescence microscopy with appropriate protein compartment markers could measure the total content of oligonucleotide in various subcellular compartments and thus answer this question. The second question is more difficult. Animal and clinical studies [1,5] have shown quite good long-term effects of ASOs and SSOs that are taken up by gymnosis, with no enhancing entity, that presumably slowly and spontaneously leak into the cytosol from an endomembrane depot. Nonetheless, the potency of ASOs and SSOs remains low, thus arguing that mobilization of the endomembrane pool may be valuable.
Summary
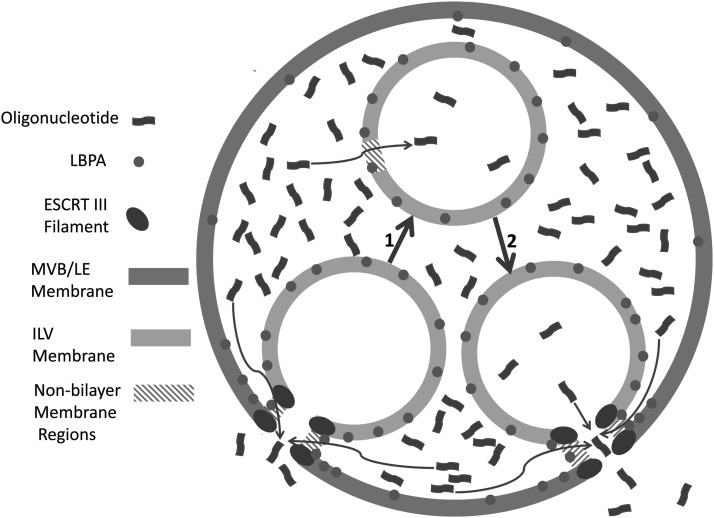
By drawing on the multiple studies of oligonucleotide trafficking, it now seems possible to generate a hypothesis about the locus and mechanism of oligonucleotide release to the cytosol (Fig. 2). Essentially, all forms of oligonucleotide delivery result in uptake by endocytosis followed by trafficking to EEs. However, productive release of oligonucleotide does not take place from this compartment despite its dynamic nature. Rather, oligonucleotides escape to the cytosol from an intermediate compartment in the trafficking pathway. This may be MVBs or hybrid EE/LE structures. The essential element is the presence of the ESCRT machinery and the formation of ILVs. ESCRT-mediated budding into the endomembrane lumen, or alternatively back fusion of ILVs to the cytosol, results in creation of unstable nonbilayer regions with transient formation of discontinuities that permit exit of oligonucleotides. Lipids that promote membrane disorder, such as LBPA, are recruited by the ESCRT machinery to the site of ILV production and contribute to the nonbilayer regions. Delivery agents such as cationic LPs or polyplexes or small molecule enhancers can accentuate this endogenous process and increase the rate and extent of oligonucleotide release by interacting with lipid or protein components of the ILV formation machinery. As noted above, there are still many unresolved questions concerning oligonucleotide trafficking. However, the tools to answer these questions are now within reach.
FIG. 2.
Model of Oligonucleotide Release from an Intermediate Endomembrane Compartment. (1) Formation of ILVs by ESCRT-mediated budding from the compartment-limiting membrane. (2) Release of ILV contents to the cytosol by back fusion of ILVs with the limiting membrane. Oligonucleotide movement across membranes takes place at discontinuities located in nonbilayer regions as indicated by curved arrows. ESCRT, endosomal sorting complex required for transport; ILV, intraluminal vesicle.
Author Disclosure Statement
R.L.J. has an equity interest in Initos Pharmaceuticals LLC a company that is developing oligonucleotide delivery agents.
References
- 1.Bennett CF, Baker BF, Pham N, Swayze E. and Geary RS. (2017). Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol 57:81–105 [DOI] [PubMed] [Google Scholar]
- 2.Bobbin ML. and Rossi JJ. (2016). RNA interference (RNAi)-based therapeutics: delivering on the Promise? Annu Rev Pharmacol Toxicol 56:103–122 [DOI] [PubMed] [Google Scholar]
- 3.Wu SY, Lopez-Berestein G, Calin GA. and Sood. AK. (2014). RNAi therapies: drugging the undruggable. Sci Transl Med 6:240ps7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kole R, Krainer AR. and Altman S. (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11:125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein CA. and Castanotto D. (2017). FDA-approved oligonucleotide therapies in 2017. Mol Ther 25:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crooke ST, Wang S, Vickers TA, Shen W. and Liang XH. (2017). Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol 35:230–237 [DOI] [PubMed] [Google Scholar]
- 7.Dowdy SF. (2017). Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol 35:222–229 [DOI] [PubMed] [Google Scholar]
- 8.Juliano RL. (2016). The delivery of therapeutic oligonucleotides. Nucleic Acids Res 44:6518–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzer C, Dirin M, Winkler AM, Baumann V. and Winkler J. (2015). Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J Control Release 203:1–15 [DOI] [PubMed] [Google Scholar]
- 10.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR. and Anderson DG. (2014). Non-viral vectors for gene-based therapy. Nat Rev Genet 15:541–555 [DOI] [PubMed] [Google Scholar]
- 11.Watts JK. and Corey DR. (2012). Silencing disease genes in the laboratory and the clinic. J Pathol 226:365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickstrom E. (2015). DNA and RNA derivatives to optimize distribution and delivery. Adv Drug Deliv Rev 87:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juliano RL, Ming X. and Nakagawa O. (2012). Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug Chem 23:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varkouhi AK, Scholte M, Storm G. and Haisma HJ. (2011). Endosomal escape pathways for delivery of biologicals. J Control Release 151:220–228 [DOI] [PubMed] [Google Scholar]
- 15.Juliano RL. and Carver K. (2015). Cellular uptake and intracellular trafficking of oligonucleotides. Adv Drug Deliv Rev 87:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittrup A. and Lieberman J. (2015). Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet 16:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty GJ. and McMahon HT. (2009). Mechanisms of endocytosis. Annu Rev Biochem 78:857–902 [DOI] [PubMed] [Google Scholar]
- 18.Goldenring JR. (2015). Recycling endosomes. Curr Opin Cell Biol 35:117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huotari J. and Helenius A. (2011). Endosome maturation. EMBO J 30:3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannes L. and Wunder C. (2011). Retrograde transport: two (or more) roads diverged in an endosomal tree? Traffic 12:956–962 [DOI] [PubMed] [Google Scholar]
- 21.Hanson PI. and Cashikar A. (2012). Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28:337–362 [DOI] [PubMed] [Google Scholar]
- 22.Scott CC, Vacca F. and Gruenberg J. (2014). Endosome maturation, transport and functions. Semin Cell Dev Biol 31:2–10 [DOI] [PubMed] [Google Scholar]
- 23.Luzio JP, Pryor PR. and Bright NA. (2007). Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632 [DOI] [PubMed] [Google Scholar]
- 24.Klumperman J. and Raposo. G. (2014). The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol 6:a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai H, Reinisch K. and Ferro-Novick S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12:671–682 [DOI] [PubMed] [Google Scholar]
- 26.Mayor S, Parton RG. and Donaldson JG. (2014). Clathrin-independent pathways of endocytosis. Cold Spring Harb Perspect Biol 6:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonifacino JS. and Neefjes J. (2017). Moving and positioning the endolysosomal system. Curr Opin Cell Biol 47:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spang A. (2016). Membrane tethering complexes in the endosomal system. Front Cell Dev Biol 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witkos TM. and Lowe M. (2017). Recognition and tethering of transport vesicles at the Golgi apparatus. Curr Opin Cell Biol 47:16–23 [DOI] [PubMed] [Google Scholar]
- 30.Kummel D. and Ungermann C. (2014). Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr Opin Cell Biol 29:61–66 [DOI] [PubMed] [Google Scholar]
- 31.Pfeffer SR. (2013). Rab GTPase regulation of membrane identity. Curr Opin Cell Biol 25:414–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henne WM, Buchkovich NJ. and Emr SD. (2011). The ESCRT pathway. Dev Cell 21:77–91 [DOI] [PubMed] [Google Scholar]
- 33.Hullin-Matsuda F, Taguchi T, Greimel P. and Kobayashi T. (2014). Lipid compartmentalization in the endosome system. Semin Cell Dev Biol 31:48–56 [DOI] [PubMed] [Google Scholar]
- 34.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, et al. (2013). Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol 31:638–646 [DOI] [PubMed] [Google Scholar]
- 35.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, et al. (2013). Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol 31:653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Tam YY, Chen S, Zaifman J, van der Meel R, Ciufolini MA. and Cullis PR. (2016). The niemann-pick C1 inhibitor NP3.47 enhances gene silencing potency of lipid nanoparticles containing siRNA. Mol Ther 24:2100–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, Manoharan M, Kirchhausen T. and Lieberman J. (2015). Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol 33:870–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch M. and Helm M. (2015). Live cell imaging of duplex siRNA intracellular trafficking. Nucleic Acids Res 43:4650–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel S, Ashwanikumar N, Robinson E, DuRoss A, Sun C, Murphy-Benenato KE, Mihai C, Almarsson O. and Sahay G. (2017). Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett 17:5711–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall A, Lachelt U, Bartek J, Wagner E. and Moghimi SM. (2017). Polyplex evolution: understanding biology, optimizing performance. Mol Ther 25:1476–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurrikoff K, Gestin M. and Langel U. (2016). Recent in vivo advances in cell-penetrating peptide-assisted drug delivery. Expert Opin Drug Deliv 13:373–387 [DOI] [PubMed] [Google Scholar]
- 42.Lindberg S, Regberg J, Eriksson J, Helmfors H, Munoz-Alarcon A, Srimanee A, Figueroa RA, Hallberg E, Ezzat K. and Langel U. (2015). A convergent uptake route for peptide- and polymer-based nucleotide delivery systems. J Control Release 206:58–66 [DOI] [PubMed] [Google Scholar]
- 43.Elouahabi A. and Ruysschaert JM. (2005). Formation and intracellular trafficking of lipoplexes and polyplexes. Mol Ther 11:336–347 [DOI] [PubMed] [Google Scholar]
- 44.Ming X, Sato K. and Juliano RL. (2011). Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes. J Control Release 153:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassane FS, Abes R, El Andaloussi S, Lehto T, Sillard R, Langel U. and Lebleu B. (2011). Insights into the cellular trafficking of splice redirecting oligonucleotides complexed with chemically modified cell-penetrating peptides. J Control Release 153:163–172 [DOI] [PubMed] [Google Scholar]
- 46.Rehman Z, Hoekstra D. and Zuhorn IS. (2013). Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano 7:3767–3777 [DOI] [PubMed] [Google Scholar]
- 47.Wang C, de Jong E, Sjollema KA. and Zuhorn. IS. (2016). Entry of PIP3-containing polyplexes into MDCK epithelial cells by local apical-basal polarity reversal. Sci Rep 6:21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazebnik M. and Pack DW. (2017). Rapid and facile quantitation of polyplex endocytic trafficking. J Control Release 247:19–27 [DOI] [PubMed] [Google Scholar]
- 49.Shukla RS, Jain A, Zhao Z. and Cheng K. (2016). Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomedicine 12:1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eltoukhy AA, Sahay G, Cunningham JM. and Anderson DG. (2014). Niemann-Pick C1 affects the gene delivery efficacy of degradable polymeric nanoparticles. ACS Nano 8:7905–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song W, Ma Z, Zhang Y. and Yang C. (2017). Autophagy plays a dual role during intracellular siRNA delivery by lipoplex and polyplex nanoparticles. Acta Biomater 58:196–204 [DOI] [PubMed] [Google Scholar]
- 52.Zhong X, Panus D, Ji W. and Wang C. (2015). Modulating polyplex-mediated gene transfection by small-molecule regulators of autophagy. Mol Pharm 12:932–940 [DOI] [PubMed] [Google Scholar]
- 53.Barnaby SN, Sita TL, Petrosko SH, Stegh AH. and Mirkin CA. (2015). Therapeutic applications of spherical nucleic acids. Cancer Treat Res 166:23–50 [DOI] [PubMed] [Google Scholar]
- 54.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, et al. (2013). Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 5:209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu XA, Choi CH, Zhang C, Hao L. and Mirkin CA. (2014). Intracellular fate of spherical nucleic acid nanoparticle conjugates. J Am Chem Soc 136:7726–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Fan C, Pei H, Shi J. and Huang Q. (2013). Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv Mater 25:4386–4396 [DOI] [PubMed] [Google Scholar]
- 57.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, et al. (2012). Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol 7:389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang L, Li J, Li Q, Huang Q, Shi J, Yan H. and Fan C. (2014). Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem Int Ed Engl 53:7745–7750 [DOI] [PubMed] [Google Scholar]
- 59.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, Worm J, Hedtjarn M, Souleimanian N, et al. (2010). Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 38:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koller E, Vincent TM, Chappell A, De S, Manoharan M. and Bennett CF. (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39:4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S, Sun H, Tanowitz M, Liang XH. and Crooke ST. (2016). Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res 44:7314–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S, Sun H, Tanowitz M, Liang XH. and Crooke ST. (2017). Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res 45:5309–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagenaar TR, Tolstykh T, Shi C, Jiang L, Zhang J, Li Z, Yu Q, Qu H, Sun F, et al. (2015). Identification of the endosomal sorting complex required for transport-I (ESCRT-I) as an important modulator of anti-miR uptake by cancer cells. Nucleic Acids Res 43:1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erazo-Oliveras A, Najjar K, Truong D, Wang TY, Brock DJ, Prater AR. and Pellois JP. (2016). The late endosome and its lipid BMP act as gateways for efficient cytosolic access of the delivery agent dfTAT and its macromolecular cargos. Cell Chem Biol 23:598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castanotto D, Lin M, Kowolik C, Wang L, Ren XQ, Soifer HS, Koch T, Hansen BR, Oerum H, et al. (2015). A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res 43:9350–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castanotto D, Lin M, Kowolik C, Koch T, Hansen BR, Oerum H. and Stein CA. (2016). Protein Kinase C-alpha is a critical protein for antisense oligonucleotide-mediated silencing in mammalian cells. Mol Ther 24:1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez-Barriga A, Nillessen B, Kranzen J, van Kessel IDG, Croes HJE, Aguilera B, de Visser PC, Datson NA, Mulders SAM, et al. (2017). Intracellular distribution and nuclear activity of antisense oligonucleotides after unassisted uptake in myoblasts and differentiated myotubes in vitro. Nucleic Acid Ther 27:144–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang XH, Sun H, Nichols JG. and Crooke ST. (2017). RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol Ther 25:2075–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, Wang Q, Hoekstra M, Kandasamy P, Kel'in AV, et al. (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136:16958–16961 [DOI] [PubMed] [Google Scholar]
- 70.Huang Y. (2017). Preclinical and clinical advances of galnac-decorated nucleic acid therapeutics. Mol Ther Nucleic Acids 6:116–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanowitz M, Hettrick L, Revenko A, Kinberger GA, Prakash TP. and Seth PP. (2017). Asialoglycoprotein receptor 1 mediates productive uptake of N-acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res 45:12388–12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, et al. (2014). Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol 32:1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alam MR, Dixit V, Kang H, Li ZB, Chen X, Trejo J, Fisher M. and Juliano RL. (2008). Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res 36:2764–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ming X, Alam MR, Fisher M, Yan Y, Chen X. and Juliano RL. (2010). Intracellular delivery of an antisense oligonucleotide via endocytosis of a G protein-coupled receptor. Nucleic Acids Res 38:6567–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alam MR, Ming X, Dixit V, Fisher M, Chen X. and Juliano RL. (2010). The biological effect of an antisense oligonucleotide depends on its route of endocytosis and trafficking. Oligonucleotides 20:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alam MR, Ming X, Fisher M, Lackey JG, Rajeev KG, Manoharan M. and Juliano RL. (2011). Multivalent cyclic RGD conjugates for targeted delivery of small interfering RNA. Bioconjug Chem 22:1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kortylewski M, Swiderski P, Herrmann A, Wang L, Kowolik C, Kujawski M, Lee H, Scuto A, Liu Y, et al. (2009). In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol 27:925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ly S, Navaroli DM, Didiot MC, Cardia J, Pandarinathan L, Alterman JF, Fogarty K, Standley C, Lifshitz LM, et al. (2017). Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res 45:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boisguerin P, Deshayes S, Gait MJ, O'Donovan L, Godfrey C, Betts CA, Wood MJ. and Lebleu B. (2015). Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv Drug Deliv Rev 87:52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lehto T, Ezzat K, Wood MJA. and El Andaloussi S. (2016). Peptides for nucleic acid delivery. Adv Drug Deliv Rev 106:172–182 [DOI] [PubMed] [Google Scholar]
- 81.Rinaldi C. and Wood MJA. (2018). Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol 14:9–21 [DOI] [PubMed] [Google Scholar]
- 82.Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL. and Lebleu B. (2006). Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J Control Release 116:304–313 [DOI] [PubMed] [Google Scholar]
- 83.Lehto T, Castillo Alvarez A, Gauck S, Gait MJ, Coursindel T, Wood MJ, Lebleu B. and Boisguerin P. (2014). Cellular trafficking determines the exon skipping activity of Pip6a-PMO in mdx skeletal and cardiac muscle cells. Nucleic Acids Res 42:3207–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ming X, Carver K, Fisher M, Noel R, Cintrat JC, Gillet D, Barbier J, Cao C, Bauman J. and Juliano RL. (2013). The small molecule Retro-1 enhances the pharmacological actions of antisense and splice switching oligonucleotides. Nucleic Acids Res 41:3673–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang B, Ming X, Cao C, Laing B, Yuan A, Porter MA, Hull-Ryde EA, Maddry J, Suto M, Janzen WP. and Juliano RL. (2015). High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic Acids Res 43:1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang L, Ariyarathna Y, Ming X, Yang B, James LI, Kreda SM, Porter M, Janzen W. and Juliano RL. (2017). A novel family of small molecules that enhance the intracellular delivery and pharmacological effectiveness of antisense and splice switching oligonucleotides. ACS Chem Biol 12:1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Castanotto D, Nam S, Horne D. and Stein C. (2017). 6BIO enhances oligonucleotide activity in cells: a potential combinatorial anti-androgen receptor therapy in prostate cancer cells. Mol Ther 25:79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Osborn MF, Alterman JF, Nikan M, Cao H, Didiot MC, Hassler MR, Coles AH. and Khvorova A. (2015). Guanabenz (Wytensin) selectively enhances uptake and efficacy of hydrophobically modified siRNAs. Nucleic Acids Res 43:8664–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilleron J, Paramasivam P, Zeigerer A, Querbes W, Marsico G, Andree C, Seifert S, Amaya P, Stoter M, et al. (2015). Identification of siRNA delivery enhancers by a chemical library screen. Nucleic Acids Res 43:7984–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Juliano RL, Wang L, Tavares F, Brown EG, James L, Ariyarathna Y, Ming X, Mao C. and Suto M. (2018). Structure-activity relationships and cellular mechanism of action of small molecules that enhance the delivery of oligonucleotides. Nucleic Acids Res 46:1601–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey JK, Shen W, Liang XH. and Crooke ST. (2017). Nucleic acid binding proteins affect the subcellular distribution of phosphorothioate antisense oligonucleotides. Nucleic Acids Res 45:10649–10671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang XH, Shen W, Sun H, Kinberger GA, Prakash TP, Nichols JG. and Crooke ST. (2016). Hsp90 protein interacts with phosphorothioate oligonucleotides containing hydrophobic 2'-modifications and enhances antisense activity. Nucleic Acids Res 44:3892–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liang XH, Shen W, Sun H, Prakash TP. and Crooke ST. (2014). TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res 42:7819–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG. and Gruenberg J. (1998). A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature 392:193–197 [DOI] [PubMed] [Google Scholar]
- 95.Wang T, Smith EA, Chapman ER. and Weisshaar JC. (2009). Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1–5 ms resolution. Biophys J 96:4122–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sudhof TC. and Rothman JE. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323:474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Renard HF, Johannes L. and Morsomme P. (2018). Increasing diversity of biological membrane fission mechanisms. Trends Cell Biol 28:274–286 [DOI] [PubMed] [Google Scholar]
- 98.Wickner W. and Rizo J. (2017). A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol Biol Cell 28:707–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bennett WF. and Tieleman DP. (2014). The importance of membrane defects-lessons from simulations. Acc Chem Res 47:2244–2251 [DOI] [PubMed] [Google Scholar]
- 100.Cullis PR, Hope MJ. and Tilcock CP. (1986). Lipid polymorphism and the roles of lipids in membranes. Chem Phys Lipids 40:127–144 [DOI] [PubMed] [Google Scholar]
- 101.Zelphati O. and Szoka FC., Jr. (1996). Mechanism of oligonucleotide release from cationic liposomes. Proc Natl Acad Sci U S A 93:11493–11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gallon M. and Cullen PJ. (2015). Retromer and sorting nexins in endosomal sorting. Biochem Soc Trans 43:33–47 [DOI] [PubMed] [Google Scholar]
- 103.Henne WM, Stenmark H. and Emr SD. (2013). Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol 5: pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dores MR. and Trejo J. (2014). Atypical regulation of G protein-coupled receptor intracellular trafficking by ubiquitination. Curr Opin Cell Biol 27:44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi T, Startchev K, Whitney AJ. and Gruenber J. (2001). Localization of lysobisphosphatidic acid-rich membrane domains in late endosomes. Biol Chem 382:483–485 [DOI] [PubMed] [Google Scholar]
- 106.Bissig C. and Gruenberg J. (2014). ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24:19–25 [DOI] [PubMed] [Google Scholar]
- 107.Edgar JR, Eden ER. and Futter CE. (2014). Hrs- and CD63-dependent competing mechanisms make different sized endosomal intraluminal vesicles. Traffic 15:197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531–534 [DOI] [PubMed] [Google Scholar]
- 109.Segura V, Perez-Aso M, Monto F, Carceller E, Noguera MA, Pediani J, Milligan G, McGrath IC. and D'Ocon P. (2013). Differences in the signaling pathways of alpha(1A)- and alpha(1B)-adrenoceptors are related to different endosomal targeting. PLoS One 8:e64996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F. and Kay MA. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541 [DOI] [PubMed] [Google Scholar]