Abstract
Lineage-specific expansion (LSE) of protein families is a widespread phenomenon in many eukaryotic genomes, but is generally more limited in bacterial genomes. Here, we report the presence of 434 genes encoding solute-binding proteins (SBPs) from the tripartite tricarboxylate transporter (TTT) family, within the 8.2 Mb genome of the α-proteobacterium Rhodoplanes sp. Z2-YC6860, a gene family over-representation of unprecedented abundance in prokaryotes. Representing over 6 % of the total number of coding sequences, the SBP genes are distributed across the whole genome but are found rarely in low-GC islands, where the gene density for this family is much lower. This observation, and the much higher sequence identity between the 434 Rhodoplanes TTT SBPs compared with the average identity between homologues from different species, is indicative of a key role for LSE in the expansion. The TTT SBP genes were found in the vicinity of genes encoding membrane components of transport systems from different families, as well as regulatory proteins such as histidine-kinases and transcription factors, indicating a broad range of functions around the sensing, response and transport of organic compounds. A smaller expansion of TTT SBPs is known in some species of the β-proteobacteria Bordetella and we observed similar expansions in other β-proteobacterial lineages, including members of the genus Comamonas and the industrial biotechnology organism Cupriavidus necator, indicating that strong environmental selection can drive SBP duplication and specialisation from multiple evolutionary starting points.
Keywords: Solute transporter, periplasmic-binding protein, Gene duplication, gene over-representation, lineage specific expansion
Impact Statement.
Intraspecific gene homology is a common evolutionary feature among prokaryotes, helping to build metabolic versatility and adaptability, but is mostly restricted in terms of gene numbers. Here, the over-representation of a gene family of unprecedented magnitude in a prokaryotic genome is described, with 434 genes belonging to the tripartite tricarboxylate transporter solute-binding proteins family in the α-proteobacterium Rhodoplanes sp. Z2-YC6860 genome. Although over-representations of smaller scale have been characterised for this family in some β-proteobacteria, we provide evidence that lineage-specific expansion, rather than horizontal gene transfer, is likely to be the major driving mechanism for this expansion, indicating that environmental selection can drive the expansion of binding proteins independently in multiple organisms.
Introduction
Gene family expansions result from events of gene duplication and horizontal gene transfer (HGT), and are major drivers for metabolic versatility and adaptation in prokaryotes and eukaryotes [1–3]. The results of high-throughput analyses of prokaryotic genomes indicate that the number of homologous genes correlates positively with genome size, possibly reflecting the versatility required from the complex environments that bacteria with large genome sizes often inhabit [1, 4–7], and that species showing similar families of homologues usually show similar metabolic features [1]. However, although the sum of genes encoding proteins that form members of gene families is significant, the number of members of individual families is usually low, with only 2 % of such families showing three or more copies [6–8], and there are very few cases where an over-representation of a single gene family is observed in a prokaryotic genome that is not related to transposon and phage insertions [6].
One of the few exceptions is the observed over-representation of histidine kinases (HPKs), reaching over 100 gene copies and representing 2.5 % of the coding sequences in some genomes [9, 10]. Although HGT seems to have played an important role in the expansion of this gene family, the largest over-representations are a consequence of gene duplication leading to lineage-specific expansions (LSE), which then results in the occurrence of ‘orphan’ HPKs (i.e. not encoded next to their cognate response regulators), that subsequently diverge through rearrangements in their N-terminal signalling domain to sense different environmental conditions [9]. In genomes where the expansion is driven by HGT, the HPK and the response regulators are duplicated together, undergoing a rapid selective pressure to co-evolve and cease ‘cross-talk’ with other systems [11].
A second example is the genome of the subsurface α-proteobacterium Geobacter sulfurreducens, which contains 111 genes encoding proteins of the cytochrome c family, representing 3 % of the total theoretical proteome [12]. This over-representation correlates with the versatile bioenergetics of this organism, which uses metal-ion-mediated electron transport to oxidize its substrates [12], although the actual function of the vast majority of these cytochromes is unknown.
A third example is the genome of the gut symbiotic Bacteroides thetaiotaomicron, which revealed the existence of 106 homologous genes to susC and 57 homologous genes to susD, encoding components of outer membrane uptake systems. In addition, 71 genes encoding glycosyltransferases are present, being involved in capsular polysaccharide biosynthesis. The over-representation of these two families is believed to relate both to the versatile uptake and utilisation of otherwise non-digestible sugars and evasion of the human immune system [10].
A final example of a large gene over-representation in bacterial genomes, relates to solute-binding proteins (SBPs) of the tripartite tricarboxylate transporters (TTT) family [13]. The TTT family are secondary transporters that use a periplasmic SBP for initial substrate recognition and binding. The TctCBA citrate uptake system from Salmonella typhimurium is the prototypical transporter of this family [14]. Each complete system is composed of three subunits: a 12-helix transmembrane protein, homologous to TctA and thought to be the solute carrier itself, a poorly-conserved four-transmembrane-domain accessory protein, homologous to TctB and of unknown function, and a periplasmic SBP, homologous to TctC and responsible for substrate specificity and initial high-affinity binding.
Genomic studies in the whooping-cough-causing agent Bordetella pertussis revealed the existence of 79 genes encoding TTT SBPs [15], and further analysis revealed that this over-representation was found in many species of the genus Bordetella, reaching 181 genes in Bordetella bronchiseptica, and in a closely related bacterium, Cupriavidus metallidurans [16]. Intriguingly, the number of genes for the membrane components did not follow the same pattern, showing no more than four representatives in Bordetella, leaving the majority of the SBPs as ‘orphans’. Later genome releases indicated that this over-representation was found also in other β-proteobacteria of the genus Advenella [17]. In this study, we further analyse the distribution of TTT SBPs in bacteria and discover a group of α-proteobacteria that have independently expanded their use of these proteins to unprecedented levels for any gene family in any currently known bacterial genome.
Results and discussion
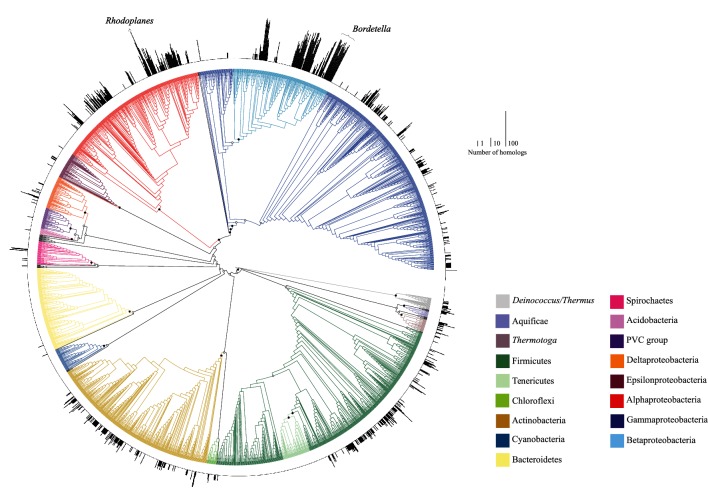
While studying the function and diversity of TTT transporters [13, 18], we analysed the number of TTT SBP homologues encoded amongst the available bacterial genomes, as previously described [13]. As of November 2017, 8049 fully assembled bacterial genomes were available in the NCBI database, belonging to 2323 different species. We used one genome per species for the analysis below. A homology search was performed using the TBLASTN tool with an e-value cut-off of 10−15, with the following set of functionally characterised TTT SBPs as query: TctC [19], BugD [20], BugE [21], Bug27 [22], TphC [23] and AdpC [18]. The resulting data were mapped onto a 16S rRNA tree to determine how the frequency of the SBPs correlated with the phylogenetic position of the organisms (Fig. 1). The known expansions of the TTT SBPs in the Bordetellae [16] can be clearly identified in the tree, as well as other expansions within the β-proteobacteria. However, examples of genomes enriched in TTT SBPs can also be seen in the α-proteobacteria, and most strikingly in the genome of Rhodoplanes sp. Z2-YC6860 (Assembly: GCA_001579845.1), where 434 TctC homologs were found to be encoded in its 8.2 Mbp genome, representing 6.2 % of the total coding sequences (CDS) (Fig. 1). Other closely related α-proteobacteria show similar but smaller enrichments, including Pseudorhodoplanes sinuspersici RIPI110T, with 99 TTT SBPs, and Bradyrhizobium icense LMTR 13T, with 43 TTT SBPs.
Fig. 1.
Distribution of TTT SBPs in the genomes of bacteria. The outer circle represents the number of TTT SBPs present in each genome, using a log2 scale. The tree was inferred using 16S rRNA sequences retrieved from the genome of each organism, and aligned using MAFFT v7 [37]. A maximum-likelihood tree was inferred using RAxML v8.2.11 [38] under the GTRCAT model, with 100 bootstrap pseudoreplicates. Bootstrap support values are indicated on nodes of major lineages when higher than 50 % (inclusive; filled circles) or lower than 50 % (open circle). Major branches are coloured as indicated in the key. Non-coloured branches are minor lineages of Bacteria.
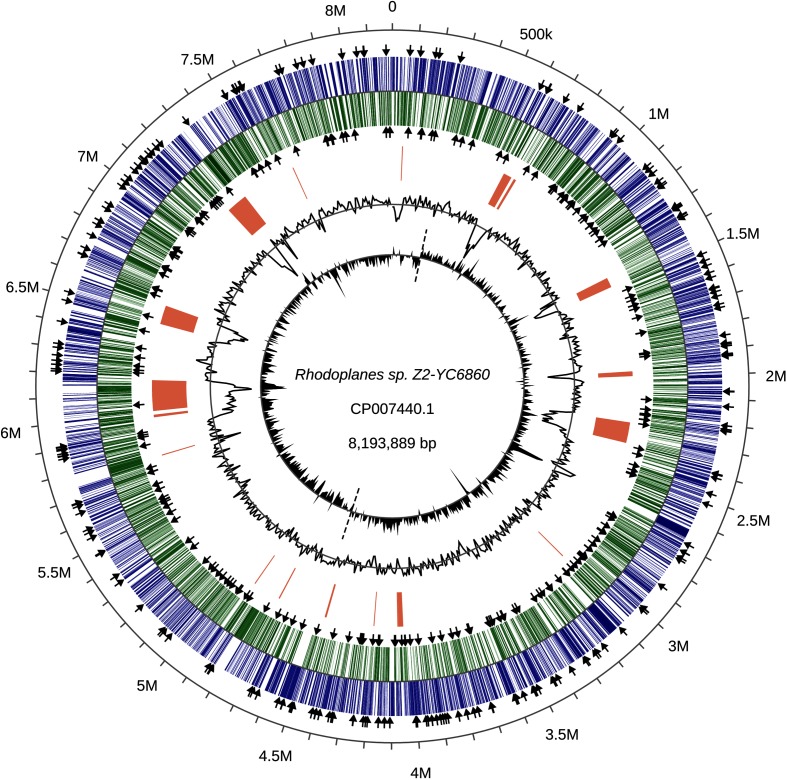
We examined the genome of Rhodoplanes sp. Z2-YC6860 in more detail to check the quality of the assembly and to investigate where the genes for the 434 TTT SBPs were located (Fig. 2). To check the assembly we plotted the GC skew (innermost ring in Fig. 2) and observed the typical skew reversal from the likely origin of replication until the likely termination site [24]. We also plotted the GC content (next innermost ring in Fig. 2) and could identify likely regions of DNA with atypically low GC contents, which were also predicted to be genomic islands using IslandViewer4 [25]. Interestingly, although the TTT SBP genes are distributed across the whole genome, the number of such genes in these low-GC regions, which comprise 9.9 % of genome length, with a GC content of 58.1 %, is only 11, which gives a ‘density’ of TTT SBP genes in these regions of 13.5 genes per Mb. The rest of the genome, in contrast, has a GC content of 64.1 %, and contains 423 TTT SBP genes, so has a ‘density’ of 57.3 TTT SBP genes per Mb. This striking tenfold difference in density indicates that recent detectable HGT is not the primary route by which these genes have been acquired as these regions in effect dilute the density of the TTT SBPs found in the rest of the genome.
Fig. 2.
Circular genome plot of the 8.193 Mb Rhodoplanes sp. Z2-YC6860 genome. The outer two tracks represent CDS on the forward strand (blue) and reverse strand (green), with black arrows indicating the location of TTT SBP genes. Orange blocks indicate genomic islands predicted by IslandViewer software [25]. The next inner circular plot represents percentage GC content calculated over a 10 kb sliding window with a range of 52.4 to 69.1% and mean of 63.5 %. The innermost circular plot represents GC skew calculated as (G−C)/(G+C) over a 10 kb sliding window. The characteristic GC skew reversal at the origin and terminus of replication is indicated by dashed lines.
Examination of the arrangement of the TTT SBPs on the genome reveals that there are 48 sets of these genes arranged in tandem arrays, while others are present in longer arrays of three, four and five genes (Table 1). Remarkably, there is a single array containing nine TTT SBPs adjacent to each other. The closely related Pseudorhodoplanes sinuspersici RIPI110T and Bradyrhizobium icense LMTR 13T also show evidence of tandem arrays, but not of this complexity. However, we cannot exclude the presence of similar organisations in closely related bacteria where genomic information is not available.
Table 1. Occurrence of TTT SBP genes as single genes or in larger arrays of genes, in the genome of Rhodoplanes sp. Z2-YC6860 and two closely related bacteria.
| Array size (number of genes) | Frequency | ||
|---|---|---|---|
| Rhodoplanes sp. Z2-YC6860 | Pseudorhodoplanes sinuspersici RIPI110T | Bradyrhizobium icense LMTR 13T | |
| 1 | 294 | 93 | 39 |
| 2 | 48 | 3 | 2 |
| 3 | 6 | – | – |
| 4 | 3 | – | – |
| 5 | 1 | – | – |
| 6 | – | – | – |
| 7 | – | – | – |
| 8 | – | – | – |
| 9 | 1 | – | – |
To investigate the mechanisms driving this massive over-representation, each of the 434 TTT SBP’s protein sequences of Rhodoplanes sp. Z2-YC6860 was searched using the blastp tool against a database comprising all TTT SBP’s identified in all 2323 bacterial genomes (see above). Self-matches were excluded and only the best hit (highest identity) was reported (Table S1, available in the online version of this article). Out of the 434 TTT SBP’s of Rhodoplanes sp. Z2-YC6860, a total of 234 sequences (53.9 %) had the best hit within the same genome. For another 98 sequences (22.6 %), the best hit was found in representatives of Rhizobacteria, which is the α-proteobacteria group that includes the genus Rhodoplanes. The best hit of the remaining 102 sequences (23.5 %) was found in species that are not within the α-proteobacteria group (Table S2). These findings suggest that gene duplications within Rhodoplanes sp. Z2-YC6860 account for more than half of this species’ TTT SBP’s, pointing to a key role of LSE in this unprecedented protein family expansion. The fact that another 22.6 % of the total genes have their best hit in other representatives of Rhizobacteria is indicative of duplication events preceding the origin of Rhodoplanes. Such best hits probably represent orthologous genes in these closely related species. The 23.5 % remaining genes with the best hit in non-α-proteobacteria species would probably have originated via HGT, although in this case it is unclear whether Rhodoplanes would have acted as donor or recipient of these genes. Note however, that a comprehensive phylogenetic analysis of the TTT SBP protein sequences across the Proteobacteria would be necessary to distinguish among these scenarios, and to exclude the hypotheses of (1) multiple gene losses in closely related groups [26], which could also produce the over-representation pattern that was exclusive to Rhodoplanes sp. Z2-YC6860, and/or (2) convergent sequence evolution, which could lead to higher sequence identity between distantly related species, therefore masking the actual mechanism underlying the origin of the gene [27].
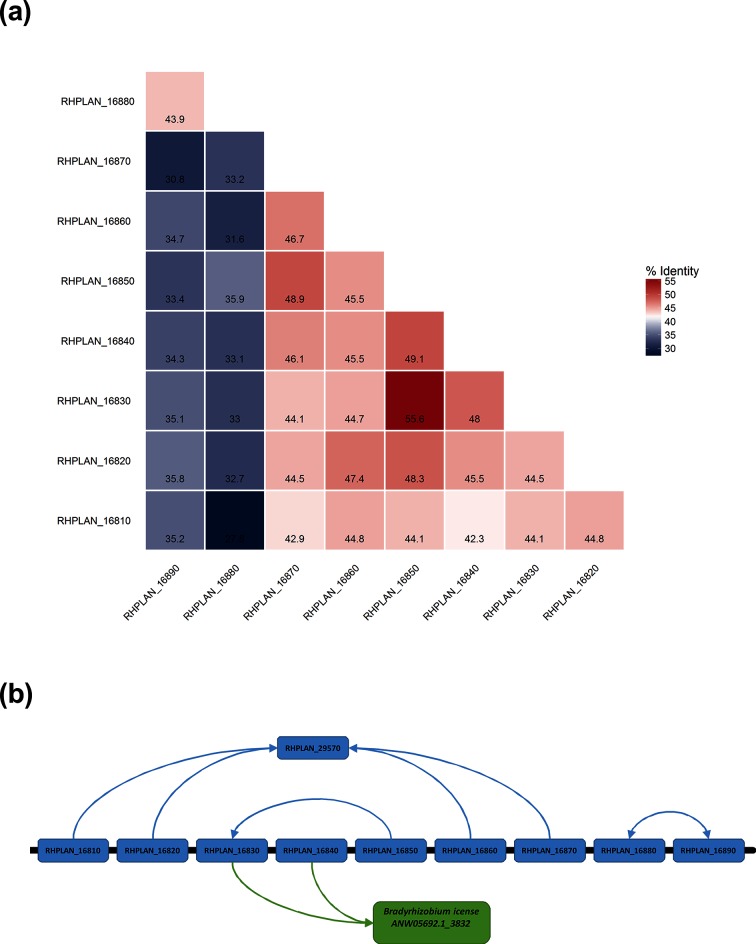
Within the unusual nine-gene array in Rhodoplanes, we examined the relationships between the proteins encoded by genes in the cluster (Fig. 3). Interestingly, with a single exception, the proteins are not more than 50 % identical to each other (Fig. 3a) and five of the nine proteins have the greatest identity to proteins outside the cluster, with RHPLAN_29570 being the closest match for four of them and the protein encoded by the gene ANW05692.1_3832 from Bradyrhizobium icense being the closest match for another two (Fig. 3b). Although an inverse correlation between cluster size and protein similarity has been previously observed in other studies [7], even this limited analysis indicates a complex pattern of gene gain and movement of genes around the chromosome.
Fig. 3.
Analysis of the relationships between the nine TTT family proteins located in the largest tandem array found in Rhodoplanes sp. Z2-YC6860. (a) Similarity matrix for amino acid sequences between proteins belonging to the nine-gene cluster. With the exception of RHPLAN_16830, which shared highest identity with RHPLAN_10480 (53.1 %), the remaining shared similarities were below 50 %. (b) Representation of highest identity for individual proteins in the cluster through blastp searches against 2323 bacterial genomes. The arrows represent the highest shared identity for each protein. Of the nine proteins in the array, only three members shared highest amino acid identity with other members of the same array, while four members shared highest identity with RHPLAN_29570, located elsewhere in the same genome. Two members were more similar to the TTT SBP ANW05692.1_3832 from Bradyrhizobium icense than to any gene inside the genome of Rhodoplanes sp. Z2-YC6860.
Finally, we considered what the physiological role of this expansion might be and noted that it appears to have occurred multiple times in different bacteria. From the genome context of the TTT SBPs we could see that many were genetically linked to known transporters. However, only nine form part of standard operons for a full TTT transporter, along with genes encoding TctA and TctB-like membrane proteins. Perhaps more surprisingly, some are linked to genes encoding subunits of other SBP-dependent transporters, with eight linked to tripartite ATP-independent periplasmic (TRAP) transporters and 41 to ATP-binding cassette (ABC) transporters. One of the proposed explanations for the over-representation of TTT SBPs in members of the genus Bordetella was that these SBPs would interact with different TTT systems, and that one membrane component would be capable of interacting with different SBPs under diverse circumstances, which could also be the case in members of the genus Rhodoplanes [16, 23]. Another possibility would be that these orphan proteins are required for processes other than transport, where the binding protein function is related to chemical sensing and signalling rather than to transport directly, in systems such as two-component systems and the chemosensory apparatus that regulates chemotaxis. For example, this sensory role has been demonstrated for the BctDECBA system of Bordetella pertussis [28, 29]. Our searches in members of the genus Rhodoplanes revealed that 14 out of the 22 identified histidine kinase genes, as well as five genes encoding proteins related to flagella motility had genes encoding TTT SBPs nearby. In addition, 70 tctC genes (16 %) had a transcription factor encoded in their immediate vicinity. The abundance of transcription factor genes around TTT SBP genes has been previously observed in the genome of Ralstonia eutropha, where 64.1 % of the 154 homologs exhibited this characteristic [30]. From the 434 TctC sequences in Rhodoplanes sp., 355 (82 %) were predicted to have a signal for periplasmic export, identified by SignalP software [31], showing that regardless of their function, they are likely to be located in the periplasm, as expected.
The proteobacterial groups we have observed to show a TTT SBP expansion all inhabit complex soil and water environments, with the exception of some species of the genus Bordetella, known for their pathogenicity, but it has been speculated whether in the latter case this over-representation was inherited from a free-living ancestor and that these genes are gradually being lost [16]. The genus Rhodoplanes was first proposed by Hiraishi and Ueda [32] to comprise pink-coloured non-sulphur purple bacteria, closely related to the members of the genus Rhodopseudomonas, but forming an exclusive cluster in 16S rRNA phylogenetic analyses. Inhabiting a wide range of complex soil and water environments, they are known to have a diverse and versatile metabolism [33–36]. We speculate that this vast array of TTT SBPs could increase this versatility by providing an evolutionary advantage in terms of sensing, response and uptake in a complex environment in a more tailored fashion [7]. Following our genomic analyses, further transcriptomic and proteomic studies would be crucial in determining how physiologically relevant these proteins are. The polyphyletic origins of TTT expansion support the hypothesis that LSE played a major role in the mechanism of the expansion, potentially following ancient small events of HGT, indicating that environmental pressure can independently drive SBP duplication and adaptation from multiple starting points given the right environmental conditions.
Conclusion
The present study describes a vast over-representation of TTT SBPs in the genome of the soil bacterium Rhodoplanes sp. Z2-YC6860, with 434 representatives. Although we cannot totally exclude the possibility that this gene family over-representation was partially driven by HGT, our findings indicate a major role of LSE in the origin of such an astonishing number of homologous genes, which may be linked to an increased metabolic versatility in this organism. Furthermore, we speculate that environmental pressure can drive SBP proliferation from multiple starting points. To the best of our knowledge, this is the most abundant gene family so far described in a prokaryotic genome.
Funding information
The present work was accomplished with funding from Brazilian funding agency CNPq (National Council for Scientific and Technological Development), though a PhD studentship in the remit of ‘Science Without Borders’ program to L. R. (248597/2013-2) and M. B. (201873/2014-1) and an BBSRC IB Catalyst grant for Project DETOX (BB/N01040X/1) to V. S., D. J. K. and G. H. T.
Conflicts of interest
The research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Data
Footnotes
Abbreviations: CDS, coding DNA sequence; HGT, horizontal gene transfer; HPK, histidine kinase; LSE, lineage-specific expansion; TTT, tripartite tricarboxylate transporters; SBP, solute-binding protein.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables are available with the online version of this article.
References
- 1.Bratlie MS, Johansen J, Sherman BT, Huang Daw, Lempicki RA, et al. Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics. 2010;11:588. doi: 10.1186/1471-2164-11-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turkarslan S, Reiss DJ, Gibbins G, Su WL, Pan M, et al. Niche adaptation by expansion and reprogramming of general transcription factors. Mol Syst Biol. 2011;7:554. doi: 10.1038/msb.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno S. Evolution by Gene Duplication. Berlin, Heidelberg: Springer; 1970. [Google Scholar]
- 4.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordero OX, Hogeweg P. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci USA. 2009;106:21748–21753. doi: 10.1073/pnas.0907584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsson J, Nylander JA, Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 2011;11:187. doi: 10.1186/1471-2148-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11:555–565. doi: 10.1101/gr.GR-1660R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, et al. A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 11.Salazar ME, Laub MT. Temporal and evolutionary dynamics of two-component signaling pathways. Curr Opin Microbiol. 2015;24:7–14. doi: 10.1016/j.mib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 13.Rosa LT, Bianconi ME, Thomas GH, Kelly DJ. Tripartite ATP-independent periplasmic (TRAP) transporters and tripartite tricarboxylate transporters (TTT): from uptake to pathogenicity. Front Cell Infect Microbiol. 2018;8:33. doi: 10.3389/fcimb.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widenhorn KA, Somers JM, Kay WW. Expression of the divergent tricarboxylate transport operon (tctI) of Salmonella typhimurium. J Bacteriol. 1988;170:3223–3227. doi: 10.1128/jb.170.7.3223-3227.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoine R, Raze D, Locht C. Genomics of Bordetella pertussis toxins. Int J Med Microbiol. 2000;290:301–305. doi: 10.1016/S1438-4221(00)80026-0. [DOI] [PubMed] [Google Scholar]
- 16.Antoine R, Jacob-Dubuisson F, Drobecq H, Willery E, Lesjean S, et al. Overrepresentation of a gene family encoding extracytoplasmic solute receptors in Bordetella. J Bacteriol. 2003;185:1470–1474. doi: 10.1128/JB.185.4.1470-1474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wübbeler JH, Hiessl S, Schuldes J, Thürmer A, Daniel R, et al. Unravelling the complete genome sequence of Advenella mimigardefordensis strain DPN7T and novel insights in the catabolism of the xenobiotic polythioester precursor 3,3'-dithiodipropionate. Microbiology. 2014;160:1401–1416. doi: 10.1099/mic.0.078279-0. [DOI] [PubMed] [Google Scholar]
- 18.Rosa LT, Dix SR, Rafferty JB, Kelly DJ. Structural basis for high-affinity adipate binding to AdpC (RPA4515), an orphan periplasmic-binding protein from the tripartite tricarboxylate transporter (TTT) family in Rhodopseudomonas palustris. Febs J. 2017;284:4262–4277. doi: 10.1111/febs.14304. [DOI] [PubMed] [Google Scholar]
- 19.Sweet GD, Somers JM, Kay WW. Purification and properties of a citrate-binding transport component, the C protein of Salmonella typhimurium. Can J Biochem. 1979;57:710–715. doi: 10.1139/o79-089. [DOI] [PubMed] [Google Scholar]
- 20.Huvent I, Belrhali H, Antoine R, Bompard C, Locht C, et al. Crystal structure of Bordetella pertussis BugD solute receptor unveils the basis of ligand binding in a new family of periplasmic binding proteins. J Mol Biol. 2006;356:1014–1026. doi: 10.1016/j.jmb.2005.11.096. [DOI] [PubMed] [Google Scholar]
- 21.Huvent I, Belrhali H, Antoine R, Bompard C, Locht C, et al. Structural analysis of Bordetella pertussis BugE solute receptor in a bound conformation. Acta Crystallogr D Biol Crystallogr. 2006;62:1375–1381. doi: 10.1107/S0907444906032653. [DOI] [PubMed] [Google Scholar]
- 22.Herrou J, Bompard C, Antoine R, Leroy A, Rucktooa P, et al. Structure-based mechanism of ligand binding for periplasmic solute-binding protein of the Bug family. J Mol Biol. 2007;373:954–964. doi: 10.1016/j.jmb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Hosaka M, Kamimura N, Toribami S, Mori K, Kasai D, et al. Novel tripartite aromatic acid transporter essential for terephthalate uptake in Comamonas sp. strain E6. Appl Environ Microbiol. 2013;79:6148–6155. doi: 10.1128/AEM.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mclean MJ, Wolfe KH, Devine KM. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/PL00006428. [DOI] [PubMed] [Google Scholar]
- 25.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzberg SL, White O, Peterson J, Eisen JA. Microbial genes in the human genome: lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 27.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 28.Antoine R, Huvent I, Chemlal K, Deray I, Raze D, et al. The periplasmic binding protein of a tripartite tricarboxylate transporter is involved in signal transduction. J Mol Biol. 2005;351:799–809. doi: 10.1016/j.jmb.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 29.Piepenbreier H, Fritz G, Gebhard S. Transporters as information processors in bacterial signalling pathways. Mol Microbiol. 2017;104:1–15. doi: 10.1111/mmi.13633. [DOI] [PubMed] [Google Scholar]
- 30.Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, et al. Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16. Nat Biotechnol. 2006;24:1257–1262. doi: 10.1038/nbt1244. [DOI] [PubMed] [Google Scholar]
- 31.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 32.Hiraishi A, Ueda Y. Rhodoplanes gen. nov., a new genus of phototrophic bacteria including Rhodopseudomonas rosea as Rhodoplanes roseus comb. nov. and Rhodoplanes elegans sp. nov. Int J Syst Bacteriol. 1994;44:665–673. doi: 10.1099/00207713-44-4-665. [DOI] [Google Scholar]
- 33.Takaichi S, Sasikala C, Ramana C, Okamura K, Hiraishi A. Carotenoids in Rhodoplanes species: variation of compositions and substrate specificity of predicted carotenogenesis enzymes. Curr Microbiol. 2012;65:150–155. doi: 10.1007/s00284-012-0139-y. [DOI] [PubMed] [Google Scholar]
- 34.Srinivas A, Sasikala C, Ramana C, Ch RV. Rhodoplanes oryzae sp. nov., a phototrophic alphaproteobacterium isolated from the rhizosphere soil of paddy. Int J Syst Evol Microbiol. 2014;64:2198–2203. doi: 10.1099/ijs.0.063347-0. [DOI] [PubMed] [Google Scholar]
- 35.Lodha TD, Srinivas A, Sasikala C, Ramana CV. Hopanoid inventory of Rhodoplanes spp. Arch Microbiol. 2015;197:861–867. doi: 10.1007/s00203-015-1112-5. [DOI] [PubMed] [Google Scholar]
- 36.Hiraishi A. Characterization of thermotolerant phototrophic bacteria, Rhodoplanes tepidicaeni sp. nov. and Rhodoplanes azumiensis sp. nov., isolated from a geothermal spring. Int J Syst Evol Microbiol. 2017;67:5038–5045. doi: 10.1099/ijsem.0.002408. [DOI] [PubMed] [Google Scholar]
- 37.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.