Summary
Cdc45 plays a critical role at the core of the eukaryotic DNA replisome, serving as an essential scaffolding component of the replicative helicase holoenzyme Cdc45-MCM-GINS (CMG) complex. A 1.66-Å-resolution crystal structure of the full-length Cdc45 protein from Entamoeba histolytica shows a protein fold similar to that observed previously for human Cdc45 in its active conformation, featuring the overall disk-like monomer shape and intimate contacts between the N- and C-terminal DHH domains. However, the E. histolytica Cdc45 structure shows several unique features, including a distinct orientation of the C-terminal DHHA1 domain, concomitant disordering of the adjacent protruding α-helical segment implicated in DNA polymerase ɛ interactions, and a unique conformation of the GINS/Mcm5-binding loop. These structural observations collectively suggest the possibility that Cdc45 can sample multiple conformations corresponding to different functional states. We propose that such conformational switch of Cdc45 may allow regulation of protein-protein interactions important in DNA replication.
Subject Areas: Biophysics, Protein Structure Aspects, Structural Biology
Graphical Abstract
Highlights
-
•
High-resolution crystal structure of a full-length Cdc45 protein
-
•
Distinct protein conformation lacking the α-helical protrusion
-
•
Unique positioning of the C-terminal DHHA1 domain possibly coupled to MCM binding
-
•
A conformational switch may regulate DNA polymerase ɛ interactions of Cdc45
Biophysics; Protein Structure Aspects; Structural Biology
Introduction
Initiation of DNA synthesis from an origin of replication begins with loading of two heterohexameric AAA+ ATPase minichromosome maintenance (Mcm) 2-7 complexes by the origin recognition complex (ORC) during the G1-phase (Bell and Dutta, 2002, Pospiech et al., 2010). At the onset of S-phase, each Mcm2-7 hexamer is activated through a series of highly regulated events that culminate in the recruitment of cell division cycle 45 (Cdc45) and Go-Ichi-Ni-San (GINS) proteins. Cdc45, Mcm2-7, and GINS together form a CMG helicase holoenzyme (Boos et al., 2012, Ilves et al., 2010, Tanaka and Araki, 2013). The two active CMG helicases separate and proceed into opposite directions, starting bidirectional unwinding of a double-stranded DNA (Vijayraghavan and Schwacha, 2012). Cdc45 and GINS remain associated with Mcm2-7 throughout DNA replication.
Recent cryoelectron microscopic (cryo-EM) studies have provided valuable information regarding the architecture of the CMG helicase and the potential mode of its DNA unwinding (Abid Ali et al., 2016, Costa et al., 2011, Georgescu et al., 2017, Sun et al., 2015, Yuan et al., 2016, Zhou et al., 2017). These studies showed that Cdc45 and GINS bind to the outer surface of the N-tier ring of the hexameric Mcm2-7 complex, bracing the flexible Mcm2/5 interface to stabilize its closed conformation (Petojevic et al., 2015). This association creates a stable platform threaded on DNA, upon which the C-tier AAA+ ATPase motor of Mcm2-7 catalyzes the translocation of the CMG complex along the DNA strand (Yuan et al., 2016). Although the precise roles of Cdc45 in the CMG helicase function are not fully understood, it likely acts as a structural scaffold and mediates interactions with other replication factors. As a distant homolog of the bacterial RecJ nuclease, Cdc45 has also been proposed to directly interact with DNA strands or facilitate interaction of other replication factors with DNA at the replication fork (Krastanova et al., 2012, Szambowska et al., 2014, Szambowska et al., 2017). The crystal structure of human Cdc45 not only confirmed its evolutionary relationship to RecJ but also revealed key differences that likely confer unique functionalities (Simon et al., 2016).
Despite the wealth of structural information available for Cdc45, a high-resolution structure of full-length Cdc45 is lacking. We sought to crystallize an intact Cdc45 protein and obtain additional structural information, to help better understand its essential function in DNA replication.
Results and Discussion
Structure of Full-Length Cdc45
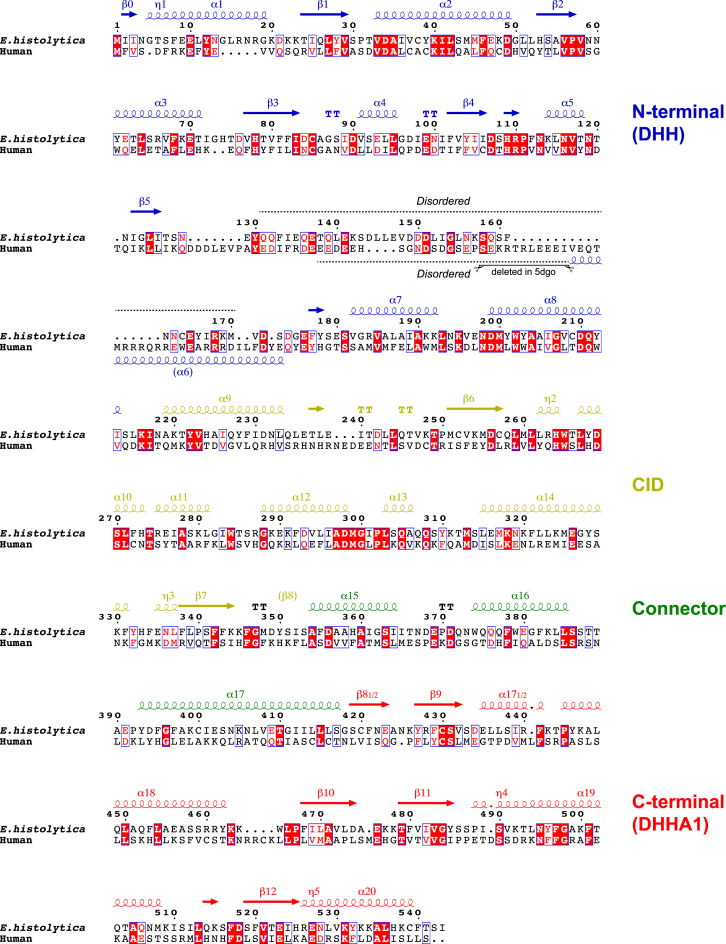
Cdc45 consists of the N-terminal DHH and C-terminal DHHA1 domains connected through an α-helical connector segment (Figure 1), which are shared structural features with the bacterial RecJ nuclease (Pellegrini, 2017, Simon et al., 2016, Yamagata et al., 2002). The CMG-interacting domain (CID) present in Cdc45 and its archaeal homolog GAN (Oyama et al., 2016), but absent from RecJ, is responsible for interactions with the rest of the CMG complex. Another unique feature of Cdc45 is the flexible insertion within the N-terminal DHH domain, comprising a low-complexity sequence rich in acidic amino acids followed by a segment with high α-helical propensity. The crystal structure of human Cdc45 was obtained by deleting a 11-amino acid stretch from this region, which likely facilitated crystallization by rendering the protein less flexible (Simon et al., 2016). We took an alternative approach, to look for a Cdc45 ortholog with a naturally shorter flexible insertion. A relatively compact 543-residue Cdc45 protein from Entamoeba histolytica, an intestinal parasite and the causative agent of amebiasis, was identified as a promising candidate for structural studies (Figure 1).
Figure 1.
Sequence Alignment between Human and Entamoeba histolytica Cdc45
Secondary structures for E. histolytica Cdc45 are shown schematically above the sequence. η and TT denote 310-helix and β-turn, respectively. The 11-residue stretch (amino acids S154 to I164) deleted to facilitate crystallization of human Cdc45 (Simon et al., 2016) is indicated. This figure was prepared using ESPript3.0 (http://espript.ibcp.fr) (Robert and Gouet, 2014).
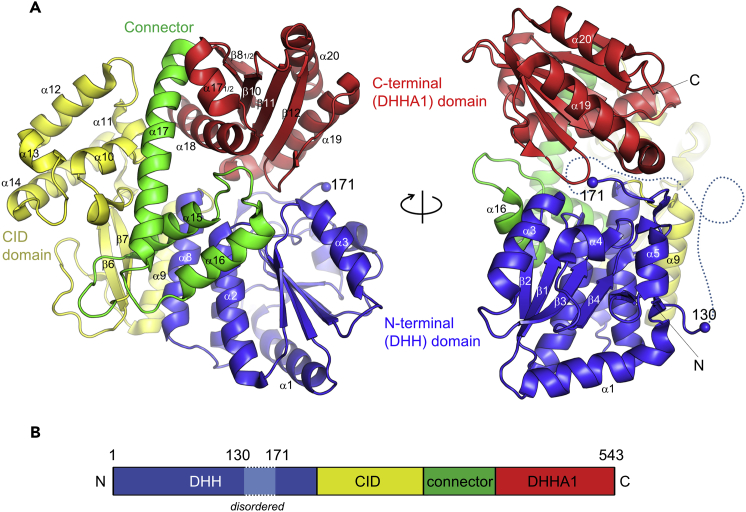
We crystallized the full-length, wild-type E. histolytica Cdc45 and solved its structure using the selenomethionine single-wavelength anomalous diffraction (SAD) phasing method (Table 1, Figure S1). The final model refined to 1.66 Å resolution includes the protein residues 1 through 542, with the exception of an internal disordered region spanning residues 131 through 170 (Figure 2). The domain organization of E. histolytica Cdc45 closely matches that of human Cdc45, and structures of all individual domains show high similarities between the two species, as expected (Figures S2 and S3). As described previously for Phe542 of human Cdc45 (Simon et al., 2016), Phe517 from the DHHA1 domain of E. histolytica Cdc45 is inserted deep into a hydrophobic pocket in the DHH domain to maintain close proximity of the two domains (Figure S4). The overall disk-like shape of E. histolytica Cdc45, as noted for the human Cdc45 structure, with a shallow cleavage between the N- and C-terminal domains makes it resemble a “Pac-Man” when viewed from the side (Figure 2A).
Table 1.
Data Collection and Refinement Statistics
| Data Collection | |
| Resolution range | 36.08–1.66 (1.72–1.66) |
| Space group | P 212121 |
| Unit cell | |
| a,b,c (Å) | 39.12 73.81 171.6 |
| Total reflections | 297,761 (28,914) |
| Unique reflections | 57,811 (5,488) |
| Multiplicity | 5.2 (5.3) |
| Completeness (%) | 96.57 (93.13) |
| I/σI | 10.5 (1.7) |
| R-merge (%) | 9.21 (83.51) |
| R-meas (%) | 10.24 (92.68) |
| R-pim (%) | 4.353 (39.31) |
| CC1/2 | 0.997 (0.675) |
| Refinement | |
| No. reflections | 57,811 (5,488) |
| Reflections for Rfree | 2,905 (292) |
| Rwork | 15.7 (27.5) |
| Rfree | 19.1 (30.0) |
| No. non-H atoms | 4,544 |
| Macromolecules | 4,084 |
| Ligands | 58 |
| Solvent | 402 |
| Protein residues | 502 |
| RMSD | |
| Bond lengths (Å) | 0.012 |
| Bond angles (˚) | 1.09 |
| Ramachandran plot | |
| Favored (%) | 98.38 |
| Allowed (%) | 1.62 |
| Outliers (%) | 0.00 |
| Average B-factor | 33.31 |
| Macromolecules | 32.22 |
| Ligands | 52.94 |
| Solvent | 41.52 |
Statistics for the highest-resolution shell are shown in parentheses.
RMSD, root-mean-square deviation.
Figure 2.
Overall Structure of E. histolytica Cdc45
(A) Ribbon model of E. histolytica Cdc45 viewed from two orthogonal orientations. Color coding for the four structural domains matches that used in Figure 1. Residues 130 and 171 on either end of the disordered region are highlighted by small blue spheres. The disordered residues (131–170) are represented by a dotted line in the right panel.
(B) A schematic diagram showing domain organization of E. histolytica Cdc45.
E. histolytica Cdc45 Shows a Unique Conformation
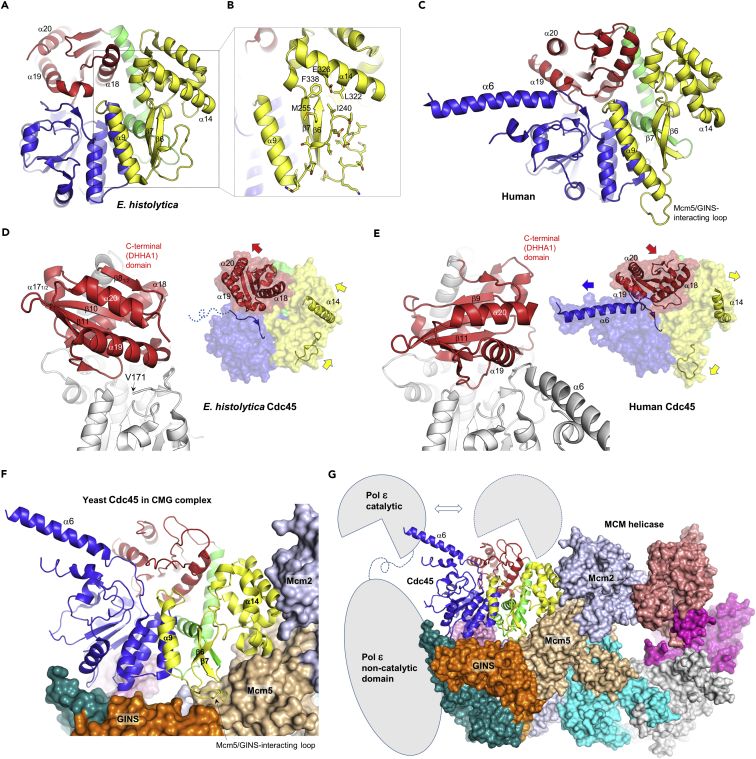
Despite overall similarity to human Cdc45, the E. histolytica Cdc45 crystal structure has several unique features (Figures 3A–3E, S5 and Video S1). First, it shows a distinct positioning of the C-terminal DHHA1 domain with respect to the rest of the molecule. When superimposed over the N-terminal 3 domains (with root-mean-square deviation of 1.4 Å over 255 Cα atoms from the DHH, CID, and connector domains), the DHHA1 domains of human and E. histolytica Cdc45 show a ∼30° rotation and translation by as much as ∼12 Å. Second, the E. histolytica Cdc45 structure does not show a “helical protrusion,” which is one of the most distinct structural features observed for human Cdc45. In the human Cdc45 crystal structure, α6, which immediately follows the low-complexity acidic stretch as mentioned above (Figure 1), protrudes away from the body of the protein (Figures 3C and 3E). The corresponding region of E. histolytica Cdc45 is disordered, and the following loop (Val171 ∼) traverses the narrow inter-domain groove between the DHH and DHHA1 domains (Figures 2A and 3D). Third, the loop between α9 and β6 in CID of E. histolytica Cdc45 (residues 233–251) takes a distinct “tucked-in” conformation where it associates with α14 (Figures 3A, 3B, and 3D). This loop conformation is stabilized by docking of Ile240 in a hydrophobic pocket lined by Met255, Leu322, and Phe338 and is accompanied by unique conformation of the surrounding structural elements in E. histolytica Cdc45, including the notably straight α14 (Figure 3B). The loop between α9 and β6 interacts with GINS subunit Psf2 and Mcm5, whereas α14 in a bent conformation interacts with Mcm2 and Mcm5, in the yeast CMG complex (Figures 3F and 3G) (Yuan et al., 2016).
Figure 3.
Structural Comparison
(A) E. histolytica Cdc45 structure.
(B) Zoomed-in view of the region highlighted by a dotted box in panel A. The loop between α9 and β6 of E. histolytica Cdc45 takes a unique “tucked-in” conformation.
(C) Human Cdc45 structure, PDB ID: 5dgo (Simon et al., 2016).
(D) E. histolytica Cdc45, with the C-terminal DHHA1 domain highlighted in red. A possible coordination between positioning of the DHHA1 domain, α14 helix, and the Mcm5/GINS-interacting loop is depicted in the right panel.
(E) Human Cdc45 with the C-terminal DHHA1 domain highlighted. Note distinct positioning of the DHHA1 domain, bent α14 helix, and untucked Mcm5/GINS-interacting loop.
(F) Yeast CMG helicase cryo-EM structure (PDB ID: 3jc6) (Yuan et al., 2016) viewed from the N-terminal face of MCM, showing extensive interactions between Cdc45 (ribbon) and Mcm2/5 and the GINS subunits (surface).
(G) A full view of the CMG helicase, with the spatial relationship between helicase and DNA polymerase ɛ shown schematically. The catalytic domain of DNA polymerase ɛ can take alternative positions, one of which is stabilized by interaction with the tip of α6 helix of Cdc45 (Zhou et al., 2017).
Active Conformation for DNA Polymerase Interactions
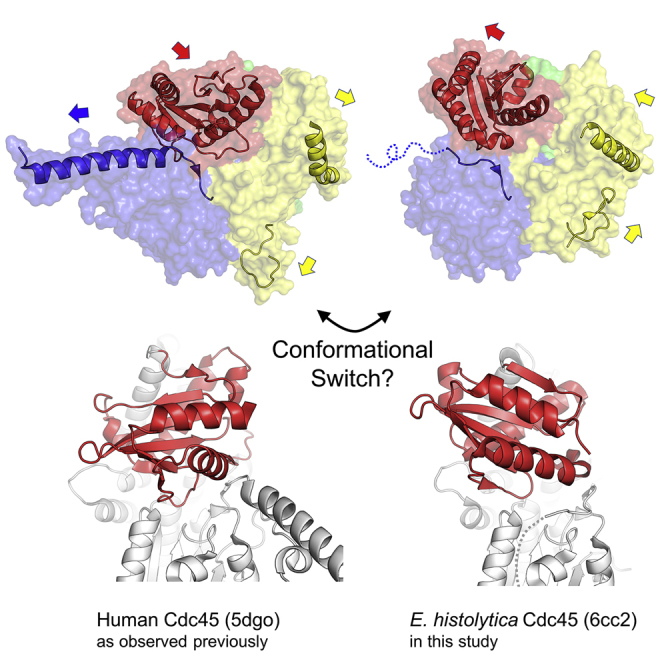
The crystal structure of human Cdc45 with a long α-helical protrusion (α6), as reported earlier (Simon et al., 2016), is similar to the cryo-EM structures of yeast or Drosophila Cdc45 in the CMG complex (Abid Ali et al., 2016, Georgescu et al., 2017, Yuan et al., 2016) (Figure S6). Chemical cross-linking studies showed that the distal end (N terminus, pointed away from the CMG core) of α6 is involved in the interaction of CMG helicase with DNA polymerase ɛ (Pol ɛ) responsible for leading-strand synthesis (Sun et al., 2015). Cryo-EM studies also suggested that the tip of α6 serves as a landing pad for the highly dynamic N-terminal catalytic domain of Pol ɛ, which was shown to undergo a large-scale swing motion, possibly corresponding to different functional states of the polymerase (Zhou et al., 2017) (Figure 3G). These observations suggest that the human Cdc45 crystal structure represents an active state of the protein, poised for functioning in the CMG complex to mediate protein-protein interactions. It is possible that the distinct conformations observed between the human and E. histolytica Cdc45 crystal structures described above simply reflect an inter-species difference: i.e., they represent divergent active conformations of Cdc45 from two different organisms. Alternatively, the observed structural differences could be due to different crystal packing.
Cdc45 Conformational Switch May Regulate DNA Replication
However, given the likely functional importance of the protruding α6 in Cdc45 from yeast, Drosophila, and humans, it is also conceivable that E. histolytica Cdc45 in its active (“on”) state would assume a conformation similar to that observed for human Cdc45. In this scenario, our E. histolytica Cdc45 crystal structure would represent a distinct functional (“off”) state of the eukaryotic Cdc45 proteins incompatible with the Pol ɛ catalytic domain interactions. In support of this hypothesis, more than half of the disordered residues (131–170) within the N-terminal DHH domain of E. histolytica Cdc45 are predicted to have an α-helical conformation by secondary structure prediction programs (e.g., PSIPRED) (McGuffin et al., 2000), suggesting that this region could take a more rigid α-helical structure under different functional contexts, for instance, when E. histolytica Cdc45 is bound to other protein factors. Such conformational change may be coupled to re-orientation of the DHHA1 domain, which displaces (pushes out) the residues nestled in the inter-domain groove between the DHH and DHHA1 domains and supports its conversion to an extended α-helical structure (Video S1). We further speculate that these structural changes might also be allosterically coupled to the conformational state of the α14 helix adjacent to the DHHA1 domain and in turn, to that of the GINS/Mcm5-binding loop (Figures 3D and 3E). Such allosteric coupling would allow Cdc45 to control positioning of the Pol ɛ catalytic domain in response to conformational state of the rest of the CMG helicase, for instance, regulating access of Pol ɛ to the leading-strand 3′ terminus to facilitate polymerase switching. Further structural and functional studies are warranted to understand the conformational flexibility of Cdc45 and its possible roles in DNA replication.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Surajit Banerjee for assistance in X-ray data collection. This work is based upon research conducted at the Advanced Photon Source (APS) Northeastern Collaborative Access Team beamlines, which are funded by the US National Institutes of Health (NIGMS P41-GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by an NIH-ORIP HEI grant (S10 RR029205). This research used resources of the APS, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357, and those of the Minnesota Supercomputing Institute. This work was supported by grants from the National Institutes of Health (NIGMS R35-GM118047 to H.A, and R01-GM074917 to A.-K.B.).
Author Contributions
A.-K.B. and H.A. conceived the project. F.K. purified the proteins and identified the crystallization condition. K.S. collected X-ray diffraction data and determined the crystal structure. K.K. performed cloning and assisted with protein purification. H.A. supervised the research and wrote the paper. All authors contributed in editing and figure preparation.
Declaration of Interests
The authors declare no competing interests.
Published: May 25, 2018
Footnotes
Supplemental Information includes Transparent Methods, six figures, and one video and can be found with this article online at https://doi.org/10.1016/j.isci.2018.04.011.
Supplemental Information
References
- Abid Ali F., Renault L., Gannon J., Gahlon H.L., Kotecha A., Zhou J.C., Rueda D., Costa A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Boos D., Frigola J., Diffley J.F. Activation of the replicative DNA helicase: breaking up is hard to do. Curr. Opin. Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M.R., Berger J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R., Yuan Z., Bai L., de Luna Almeida Santos R., Sun J., Zhang D., Yurieva O., Li H., O'Donnell M.E. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. USA. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I., Petojevic T., Pesavento J.J., Botchan M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krastanova I., Sannino V., Amenitsch H., Gileadi O., Pisani F.M., Onesti S. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 2012;287:4121–4128. doi: 10.1074/jbc.M111.285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Oyama T., Ishino S., Shirai T., Yamagami T., Nagata M., Ogino H., Kusunoki M., Ishino Y. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res. 2016;44:9505–9517. doi: 10.1093/nar/gkw789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L. Structural insights into Cdc45 function: was there a nuclease at the heart of the ancestral replisome? Biophys. Chem. 2017;225:10–14. doi: 10.1016/j.bpc.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petojevic T., Pesavento J.J., Costa A., Liang J., Wang Z., Berger J.M., Botchan M.R. Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc. Natl. Acad. Sci. USA. 2015;112:E249–E258. doi: 10.1073/pnas.1422003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech H., Grosse F., Pisani F.M. The initiation step of eukaryotic DNA replication. Subcell. Biochem. 2010;50:79–104. doi: 10.1007/978-90-481-3471-7_5. [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A.C., Sannino V., Costanzo V., Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat. Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Shi Y., Georgescu R.E., Yuan Z., Chait B.T., Li H., O'Donnell M.E. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szambowska A., Tessmer I., Kursula P., Usskilat C., Prus P., Pospiech H., Grosse F. DNA binding properties of human Cdc45 suggest a function as molecular wedge for DNA unwinding. Nucleic Acids Res. 2014;42:2308–2319. doi: 10.1093/nar/gkt1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szambowska A., Tessmer I., Prus P., Schlott B., Pospiech H., Grosse F. Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res. 2017;45:3217–3230. doi: 10.1093/nar/gkw1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013;5:a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S., Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell. Biochem. 2012;62:113–134. doi: 10.1007/978-94-007-4572-8_7. [DOI] [PubMed] [Google Scholar]
- Yamagata A., Kakuta Y., Masui R., Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl. Acad. Sci. USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Bai L., Sun J., Georgescu R., Liu J., O'Donnell M.E., Li H. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.C., Janska A., Goswami P., Renault L., Abid Ali F., Kotecha A., Diffley J.F.X., Costa A. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc. Natl. Acad. Sci. USA. 2017;114:4141–4146. doi: 10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.