Abstract
Recent evidence suggests that dysregulation of iron regulatory factors may play essential roles in cancer pathophysiology. Six-transmembrane epithelial antigen of prostate 3 (STEAP3) is a metalloreductase, which is vital for cellular iron uptake and homeostasis. However, the clinical significance and function of STEAP3 in the development of human gliomas remain unclear. Through analysis of publicly available databases, we found that STEAP3 was highly expressed in malignant gliomas, especially in the mesenchymal glioma molecular subtype and isocitrate dehydrogenase 1/2 (IDH1/2) wild-type gliomas. Expression levels of STEAP3 in gliomas correlated inversely with patient overall survival (OS) and served as an independent prognostic marker by multivariate Cox regression analysis. In functional assays performed with RNA knockdown, loss of STEAP3 attenuated aggressive phenotypes in glioma cells, including cell proliferation, invasion, and sphere formation in vitro and tumor growth in vivo. Finally, STEAP3 drives these activities by inducing mesenchymal transition, promoting transferrin receptor (TfR) expression, and activating STAT3-FoxM1 axis signaling. Taken together, these results indicate that STEAP3 functions as an oncogenic mediator in glioma progression and is thus a potential therapeutic target for the treatment of the disease.
Abbreviations: FoxM1, forkhead box protein M1; STAT3, signal transducer and activator of transcription 3; SOX2, SRY-related high-mobility-group (HMG)-box protein-2
Introduction
Glioblastoma (GBM) is the most aggressive, deadly, and common brain malignancy occurring in adults [1]. Despite advances in surgical technique, radiotherapy, and chemotherapy, the median survival for GBM patients has remained at a mere 14–15 months [2]. The major challenge in treating GBM is infiltration and relapse, which are presumed to be associated with the process of epithelial-mesenchymal transition and the acquisition of stem cell-like properties [3], such as self-renewal and invasiveness [4]. Therefore, the discovery of new therapeutic molecular targets and pathways which expose vulnerabilities in GBM is desperately needed.
Many biological processes are affected in tumor progression. Among them interestingly is iron metabolism. Iron has specific functions necessary for cell viability/growth. It is indispensable for the synthesis/function of proteins or enzymes that regulate respiratory complexes, DNA, hemesynthesis, mitosis, and epigenetic modifications, all of which are abnormal in cancer [5], [6]. Iron uptake and dependence are in fact increased in glioma stem cells (GSCs). In addition, it has been reported that iron is an important regulator of EMT and metastasis in human cancers [7], [8].
As a member of iron regulatory protein family, STEAP3 functions as a ferrireductase which reduces ferric iron to ferrous iron in endosomes [9]. Under normal conditions, iron exists in two forms, ferrous (Fe2+) and ferric (Fe3+). Fe3+ binds to transferrin in order to form holo-transferrin, which can then be taken up by TfR located on the cell membrane. The endosome containing the transferrin-TfR-1 complex becomes acidified, which accelerates release of ferric iron from holo-transferrin [10]. Importantly, Fe3+ must be reduced to the Fe2+ form before it can be delivered out into the cytoplasm by the divalent metal transporter-1 (DMT1) [11] or transient receptor potential (TRP) protein TRPML [12]. Thus, STEAP3 has a critical role in iron uptake.
Recently, dysregulation of the STEAP family (STEAP1, STEAP2, STEAP3, and STEAP4) has been observed in various cancers. STEAP1 is highly expressed in many human malignancies, including prostate, lung, bladder, colon, ovarian, and Ewing's sarcoma cancers [13], [14], [15]. Furthermore, STEAP1 has also been identified as an intriguing target for antibody therapy in multiple solid human tumors [15]. STEAP2 has been shown to contribute to the regulation of cell proliferation, cell cycle progression, and apoptosis in prostate cancer [16]. Finally, several phosphorylation sites within the STEAP4 protein have been associated with its functions and signaling pathways in cancer cells [17].
However, the molecular pathways that require iron metabolism and drive malignant transition and acquisition of stemness of glioma cells remain to be elucidated. In this study, we investigated the role of STEAP3 in human glioma. Expression of STEAP3 was examined in primary tumor samples and correlated with clinical outcome. Functional studies were performed in vitro and in vivo to investigate the role of STEAP3 in migration, invasion, proliferation, stemness, and tumor progression. Our results indicate that iron metabolism and STEAP3 should be further investigated as therapeutic targets for the treatment of human glioma.
Methods
Ethics Statement
The research strategy was approved by the Research Ethics Committee of Shandong University and the Ethics Committee of Qilu Hospital (Shandong, China). All experiments were performed in accordance with the relevant guidelines and regulations, and written informed consent was obtained from all patients. The Institutional Animal Care and Use Committee (IACUC) of Shandong University approved all surgical interventions and post-operative animal care.
Clinical Specimens and Database Searches
Archived paraffin embedded glioma tissues (WHO grade II-IV) were collected from patients (n = 60) who underwent surgery in the Department of Neurosurgery, Qilu Hospital of Shandong University. Normal brain tissue samples (n = 5) were taken from trauma patients who underwent partial resection of normal brain for decompression treatment for severe head injuries. The Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (http://cancergenome.nih.gov) were mined for relevant molecular data.
Immunohistochemistry (IHC)
Sections (4 μm) were cut from formalin-fixed, paraffin-embedded tissues of WHO grade II – IV gliomas, deparaffinized, rehydrated in an alcohol series, and pretreated with high-temperature antigen retrieval to 98 °C in 0.01 M citrate buffer for 20 min. Endogenous HRP activity was quenched with 3% H2O2 diluted in ethanol for 30 min at room temperature. Sections were blocked with 10% normal goat serum (Beyotime, China), incubated with anti-STEAP3 (Abcam, UK; dilution 1: 500) and anti-TfR antibodies (Abcam, UK; dilution 1: 800) at 4 °C overnight, and rinsed with physiological phosphate buffered saline (PBS) three times for five minutes each. Detection was performed at 37 °C for 30 minutes with a goat anti-rabbit secondary antibody and HRP conjugated to a polymer according to the manufacturer's instructions. Signal was developed with 3,3′-diaminobenzidine (DAB; Beyotime, China), and slides were counterstained with hematoxylin. For negative controls, sections were incubated with normal goat serum instead of the primary antibody. Staining of cancer cells was scored as follows: 0, no staining; 1, weak staining in <50% of cells; 2, weak staining in ≥50% of cells; 3, strong staining in <50% of cells; and 4, strong staining in ≥50% of cells.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
Analysis for genes correlated with STEAP3 expression was performed on the TCGA GBM dataset with MATLAB software (MathWorks; Natick, MA, USA). Positively and negatively correlated genes (P < .01) were analyzed using the DAVID web tool (http://david.abcc.ncifcrf.gov/home.jsp) to identify the biological processes and KEGG signaling pathways associated with STEAP3 expression in gliomas.
Cell Culture
Patient-derived GBM#01 GSCs were isolated from a GBM surgical specimen in the Department of Neurosurgery, Qilu Hospital. Patient-derived GBM#P3 GSCs were kindly provided by Professor Rolf Bjerkvig, Department of Biomedicine, University of Bergen, Norway. GSCs were cultured in serum-free Neurobasal medium (Gibco, USA) supplemented with 2% B27 Neuro Mix (Thermo Fisher Scientific, USA), 20 ng/mL epidermal growth factor (EGF; Thermo Fisher Scientific, USA), and 10 ng/mL basic fibroblast growth factor (bFGF; PeproTech, USA). Tumor spheres were split using accutase (Thermo Fisher Scientific, USA) to expand GSCs.
Gene Knockdown and Ectopic Expression
Stable knockdown of STEAP3 was generated by transducing an sh-STEAP3 lentiviral expression construct in cells (Genechem, China). The shRNA sequence used was the following: 5′-GCTTCTATGCCTACAACTT-3′. For ectopic expression of STEAP3, the full length ORF of the gene was cloned into pENTER vectors. Transfection was performed with Lipofectamine 3000 (Life Technologies, Carlsbad, CA). Cell cultures stably expressing STEAP3 were obtained after selection with puromycin (Life Technologies) for at least 1 week. STEAP3 siRNA and TfR siRNA were purchased from Riobio Co. Ltd. (Guangzhou, China) and transfected into glioma cells using Lipofectamine 3000. The siRNA sequences used were the following: STEAP3: 5′-GCUUCUAUGCCUACAACUU-3′ and 5′-GCCAGAACAAGUUCUUCAA-3′; TfR#1: 5′-GGUAGUUCAAUACCAGUUA-3′.
Western blot analysis
Protein lysates were prepared from human or mouse glioma tissue, 10 to 12 samples for each experimental group, and lysed for 30 min in RIPA buffer (Beyotime, China) supplemented with a protein inhibitor cocktail. Protein concentrations were determined using the BCA assay according to the manufacturer's instructions (Beyotime, China). Protein lysates (20 μg) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, China). Membranes were blocked for 1 h in Tris Buffered Saline with Tween 20 (TBS-T, 10 mM Tris, 150 mM NaCl, 0.1% Tween 20) containing 5% bovine serum albumin (BSA; Beyotime, China), and incubated overnight at 4 °C with primary antibody followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Beyotime; China; dilution 1: 5000) dissolved in antibody dilution buffer (Beyotime; China) for 1 h at room temperature. Rinses were performed in between incubations with tris buffered saline with tween 20 (TBS-T, 10 mM Tris, 150 mMNaCl, 0.1% Tween 20). Proteins were visualized with chemiluminescence (Bio-Rad, USA) according to the manufacturer's protocol. The following primary antibodies were used: CDH2 (Cell Signaling Technology, USA; dilution 1: 1000), Snail (Cell Signaling Technology, USA; dilution 1: 1000), Slug (Cell Signaling Technology, USA; dilution 1: 1000), MMP-2 (Cell Signaling Technology, USA; dilution 1: 1000), GAPDH (Santa Cruz, USA; dilution 1: 2000), STEAP3 (Abcam, UK; dilution 1: 500), TfR (Abcam, UK; dilution 1: 1000), Ferritin (Abcam, UK; dilution 1: 1000), FoxM1 (Abcam, UK; dilution 1: 1000), STAT3 (Abcam, UK; dilution 1: 1000), and p-STAT3 (Abcam, UK; dilution 1: 1000). GAPDH was used as a control for protein loading. Data shown are representative of at least three independent biological replicates.
3D Tumor Spheroid Invasion Assay
Glioma spheroids were generated by incubating cells in spheroid formation matrix for 96 h in a 3D culture qualified 96-well spheroid formation plate. Spheroids were then embedded into the invasion matrix (Trevigen; Gaithersburg, MD; USA) composed of basement membrane proteins in a 96-well plate. Glioma spheroids were photographed under bright field microscopy (Nikon; Tokyo, Japan). The spheroid at 0 h was used as a reference point for measurement of the distance invaded by sprouting cells.
Co-Immunoprecipitation (Co-IP)
Cells were lysed in IP lysis buffer (Pierce/Thermo Fisher Scientific, USA). Immunoprecipitation was performed with cell lysates (20 μg) in the presence of antibodies (1–5 mg) and A/G agarose beads (Pierce/Thermo Fisher Scientific, USA) overnight at 4 °C with constant agitation. Controls were samples incubated with agarose beads after immunoprecipitation with a control immunoglobulin. The immunoprecipitated complexes were then rinsed with lysis buffer three times. Targeted proteins were eluted by heating the mixture to 99 °C for 10 min in protein loading buffer. One tenth of the eluate was retained for western blotting confirmation of the pull-down, and the remainder of the eluate was used to identify the proteins in the immune complexes.
Immunofluorescence (IF)
Transfected cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.4% Triton X-100 for 10 min. Cells were incubated in 5% normal goat serum for 60 min at room temperature. The serum was drained off, and the slides were blot-dried. Coverslips were incubated with primary antibodies against STEAP3 (Abcam, UK; dilution 1: 100), TfR (Abcam, UK; dilution 1: 100), Nestin (Santa Cruz, USA; dilution 1: 200), and SOX2 (Abcam, UK; dilution 1: 100) at 4 °C overnight followed by incubation with Alexa-conjugated secondary antibody (Abcam, UK; dilution 1: 800) for 1 h. Nuclei were counterstained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, Sigma-Aldrich, USA; working dilution 1: 1000) for 5 min at room temperature and mounted in 50% glycerin mounting solution. Rinses in between steps were performed with phosphate buffered saline (PBS). Representative images of STEAP3 and TfR were acquired in confocal microscopy (LSM780, Zeiss). Representative images of Nestin and SOX2 were viewed with a Nikon inverted fluorescence microscope.
Cell Proliferation Assay and Tumor Sphere Formation Assay
Cell growth was assessed using the Luminescent Cell Viability Assay kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. For tumor sphere formation assays, GSCs transfected with si-Ctrl and si-STEAP3 were seeded into 12-well plates at a density of 1000 cells/well and cultured in Neurobasal medium with B27 supplement and growth factors. Tumor spheres were photographed under bright field microscopy (Nikon).
InVitro Extreme Limiting Dilution Assay
GSCs were seeded into a 96-well plate at a density of 1 to 50 cells/well with eight replicates for each concentration. After ten days, tumor spheres in each well were counted, and the sphere formation efficiency was calculated using extreme limiting dilution analysis as previously described [18].
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from cells using RNAiso (Takara, Japan) according to the manufacturer's protocol. Reverse transcription was conducted using the PrimeScript™ RT reagent kit (Takara, Japan). qRT-PCR was performed with SYBR premix Ex Taq (Takara, Japan) on the CFX96 Real Time PCR Detection System (Roche 480II; Berlin, Germany). GAPDH mRNA was used to normalize mRNA expression, and the results are representative of at least three independent experiments. The sequences of the primers used are shown in Supplementary Table S1.
Implantations
Glioma cells (1 × 106) were infected with sh-STEAP3 or sh-Ctrl lentivirus and implanted stereotactically into the brains of 4-week-old nude mice as previously described (SLAC laboratory animal Center; Shanghai, China). Animals that showed symptoms such as severe hunchback posture, apathy, messy fur, decreased motion or activity, dragging legs, or drastic loss of body weight were sacrificed by cervical dislocation. After death, tumor tissues were formalin-fixed, paraffin-embedded, and sectioned.
Statistical Analysis
Survival curves were estimated by the Kaplan–Meier method and compared using the log-rank test. The cut-off level was set at the median value of STEAP3 expression levels. Expression patterns of STEAP3 in different glioma subtypes were determined using data extracted from publicly available databases. A two-tailed χ2 test was used to determine the association between STEAP3 expression and clinicopathological characteristics. The Pearson correlation was applied to evaluate the linear relationship between gene expression levels. The one-way ANOVA test or t-test was used for all other data comparisons using GraphPad Prism software program (GraphPad, La Jolla, CA, USA). Data for each treatment group were represented as mean±S.E.M. and compared with other groups for significance by one-way ANOVA followed by Bonferroni's post hoc test (multiple comparison tests). All tests were two-sided, and P-values < .05 were considered to be statistically significant.
Results
STEAP3 Expression is Elevated in Primary Human GBM Specimens
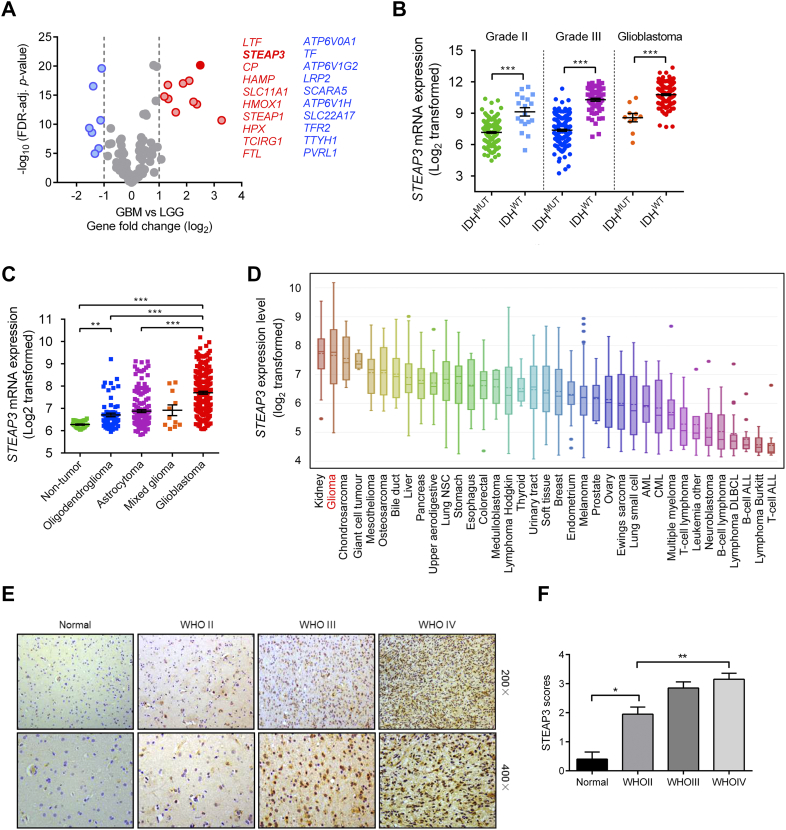
We used a bioinformatics approach to investigate the expression of iron metabolism-related genes in GBM and low-grade glioma (LGG) samples from the TCGA. We first generated a list of 121 genes involved in the biology of iron metabolism using the following Molecular Signature Gene Sets: GO_IRON_ION_HOMEOSTASIS, GO_CELLULAR_IRON_ION_HOMEOSTASIS, GO_IRON_ION_TRANSPORT, and REACTOME_IRON_UPTAKE_AND_TRANSPORT. Through analysis of the differences in mRNA expression levels (log2 transformed) between TCGA GBM and LGG samples, STEAP3 emerged as one of the top three genes (LTF, STEAP3, and CP) with increased expression in GBM (Figure 1A). We further examined the association of STEAP3 expression with GBM by incorporating IDH mutation status in our analysis of the TCGA clinical data. IDH mutated gliomas in general expressed STEAP3 at lower levels than their IDH wild-type counterparts (WHO grade II (P < .001); WHO grade III (P < .001); and GBM (P < .001) patients; Figure 1B). Analysis of the Rembrandt database demonstrated that STEAP3 expression was also higher in multiple histological subtypes of gliomas, relative to non-neoplastic samples, and increased with increasing tumor grade (Figure 1C). Finally, data derived from the Cancer Cell Line Encyclopedia indicated that glioma cell lines possess higher expression of STEAP3 than most cancer cells lines derived from other lineages (Figure 1D).
Figure 1.
STEAP3 expression is elevated in primary human GBM specimens. (A) Fold-change (log2) in mRNA levels of iron regulatory associated genes based on tumor pathology, GBM or LGG. Data was obtained from the TCGA dataset. (B) Analysis of STEAP3 mRNA levels (log2) in WHO grade II, grade III, and grade IV gliomas from the TCGA as a function of IDH mutation status. (C) STEAP3 mRNA expression (log2) in non-tumor brain samples and different histological subtypes of gliomas from the Rembrandt dataset. (D) STEAP3 mRNA expression (log2) across cancer cell lines from different tissues of origin. Glioma cell lines are highlighted in red. (E) Representative images of IHC staining for STEAP3 in normal brain and different pathological grades of astrocytomas showing cytoplasmic positivity. Magnification: 200×, upper panel; 400×, lower panel. (F) Scores for IHC staining represented in bar graphs. Five random fields from each section were counted. Data is shown as the mean ± the standard error of the mean (SEM) for each group. *P < .05, **P < .01, ***P < .001.
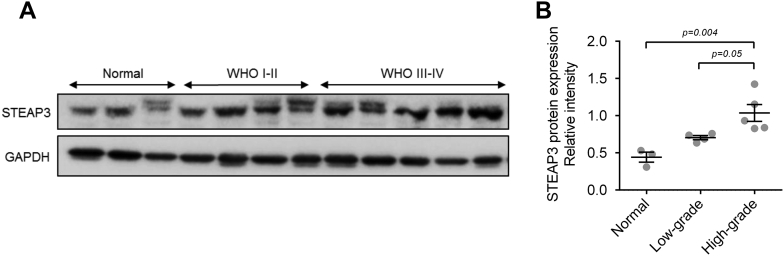
IHC staining of STEAP3 in tissue samples from primary gliomas WHO grade II-IV patients (n = 60) and normal brain (n = 5) demonstrated that protein levels were more often increased in GBM samples, while expression was low/undetectable in normal brain tissues (grade II, 8/20, 40%; grade III, 14/20, 70%; GBM, 17/20, 85%; Fig 1E and F; Table. 1). Western blot analysis of lysates prepared from normal brain (n = 3), LGG (n = 4), and high-grade gliomas (HGG, n = 5) also demonstrated that STEAP3 protein levels were increased in gliomas (Supplementary Figure S1). Taken together, the expression of STEAP3 was up-regulated and positively correlated with increasing grade in gliomas based on results from in silico experiments and an independent set of primary glioma specimens.
Table 1.
Correlations of STEAP3 expression with preoperative clinicopathological features in glioma patients
| Variables | No. of Cases |
STEAP3 Expression |
P | |
|---|---|---|---|---|
| Low | High | |||
| Age (year) | ||||
| <55 | 34 | 14 | 20 | .832 |
| ≥55 | 26 | 10 | 16 | |
| Sex | ||||
| Male | 27 | 11 | 15 | .621 |
| Female | 33 | 13 | 23 | |
| Tumor size | ||||
| <4 cm | 35 | 14 | 21 | 1 |
| ≥4 cm | 25 | 10 | 15 | |
| Cystic change | ||||
| Absent | 27 | 15 | 12 | .525 |
| Present | 33 | 22 | 12 | |
| Edema | ||||
| None to mild | 37 | 16 | 21 | .515 |
| Moderate to severe | 23 | 8 | 15 | |
| WHO grade | ||||
| II | 20 | 12 | 8 | |
| III + IV | 40 | 9 | 31 | .004 |
Supplementary Figure S1.
Expression of STEAP3 is elevated in high grade gliomas.
High STEAP3 Expression is a Prognostic Marker for Poor Clinical Outcome in Glioma Patients
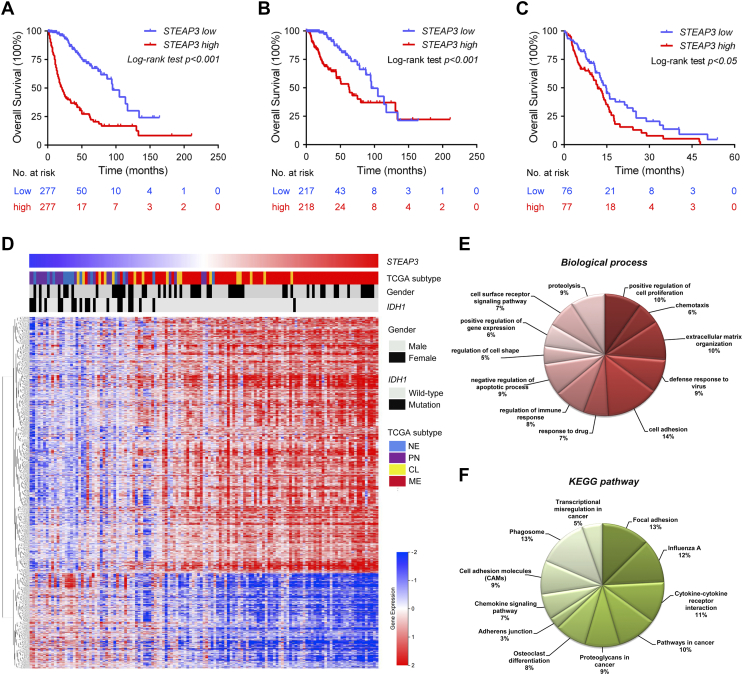
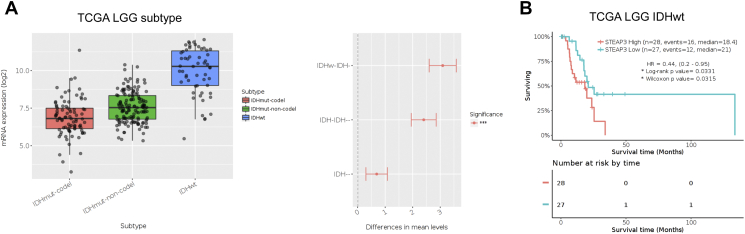
To examine the value of the gene as a prognostic marker, Kaplan–Meier survival curves were generated based on median values of STEAP3 expression in gliomas in publicly available databases. In the analysis of the TCGA data including all glioma pathologies, patients with higher STEAP3 expression levels exhibited significantly shorter overall survival (OS) than patients with lower STEAP3 expression (P < .001; Figure 2A). Shorter OS was also associated with higher STEAP3 expression in the analysis performed on LGG and GBM patients independently (P < .001 and P < .05, respectively; Figure 2, B and C). We further evaluated the expression pattern and prognostic significance of STEAP3 in different LGG molecular subtypes. Higher STEAP3 levels were more common in IDH wild-type LGGs compared to IDH mutated LGGs, including both chromosome 1p/19q intact and co-deleted cases (Supplementary Figure S2A). STEAP3 was also found to be negatively correlated with OS in IDH wild-type LGG (P < .05; Supplementary Figure S2B). Finally, STEAP3 expression was validated as an independent prognostic marker in multivariate Cox regression analysis of TCGA (HR = 1.177, 95% CI = 1.034 to 1.339, P = .013; Supplementary Table S2) and CGGA (HR = 1.617, 95% CI = 1.617 to 2.499, P = .030; Supplementary Table S3) data.
Figure 2.
High STEAP3 expression is a prognostic marker for poor clinical outcome in glioma patients. Kaplan–Meier analysis of patient survival data based on high vs low expression of STEAP3 in all gliomas (A), LGG (B), and GBM (C). The median value of STEAP3 expression was used as the cutoff in indicated databases. P-values were obtained from the log-rank test. (D) Correlation analysis was performed with TCGA GBM mRNA microarray data to obtain a set of STEAP3 associated genes. Cluster analysis was performed on STEAP3-associated genes based on STEAP3 expression. The resultant heatmap shows relative expression levels of STEAP3-associated genes in individual glioma cases where red is higher expression and blue is lower expression. (E) Biological processes and (F) Pathway analysis performed using the set of STEAP3-associated genes. Results are based the KEGG and GO databases.
Supplementary Figure S2.
STEAP3 prefers to express in IDHwt of gliomas in which predicts a poor prognosis.
Biological Process and Pathway Analysis of STEAP3 Potential Function
Cluster analysis was performed on the expression data of GBM patients to identify gene expression patterns correlating with STEAP3 (Figure 2D). High expression of STEAP3 was strongly associated with IDH wild-type GBM, while low expression of STEAP3 was tended to correlate with IDH mutated GBM (Figure 2D). Genes positively associated with STEAP3 (P < .01) were subjected to GO and KEGG analysis to identify function. GO analysis indicated that STEAP3 was strongly associated with several biological processes, including cell adhesion, promotion of cell proliferation, extracellular matrix organization, proteolysis, and regulation of the immune response (Figure 2E). In KEGG analysis, the up-regulated genes were enriched in pathways related to focal adhesion, phagosomes, cytokine-cytokine receptor interactions, proteoglycans in cancer, and cell adhesion molecules (CAMs; Figure 2F).
STEAP3 Promotes GBM Cell Growth and GSC Self-Renewal
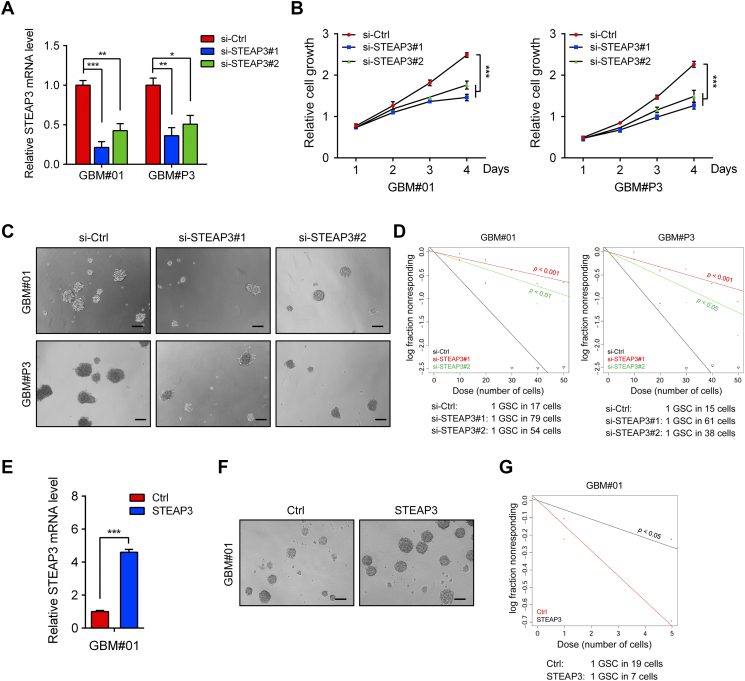
To investigate whether the gene has an oncogenic role in human glioma, we performed STEAP3 knockdown experiments with two siRNA in GBM#01 and GBM#P3 GSCs and assessed cell growth in vitro. qRT-PCR and western blot analyses demonstrated that both siRNAs efficiently knocked down STEAP3 at the mRNA and protein levels (Figs 3A, 4F). Knockdown with si-STEAP3 significantly reduced cell growth in both GBM#01 and GBM#P3 GSC compared to the si-Ctrl group (P < .001, si-Ctrl vs si-STEAP3, respectively; Figure 3B).
Figure 3.
STEAP3 promotes GBM cell growth and GSC self-renewal. (A) STEAP3 knockdown efficiency as assessed by qRT-PCR in GBM#01 and GBM#P3 GSCs transfected with si-Ctrl or two different siRNAs targeting STEAP3 (si-STEAP3#1 and #2). GAPDH was used as the internal normalization control. (B) Cell growth curves generated for GBM#01 and GBM#P3 cells transfected in vitro with si-Ctrl and si-STEAP3 from data obtained using the Luminescent Cell Viability Assay kit. Measurements (luminescence) was obtained at days 1, 2, 3, and 4. (C) Representative images of tumor sphere formation for GBM#01 and GBM#P3 GSCs transfected with si-Ctrl and si-STEAP3. Scale bar = 100 μm. (D) Extreme limiting dilution assay performed with GBM#01 and GBM#P3 GSCs. GSCs were transfected with si-Ctrl and si-STEAP3, and analysis was performed 10 days after transfection. Stem cell frequency was calculated using ELDA. (E) Ectopic expression of STEAP3 in GBM#01 (GBM#01-STEAP3) confirmed by qRT-PCR. GAPDH was used as an internal normalization control. GBM#01-STEAP3 in tumor sphere formation assays (F) and ELDA (G). Data are shown as the mean ± SEM from three independent experiments. *P < .05; **P < .01; ***P < .001.
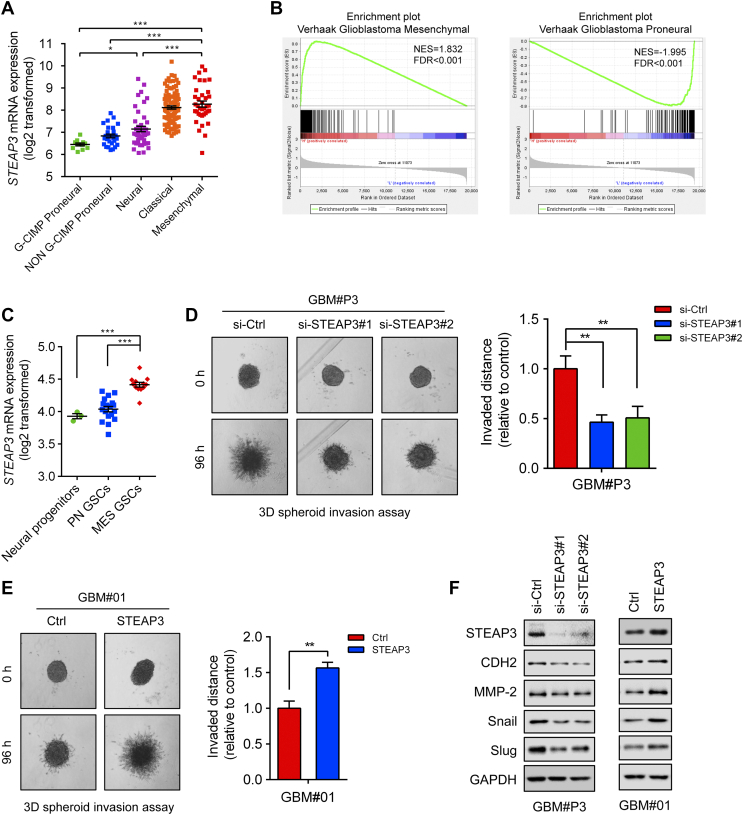
Figure 4.
STEAP3 is enriched in the GBM mesenchymal molecular subtype and promotes cell invasive potential. (A) STEAP3 mRNA expression in different molecular subtypes (CpG island methylator phenotype (G-CIMP) proneural, non–G-CIMP proneural, neural, classical, mesenchymal) from the TCGA GBM microarray dataset. (B) GSEA enrichment analysis of mesenchymal and proneural signatures (Verhaak) in STEAP3highvs STEAP3low samples in the TCGA GBM dataset. Normalized enrichment score (NES) and FDR are shown for each plot. (C) STEAP3 mRNA expression in neural progenitor cells (n = 3), PN GSCs (n = 18), and MES GSCs (n = 12) in microarray data. (D) Representative images of spheroids in 3D invasion assay for GBM#P3 GSCs transfected with si-Ctrl and si-STEAP3 and evaluated at 0 h and 96 h. The distance of invading cells from the tumor spheres was determined after 96 h. (E) 3D invasion assay at 96 h for GBM#01 GSCs overexpressing STEAP3. (F) Western blot for protein levels of key factors involved in mesenchymal transition in lysates (20 μg) prepared from GBM#P3 and GBM#01 GSCs. GAPDH was used as a loading control. Data are shown as the mean ± SEM from three independent experiments. *P < .05; **P < .01; ***P < .001.
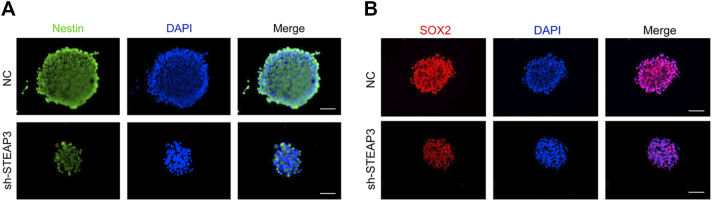
To investigate whether loss of STEAP3 influences stem cell–associated properties, we performed tumor sphere formation and extreme limiting dilution assays (ELDA) with GBM#01- and GBM#P3-si-STEAP3 GSCs. STEAP3 knockdown resulted in a remarkable decrease in GSC sphere number (Figure 3C), as well as in the frequency of sphere formation (Figure 3D) in both GBM#01 and GBM#P3 GSCs. The relationship between STEAP3 and the property of stemness was further confirmed in immunofluorescence staining for Nestin and SOX2; both proteins were decreased in GBM#01 tumor spheres (Supplementary Figure S3).
Supplementary Figure S3.
Silencing of STEAP3 down-regulated expression of Nestin and SOX2.
In the converse experiment, where we overexpressed STEAP3 in GBM#01 GSC (Figures 3E and 4F), tumor sphere formation and ELDA analysis demonstrated that the gene promoted cell growth and stem cell–associated properties in GBM#01 GSC cells (Figure 3, F and G). These data indicated that STEAP3 promoted cell proliferation and self-renewal in GBM#01 and GBM#P3 GSCs.
STEAP3 is Enriched in the GBM Mesenchymal Molecular Subtype and Promotes Cell Invasive Potential
Human gliomas have been molecularly categorized into different subtypes: classical, mesenchymal, proneural, and neural. Classical and mesenchymal subtypes are associated with poorer survival outcomes relative to proneural and neural subtypes which are often IDH1/2 mutated [19], [20], [21]. In the TCGA dataset, increased STEAP3 levels were associated with the mesenchymal molecular subtype compared to the proneural CpG island methylator phenotype (G-CIMP) or non-G-CIMP molecular subtypes (Figure 4A). Similar results were observed through Gene Set Enrichment Analysis (GSEA) in GBM patients (Figure 4B). To further characterize the pathophysiological role of STEAP3 in GSCs of different GBM molecular subtypes, we analyzed microarray data GSE67089 from neural progenitor stem cells (n = 3), mesenchymal (MES) GSC (n = 12) and proneural (PN) GSC (n = 18). STEAP3 expression in MES GSCs was elevated compared to neural progenitor or PN GSCs (P < .001; Figure 4C).
It is well known that the epithelial–mesenchymal transition process plays an essential role in the invasion and metastasis of diverse cancers [22], [23], [24], [25]. Targeting molecules that mediate the invasive properties of GBM is one therapeutic strategy for the treatment of the disease. To test whether STEAP3 might drive cell invasion, GBM#P3 GSCs, a highly invasive cell type, were evaluated in a 3D collagen spheroid invasion assay. GSCs were transfected with siRNAs (si-Ctrl and si-STEAP#1 and #2) and seeded into wells of 96-well plates coated with Matrigel. The invaded distance of cells from the tumor spheroid after 96 h was significantly reduced in GBM#P3 treated si-STEAP3 cells relative to controls (P < .01, Figure 4D). In contrast, overexpression of STEAP3 in GBM#01 GSCs, a less invasive cell type, promoted tumor invasion (P < .01; Figure 4E).
Because high STEAP3 expression was associated with the GBM mesenchymal molecular subtype, we explored whether molecular markers involved in the mesenchymal transition were correspondingly regulated. Western blot analysis revealed that knockdown of STEAP3 led to decreased expression of molecular markers of the mesenchymal transition, such as CDH2, Snail, Slug, and matrix metalloproteinase-2 (MMP-2). Overexpression of STEAP3 however led to increased expression of these markers (Figure 4F). Altogether, our findings indicated that STEAP3 might promote invasion of human glioma cells by inducing the mesenchymal transition.
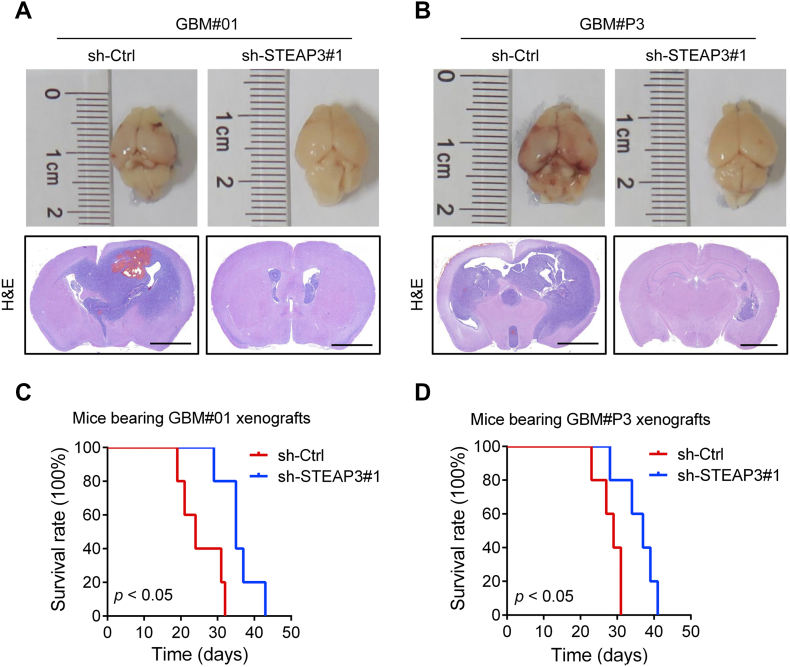
Knockdown of STEAP3 Inhibits Tumor Growth in An Orthotopic Mouse Model
To further elucidate the role of STEAP3 in the development of human glioma, tumor growth was examined in an orthotopic mouse model established with GBM#01 or GBM#P3 cells transduced with sh-Ctrl (n = 5) or sh-STEAP3#1 (n = 5). Knockdown of STEAP3 dramatically suppressed tumor growth and prolonged OS of tumor-bearing mice (P < .05, sh-Ctrl vs sh-STEAP3#1; Figure 5, A–D).
Figure 5.
Knockdown of STEAP3 inhibits tumor growth in an orthotopic mouse model. (A and B) Representative Images of mouse brains implanted with GBM#01 or GBM#P3 GSCs transduced with sh-Ctrl (n = 5) or sh-STEAP3#1 (n = 5). Representative H&E staining from indicated groups. Scale bar = 2 mm. (C and D) Survival analysis for animals implanted with GBM#01- or GBM#P3-sh-Ctrl or -sh-STEAP3#1 GSCs (P < .05 by the log-rank test).
STEAP3 Activates the TfR-STAT3 Pathway in GBM Cells In Vitro
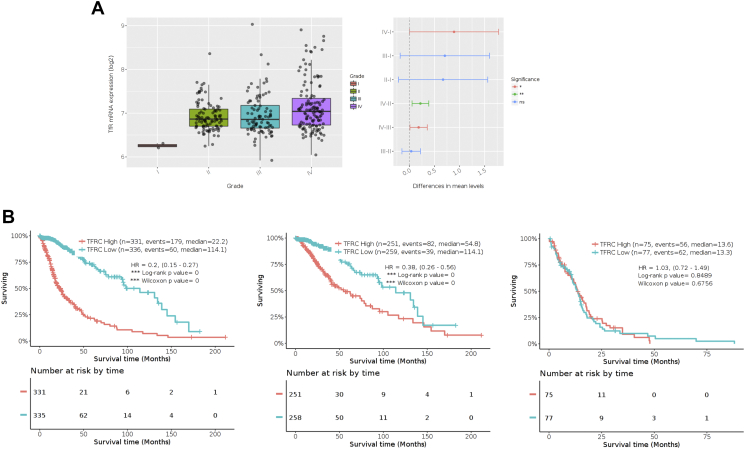
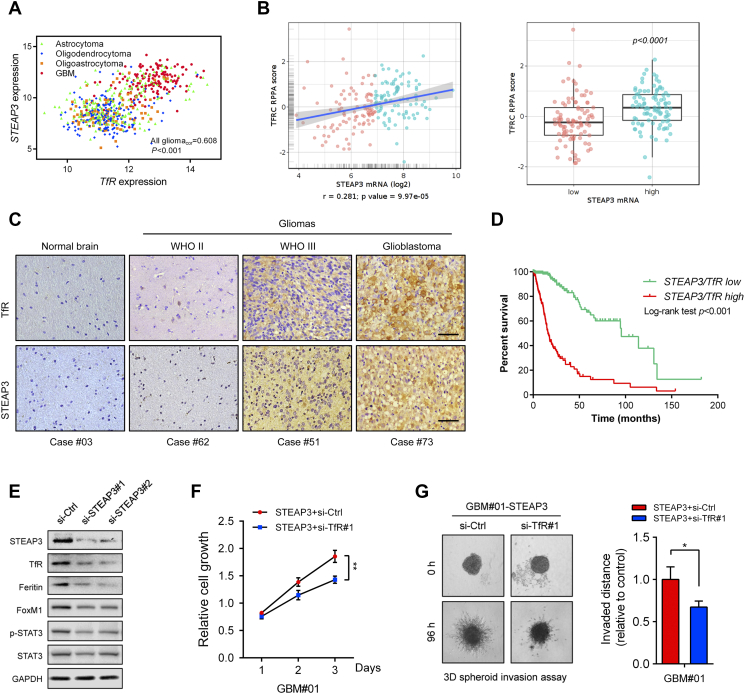
STEAP3 is a member of the transmembrane ferrireductases, which exert their function by reducing ferric iron to ferrous iron in endosomes. Intriguingly, it has been demonstrated that STEAP1 and STEAP2 colocalize with transferrin receptor (TfR) [24]. Therefore, we investigated the relationship between STEAP3 and TfR in gliomas. First, TfR was observed to be highly expressed in GBM compared to LGG (Supplementary Figure S4). Correlation analysis between TfR and STEAP3 in glioma samples from the TCGA database revealed a strong linear association between these two mRNAs (Pearson = 0.608, P < .001; Figure 6A). We also found TfR to be one of the top five proteins (annexin.1, p62.LCK.ligand, PAI.1, stathmin, TfR) with increased expression in STEAP3high GBM patients compared to STEAP3low GBM patients based on TCGA RPPA data (P < .0001; Figure 6B, supplementary Table S5). Moreover, intensity of IHC staining was comparable for TfR and STEAP3 in different cases of normal brain and glioma specimens (Figure 6C).
Supplementary Figure S4.
Elevated TfR in glioma patients predicted a poor prognosis.
Figure 6.
STEAP3 activates the TfR-STAT3 pathway in GBM cells in vitro. (A) Correlation between STEAP3 and TfR mRNA expression in gliomas from the TCGA dataset. The statistical significance of correlation was evaluated using a linear regression model (TCGA all gliomacor = 0.608, P < .001). (B) Correlation between STEAP3 mRNA and TfR protein levels in TCGA GBM patients (r = 0.281, P = 9.97e-05). TfR protein levels were increased in the STEAP3high group (P < .001). (C) Representative images of paired IHC staining for TfR and STEAP3 in individual cases of normal brain and different pathological grades of astrocytoma. Scale bar = 100 μm. (D) Kaplan–Meier curve analysis of survival data from patients based on low/high co-expression of STEAP3/TfR. P-values were obtained from the log-rank test. (E) Western blot analysis of TfR, Ferritin, FoxM1, STAT3 and phosphorylated-STAT3 in GBM#P3 GSCs transfected with si-Ctrl and si-STEAP3. GAPDH was used as a loading control. (F) Growth curves generated for GBM#01-STEAP3 GSCs transfected with si-Ctrl and si-TfR in vitro. Measurements were obtained at days 1, 2, and 3 using the Luminescent Cell Viability Assay kit. (G) 3D invasion assay at 96 h for GBM#01-STEAP3 transfected with TfR knockdown siRNA. Data are shown as the mean ± SEM from three independent experiments. *P < .05; **P < .01; ***P < .001.
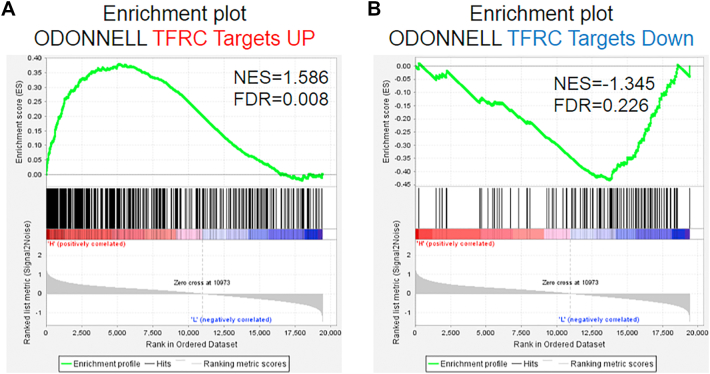
In analysis of the TCGA datasets, patients with TfRhigh/STEAP3high exhibited significantly poorer OS than patients with TfRlow/STEAP3low (P < .001; Figure 6D). GSEA furthermore revealed that STEAP3 expression levels positively correlated with TfR-activated gene signatures (Supplementary Figure S5), suggesting that the TfR pathway might mediate the pro-tumor effects of STEAP3.
Supplementary Figure S5.
Gene set enrichment analysis for TFRC.
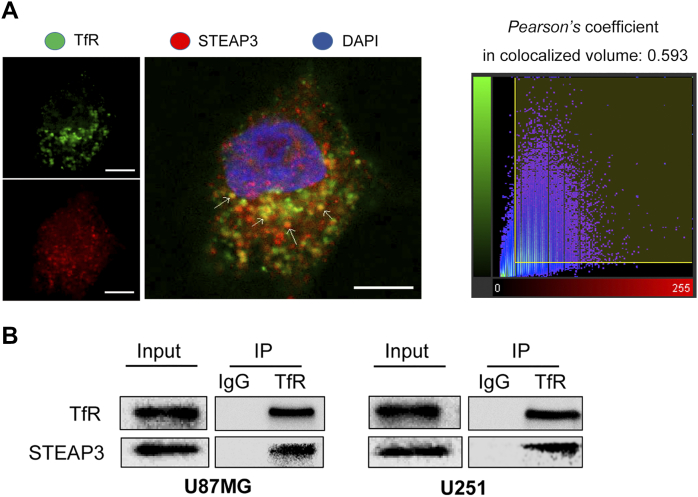
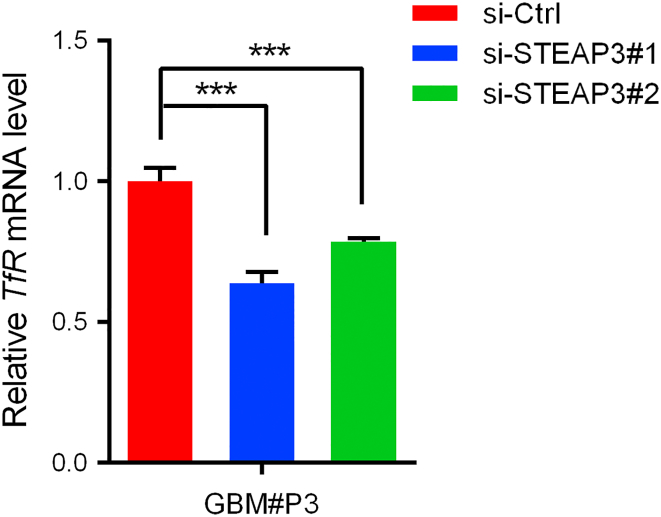
Several additional results demonstrated that a physical as well as functional relationship exists between STEAP3 and TfR in human glioma cells. First, immunofluorescence staining acquired through confocal microscopy demonstrated that STEAP3 co-localized with TfR in U251 glioma cells (Pearson's coefficient in co-localized volume = 0.593, Supplementary Figure S6A). Co-IPs confirmed these results. In lysates prepared from glioma cell lines, STEAP3 was found in pull-downs performed with TfR antibody (Supplementary Figure S6B). Second, siRNA knockdown of STEAP3 led to decreased expression of TfR and Ferritin in GBM#P3 GSCs (Figure 6E; Supplementary Figure S7). Third, a previous study reported that depleting Ferritin disrupted mitotic progression in cancer stem cells through the STAT3-FoxM1 regulatory axis [25]. We therefore investigated whether STEAP3 knockdown influenced protein levels of STAT3 and FoxM1. STEAP3 siRNA knockdown led to decreased FoxM1 and phosphorylated STAT3, as well as total STAT3 protein levels (Figure 6E).
Supplementary Figure S6.
STEAP3 forms a complex with TFR in U251 and U87MG cells.
Supplementary Figure S7.
si-RNAs induced knockdown efficiency of STEAP3 were examined by qRT-PCR.
Finally, we used siRNA knockdown of TfR to characterize its functional relationship with STEAP3. Si-RNA-induced knockdown of TfR significantly abrogated STEAP3 induced cell proliferation and invasion in GBM#01-STEAP3 GSCs (Figure 6, F and G, Supplementary Figure S8). These results indicated that STEAP3 might contribute to cancer progression by activating TfR and the downstream Ferritin-STAT3 pathway.
Supplementary Figure S8.
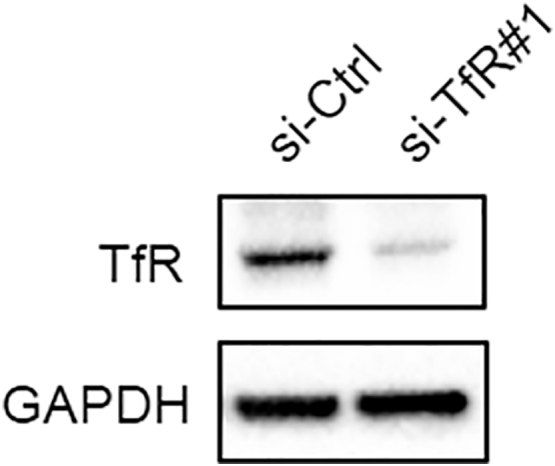
si-TfR1 induced knockdown of TFR was examined by western blot.
Discussion
In recent years, cellular iron metabolism has emerged as a unique pathway markedly altered in cancer cells. Importantly, iron metabolism contributes to the activation of a number of biological processes, including proliferation, EMT, and TGFβ and Wnt growth factor signaling [8]. Here, we found that STEAP3, an enzyme involved in iron metabolism, was one of the top three genes with increased expression in GBM compared to LGG, correlated with poor clinical prognosis. Silencing STEAP3 in glioma cell cultures attenuated many biological processes associated with cancer development, including tumor growth and invasion, as well as mesenchymal transition. Furthermore, stem cell-like properties, such as tumor sphere formation and expression of stem cell markers, were also inhibited in human glioma cells transfected with si-STEAP3. Finally, we demonstrated that STEAP3 colocalizes with TfR, a receptor which is critical for the malignant behavior of GSCs [25]. Through our study, STEAP3 emerged as an important protein that induces mesenchymal transition and stem-like traits in glioma cells and thus, as a potential therapeutic target.
STEAP3 is highly expressed in hematopoietic tissues, which support important physiologic functions associated with iron metabolism, especially in erythroid precursors. The protein has a 6-transmembrane domain at the COOH-terminal region and a cytoplasmic N-terminal oxidoreductase domain, which is critical for iron and copper uptake [6]. Iron uptake can be facilitated by STEAP3, which reduces endosomal ferric iron bound to transferrin to the ferrous form in erythroid cells [26]. Recently, the STEAP3 gene has been reported to be expressed in multiple malignant cell types, including prostate, colorectal, and lung cancers [27]. Altered iron states do occur in glioma cells through changes in iron uptake, which may contribute to rapid proliferation characteristic of aggressive tumor types [28]. Thus, increased STEAP3 may be necessary to sustain high proliferation rates in aggressive human gliomas.
Intriguingly, iron uptake and dependence have been shown to be enhanced in GSCs which are thought to be the cells fundamental to the development and recurrence of malignancies. GSCs exhibit properties of normal stem cells and like a stem cell, have the capacity to generate all cell types that make up a tumor tissue [29]. Thus, GSCs retain unlimited potential to drive tumor cell growth. Importantly, cellular iron has been shown to contribute to the stem cell-like phenotype. First, cells acquire stem-like features by undergoing the process of EMT during glioma progression, and cellular iron has been shown to have a role in regulating EMT. In addition, GSCs release several cytokines and chemokines crucial for tumor migration and invasion in EMT. Cellular iron is also known to contribute to the activation of transforming growth factor (TGFβ) and Wnt pathways which are two major signaling pathways inducing EMT [30], [31], [32].
Finally, we demonstrated that STEAP3 activates the TfR-STAT3 pathway in GBM, and that knockdown of TfR significantly influences the impact of STEAP3 overexpression on malignant phenotypes in GSC. TfR and ferritin are two key iron regulators, which are also important for the propagation of GSCs and tumor development in vivo [25]. TfR is highly expressed in many human malignancies, such as leukemia, lymphoma, glioma, and bladder, breast, and lung cancers. These findings indicate that increases in iron are needed in tumor cells [33]. In addition, TfR has been identified as an important therapeutic target or delivery tool in the treatment and imaging of gliomas [34], [35]. Here, we found that STEAP3 interacts directly with TfR like other family members STEAP1 and STEAP2 [23]. In addition, STEAP3 is important for optimal TfR1 internalization in erythroid cells [24]. These results suggest that STEAP3 might contribute to cancer progression through interaction with TfR and regulation of downstream Ferritin-FoxM1-STAT3 signaling. However, further investigation is necessary to elucidate the role of the STEAP3-TfR complex in the development of human glioma and downstream molecular pathways in order to provide new effective therapeutic targets of GBM.
In summary, STEAP3 is overexpressed in human glioma and portends a poor clinical outcome in patients. Knockdown experiments indicated that the protein contributes to the efficiency of several biological processes critical to the maintenance of the tumorigenic phenotype. We have identified a regulatory role for STEAP3 in some signaling pathways, but there may be others. Many signaling pathways are implicated in the biology of GSCs, including those involving notch, SHH, VEGF, STAT3, and BMP [36]. Therefore, STEAP3 holds promise in the treatment of human GBM as a molecular target which may have implications for specifically eliminating GSCs.
Conclusions
In our study, we found that STEAP3 not only promotes malignant progression of human glioma but is also a prognostic marker of glioma patients. Remarkably, a link between STEAP3 and TfR as well as the downstream FoxM1-STAT3 signaling was also revealed. This physical and functional interaction might serve as a mechanism underlying STEAP3-mediated glioma progression. In summary, STEAP3 provides a novel therapeutic target in the treatment of human glioma.
The following are the supplementary data related to this article.
Gene primers.
Univariate and multivariate Cox regression of STEAP3 expression for overall survival in TCGA glioma patients (n=667)
Univariate and multivariate Cox regression of STEAP3 expression for overall survival in CGGA glioma patients (n=325)
Datasets summary.
Top 10 proteins with increased expression in STEAP3high GBM patients compared with STEAP3low GBM patients according to TCGA RPPA data
STR profile information
Author Contributions
X.L. and J.W. conceived and designed the experiments; M.H, R.X., Y.X., C.Z., Y.W., X.Z. and N.Y. performed the experiments; S.W., S.N. and J.J. analyzed the data; B.H., A.C., Q.Z., W.L. and D.Z. contributed reagents/materials/analysis tools. All authors were involved in writing the paper.
Acknowledgements
This work was supported by the Natural Science Foundation of China Grant (81572487, 81702474, 81701329, 81472353), the Special Foundation for Taishan Scholars (ts20110814, tshw201502056, and tsqn20161067), the Department of Science & Technology of Shandong Province (2015ZDXX0801A01, 2017CXGC1502 and 2015GSF118061), the Shandong Provincial Natural Science Foundation (ZR2017MH116 and ZR2017MH015), the Fundamental Research Funds of Shandong University (2016JC019), the Science Foundation of Qilu Hospital of Shandong University (2017QLQN02 and 2017QLQN03), the Jinan Science and Technology Bureau of Shandong Province (201704096 and 201704124), and the University of Bergen and the K.G. Jebsen Brain Tumor Research Centre.
Contributor Information
Xingang Li, Email: lixg@sdu.edu.cn.
Jian Wang, Email: jian.wang@biomed.uib.no.
References
- 1.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, Mclendon RE. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Paw I, Carpenter RC, Watabe K. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane DJR, Mills TM, Shafie NH. Expanding horizons in iron chelation and the treatment of cancer: Role of iron in the regulation of ER stress and the epithelial–mesenchymal transition. Biochim Biophys Acta. 2014;1845:166–181. doi: 10.1016/j.bbcan.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13:342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishitani S, Noma K, Ohara T. Iron depletion-induced downregulation of N-cadherin expression inhibits invasive malignant phenotypes in human esophageal cancer. Int J Oncol. 2016;49:1351–1359. doi: 10.3892/ijo.2016.3640. [DOI] [PubMed] [Google Scholar]
- 8.Lui GYL, Zaklina K, Vera R. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget. 2015;6:18748–18779. doi: 10.18632/oncotarget.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohgami RS, Campagna DR, Greer EL. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nature Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentze MW, Muckenthaler MU, Galy B. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Fleming MD, Trenor CC, Su MA. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nature Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 12.Dong XP, Cheng X, Mills E. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CH, Chen SL, Sung WW. The Prognostic Role of STEAP1 Expression Determined via Immunohistochemistry Staining in Predicting Prognosis of Primary Colorectal Cancer: A Survival Analysis. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodeberg DA, Nuss RA, Elsawa SF. Recognition of six-transmembrane epithelial antigen of the prostate-expressing tumor cells bypeptide antigen-induced cytotoxic T lymphocytes. Clin Cancer Res. 2005;11:4545–4552. doi: 10.1158/1078-0432.CCR-04-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Challita-Eid PM, Morrison K, Etessami S. Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res. 2007;67:5798–5805. doi: 10.1158/0008-5472.CAN-06-3849. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Jin Y, Arnoldussen YJ. STAMP1 is both a proliferative and an antiapoptotic factor in prostate cancer. Cancer Res. 2010;70:5818–5828. doi: 10.1158/0008-5472.CAN-09-4697. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, Chiba J. STEAP4 regulates focal adhesion kinase activation and CpG motifs within STEAP4 promoter region are frequently methylated in DU145, human androgen-independent prostate cancer cells. Int J Mol Med. 2009;24:599–604. doi: 10.3892/ijmm_00000270. [DOI] [PubMed] [Google Scholar]
- 18.Alvarado AG, Turaga SM, Sathyan P. Coordination of self-renewal in glioblastoma by integration of adhesion and microRNA signaling. Neuro Oncol. 2016;18:656–666. doi: 10.1093/neuonc/nov196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips HS, Kharbanda S, Chen R. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Verhaak RG, Hoadley KA, Purdom E. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao P, Joshi K, Li J. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani SA, Guo W, Liao MJ. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufhold S, Garbán H, Bonavida B. Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J Exp Clin Cancer Res. 2016;35:1–14. doi: 10.1186/s13046-016-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohgami RS, Campagna DR, Mcdonald A. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schonberg D, Miller T, Wu Q. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28:441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohgami RS, Campagna DR, Greer EL. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubert RS, Vivanco I, Chen E. STEAP: A prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci U S A. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khiroya Heena, Moore Jasbir S, Ahmad Nabeel. IRP2 as a potential modulator of cell proliferation, apoptosis and prognosis in nonsmall cell lung cancer. Eur Respir J. 2017;49 doi: 10.1183/13993003.00711-2016. [DOI] [PubMed] [Google Scholar]
- 29.Stopschinski BE, Beier CP, Beier D. Glioblastoma cancer stem cells-from concept to clinical application. Cancer Lett. 2012;338:32–40. doi: 10.1016/j.canlet.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Zhang D, Yue F. The iron chelators Dp44mT and DFO inhibit TGF-beta-induced epithelial-mesenchymal transition via up-regulation of N-Myc downstream-regulated gene 1 (NDRG1) J Biol Chem. 2012;287:17016–17028. doi: 10.1074/jbc.M112.350470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song S, Christova T, Perusini S. Wnt inhibitor screen reveals iron dependence of beta-catenin signaling in cancers. Cancer Res. 2011;71:7628–7639. doi: 10.1158/0008-5472.CAN-11-2745. [DOI] [PubMed] [Google Scholar]
- 32.Roberts AB, Wakefield LM. The two faces of transforming growth factor β in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniels TR, Delgado T, Rodriguez JA. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Recht LD, Griffin TW, Raso V. Potent cytotoxicity of an antihuman transferrin receptor-ricin A-chain immunotoxin on human glioma cells in vitro. Cancer Res. 1990;50:6696–6700. [PubMed] [Google Scholar]
- 35.Dixit S, Novak T, Miller K. Transferrin receptor-targeted theranostic gold nanoparticles for photosensitizer delivery in brain tumors. Nanoscale. 2014;7:1782–1790. doi: 10.1039/c4nr04853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebelt BD, Shingu T, Xin Z. Glioma stem cells: signaling, microenvironment, and therapy. Stem Cells Int. 2016;2016(7849890) doi: 10.1155/2016/7849890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene primers.
Univariate and multivariate Cox regression of STEAP3 expression for overall survival in TCGA glioma patients (n=667)
Univariate and multivariate Cox regression of STEAP3 expression for overall survival in CGGA glioma patients (n=325)
Datasets summary.
Top 10 proteins with increased expression in STEAP3high GBM patients compared with STEAP3low GBM patients according to TCGA RPPA data
STR profile information