Abstract
Bordetella pertussis, the causative agent of whooping cough, has experienced a resurgence in the past 15 years, despite the existence of both whole-cell and acellular vaccines. Here, we performed whole genome sequencing analysis of 149 clinical strains, provided by the National Institute of Infectious Diseases (NIID), Japan, isolated in 1982–2014, after Japan became the first country to adopt acellular vaccines against B. pertussis. Additionally, we sequenced 39 strains provided by the Konan Kosei Hospital in Aichi prefecture, Japan, isolated in 2008–2013. The genome sequences afforded insight into B. pertussis genome variability and population dynamics in Japan, and revealed that the B. pertussis population in Japan was characterized by two major clades that divided more than 40 years ago. The pertactin gene was disrupted in about 20 % of the 149 NIID isolates, by either a deletion within the signal sequence (ΔSS) or the insertion of IS element IS481 (prn :: IS481). Phylogeny suggests that the parent clones for these isolates originated in Japan. Divergence dating traced the first generation of the pertactin-deficient mutants in Japan to around 1990, and indicated that strains containing the alternative pertactin allele prn2 may have appeared in Japan around 1974. Molecular clock data suggested that observed fluctuations in B. pertussis population size may have coincided with changes in vaccine usage in the country. The continuing failure to eradicate the disease warrants an exploration of novel vaccine compositions.
Keywords: Bordetella pertussis, pertactin, phylogenomics, acellular vaccine, epidemiology, Japan
Data Summary
1. The genome sequence reads of all 188 B. pertussis strains were deposited in the European Nucleotide Archive at www.ebi.ac.uk/ under project number PRJEB18624.
2. All supporting data are accessible as supplementary tables. Data include: (1) metadata, sequencing statistics and typing results on 188 Japanese B. pertussis strains (Table S1, available in the online version of this article); (2) metadata and typing results on 943 B. pertussis strains from this study and the literature (Table S2); (3) sequences and accession numbers of major B. pertussis antigen alleles utilized during in silico phenotyping of sequenced strains (Table S3); (4) all SNPs detected among the 188 contemporary Japanese B. pertussis isolates and strain Tohama I (Table S4); (5) SNPs overrepresented in isolates with prn :: IS481 and/or prnΔSS mutations (Table S5); and (6) rates of genetic variants observed for the orthologous groups of core genes from 188 B. pertussis isolates from Japan, 1982–2014, and Tohama I (Table S6).
Impact Statement.
Japan was the first country to adopt acellular vaccines against Bordetella pertussis, the causative agent of whooping cough. After switching from whole-cell to acellular vaccines, a resurgence in the incidence of pertussis in many Western countries, including Japan, was observed. Whole genome sequencing of B. pertussis strains harvested across this country identified the B. pertussis population dynamics in this environment. In addition to the existence of strains belonging to two different clades that diverged over 40 years ago, clonal spread of pertactin-negative strains was observed in Japan. Our data revealed that these isolates may have arisen in Japan around 1990. We also show a potential direct impact of changes in vaccine usage on the B. pertussis population size in the country, and present data on highly variant and very stable bacterial proteins that may aid the development of novel vaccine formulations to better fight the disease.
Introduction
Bordetella pertussis is a non-motile, aerobic, Gram-negative coccobacillus that causes whooping cough (pertussis) in humans. Pertussis is a highly contagious disease of the respiratory tract that, in rare cases, can be fatal, particularly in neonates. Fortunately, vaccines have been developed that reduce disease impact and limit spread of the bacterium in the human population.
More than 100 years ago, whole-cell vaccines (WCVs) against B. pertussis were licensed in the USA [1], and were subsequently adopted in many industrialized countries as a combinatorial mixture with diphtheria and tetanus toxoids. In the mid-1970s, increasing reports of adverse reactions to WCV preparations in Japan resulted in a government decision to temporarily halt mass B. pertussis vaccination in 1975 and to increase vaccination age after reintroduction of the programme [2]. As a consequence, the development of safer vaccination alternatives was prioritized, and in 1981 Japan was the first country to develop acellular pertussis vaccines (ACVs) and to adopt these for use in the general population [2, 3]. The ACVs invariably contained inactivated pertussis toxin and filamentous haemagglutinin, and were often supplemented with fim2-encoded fimbriae and pertactin, the latter an outer membrane protein that promotes adhesion to human tracheal epithelial cells of the host [4]. ACVs helped regain control over pertussis disease in Japan [5], and were shown to be as protective as whole cell-based vaccine preparations [6].
Despite growing vaccine coverage, worldwide incidence of pertussis has increased in recent years. Possible explanations for this resurgence include (i) genetic changes in circulating B. pertussis strains (see below), (ii) waning of vaccine-induced immunity, which may be rectifiable by repeated vaccination [7–11], (iii) increased awareness and reporting of pertussis cases, and (iv) an improved diagnosis of pertussis disease (summarized in [12]). In addition, studies in baboons suggest that ACVs, while protecting against disease, have limited impact, if any, on infection with and transmission of B. pertussis [13, 14].
Both pre-dating and after ACV development, genomic variations have been observed for ACV-targeted antigens [9, 15–17], some of which may be the result of vaccine-induced selective pressure. A mutation in the promoter region of the operon encoding the pertussis toxin, ptxP3, resulted in production of elevated levels of the toxin [18], and now predominates in many countries where ACVs have been deployed [19–22]. Similarly, the non-vaccine pertactin allele prn2 has become prevalent in many industrialized countries, including Australia, Japan, the UK, the Netherlands, Canada, Austria and the USA [20, 22–27].
Of particular interest is the reported increase in B. pertussis isolates not expressing pertactin at all (Prn−), observed in many countries that have adopted ACVs [21, 28–35]. The rate of Prn− isolates significantly correlates with vaccine use in the USA [33]. In Japan, the first known pertactin-deficient (Prn−) B. pertussis strain was harvested in 1997, marking the beginning of an increase of the fraction of pertactin-negative strains in the Japanese B. pertussis population to over 30 % during the first decade of the 21st century [36, 37]. The loss of pertactin by some B. pertussis strains does not influence disease severity [38, 39]. However, these isolates show increased fitness and/or prolonged infection times in animal host populations immunized with ACVs [40, 41]. Notably, an increased prevalence of Prn– strains is often reported in countries that use pertactin-comprising ACVs for general vaccination [21, 28–30, 32, 35]. However, a recent study revealed a surprising decrease in B. pertussis Prn– frequency in Japan in the past 3 years [42].
Many B. pertussis genomes have been sequenced and deposited in public repositories. Genomes of the strains harvested during the pertussis epidemic in Australia of 2008–2012 revealed microevolution events, predominant prn2 alleles and mixing of strains from a concurrent epidemic in the UK [43]. Genome sequencing performed on strains of that UK outbreak revealed that the genes encoding antigens present in ACVs may evolve at a faster rate than other surface proteins of the bacterium, potentially suggesting vaccine-induced selective pressure [25]. Genome sequencing confirmed clonal expansion of B. pertussis on four occasions in the Netherlands [26] and suggested differences in the mutation rates of the bacterium depending on vaccination coverage and method in China, the Netherlands and Finland [44]. An extensive study examined and characterized the global population structure of B. pertussis, sequencing over 300 isolates, harvested worldwide between 1920 and 2010 [16]. This study included 17 strains collected in Japan between 1988 and 2007, but seemed not to sample any Prn− isolates.
The aim of this study was to further monitor and analyse the B. pertussis population dynamics in Japan as the first country to adopt ACV vaccination. To do this, a set of representative B. pertussis clinical isolates collected between 1982 and 2014 was analysed by whole-genome sequencing. Our hope was to pinpoint the selection of the first generation of Prn− strains, and to put these into an evolutionary context of currently circulating Prn– and Prn+ strains, in Japan.
Methods
Selection of B. pertussis clinical isolates
A set of Japanese B. pertussis isolates was obtained from the Konan Kosei Hospital (n=39), representing all B. pertussis cases in that hospital between 2008 and 2013. In addition, isolates were procured from the National Institute of Infectious Diseases of Japan (NIID; n=151, genome sequences were obtained from 149 of these), representing a random pool of clinical isolates collected from various locations in Japan between 1982 and 2014. A summary of strain characteristics is given in Table 1, while a more detailed description of every strain and associated metadata is given in Table S1. The isolates originated from 29 different locations across all five major islands of the country (Hokkaido, Honshu, Shikoku, Kyushu and Okinawa).
Table 1. Summary of Japanese B. pertussis strains investigated by whole genome sequencing.
| Period | Total no. of isolates* | No. of locations | No. of ∆prn isolates | ∆prn mechanisms | Identified gene alleles | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| prn | ptxP | ptxA | fim2 | fim3 | fhaB | |||||
| 1981–1985 | 9 | 2 | 0 | mRNA↓† | 1 | 1 | 2 | 1 | 1 | 1 |
| 1986–1990 | 13 | 5 | 0 | – | 1 | 1, 8 | 1, 2 | 1 | 1 | 1 |
| 1991–1995 | 18 | 7 | 0 | – | 1, 2 | 1, 3 | 1, 2 | 1 | 1 | 1 |
| 1996–2000 | 42 | 11 | 8 | prn∆SS, prn :: IS481 | 1, 2 | 1, 3 | 1, 2 | 1 | 1 | 1 |
| 2001–2005 | 23 | 16 | 10 | prn∆SS, prn :: IS481 | 1, 2 | 1, 3, 8 | 1, 2 | 1 | 1, 2 | 1 |
| 2006–2010 | 43 (19) | 15 | 23 | prn∆SS, prn :: IS481 | 1, 2 | 1, 3 | 1, 2 | 1 | 1, 2, 4 | 1 |
| 2011–2014 | 40 (20) | 12 | 6 | prn∆SS | 1, 2 | 1, 3 | 1, 2 | 1 | 1, 2, 4 | 1 |
*The number of isolates from Konan Kosei is shown in parentheses.
†One isolate exhibited reduced prn mRNA production.
Genome sequencing and assembly
Whole genome sequencing and assembly were performed by BaseClear B.V. Paired-end sequence reads (2×125 bp) were generated using the Illumina HiSeq2500 system. FASTQ sequence files were generated using the Illumina Casava pipeline version 1.8.3. Initial quality assessment was based on data passing the Illumina Chastity filtering. Subsequently, reads containing adapters and/or PhiX control signal were removed using an in-house filtering protocol. The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.10.0. The quality of the FASTQ sequences was enhanced by trimming off low-quality bases using the ‘Trim sequences’ option of Qiagen’s CLC Genomics Workbench version 7.5 or 8.0. The quality-filtered sequence reads were assembled into contigs, using SPAdes v3.10.1 with default settings, including the careful mode. Final genome coverage and contig numbers are shown in Table S1.
The genome sequence reads of all 188 B. pertussis strains were deposited in the European Nucleotide Archive at www.ebi.ac.uk/ under project number PRJEB18624.
Phylogenetic reconstruction
Illumina sequence reads or complete genomes of all sequenced B. pertussis isolates from this study, as well as qualifying B. pertussis genome sequences available from GenBank and those deposited from Sealey and others [25] in the European Nucleotide Archive ENA (755 GenBank and ENA genomes, access date 9 Febuary 2018) were mapped against the closed genome of B. pertussis Tohama I [45], using Snippy with default settings [46]. Genomes were only included when coverage was sufficient (defined as fewer than 300 kb with a coverage of <10) and when isolation location and date were provided. Typing data were extracted from various reference sources [16, 25, 45, 47–52] or inferred from sequenced genomes (Table S2). Phylogenetic trees with and without inclusion of the 755 worldwide isolates were reconstructed using FastTree2 [53] with a generalized time-reversible (GTR) model with gamma correction on the resulting core genome alignment. The resulting trees were rooted using the genome of Bordetella bronchiseptica MO149 [54] prior to visualization using iTOL [55].
In silico typing and phenotypic analysis of isolates
For in silico typing, alleles of the genes ptxA, ptxP, fim2, fim3, prn and fhaB were aligned against assembled whole genome sequences of the isolates using blast +2.4.0 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST). A gene was assigned an allele if a 100 % match was found against the polymorphic typing region, as previously defined [56, 57]. Apparent mutations were manually verified from the sequence alignments. The numbers of alleles considered were as follows: prn, 17 alleles, ptxP, 11 alleles, ptxA, 3 alleles; fim2, 2 alleles; fim3, 6 alleles; fhaB, 2 alleles. Alelle sequences and accession numbers are shown in Table S3. We used assembly data for this analysis, as well as for SNP and gene variant detection (below), because the use of mapped reads may disable detection of insertions and deletions.
Phenotypic analyses for expression of Prn, Fim2 and Fim3 were performed as previously described [36], using immunoblotting of 1 µg protein samples per lane (Prn) and whole-cell ELISA techniques (Fim2, Fim3).
Divergence dating and recombination analysis
A core genome alignment was created on the isolates from this study using Parsnp v1.2 [58], and SNPs were detected from the alignment. SNPs closer than 1 kb to each other were excluded to remove rapidly evolving regions or regions that are under selective pressure which could affect the temporal signal. This approach resulted in removal of 400 SNPs from consideration. Pearson correlation between root to tip distance of the phylogenetic tree and isolate date was significant (P<0.0001, R2=0.38), suggesting a strong temporal signal in the SNP data. Divergence dating was performed in beast [59], using the isolation dates as tip dates in the phylogenetic tree with a GTR model of evolution. An exponential clock model was used in combination with a Bayesian skyline demographic model with four groups, essentially as previously described [16]. A Markov chain was run for 100 000 000 generations, with parameter values sampled every 10 000 generations. The chain was checked for expected sample sizes (ESSs)>200 using Tracer (http://tree.bio.ed.ac.uk/software/tracer/), with the first 10 000 000 chains removed [59]. A Bayesian skyline plot was drawn in Tracer with dates between 1950 and 2014. A maximum clade credibility tree was computed with TreeAnnotator, keeping tree heights intact and with a burn-in of 10 000 000 chains. beast results were visualized using Tracer and Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
Detection of SNPs, and association of SNPs with Prn+ and Prn– B. pertussis isolates
Complete genome sequences of B. pertussis isolates were compared using Parsnp v1.2 [58]. Presence and absence of core genome SNPs, as identified by Parsnp, were tested for their prevalence in Prn− and Prn+ strains using Fisher's exact 2×2 contingency tests.
Rates of genetic variants
Proteins were clustered into orthologous groups (OGs) using Roary [60] with default settings. Prior to clustering, genes containing transposase sequences were removed using blast against genes coding for transposases [61], to prevent spurious clustering of proteins on these sequences. Amino acid and nucleotide sequences of these OGs were compared and every unique gene or protein sequence was assigned an allele number. Unique numbers of variants per OG were counted and summarized. Truncated genes shorter than 90 % of their supposed length were removed from the analysis. The computed variant numbers per OG (minus 1) were finally divided by the length of the represented gene to generate a density value that allows cross-comparisons between genes.
Note that a genetic variant is based on the entire ORF sequence, while prn, ptxP, ptxA, fim2, fim3 and fhaB alleles only pertain to the typing regions defined for each gene. Consequently, there can be more genetic variants than alleles for these genes.
Results and discussion
Phylogenetic analysis of pertussis disease in Japan reveals two major clades
To gain insight into the B. pertussis phylogeny and population dynamics in the country that first adopted ACVs, we sequenced the genomes of 149 clinical B. pertussis isolates observed across Japan between 1982 and 2014. These strains were provided by the NIID. In addition, we sequenced the genomes of 39 isolates from the Konan Kosei Hospital, a local healthcare facility in Aichi prefecture in Japan, with isolation dates from 2008 to 2013. Overall, about 1.44 million reads (sd±0.3) were obtained per isolate, resulting in an average genome coverage of 88.2× (sd±18.3). Core genome alignment, which included 3 552 899 bp DNA, revealed a total of 1272 SNPs between the 188 isolates and B. pertussis strain Tohama I, the strain used for preparation of the ACVs that are utilized in Japan.
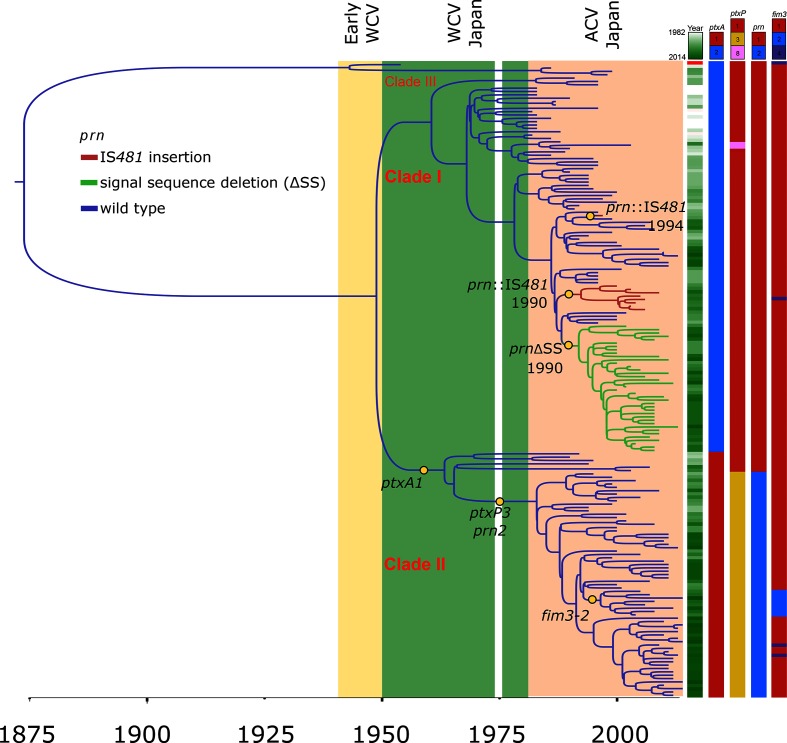
Phylogenetic reconstruction based on 872 SNPs identified in the core genome present in all sequenced strains revealed that two clades of B. pertussis represent the primary causes of pertussis disease in Japan from 1982 to 2014 (Fig. 1). This analysis excluded SNPs that were within 1 kb of each other. The clades are characterized by defined differences in the alleles of major antigens of the bacterium. The first clade consists of 112 isolates predominated by exhibition of pertussis toxin allele ptxA2, pertussis toxin promoter allele ptxP1 and pertactin allele prn1. The vast majority of the 73 strains forming the second clade contain the ptxA1 : ptxP3 : prn2 combination. Median isolation dates reveal the first clade (median isolation year 2000) to contain overall older strains than the second clade (median isolation year 2009), indicating an overall increase in prevalence of strains containing the prn2 and ptxP3 alleles. A closer look at the temporal prevalence of different alleles of the prn and ptxA genes and ptxP confirms this observation (Fig. 2a–c).
Fig. 1.
Phylogeny of B. pertussis isolates in Japan. Shaded regions indicate time periods of WCV and ACV use, with the notable gap in vaccine use in 1975. Isolation year, alleles of ptxP, ptxA and prn, as well as pertactin deficiency is indicated for each isolate on the right. The clustered isolates of two variants of pertactin-deficient strains are highlighted in red (prn :: IS481) and green (ΔSS). Notable nodes are illustrated and beast-based divergence dating results are shown for the pertactin-deficient variants.
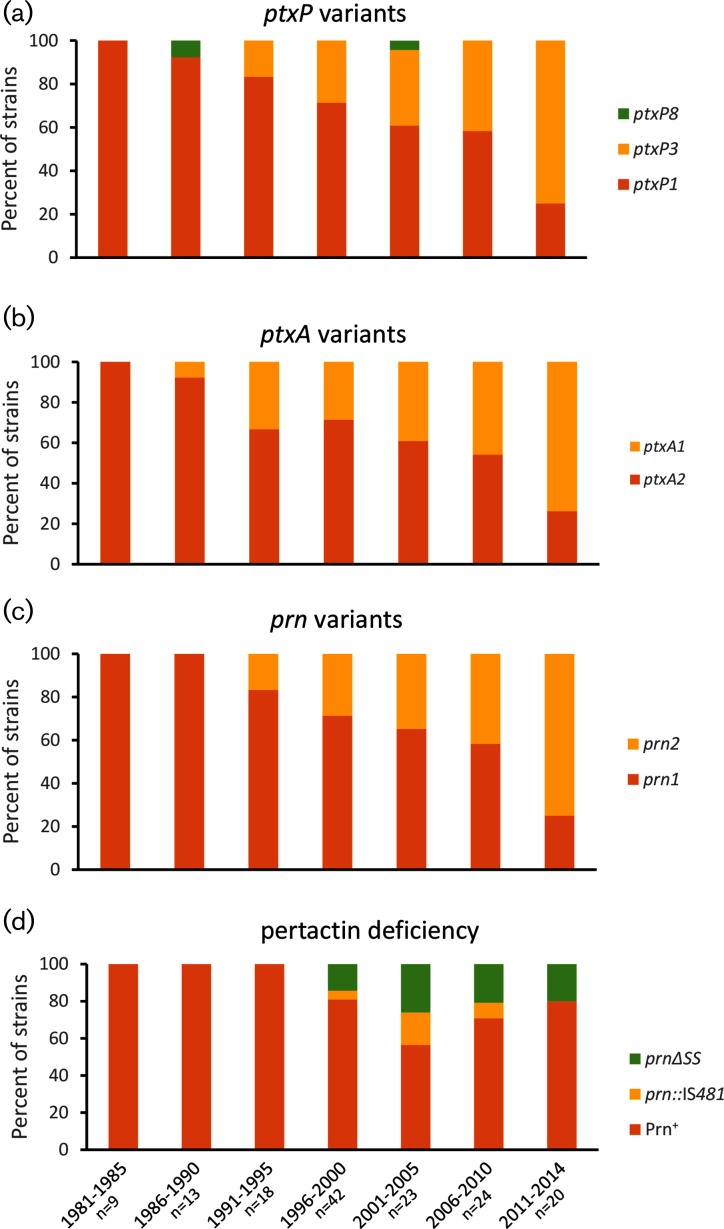
Fig. 2.
Changes in B. pertussis allele frequencies of ptxP, ptxA and prn as well as pertactin deficiency rates over time in Japan, 1982–2014. Konan Kosei isolates were excluded from the calculation.
A minor third clade was also identified, which consisted of reference isolate B. pertussis Tohama I (ptxA1 : ptxP1 : prn1) and three additional isolates (BP56, BP121 and BP194), all harvested before 2000. The population structure in Japan broadly mirrors observations made in other countries. However, we noted one minor distinction. A separate clonally expanding clade that included ptxA1 : ptxP3 : prn2 strains with the fim3-2 allele was detected in the UK and the Netherlands in the early 2000s [25, 26]. A small fim3-2 clade of eight isolates with the same antigenic combination was also present in Japan, but has not expanded to the same predominance and clusters deep within clade II (Figs 1 and 3).
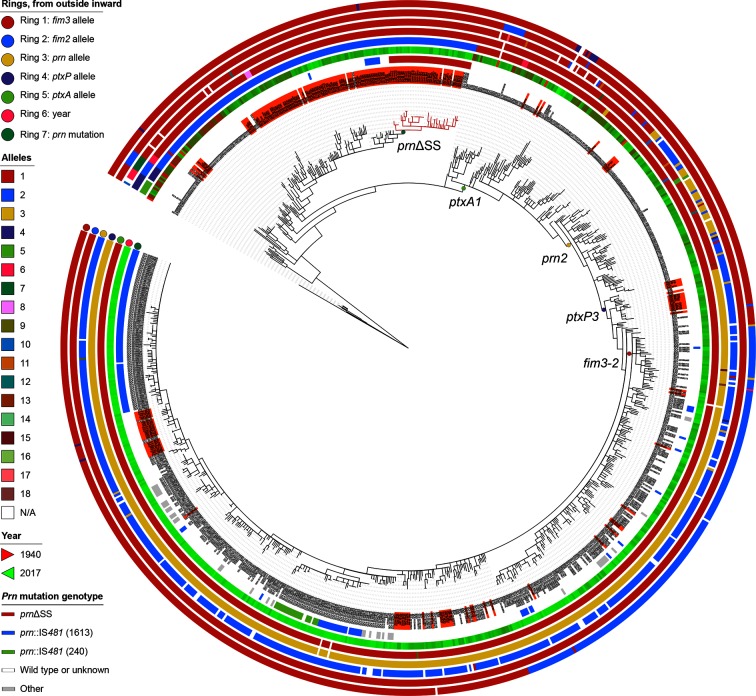
Fig. 3.
Phylogenetic tree of B. pertussis isolates in Japan, contextualized with worldwide isolates. Japanese isolates are on an orange background. Worldwide isolates are those investigated in previous studies [16, 25] and all other qualifying genome sequences deposited in GenBank by 9 February 2018 (see Methods). Alleles of fim3, fim2, prn, ptxP and ptxA and isolation year are colour-coded, as indicated on the left. The variants of pertactin deficiency are highlighted in blue and green (prn :: IS481), red (ΔSS) and grey (other). Notable nodes are annotated. Circles represent, from outermost inwards, fim3, fim2, prn, ptxP and ptxA allele status, followed by isolation year of each strain and prn mutation status. Distances in the tree were adjusted to sites per core genome and square root transformed to enhance resolution at short phylogenetic distances.
Comparison with previously sequenced isolates and worldwide context
Extensive earlier work sequenced B. pertussis genomes obtained worldwide, to gain insight into evolutionary patterns on a global scale [16]. This study included 17 strains harvested from Japan between 1988 and 2007 and pertactin deficiency had not been recognized in the genome assemblies for any of these strains. However, an earlier report had suggested lack of pertactin in one of these isolates, strain BP310 [36]. We therefore re-sequenced nine of these 17 strains (BP22, BP120, BP128, BP121, BP31, BP56, BP119, BP227, BP310), and confirmed a ΔSS pertactin phenotype in strain BP310. After assembly of the reads from all 346 isolates included in the earlier study [16], previously unnoticed ΔSS pertactin genotypes were also noted in two further strains, B050, harvested in 2007 in Australia, and B107, a 2002 isolate from Hong Kong. A phylogenetic placement of our panel of Japanese isolates with the strains sequenced as part of the worldwide investigation in 2014 provided a worldwide context for the Japanese isolates. This context was expanded to include all other qualifying B. pertussis genome sequences in GenBank and ENA (see Methods, Fig. 3), for a total of 943 genomes. Among other observations, the tree indicated that three additional isolates, harvested in 2010–2012 in the USA, clustered with the Japanese prnΔSS isolates and were possibly introduced into North America from Japan.
The Japanese isolates populated all major branches of the global tree, indicating that the parameters used for tree reconstruction minimized any systematic difference introduced by the distinct sequencing approaches for the different strains. However, no Japanese isolate was identified that populated the intermediate evolutionary clade after the appearance of the prn2 allele but pre-dating the introduction of the ptxP3 mutation, i.e. ptxP1 : prn2.
We observed strong geographical clustering of the Japanese ptxA2 isolates, suggesting that they had evolved there for a prolonged amount of time and remained clonal. This was unexpected, given the lack of geographical clustering of worldwide B. pertussis isolates observed elsewhere [16]. However, the tree intimated a few Japanese isolates to have been introduced into the country from elsewhere, by placing these in close proximity to older strains from the worldwide panel. These ‘island’ clones are exemplified by BP28, a strain that closely resembles B250, a Swedish isolate harvested in 1970, and isolates BP227 and BP316, which are similar to B014, a strain harvested in 1971 in Australia, and two European strains isolated in 1968 (B071 from Denmark and B221 from Poland). Conversely, a few strains found in the global strain panel had very close relatives in the Japanese strain panel, such as the previously mentioned three USA strains clustering with the prnΔSS isolates.
Three isolates from our study, BP56, BP194 and BP121, two Japanese isolates from the worldwide panel (B133 and B135, harvested in the late 1990s), and one additional Japanese isolate from 1950 (GCA_002083095) clustered in an outgroup with the model isolate Tohama I, and represent a remote branch that has become extremely rare and possibly extinct in modern Japan. On a worldwide scale, this small cluster included a few other older strains, such as J042, an isolate from 1947 isolated in the USA, as well as B068 and GCA_001605275, slightly more distant Chinese isolates from the 1950s.
Allele typing
In silico typing was performed on all sequenced isolates for major antigens of B. pertussis, i.e. ptxA, fim2, fim3, prn and fhaB, as well as the pertussis toxin promoter region ptxP. Results are shown in Table 1 and, in single-strain resolution, in Table S1. For ptxA, two types were found, ptxA1 (n=73) and ptxA2 (n=115), separated along clade lines. For ptxP, three types were found, ptxP1 (n=119), ptxP3 (n=67) and ptxP8 (n=2). All ptxP3 isolates and 6/119 ptxP1 isolates constituted clade II of the phylogenetic tree shown in Fig. 1, while two ptxP1 strains clustered with the type strain Tohama I in the small and possibly extinct clade III. The remaining 111 ptxP1 strains and the two ptxP8 isolates formed clade I. All strains contained fim2-1 (n=188). For fim3, three types were found, fim3-1 (n=177), fim3-2 (n=8) and fim3-4 (n=3). For fhaB, only fhaB-1 was detected (n=188).
Based on previously defined amino acid repeat variations in pertactin sequences [57], two prn types were genotypically identified in the strain panel, prn1 (n=121) and prn2 (n=67). Among the prn1 strains, 38 had a deletion in their 5′ signal sequence-encoding region (ΔSS) and nine were interrupted by an IS481 element (prn :: IS481) in their prn gene.
Allelic shifts and deficiency of pertactin
A decline in prn1 frequency, paralleling ascending ptxA1 and ptxP3 prevalence, was observed in existing isolates in Japan, a shift that experienced its biggest momentum at around 2010 (Fig. 2a, b). These events mirror similar shifts toward alternative antigen alleles observed earlier on a worldwide scale, primarily in the industrialized world with high vaccine coverage. The start of this worldwide shift pre-dated use of ACV preparations and suggested exertion of selective immune pressure on pertactin by WCVs. Consequently, allelic variation of B. pertussis antigens probably contributed to survival of B. pertussis after WCV adoption, as previously championed [16].
The vaccine strain Tohama I expresses prn1 and ptxA2, and the recent shift favours strains expressing antigen variants not included in the ACV preparations customary in Japan. Evidence of increasing incidence of pertactin variants not represented in vaccine preparations had been presented before (exemplified by [17, 62]), and similar changes had been described, beginning in the WCV era, on a worldwide scale [16]. However, this observation may be restricted to countries with high vaccine coverage, as countries with lower, or delayed, vaccine coverage have not (yet) experienced similar predominance of prn2 isolates [44, 63], or are apparently devoid of prn2 strains [64]. Notably, an allele shift to prn2:ptxA1 resulted in prolonged survival in murine test animals treated with an ACV derived from the prn1 isolate Tohama I [65].
Divergence dating using beast on a core genome alignment (see Methods) places the divergence node between prn2 and prn1 strains in Japan at around 1974, well within the period of WCV usage, but pre-dating the development of ACVs against B. pertussis. This suggests that WCVs induce pertactin-selective immune pressure. Allelic variation of pertactin has been observed in many countries prior to introduction of ACVs, during periods of high WCV coverage [66–68].
Pre-dating the decline in prn1 predominance, pertactin-deficient isolates appeared in the mid-1990s in Japan. In our strain panel, this deficiency occurred exclusively in prn1 alleles, in two ways. The first consists of an integration of the insertion sequence IS481 (in the forward orientation) at nucleotide position 1598 of the prn gene (equivalent to nucleotide position 1613 in prn2). The second consists of a deletion of nucleotides 26–109 in the signal sequence of the prn gene. Both disruption of the gene by IS481 and signal sequence deletion have been previously observed and described in Japanese B. pertussis Prn– isolates [37], and IS481 insertions, although in different locations within the gene, were also observed elsewhere [30, 32, 34]. In our strain selection, divergence dating estimates the divergence of the prnΔSS cluster in the phylogenetic tree depicted in Fig. 1 to have occurred in 1990 [95 % confidence interval (CI): 1989–1993]. In the same year, a prn :: IS481-containing mutant cluster may have first appeared (95 % CI: 1989–1995). A lineage that included a third single prn :: IS481 isolate, BP118, was predicted to have diverged from an intact ancestor around 1994 (95 % CI: 1991–1997). Such pertactin-deficient isolates reached a maximal frequency in Japan of over 40 % in the early 2000s (Fig. 2d). The phylogenetic tree including the worldwide strain collection (Fig. 3) does not detect a close ancestor to the Japanese prnΔSS isolates in the worldwide collection, and therefore suggests this cluster to have arisen in Japan. Similarly, no close relative of the prn :: IS481 strains isolated in Japan was detected in the panel of worldwide isolates.
However, Prn– isolates failed to take over the B. pertussis population in Japan, and a recent study reveals a decrease in B. pertussis Prn– frequency in Japan to below 10 % in the past 3 years [42]. One reason for this drop may be the rising predominance of prn2-containing isolates, many of which have close relatives elsewhere in the world (illustrated in Fig. 3). However, this drop in pertactin-negative B. pertussis strains may also be caused, in part, by the wide introduction of new ACV preparations in Japan in 2012 that do not contain pertactin [42]. It remains unclear why Prn− isolates thrive to various degrees in different territories, exemplified by their rapidly increasing prevalence in Australia [32, 43] and the USA [69, 70], but continued low frequency in neighbouring Canada and the UK [71]. One explanation assumes that differential vaccine preparations and immunization regimens impose different levels of immune pressure on pertactin, dictating a varying need for and availability of possible escape routes.
While more than a dozen genetic disruption mechanisms of pertactin have been identified, including alternative IS elements (such as IS1002), SNPs, premature stop codons and various deletions [21, 32, 70, 72], Prn− strains fail to fully outcompete Prn+ isolates. As a case in point, only a single Prn− strain has been identified in a recent study that analysed 95 isolates harvested mostly between 2000 and 2012 in the UK, a country that switched from WCV to ACV for primary immunizations in October 2004 [25, 73]. Therefore, pertactin deficiency in itself, while perhaps initially providing an evolutionary benefit, may not carry sufficient advantages to eradicate other B. pertussis isolates, particularly those with a non-vaccine-type prn allele, e.g. prn2, and/or increased pertussis toxin production afforded by the ptxP3 mutation.
Gene variants and SNPs present in isolates with disrupting mutations in the pertactin gene
Genome-wide comparison of gene variants in Prn+ and Prn− isolates identified several gene differences present in isolates without functional pertactin. The majority of these associations probably represent hitchhiking mutations that occurred during acquisition of pertactin deficiency, and were clonally retained. Among gene variants, the most significant association was detected for an 11 bp deletion in gene BP0310 (position 315 417–315 427 in the genome of strain Tohama I), encoding a putative acyl-CoA-dehydrogenase, resulting in a truncated protein (data not shown). This variant was absent from the prn :: IS481 isolates and from the vast majority of Prn+ strains but was found in all prnΔSS isolates and in only four Prn+ isolates (BP1, BP9, BP61 and BP156). The latter four isolates cluster with the prnΔSS isolates on the phylogenetic tree, perhaps indicating that this deletion preceded the deletion of the signal sequence in prn.
Table S4 shows all SNPs identified in this study among all 188 isolates, compared to strain Tohama I, and Table S5 illustrates the results of an analysis of SNP differences between Prn+ and Prn− strains of the ptxA2 : ptxP1 : prn1 clade. We identified a non-synonymous SNP present exclusively and consistently in all prnΔSS strains in gene BP0534, encoding a putative enoyl-CoA hydratase/isomerase. No SNPs were found that were present in all prn :: IS481 Prn− isolates but not in Prn+ ptxA2 : ptxP1 : prn1 isolates. However, SNPs in BP0332 and BP1640, both encoding proteins of unknown function, were detected in all but one prn :: IS481 Prn− isolate, and nowhere else. The prn :: IS481 Prn− isolate lacking this SNP was BP118, a strain that clustered separately from all other prn :: IS481 isolates on the phylogenetic trees depicted in Figs 1 and 3. The functional effects of the detected SNPs and gene variants on bacterial pathogenicity, growth and fitness are unknown but may be verified with additional future experiments.
Local B. pertussis population dynamics may differ profoundly from national trends
The 38 strains that contained a deletion of the signal sequence of the pertactin gene were deficient in pertactin production and clustered together on the phylogenetic tree, indicating a clonal spread of the isolate. These strains had been harvested over the course of 13 years, between 2000 and 2013. A total of 18 of these strains had been provided by the Konan Kosei Hospital. Among all B. pertussis isolates reported in this hospital, prevalence of this clone reached over 90 % in 2008 and 100 % in 2009, while, according to our strain panel, the same clone represented only around 16 % of B. pertussis isolates found in the rest of the country at that time. After 2009, the isolate’s prevalence in Konan fell back to approximately national levels. This observation suggests that this particular prnΔSS isolate prevailed and persisted over at least 2 years within the Konan community. It is therefore likely that the overall fitness of this strain was at least as good as that of standard Prn+ isolates that existed in the vaccinated population of Konan at the same time.
Rates of genetic variants
We performed an examination of the rates of genetic variants (nucleotide variant rates per base pair) observed in the core genome among the sequenced B. pertussis isolates from Japan. Table S6 shows all variation rates for each B. pertussis Tohama I gene included in the core genome. The analysis confirmed a higher overall rate of variants for the nine genes predicted to encode ACV-targeted proteins (fhaB, fim2, fim3, prn, ptxA–E), compared to all genes (0.0014 versus 0.0006 variants/bp). A summary of the observed overall rates and the amino-acid-changing rates for the functional categories of the B. pertussis Tohama I core genes is depicted in Table 2. The analysis suggests that genes targeted by ACVs do indeed mutate more frequently than coding sequences in most other gene categories, an effect that had been previously described [25]. They also mutate, on average, more frequently than genes encoding proteins predicted to be located extracellularly (20 genes, 0.0006 variants/bp) or in the outer membrane (70 genes, 0.0010 variants/bp). In contrast to previous observations [16], this trend of elevated mutation frequency does not extend to a similar degree from the ACV genes to other virulence-associated genes in our strain panel. Perhaps this can be explained in part by the early adoption of ACVs in Japan, thereby focusing pressures onto the vaccine-targeted proteins earlier than in other countries.
Table 2. Rates of genetic variants observed in functional gene categories in Japanese B. pertussis isolates.
| Functional category* | Number of genes | Median gene length | Non-silent variant rate† | Total variant rate‡ |
|---|---|---|---|---|
| Fatty acid metabolism | 21 | 936 | 0.0002 | 0.0004 |
| Small molecule degradation | 107 | 1068 | 0.0003 | 0.0004 |
| Macromolecule degradation | 46 | 1234.5 | 0.0003 | 0.0004 |
| Cell division | 18 | 1014 | 0.0003 | 0.0004 |
| Ribonucleotide biosynthesis | 27 | 1080 | 0.0004 | 0.0004 |
| Macromolecule synthesis/modification | 194 | 1068 | 0.0004 | 0.0005 |
| Cell processes | 62 | 927 | 0.0004 | 0.0005 |
| Cofactor biosynthesis | 86 | 846 | 0.0004 | 0.0005 |
| Ribosome constituents | 55 | 384 | 0.0004 | 0.0005 |
| Amino acid biosynthesis | 91 | 1134 | 0.0004 | 0.0005 |
| Adaptation | 48 | 563.5 | 0.0005 | 0.0005 |
| Central/intermediary metabolism | 96 | 1016.5 | 0.0004 | 0.0005 |
| Energy metabolism | 119 | 1056 | 0.0004 | 0.0005 |
| Transport/binding proteins | 326 | 970.5 | 0.0004 | 0.0006 |
| Miscellaneous | 342 | 886.5 | 0.0005 | 0.0006 |
| Conserved hypothetical | 419 | 681 | 0.0005 | 0.0006 |
| Cell surface | 571 | 951 | 0.0005 | 0.0006 |
| Protection responses | 24 | 639 | 0.0006 | 0.0006 |
| Regulation | 270 | 829.5 | 0.0007 | 0.0008 |
| Virulence-associated genes | 89 | 1095 | 0.0007 | 0.0009 |
| Unknown | 134 | 463.5 | 0.0008 | 0.0010 |
| ACV genes | 9 | 681 | 0.0012 | 0.0014 |
| Phage-related or transposon-related | 21 | 648 | 0.0013 | 0.0016 |
| All genes | 3168 | 879 | 0.0005 | 0.0006 |
*Functional categories are from a previous source [16].
†The variant rate was calculated as (sum of amino acid-changing variants of genes in category – number of genes in category)/(sum of gene length of all genes in category, in bp).
‡The variant rate was calculated as (sum of nucleotide variants of genes in category – number of genes in category)/(sum of gene length of all genes in category, in bp).
A number of genes encoding proteins that localize close to the bacterial surface, or whose localization has not been characterized in detail as yet, display a higher than average genetic variation rate, suggesting that these may have experienced high antigenic pressure. Among the 15 genes that encode seven or more amino acid variants in our strain panel were fhaB (11 variants) and two autotransporters, tcfA (seven variants) and sphB2 (31 variants). Further investigation may be warranted to better understand the observed sequence variation in these genes.
The genetic variant analysis also revealed 33 outer membrane proteins, 17 of which are known to be under Bvg control, which displayed only one genetic variant in the 188 Japanese strains (Table S6). These proteins may be of some interest for future strategies to combat pertussis, although strong sequence conservation may be indicative of low immune pressure. Among these are components of several TonB-dependent iron uptake systems, including the putative haem receptor HemC and putative ferric siderophore receptors encoded by the bfr operon [74]. The protein that suppresses the bactericidal activity of the complement, BrkA [75], and the putative fimbrial usher protein FimC [76] were also among the gene products with no observed variations. Seven of these genes, including bfrH and the gene encoding the outer membrane protein OmpA, were also conserved in 343 worldwide strains [16].
A normalized comparison of previously reported SNP densities from isolates stemming predominantly from the WCV era [16] with the ACV-era variant rates observed in the Japanese strain panel did not reveal significant differences in the propensity for genetic variation in the nine genes encoding ACV-targeted proteins (data not shown).
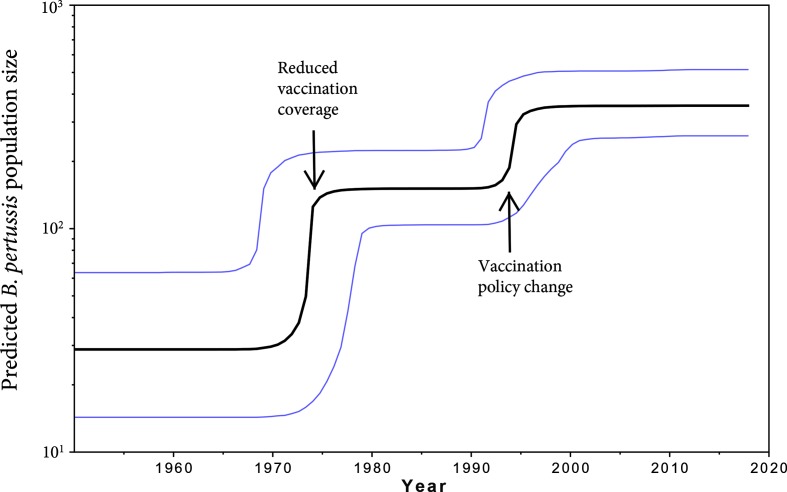
Temporal fluctuations of the B. pertussis population size in Japan
Based on the molecular clock data analysis as determined by beast, fluctuations in the B. pertussis effective population size per year relative to the sample data were inferred from the fluctuations in the number of mutations in the B. pertussis genomes sequenced. For this analysis, the isolates obtained from Konan Kosei Hospital were excluded. As illustrated in Fig. 4, two increases in effective population size were predicted between 1950 and 2012 – one around 1974, and another increase in the mid-1990s. The first increase coincides with a public debate on vaccine safety following two infant deaths within 24 h after WCV administration in the winter of 1974/75, which led to a 2-month temporary suspension of the use of the vaccine and a subsequent raise in age from 3 months to 2 years at which the primary vaccine dose was to be administrated [77]. These events may explain the inferred increase in B. pertussis population size. The second increase coincides with a change in the Preventive Vaccination Law in 1994 in Japan where mass vaccination in regional Public Health Centers was replaced by individual private inoculation upon recommendation, while at the same time the recommended age for the primary series of ACV inoculation was changed from 2 years back to 3–12 months [78, 79]. Consistent with our prediction of an increased B. pertussis population size in Japan after temporary cessation and/or a lower vaccination rate, previous studies have detected the reverse effect on population diversity after introduction of vaccine programmes [73, 80, 81]. However, because genome sequencing has higher resolution than the typing methods employed in those studies, the magnitudes of these effects are difficult to compare.
Fig. 4.
Bayesian Skyline Plot of B. pertussis in Japan, excluding Konan Kosei isolates. Fluctuations in the B. pertussis effective population size per year relative to the sample data were inferred from the fluctuations in the number of mutations in the B. pertussis genomes sequenced. The 95 % CIs are in blue. The predictions are based on the molecular clock data analysis as determined by beast (see Methods). Coinciding events that may affect population size are marked by arrows.
Notably, despite the beast-predicted increase and subsequent stabilization of the effective population size of B. pertussis in the mid-1990s in Japan, the actual case numbers reported to the NIID declined in the late 1990s [82, 83]. ACVs may therefore effectively protect against developing pertussis disease, but may not prevent asymptomatic carriage and circulation of the bacterial strains, as observed in vaccination studies in baboons [13].
In Japan, as in other parts of the world, pertussis has experienced a worldwide resurgence that cannot be attributed to lower vaccine coverage alone. The rising percentage of pertussis cases in Japanese teenagers or adults [84] suggests waning immunity to be one contributing factor, coupled with a vaccination schedule that incorporates a booster at an early age (2 years) but lacks a secondary immunization for adolescents, as championed in the USA [2]. Recent studies revealed that the level of decrease in vaccine effectiveness against pertussis disease depends on both the specific vaccine antigen and the subject’s age [85]. As a short-term solution, the efficacy of existing vaccines may be improved by a change in vaccination booster regimens, by an expansion of maternal immunizations or by improving existing ACV antigens. However, novel targets for future ACV approaches, including a potential change in adjuvants, may be desirable [86, 87].
Conclusions
A focused genome analysis of almost 200 B. pertussis strains prevalent in the last 35 years in Japan revealed the existence of two major phylogenetic clades that coexist in the country. A shift in the population occurred in the past 10 years where isolates of the clade characterized by the ptxA1 : ptxP3 : prn2 genotype began to dominate the B. pertussis population in Japan over isolates of the ptxA2 : ptxP1 : prn1 genotype. This shift coincides with an increase in pertussis cases in the country.
Despite a lack of evidence for a direct effect of ACVs on the genotype shift of the B. pertussis population in Japan, this study noted a high rate of genetic variants for the genes targeted in the current ACV preparations, compared with other cell surface proteins. This observation confirms a certain degree of selective evolutionary pressure exerted by the ACVs.
Pertactin deficiency appeared around 1990 on at least two separate occasions, and probably again in the mid-1990s, in Japan, after ACVs had been adopted. Pertactin-deficient prn1 isolates continue to be observed in Japan, although with decreasing prevalence since 2005.
Genetic variant analysis revealed a number of candidate proteins that are part of the B. pertussis core genome and may resist genetic variation, a subset of which consists of outer membrane proteins. In addition, several genes were identified that have a relatively high variant frequency, perhaps suggesting good antigenic properties. Such information may help in selecting candidate targets for a more effective pertussis vaccine.
Data bibliography
Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, et al. ENA SRA PRJEB2274 (2015).
Funding information
Writing and editorial support was funded by Janssen Vaccines and Prevention B.V. and provided by APEX Think Corporation, Fallbrook, CA, USA.
Conflicts of interest
AZ is an employee of Utrecht University, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, in Utrecht, the Netherlands, providing services for Janssen Vaccines and Prevention B.V., Leiden, the Netherlands. JP and JG are employees of Janssen Vaccines and Prevention B.V. (former Crucell Holland B.V.), part of the Janssen Pharmaceutical Companies of Johnson and Johnson, Department of Bacterial Vaccines Discovery and Early Development, in Leiden, the Netherlands. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in this paper, apart from those disclosed.
Acknowledgements
The authors acknowledge the contribution of Tomomi Kimura, Peter Hermans, Selma Wiertsema and Sander Fortanier for supporting the initiation and execution of the study.
Supplementary Data
Footnotes
Abbreviations: ACV, acellular vaccine; IS, insertion sequence; NIID, National Institute of Infectious Diseases, Japan; OG, orthologous group; Prn, pertactin; SS, signal sequence; WCV, whole-cell vaccine; CI, confidence interval.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables are available with the online version of this article.
References
- 1.CDC Epidemiology and Prevention of Vaccine-Preventable Diseases. 2015. www.cdc.gov/vaccines/pubs/pinkbook/pert.html#vaccines accessed 10/03/2016.
- 2.Watanabe M, Nagai M. Acellular pertussis vaccines in Japan: past, present and future. Expert Rev Vaccines. 2005;4:173–184. doi: 10.1586/14760584.4.2.173. [DOI] [PubMed] [Google Scholar]
- 3.Sato Y, Kimura M, Fukumi H. Development of a pertussis component vaccine in Japan. Lancet. 1984;323:122–126. doi: 10.1016/S0140-6736(84)90061-8. [DOI] [PubMed] [Google Scholar]
- 4.Kato T. Pertussis vaccine in Japan. J Infect Chemother. 1999;5:185–189. doi: 10.1007/s101560050032. [DOI] [PubMed] [Google Scholar]
- 5.Kuno-Sakai H, Kimura M. Safety and efficacy of acellular pertussis vaccine in Japan, evaluated by 23 years of its use for routine immunization. Pediatrics International. 2004;46:650–655. doi: 10.1111/j.1442-200x.2004.01970.x. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y, Sato H. Development of acellular pertussis vaccines. Biologicals. 1999;27:61–69. doi: 10.1006/biol.1999.0181. [DOI] [PubMed] [Google Scholar]
- 7.Hara M, Fukuoka M, Tashiro K, Ozaki I, Ohfuji S, et al. Pertussis outbreak in university students and evaluation of acellular pertussis vaccine effectiveness in Japan. BMC Infect Dis. 2015;15:45. doi: 10.1186/s12879-015-0777-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horiba K, Nishimura N, Gotoh K, Kawaguchi M, Takeuchi S, et al. Clinical manifestations of children with microbiologically confirmed pertussis infection and antimicrobial susceptibility of isolated strains in a regional hospital in Japan, 2008–2012. Jpn J Infect Dis. 2014;67:345–348. doi: 10.7883/yoken.67.345. [DOI] [PubMed] [Google Scholar]
- 9.Mooi FR, van der Maas NAT, de Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation – two sides of the same coin. Epidemiol Infect. 2014;142:685–694. doi: 10.1017/S0950268813000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheridan SL, Frith K, Snelling TL, Grimwood K, McIntyre PB, et al. Waning vaccine immunity in teenagers primed with whole cell and acellular pertussis vaccine: recent epidemiology. Expert Rev Vaccines. 2014;13:1081–1106. doi: 10.1586/14760584.2014.944167. [DOI] [PubMed] [Google Scholar]
- 11.Klein NP, Bartlett J, Fireman B, Aukes L, Buck PO, et al. Waning protection following 5 doses of a 3-component diphtheria, tetanus, and acellular pertussis vaccine. Vaccine. 2017;35:3395–3400. doi: 10.1016/j.vaccine.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 12.He Q, Mertsola J. Factors contributing to pertussis resurgence. Future Microbiol. 2008;3:329–339. doi: 10.2217/17460913.3.3.329. [DOI] [PubMed] [Google Scholar]
- 13.Warfel JM, Zimmerman LI, Merkel TJ. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc Natl Acad Sci USA. 2014;111:787–792. doi: 10.1073/pnas.1314688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warfel JM, Zimmerman LI, Merkel TJ. Comparison of three whole-cell pertussis vaccines in the baboon model of pertussis. Clin Vaccine Immunol. 2016;23:47–54. doi: 10.1128/CVI.00449-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mooi FR, He Q, van Oirschot H, Mertsola J. Variation in the Bordetella pertussis virulence factors pertussis toxin and pertactin in vaccine strains and clinical isolates in Finland. Infect Immun. 1999;67:3133–3134. doi: 10.1128/iai.67.6.3133-3134.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bart MJ, Harris SR, Advani A, Arakawa Y, Bottero D, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio. 2014;5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, et al. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooi FR, van Loo IHM, van Gent M, He Q, Bart MJ, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loconsole D, de Robertis AL, Morea A, Metallo A, Lopalco PL, et al. Resurgence of pertussis and emergence of the Ptxp3 toxin promoter allele in South Italy. Pediatr Infect Dis J. 2018;37:1. doi: 10.1097/INF.0000000000001804. [DOI] [PubMed] [Google Scholar]
- 20.Octavia S, Sintchenko V, Gilbert GL, Lawrence A, Keil AD, et al. Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008–2010. J Infect Dis. 2012;205:1220–1224. doi: 10.1093/infdis/jis178. [DOI] [PubMed] [Google Scholar]
- 21.Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison MJ, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol. 2014;21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuel M, Jamieson FB, Tang P, Brown S, Farrell D, et al. Genetic analysis of Bordetella pertussis in Ontario, Canada reveals one predominant clone. Int J Infect Dis. 2013;17:e413. doi: 10.1016/j.ijid.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Kodama A, Kamachi K, Horiuchi Y, Konda T, Arakawa Y. Antigenic divergence suggested by correlation between antigenic variation and pulsed-field gel electrophoresis profiles of Bordetella pertussis isolates in Japan. J Clin Microbiol. 2004;42:5453–5457. doi: 10.1128/JCM.42.12.5453-5457.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidtke AJ, Boney KO, Martin SW, Skoff TH, Tondella ML, et al. Population diversity among Bordetella pertussis isolates, United States, 1935–2009. Emerg Infect Dis. 2012;18:1248–1255. doi: 10.3201/eid1808.120082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sealey KL, Harris SR, Fry NK, Hurst LD, Gorringe AR, et al. Genomic analysis of isolates from the United Kingdom 2012 pertussis outbreak reveals that vaccine antigen genes are unusually fast evolving. J Infect Dis. 2015;212:294–301. doi: 10.1093/infdis/jiu665. [DOI] [PubMed] [Google Scholar]
- 26.van Gent M, Bart MJ, van der Heide HGJ, Heuvelman KJ, Mooi FR. Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One. 2012;7:e46407. doi: 10.1371/journal.pone.0046407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner B, Melzer H, Freymüller G, Stumvoll S, Rendi-Wagner P, et al. Genetic variation of Bordetella pertussis in Austria. PLoS One. 2015;10:e0132623. doi: 10.1371/journal.pone.0132623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barkoff A-M, Mertsola J, Guillot S, Guiso N, Berbers G, et al. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol. 2012;19:1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegerle N, Paris A-S, Brun D, Dore G, Njamkepo E, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;18:E340. doi: 10.1111/j.1469-0691.2012.03925.x. [DOI] [PubMed] [Google Scholar]
- 30.Queenan AM, Cassiday PK, Evangelista A. Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med Overseas Ed. 2013;368:583–584. doi: 10.1056/NEJMc1209369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden KE, Williams MM, Cassiday PK, Milton A, Pawloski L, et al. Molecular epidemiology of the pertussis epidemic in Washington State in 2012. J Clin Microbiol. 2014;52:3549–3557. doi: 10.1128/JCM.01189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis. 2014;20:626–633. doi: 10.3201/eid2004.131478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin SW, Pawloski L, Williams M, Weening K, Debolt C, et al. Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage. Clin Infect Dis. 2015;60:223–227. doi: 10.1093/cid/ciu788. [DOI] [PubMed] [Google Scholar]
- 34.Quinlan T, Musser KA, Currenti SA, Zansky SM, Halse TA. Pertactin-negative variants of Bordetella pertussis in New York State: a retrospective analysis, 2004–2013. Mol Cell Probes. 2014;28:138–140. doi: 10.1016/j.mcp.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Zeddeman A, van Gent M, Heuvelman CJ, van der Heide HG, Bart MJ, et al. Investigations into the emergence of pertactin-deficient Bordetella pertussis isolates in six European countries, 1996 to 2012. Euro Surveill. 2014;19:20881. doi: 10.2807/1560-7917.ES2014.19.33.20881. [DOI] [PubMed] [Google Scholar]
- 36.Miyaji Y, Otsuka N, Toyoizumi-Ajisaka H, Shibayama K, Kamachi K. Genetic analysis of Bordetella pertussis isolates from the 2008–2010 pertussis epidemic in Japan. PLoS One. 2013;8:e77165. doi: 10.1371/journal.pone.0077165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsuka N, Han H-J, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One. 2012;7:e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodilis H, Guiso N. Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg Infect Dis. 2013;19:471–474. doi: 10.3201/eid1903.121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke M, Mcintyre PB, Blyth CC, Wood N, Octavia S, et al. The relationship between Bordetella pertussis genotype and clinical severity in Australian children with pertussis. J Infect. 2016;72:171–178. doi: 10.1016/j.jinf.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Hegerle N, Dore G, Guiso N. Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine. Vaccine. 2014;32:6597–6600. doi: 10.1016/j.vaccine.2014.09.068. [DOI] [PubMed] [Google Scholar]
- 41.Safarchi A, Octavia S, Luu LDW, Tay CY, Sintchenko V, et al. Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine. 2015;33:6277–6281. doi: 10.1016/j.vaccine.2015.09.064. [DOI] [PubMed] [Google Scholar]
- 42.Hiramatsu Y, Miyaji Y, Otsuka N, Arakawa Y, Shibayama K, et al. Significant decrease in pertactin-deficient Bordetella pertussis isolates, Japan. Emerg Infect Dis. 2017;23:699–701. doi: 10.3201/eid2304.161575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safarchi A, Octavia S, Wu SZ, Kaur S, Sintchenko V, et al. Genomic dissection of Australian Bordetella pertussis isolates from the 2008–2012 epidemic. J Infect. 2016;72:468–477. doi: 10.1016/j.jinf.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Liu B, Gröndahl-Yli-Hannuksila K, Tan Y, Feng L, et al. Whole-genome sequencing reveals the effect of vaccination on the evolution of Bordetella pertussis. Sci Rep. 2015;5:12888. doi: 10.1038/srep12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowden KE, Weigand MR, Peng Y, Cassiday PK, Sammons S, et al. Genome structural diversity among 31 Bordetella pertussis isolates from two recent U.S. whooping cough statewide epidemics. mSphere. 2016;1:e00036-16. doi: 10.1128/mSphere.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2015. https://github.com/tseemann/snippy accessed 20 September, 2017.
- 47.Weigand MR, Peng Y, Loparev V, Batra D, Bowden KE, et al. The history of Bordetella pertussis genome evolution includes structural rearrangement. J Bacteriol. 2017;199:e00806-16. doi: 10.1128/JB.00806-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bart MJ, van der Heide HGJ, Zeddeman A, Heuvelman K, van Gent M, et al. Complete genome sequences of 11 Bordetella pertussis strains representing the pandemic ptxP3 lineage. Genome Announc. 2015;3:e01394-15. doi: 10.1128/genomeA.01394-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bart MJ, Zeddeman A, van der Heide HGJ, Heuvelman K, van Gent M, et al. Complete genome sequences of Bordetella pertussis isolates B1917 and B1920, representing two predominant global lineages. Genome Announc. 2014;2:e01301-14. doi: 10.1128/genomeA.01301-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J, Zhang Y, Buboltz AM, Zhang X, Schuster SC, et al. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics. 2012;13:545. doi: 10.1186/1471-2164-13-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park J, Chen C, Harvill ET, Dudley EG, Zhang Y. Diversity of secretion systems associated with virulence characteristics of the classical bordetellae. Microbiology. 2015;161:2328–2340. doi: 10.1099/mic.0.000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gates I, DuVall M, Ju H, Tondella ML, Pawloski L, et al. Development of a qualitative assay for screening of Bordetella pertussis isolates for pertussis toxin production. PLoS One. 2017;12:e0175326. doi: 10.1371/journal.pone.0175326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- 55.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan W-F, Maharjan RP, Reeves PR, Sintchenko V, Gilbert GL, et al. Rapid and accurate typing of Bordetella pertussis targeting genes encoding acellular vaccine antigens using real time PCR and High Resolution Melt analysis. J Microbiol Methods. 2009;77:326–329. doi: 10.1016/j.mimet.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 57.Muyldermans G, Pierard D, Hoebrekx N, Advani R, van Amersfoorth S, et al. Simple algorithm for identification of Bordetella pertussis pertactin gene variants. J Clin Microbiol. 2004;42:1614–1619. doi: 10.1128/JCM.42.4.1614-1619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tizolova A, Guiso N, Guillot S. Insertion sequences shared by Bordetella species and implications for the biological diagnosis of pertussis syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:89–96. doi: 10.1007/s10096-012-1718-3. [DOI] [PubMed] [Google Scholar]
- 62.Fry NK, Neal S, Harrison TG, Miller E, Matthews R, et al. Genotypic variation in the Bordetella pertussis virulence factors pertactin and pertussis toxin in historical and recent clinical isolates in the United Kingdom. Infect Immun. 2001;69:5520–5528. doi: 10.1128/IAI.69.9.5520-5528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.du Q, Wang X, Liu Y, Luan Y, Zhang J, et al. Direct molecular typing of Bordetella pertussis from nasopharyngeal specimens in China in 2012–2013. Eur J Clin Microbiol Infect Dis. 2016;35:1211–1214. doi: 10.1007/s10096-016-2655-3. [DOI] [PubMed] [Google Scholar]
- 64.Galit SR, Otsuka N, Furuse Y, Almonia DJ, Sombrero LT, et al. Molecular epidemiology of Bordetella pertussis in the Philippines in 2012–2014. Int J Infect Dis. 2015;35:24–26. doi: 10.1016/j.ijid.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Komatsu E, Yamaguchi F, Abe A, Weiss AA, Watanabe M. Synergic effect of genotype changes in pertussis toxin and pertactin on adaptation to an acellular pertussis vaccine in the murine intranasal challenge model. Clin Vaccine Immunol. 2010;17:807–812. doi: 10.1128/CVI.00449-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caro V, Elomaa A, Brun D, Mertsola J, He Q, et al. Bordetella pertussis , Finland and France. Emerg Infect Dis. 2006;12:987–989. doi: 10.3201/eid1206.051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber C, Boursaux-Eude C, Coralie G, Caro V, Guiso N. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J Clin Microbiol. 2001;39:4396–4403. doi: 10.1128/JCM.39.12.4396-4403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poynten M, Mcintyre PB, Mooi FR, Heuvelman KJ, Gilbert GL. Temporal trends in circulating Bordetella pertussis strains in Australia. Epidemiol Infect. 1999;132:185–193. doi: 10.1017/S095026880300164X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Breakwell L, Kelso P, Finley C, Schoenfeld S, Goode B, et al. Pertussis vaccine effectiveness in the setting of pertactin-deficient pertussis. Pediatrics. 2016;137:5. doi: 10.1542/peds.2015-3973. [DOI] [PubMed] [Google Scholar]
- 70.Vodzak J, Queenan AM, Souder E, Evangelista AT, Long SS. Clinical manifestations and molecular characterization of pertactin-deficient and pertactin-producing Bordetella pertussis in children, Philadelphia 2007–2014. Clin Infect Dis. 2017;64:60–66. doi: 10.1093/cid/ciw632. [DOI] [PubMed] [Google Scholar]
- 71.Shuel M, Lefebvre B, Whyte K, Hayden K, de Serres G, et al. Antigenic and genetic characterization of Bordetella pertussis recovered from Quebec, Canada, 2002–2014: detection of a genetic shift. Can J Microbiol. 2016;62:437–441. doi: 10.1139/cjm-2015-0781. [DOI] [PubMed] [Google Scholar]
- 72.Weigand MR, Peng Y, Cassiday PK, Loparev VN, Johnson T, et al. Complete genome sequences of Bordetella pertussis isolates with novel pertactin-deficient deletions. Genome Announc. 2017;5:e00973-17. doi: 10.1128/genomeA.00973-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Litt DJ, Neal SE, Fry NK. Changes in genetic diversity of the Bordetella pertussis population in the United Kingdom between 1920 and 2006 reflect vaccination coverage and emergence of a single dominant clonal type. J Clin Microbiol. 2009;47:680–688. doi: 10.1128/JCM.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brickman TJ, Anderson MT, Armstrong SK. Bordetella iron transport and virulence. Biometals. 2007;20:303–322. doi: 10.1007/s10534-006-9031-1. [DOI] [PubMed] [Google Scholar]
- 75.Barnes MG, Weiss AA. BrkA protein of Bordetella pertussis inhibits the classical pathway of complement after C1 deposition. Infect Immun. 2001;69:3067–3072. doi: 10.1128/IAI.69.5.3067-3072.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, et al. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/S0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 77.Noble GR, Bernier RH, Esber EC, Hardegree MC, Hinman AR, et al. Acellular and whole-cell pertussis vaccines in Japan. Report of a visit by US scientists. JAMA. 1987;257:1351–1356. [PubMed] [Google Scholar]
- 78.Nakayama T. Vaccine chronicle in Japan. J Infect Chemother. 2013;19:787–798. doi: 10.1007/s10156-013-0641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Infectious Disease Surveillance Center, Japan Infectious Agents Surveillance Report: Pertussis, Japan, 1982–1996. 1997. http://idsc.nih.go.jp/iasr/18/207/tpc207.html accessed 02/27/2017.
- 80.van Loo IHM, Mooi FR. Changes in the Dutch Bordetella pertussis population in the first 20 years after the introduction of whole-cell vaccines. Microbiology. 2002;148:2011–2018. doi: 10.1099/00221287-148-7-2011. [DOI] [PubMed] [Google Scholar]
- 81.van Loo IHM, van der Heide HGJ, Nagelkerke NJD, Verhoef J, Mooi FR. Temporal trends in the population structure of Bordetella pertussis during 1949–1996 in a highly vaccinated population. J Infect Dis. 1999;179:915–923. doi: 10.1086/314690. [DOI] [PubMed] [Google Scholar]
- 82.Infectious Disease Surveillance Center, Japan Infectious Agents Surveillance Report: Pertussis, Japan, 2005–2007. 2008. http://idsc.nih.go.jp/iasr/29/337/tpc337.html accessed 02/27/2017.
- 83.National Institute of Infectious Diseases, Japan NESID Annual Surveillance Data Sentinel-Reporting Diseaes 2014-2. 2015. www.nih.go.jp/niid/en/survei/2085-idwr/ydata/6054-report-eb2014-2.html accessed 02/27/2017.
- 84.National Institute of Infectious Diseases, Japan Pertussis, Japan, 2008-2011. Infectious Agents Surveillance Report 2012. p. 321-332. www.niid.go.jp/niid/en/iasren/865-iasr/3027-tpc394.html accessed 02/27/2017.
- 85.van Twillert I, Bonačić Marinović AA, Kuipers B, van Gaans-van den Brink JAM, Sanders EAM, et al. Impact of age and vaccination history on long-term serological responses after symptomatic B. pertussis infection, a high dimensional data analysis. Sci Rep. 2017;7:40328. doi: 10.1038/srep40328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dorji D, Mooi F, Yantorno O, Deora R, Graham RM, et al. Bordetella pertussis virulence factors in the continuing evolution of whooping cough vaccines for improved performance. Med Microbiol Immunol. 2018;207 doi: 10.1007/s00430-017-0524-z. [DOI] [PubMed] [Google Scholar]
- 87.Geurtsen J, Fae KC, van den Dobbelsteen GPJM. Importance of (antibody-dependent) complement-mediated serum killing in protection against Bordetella pertussis. Expert Rev Vaccines. 2014;13:1229–1240. doi: 10.1586/14760584.2014.944901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.