Abstract
Tractable human tissue-engineered 3D models of cancer that enable fine control of tumor growth, metabolism, and reciprocal interactions between different cell types in the tumor microenvironment promise to accelerate cancer research and pharmacologic testing. Progress to date mostly reflects the use of immortalized cancer cell lines, and progression to primary patient-derived tumor cells is needed to realize the full potential of these platforms. For the first time, we report endothelial sprouting induced by primary patient tumor cells in a 3D microfluidic system. Specifically, we have combined primary human clear cell renal cell carcinoma (ccRCC) cells from six independent donors with human endothelial cells in a vascularized, flow-directed, 3D culture system (“ccRCC-on-a-chip”). The upregulation of key angiogenic factors in primary human ccRCC cells, which exhibited unique patterns of donor variation, was further enhanced when they were cultured in 3D clusters. When embedded in the matrix surrounding engineered human vessels, these ccRCC tumor clusters drove potent endothelial cell sprouting under continuous flow, thus recapitulating the critical angiogenic signaling axis between human ccRCC cells and endothelial cells. Importantly, this phenotype was driven by a primary tumor cell–derived biochemical gradient of angiogenic growth factor accumulation that was subject to pharmacological blockade. Our novel 3D system represents a vascularized tumor model that is easy to image and quantify and is fully tunable in terms of input cells, perfusate, and matrices. We envision that this ccRCC-on-a-chip will be valuable for mechanistic studies, for studying tumor-vascular cell interactions, and for developing novel and personalized antitumor therapies.
Abbreviations: ccRCC, clear cell renal cell carcinoma; Ct, cycle threshold; EGM, endothelial growth medium
Introduction
Cancer of the kidney and renal pelvis is the eighth most common malignancy in the United States, with a 5-year survival of less than 12% when diagnosed as metastatic disease [1]. Approximately 85% to 90% of kidney cancers are derived from the proximal tubular epithelial cells of the renal cortex and are classified as renal cell carcinoma (RCC) [2], [3]. Of these, RCC of the clear cell type (ccRCC) accounts for approximately 75% of cases. Most sporadic ccRCCs in humans carry inactivating mutations in the VHL tumor suppressor gene, which lead to constitutive stabilization of the hypoxia inducible transcription factors HIF-1α and HIF-2α [4], [5]. These transcription factors in turn activate a canonical cellular response which includes metabolic reprogramming towards aerobic glycolysis and secretion of numerous factors, including VEGFA, that promote angiogenesis [6], [7]. If caught at an early stage, ccRCC is potentially curable, whereas advanced disease remains difficult to treat. Therapy relies in part on inhibition of the potent VEGFA-driven angiogenic signaling axis, but the development of resistance is common, owing in part to the upregulation of alternate angiogenic factors [8], [9], [10]. While there is a need to identify more effective therapeutic strategies, current research tools to study ccRCC have limitations, and patient-specific human models are needed to develop precision therapies.
Most kidney cancer research to date has utilized primary cultures or long-standing tumor cell lines grown in 2D monolayers, or animal tumor models. The major drawback of conventional 2D tumor cultures in vitro is the failure to maintain the primary tumor phenotype and complexity over time [11]. A recent study created primary human ccRCC 3D organoids within decellularized renal extracellular matrix or synthetic polysaccharide scaffolds and demonstrated that the tumor cells sustained the original tumor immunohistochemical and gene expression features longer than conventional 2D monolayer cultures [12]. However, this system did not incorporate endothelial cells or study the effects of directional flow/perfusion on tumor phenotypes.
Though rodent models have much improved our understanding of ccRCC, they still fail to completely capture key features of the human disease. For example, patients with von Hippel–Lindau disease inherit one defective copy of the VHL gene and develop ccRCC at very high rates following sporadic inactivation of the remaining allele in kidney tubular epithelial cells [13]. In contrast, the Vhl-null mouse develops tubular hyperplasia but not frank carcinoma [14], [15], [16]. Mice transgenic for a constitutively active form of HIF-1α appear to develop cystic ccRCC in situ but only at low frequency [17]. In fact, only after inactivation of the tumor suppressor genes Trp53 and Rb together with Vhl do mice spontaneously develop ccRCC [18]. Even with the long latency associated with spontaneous tumor development in these systems, spontaneous animal ccRCC tumor models still do not capture the complete molecular and phenotypic complexity of the human disease.
Recent studies using primary ccRCC xenografts (“tumorgrafts”) in mice [19], [20], [21] or on chicken chorioallantoic membranes [22] have permitted the examination of patient-to-patient variability in drug sensitivity. However, the use of mice for serial transplantation of xenografts is expensive, time consuming, and labor intensive. Furthermore, not all patients' tumors will engraft successfully into mice [20], [21]. Lastly, it is difficult to mechanistically dissect human-specific tumor-vascular and other cell-cell reciprocal interactions in these xenograft systems.
Thus, 3D tissue-engineered, patient-specific culture systems that efficiently recapitulate the in vivo tumor phenotype would overcome many of these challenges and provide a tractable tool for mechanistic and pharmacological studies. In this regard, the well-defined cell types in the ccRCC microenvironment (tumor, vascular endothelium, and circulating immune cells) and the typical lack of a prominent stromal response make this an ideal tumor type for modeling in microphysiological culture systems. Here, we aimed to reconstruct the 3D vascular microenvironment of ccRCC by incorporating primary tumor cell clusters into the matrix surrounding engineered human vessels subjected to continuous flow. We have previously used this framework to develop a microphysiological system for studies of human renal proximal tubule function [23]. We demonstrate that this platform reproduces the ability of ccRCC to stimulate angiogenic sprouting and provides a basis for studies of pharmacological blockade. We envision that this approach will also prove useful for studying tumor-endothelial cell interactions, testing the efficacy of novel antitumor agents on a patient-specific basis, and exploring the mechanisms of transformation.
Materials and Methods
Unless otherwise stated, all media and reagents were purchased from Thermo Fisher Scientific (Waltham, MA).
Primary Culture of ccRCC and Normal-Adjacent Renal Cortex
The research protocol to access tumor and normal-adjacent tissue from patients undergoing nephrectomies for renal masses at the University of Washington Medical Center was approved by the Institutional Review Board (File 44773). Informed consent to use leftover tissue was obtained. A final pathologist's diagnosis of ccRCC was confirmed for all cases included in this study (Supplementary Table S1). Tissues were procured by NWBioTrust (Seattle, WA) immediately following surgery in HBSS containing penicillin-streptomycin and stored at 4°C prior to transfer and processing within 24 hours. Tissues were minced to a slurry with a sterile blade and disrupted in RPMI containing 0.2 mg/ml Liberase-Thermolysin and 100 U/ml DNase I (Roche, Mannheim, Germany) with shaking at 37°C for 30 minutes and vortexing every 10 minutes. Digests were stopped with RPMI and 10% heat-inactivated FBS; strained through a 40-μm strainer; diluted with an equal volume of Dulbecco's PBS (Corning Life Sciences, Tewksbury, MA); collected at 370×g for 3 minutes; resuspended in primary culture media consisting of RPMI, 10% heat-inactivated FBS, 1× antibiotic-antimycotic supplemented with insulin, transferrin, sodium selenite, BSA, and linoleic acid (ITS+ media supplement, Sigma Aldrich, St. Louis, MO); and cultured at 37°C 5% CO2. Initial PBS wash and media change were performed within 1 to 2 days and thereafter every 3 to 4 days.
Gene Expression
Gene expression values from RNA sequencing data from primary ccRCC and normal renal cortex tissues were obtained from The Cancer Genome Atlas expression browser [24]. For real-time gene expression assays, RNA was extracted using RNeasy Plus kits (Qiagen, Valencia, CA). The cDNA was synthesized using random hexamers and the Superscript III First-Strand Synthesis Kit and was used to seed real-time SYBR Green or Taqman PCR reactions for each gene (Supplementary Table S2) using standard cycling conditions for the Applied Biosystems 7900HT thermocycler. The presence of a single peak in the dissociation curve analysis was confirmed for all SYBR Green assays. Cycle threshold (Ct) values were determined using the Applied Biosystems Sequence Detection Software. All qPCRs were performed in triplicate except for those with samples H24 and H25, which were performed in duplicate. The coefficient of variance of the mean Ct values for all assays was less than 2.5%. Relative quantification was calculated as 2–delta Ct, where delta Ct values were obtained by subtracting the control gene mean Ct value from the target gene mean Ct value using the control gene HMBS, which was previously shown to be stable in comparisons of primary RCC and normal kidney tissues [25].
Luminex Assays
Media was harvested from 24-hour adherent cultures of ccRCC or normal-adjacent renal cortex and centrifuged at 900×g for 3 minutes at 4°C to remove cells and debris. Supernatants were assayed for levels of secreted ANGPTL4 using a Luminex bead-based multiplex assay (R&D Systems, Minneapolis, MN), a Luminex 200 instrument (Luminex Corporation, Austin, TX), and BioPlex version 5.0 software (Bio-Rad Laboratories, Hercules, CA). Values were normalized for differences in cell number based on RNA yields determined by quantitative RT-PCR for the endogenous control gene HMBS.
Transwell Migration Assays
ccRCC and normal-adjacent renal cortex cells were seeded in 24-well plates in 750 μl endothelial basal medium (cat. no. CC-3121, Lonza, Walkersville, MD) containing 0.1% BSA and incubated for 6 hours at 37°C. HUVEC-2 cells (Corning Life Sciences) were cultured in endothelial growth medium (EGM, cat no. CC-4133, Lonza), washed three times with PBS, harvested using enzyme-free cell dissociation buffer, and added as 2×105 cells/mL in 0.25 ml endothelial basal medium and 0.1% BSA to the top chambers of 3-μm pore size, fibronectin-coated BioCoat Fluoroblok cell culture inserts (Corning Life Sciences). After 16 hours, inserts were removed, dipped in HBSS to remove media, and stained with 5 μg/ml calcein AM (Corning Life Sciences) in HBSS for 90 minutes at 37°C. Inserts were then transferred to another plate with fresh HBSS, and bottom fluorescence intensity measurements were obtained at excitation/emission 485/535 nm (peak calcein excitation/emission = 494/517 nm) using a 2014 Envision plate reader (Perkin Elmer, Waltham, MA).
ccRCC-Induced Sprouting in Microphysiological Devices
ccRCC and normal-adjacent renal cortex cells were aggregated into clusters overnight in 40-μl hanging drops with 80% primary cell media diluted in RPMI containing methylcellulose (0.24% final concentration). Each drop contained 400 cells for all samples except B44 tumor and B46 tumor, which contained 500 cells. Clusters were harvested, washed with PBS, resuspended gently with trimmed P-200 tips in 30 μl of EGM, and mixed gently at 2000 clusters/ml in a collagen I solution prepared following a standard protocol [26]. Briefly, a type I collagen solution was prepared by incubating rat tail tendons in 0.1% acetic acid, cleared by centrifugation, lyophilized, and resuspended in 0.1% acetic acid with agitation at a stock concentration of 15 mg/ml. This solution was further diluted on ice to 6 mg/ml in EGM and 1× M199 media and neutralized by adding drops of 1 M NaOH immediately prior to resuspending clusters. Single-channel Nortis microphysiological devices (model SCC-001, Nortis, Woodinville, WA) were washed with 100% ethanol, dried, and chilled at 4°C. For each device, approximately 150 μl of this solution was injected using a chilled 1-ml syringe and 18-gauge blunt needle until the extracellular matrix chamber was filled without bubbles. The matrix valves were sealed, and collagen was polymerized for 20 minutes at 4°C, 1 hour at room temperature, and 3 hours in a humidified 37°C incubator prior to extracting mandrel rods (polyimide-coated fused silica fibers) to form lumens. Preflow was initiated through the lumens with a syringe pump using EGM at 3 μl/min for 30 minutes. A single cell suspension of HUVEC-2 cells (passed through a 40-μm cell strainer) in EGM was then injected with a Hamilton syringe as 2.5 μl at 107 cells/ml into the lumens and allowed to adhere for 15 minutes at 37°C prior to initiating directional flow at 0.5 μl/min for 5 days with fresh media supplied at day 3. Flow was performed with EGM in all experiments with the exception of B58 tumor, which also contained 0.05% dimethyl sulfoxide. For VEGF inhibition experiments, 200 ng/ml soluble recombinant human VEGFR2-Fc chimera (cat. no. 357-KD-050, R&D Systems) or a vehicle control (0.0002% BSA) was included.
Extraction of ccRCC Cells from Collagen Gels
ccRCC clusters were resuspended in 6 mg/ml collagen I at 2000 clusters/ml as described above. After incubation of the collagen gels at 37°C for 24 hours in EGM, they were minced with a blade and digested with RPMI containing 0.2 mg/ml Liberase-Thermolysin and 100 U/ml DNase I with shaking at 37°C for 30 minutes and vortexing every 10 minutes. Digests were stopped with cold RPMI and 10% heat-inactivated FBS, and cells were collected at 2400×g for 3 minutes at 4°C and snap-frozen in liquid nitrogen prior to RNA extraction.
Immunofluorescence, Confocal Microscopy, and Quantification of Sprouting
Devices were fixed with 4% formaldehyde in PBS for 60 minutes at 2 μl/min and permeabilized and blocked with 5% BSA and 0.1% Triton X-100 in PBS for 120 minutes at 20 μl/min. A primary rabbit anti-CD31 antibody in 2% BSA and PBS (1:100, cat. no. ab28364, Abcam, Cambridge, MA) was perfused for 35 minutes at 10 μl/min, and then devices were incubated with the valves closed overnight followed by washing with PBS for 120 minutes at 20 μl/min. The following fluorescent probes were then added to 2% BSA and PBS and perfused for 55 minutes at 10 μl/min followed by washing as above: EPCAM-PE (1:50, cat. no. 130-091-253, Miltenyi Biotec, Auburn, CA); goat anti–rabbit-488, (1:150, cat. no. A11008); Phalloidin-647 (1:100, cat. no. A22287), and 33 μg/ml Hoechst 33342. Confocal imaging was performed using a Nikon A1R microscope (Nikon Instruments, Melville, NY). The method for manual quantification of sprouting frequency in confocal z-stacks was adapted with modification from Nagao et al. [27]. Specifically, confocal z-stack volume projections were generated using Nikon Elements software, and the number of 50×50×400-μm regions lateral to the central vessel that contained at least one CD31+ sprout originating from the vessel wall were counted. Sprouting in chips containing cell clusters is presented relative to the amount of sprouting observed in collagen-only chips. Confocal images were processed using Fiji software [28]. Surface rendering and movies were constructed using Imaris software (Bitplane, Zurich, Switzerland).
COMSOL Modeling
The concentration field of growth factors secreted from the tumor clusters was simulated with COMSOL Multiphysics software, package version 5.0 [29], in 3D matrices around the vessel tubes. The outer boundary of the gel, the thickness of the gel, and the relative location of vessels in the gel were selected based on the manufacturer's specifications for the Nortis model SCC-001 single-channel chip. The predefined Navier-Stokes and diffusion-reaction equations in COMSOL were used for fluid flow and growth factor concentration in the tube and interstitial space. The stationary solver for laminar flow was chosen for the Navier-Stokes equation. The fluid properties were defined as viscosity of 1×10−3 Pa⋅s and density of 1×103 kg/m3 for water. The inlet boundary conditions were laminar flow with constant flow of 0.5 μl/min with vessel tube diameter of 0.125 mm, and the outlet boundary conditions were laminar flow with zero pressure. The secretion rate of tumor cells in the matrix was estimated to be 5×10−4 mol/m3⋅s [30], [31].
Statistics
For RNA sequencing data from TCGA, the differences in gene expression values between tumor and normal samples were evaluated using a two-tailed t test. For sprouting and quantitative RT-PCR assays, within each patient, differences in sprouting quantification or gene expression values between tumor and normal, or 2D and 3D, or vehicle and VEGFR2-Fc were calculated after log transforming the data. The mean of these differences across patients was computed, and the null hypothesis that the mean difference is equal to zero was tested using a one-sample, two-tailed t test.
Results
Angiogenic Factors are Upregulated in Primary Cultures of Human ccRCC
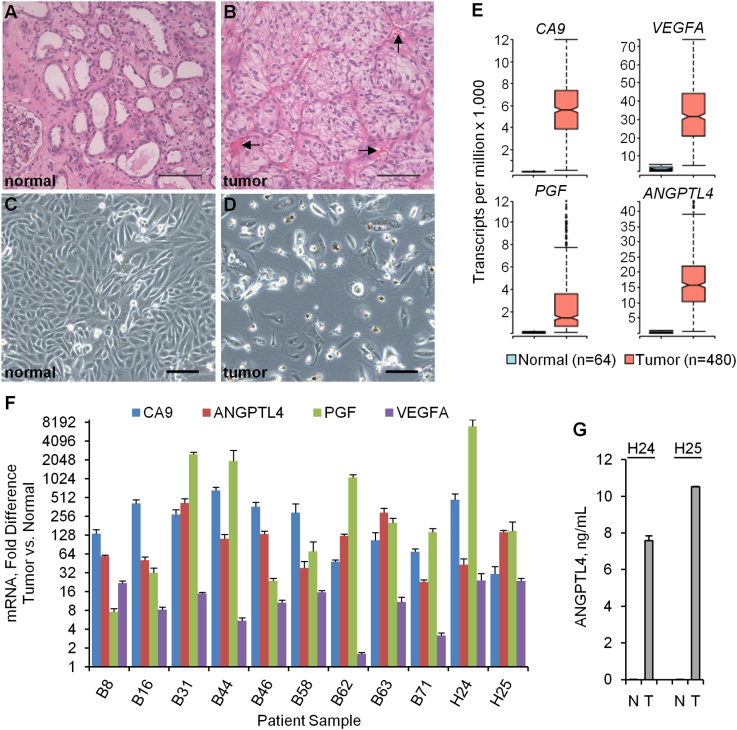
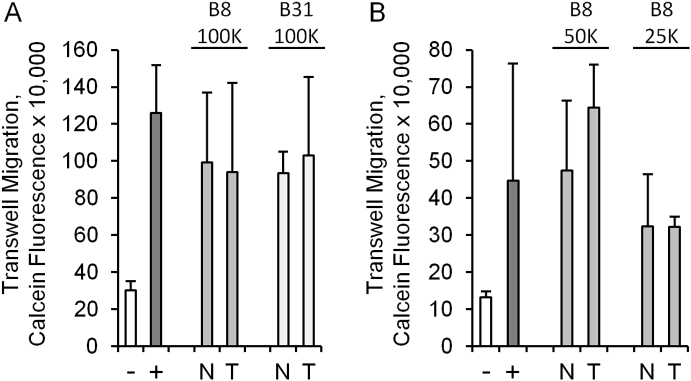
The typical histopathological features of ccRCC and normal-adjacent renal cortex are shown in Figure 1, A and B. Tumor sections were characterized by nests of epithelial cells with a clear cytoplasm and distinct cell membrane surrounded by extensive vascular networks. Overall, primary cultures established from normal-adjacent renal cortex proliferated much more rapidly (doubling time of 1-2 days) than their tumor cell counterparts, which either were static or required weeks for limited expansion in 2D monolayer cultures, as shown previously [12]. In total, we studied 11 primary cultures of ccRCC that were devoid of contaminating colonies of normal renal cortex cells (Figure 1, C and D). The ccRCC phenotype of tumor cells in these cultures compared to separate cultures of normal-adjacent renal cortex performed in parallel was confirmed by the upregulation of the renal cell carcinoma antigen, CA9, as well as the angiogenic genes ANGPTL4, PGF, and VEGFA, which are frequently upregulated in ccRCC specimens in The Cancer Genome Atlas database (Figure 1E), with donor-to-donor variation (Figure 1F). Likewise, at the protein level, ANGPTL4 was observed at higher concentrations in supernatants derived from the ccRCC cultures (Figure 1G). As a first step to studying primary tumor-endothelial cell interactions, we tested whether primary ccRCC cells could induce migration of HUVECs in a conventional 3-μm Transwell migration assay. While there was minimal HUVEC migration through the Transwell pores in the absence of a chemotactic stimulant (serum), both tumor and normal cell 2D monolayers induced similar levels of migration regardless of variation in cell seeding density (Figure 2). We next developed a novel assay system evaluating not only migration but also extracellular matrix invasion and sprouting in a novel 3D culture platform.
Figure 1.
Angiogenic factors are upregulated in primary cultures of human ccRCC. (A and B) Hematoxylin and eosin staining of formalin-fixed, paraffin-embedded normal-adjacent renal cortex (normal) and ccRCC (tumor). Arrows indicate examples of vascular networks. (C and D) Bright-field micrographs of primary cultures derived from normal-adjacent renal cortex and ccRCC. Scale bar, 100 μm for all image panels. (E) Gene expression levels of the renal cell carcinoma antigen, CA9, and the angiogenic factors ANGPTL4, PGF, and VEGFA in ccRCC versus normal-adjacent renal cortex tissues from the TCGA RNA expression data. Box, 25th-75th percentiles; middle notch, median; whiskers, 1.5 times the interquartile range; open circles, values outside of the interquartile range. The top and bottom 5% of values have been omitted for clarity of presentation. For all four genes, P < .0001 for comparison of the expression levels in tumor versus normal samples. (F) Gene expression levels determined by quantitative RT-PCR in primary cultures of ccRCC versus normal-adjacent renal cortex. Error bars represent the standard deviation of triplicate reactions for all samples except H24 and H25 (duplicate reactions). For all four genes, P < .0001 for comparison of the expression levels in tumor versus normal across all patients. G) Secreted protein concentrations of ANGPTL4 measured by Luminex in culture supernatants from normal-adjacent renal cortex (N) or ccRCC cells (T). Error bars represent the average deviation of duplicate determinations. The P value is < .01 for the comparison of ANGPTL4 levels in tumor versus normal cultures across both patients.
Figure 2.
Traditional Transwell assays do not recapitulate ccRCC-specific effects on endothelial cell migration. (A and B) Migration of HUVEC cells through 3-μm pores to the bottom of Transwell inserts (calcein fluorescence) after 16 hours in the presence of RPMI media containing no chemoattractant (−), 5% FBS (+), normal-adjacent renal cortex cells (N), or tumor cells (T). The patient samples and number of normal and tumor cells are shown above the graphs (differences between tumor and normal are not significant). Error bars are the standard deviation of triplicate Transwell inserts.
Primary ccRCC But Not Normal-Adjacent Renal Cortex Induces HUVEC Sprouting in a Flow-Directed 3D Microphysiological System
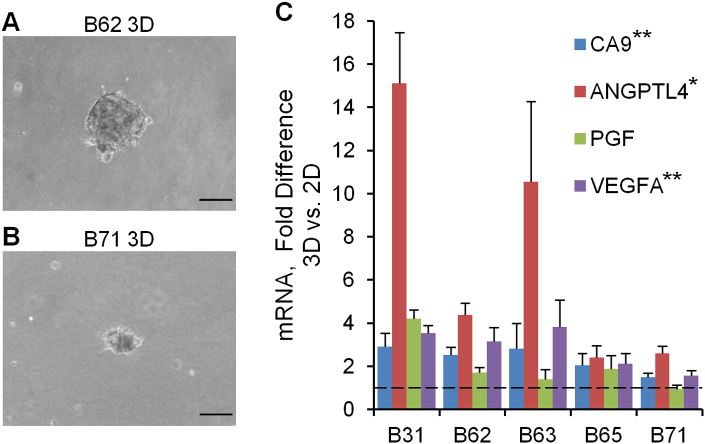
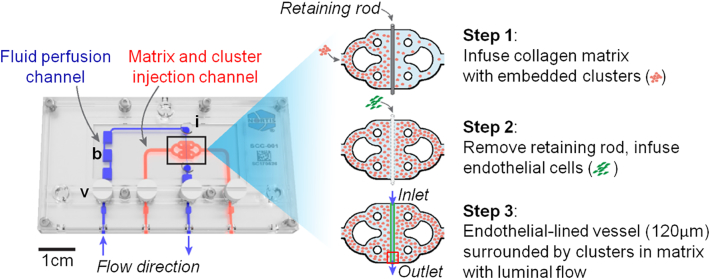
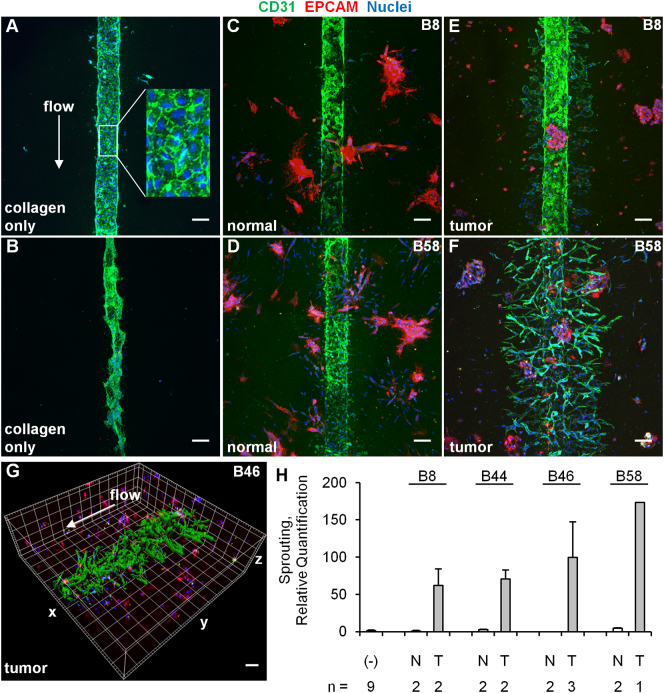
There is increasing interest in developing 3D tumor models that more closely replicate the in vivo tumor phenotype than conventional 2D monolayers [32], [33], [34].To incorporate primary ccRCC cells into 3D structures, we formed tumor clusters and resuspended them in type I collagen. Consistent with the prior ccRCC-only organoid study [12], we observed that the expression of key ccRCC markers was increased at the mRNA level even within 24 hours of tumor cell placement in 3D clusters compared to 2D monolayer cultures (Figure 3). To incorporate the tumor clusters into a 3D microphysiological system, we used a tissue chip from Nortis, Inc. (Woodinville, WA), which we have previously utilized successfully to study 3D tubular epithelial cell physiology and pharmacology [23]. A previous study demonstrated the utility of this system for modeling various types of human vasculature that displayed many features of in vivo vasculature including the deposition of basement membrane proteins, cell-cell junction formation, sprouting angiogenesis, paracellular permeability, and cell migration across vascular barriers [35]. Key features include a controlled rate of directional media flow through a cell-lined cylindrical lumen surrounded by a customizable extracellular matrix within a sealed system. Seeding the chips consisted of 1) infusing collagen with embedded clusters into the matrix channel and allowing the collagen to polymerize; 2) removing a retaining rod from the collagen to form a lumen and infusing the lumen with HUVECs; and 3) establishing directional fluid flow of medium through the hollow endothelial cell-lined lumen (Figure 4). The entire length of the endothelial-lined engineered vessel can be imaged, including the inlet, middle, and outlet regions. Tumor clusters in collagen from 4 independent donors were successfully delivered to 15 chips that were subsequently seeded with HUVECs in the central lumen of the chip in 6 independent experiments. As controls, we also injected 18 chips with collagen only and 8 chips with collagen containing clusters of normal-adjacent renal cortex cells. Since the matrix surrounding the central lumen is transparent, standard immunofluorescent staining and confocal microscopy were performed for select devices in each condition after 5 days of flow, and representative images are shown in Figure 5, A-F. HUVECs in devices containing only collagen formed vessel walls with cell-cell junctions but exhibited minimal sprouting (Figure 5A), and cell loss from the lumen frequently resulted in atrophy of the vascular tube near the outlet (Figure 5B). Similarly, when stabilized in the presence of clusters formed from normal-adjacent renal cortex, endothelial sprouting remained minimal (Figure 5, C and D). In contrast, robust endothelial sprouting was observed in all devices containing tumor cells from all patients examined (Figure5, E-H). Unlike the collagen-only chips, no atrophy of the vessels was observed, suggesting that factors produced by the cell clusters may counteract endothelial cell loss. Inclusion of the solvent dimethyl sulfoxide at 0.05%, a concentration compatible with drug studies (i.e., as a vehicle-only control), did not compromise the ability of HUVEC to sprout in response to ccRCC (Figure 5F). Sprouting was characterized by maintenance of the central lumen, and no retraction was observed after as many as 7 days of culture (Figure 5G and Supplementary Movie S1). Thus, this customizable culture chip system rapidly recapitulates the ability of ccRCC to stimulate angiogenic sprouting in a format that is simple to detect, image, and quantify.
Figure 3.
Upregulation of angiogenic growth factors in 3D tumor clusters. (A and B) Bright-field micrographs of tumor cells in 3D tumor clusters after 24 hours of culture. Similar clusters were observed for all samples except B63, which were dispersed upon resuspension in the collagen. (C) Gene expression levels determined by quantitative RT-PCR. Error bars are the standard deviation of triplicate reactions. Scale bar, 100 μm. Asterisks indicate P < .01 (**) or P < .05 (*).
Figure 4.
Steps in seeding microphysiological devices. Nortis single-channel microphysiological system containing ports for extracellular matrix collagen (red) and luminal flow (blue). b, bubble trap; i, injection port; v, valve. The red box shows the size of the region imaged (1024×1024 μm) in the panels in Figure 5 (the box is situated over the outlet region).
Figure 5.
Primary ccRCC cells but not normal-adjacent renal cortex–derived cells induce HUVEC sprouting in a flow-directed 3D microphysiological system. (A-F) Confocal z-stack projections (400-μm total thickness) of vessels formed from HUVECs in the presence of collagen only, collagen containing clusters of normal-adjacent renal cortex cells (normal), or ccRCC cells (tumor). HUVECs are labeled with the endothelial marker CD31, and the normal and tumor cells are labeled with the epithelial cell marker EPCAM. The white arrow represents the direction of flow through the lumen, which is in the same direction in all panels. Flow was maintained for 5 days. Scale bar, 100 μm. The enlarged inset in panel A is 100 μm wide. (G) 3D surface rendering of tumor-induced sprouting in a chip exposed to flow for 7 days. Each line in the grid of boxes represents 100 μm. (H) Quantification of the region containing sprouting from HUVEC vessels grown in the presence of collagen only (−), normal-adjacent renal cortex (N), or ccRCC tumor cells (T). The number of chips for which confocal staining and quantification were performed for each condition is shown below the graph. Error bars are the average deviation. The P value is < .01 for the comparison of sprouting between tumor versus normal across all patients.
Tumor-Induced Endothelial Sprouting at the Outlet Region of the ccRCC-on-a-Chip is Consistent with a Biochemical Gradient Model of Angiogenic Growth Factor Accumulation
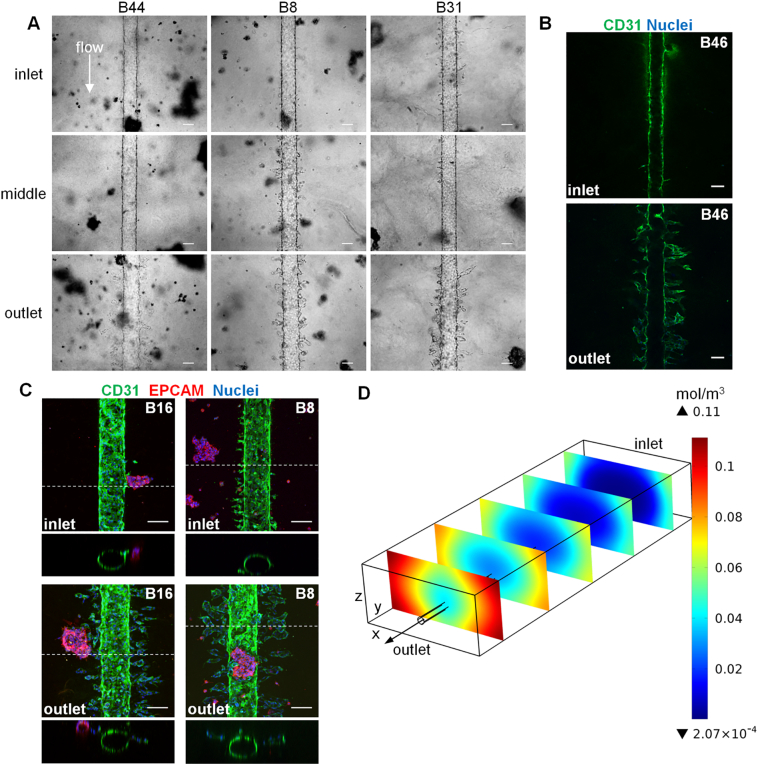
The use of tumor clusters provided the opportunity to assess if sprouting occurred directionally towards these point sources or uniformly throughout the matrix region. Despite the uniform distribution of tumor clusters throughout the entire extracellular matrix collagen surrounding the vessel, prominent endothelial sprouting occurred primarily at the outlet region in all tumor-containing devices. This was apparent both in bright-field (Figure 6A) and immunofluorescence images (Figure 6B) from chips containing clusters derived from all patient-derived primary ccRCC cultures. Endothelial sprouts tapered off in length going from the outlet to the middle region and were rarely observed at the inlet region. Close examination of regions where tumor clusters were in close proximity to the HUVEC lumen confirmed that sprouting was uniform in all directions at the outlet and not just limited to directional migration towards the clusters (Figure 6C). Computational simulation modeling [29] of laminar flow in permeable endothelial channels coupled with a diffusion reaction of growth factors produced by clusters in the bulk matrix predicted angiogenic growth factor accumulation and higher concentration at the outlet of the device compared to other regions (Figure 6D). Taken together, these results support a biochemical gradient model of angiogenic growth factor accumulation that favors sprouting at the outlet region of the flow-directed microphysiological system.
Figure 6.
ccRCC-induced sprouting primarily at the outlet region of the Nortis 3D device is consistent with COMSOL modeling. (A) Bright-field micrographs of HUVEC vessels (day 5) at the inlet, middle, and outlet regions of chips containing tumor clusters formed from ccRCC cultures. (B) Confocal cross-section of HUVEC vessel at the inlet versus outlet region of a chip containing tumor clusters from patient B46 (day 7). (C) Confocal volume projections (300-μm total thickness) at the outlet and inlet regions of chips that were reimaged at 20× zoom (compare to original 10× imaging of the same chips in Figures 5 and 7). The dotted line indicates the position of the orthogonal view shown below each volume projection. Scale bar, 100 μm. (D) COMSOL multiphysics model of flow dynamics demonstrating an angiogenic growth factor concentration gradient favoring sprouting closer to the outlet region of the Nortis 3D device.
Pharmacological Blockade of ccRCC-Induced HUVEC Sprouting
Clinically, the production of angiogenic factors such as VEGFA has been exploited to interrupt the ccRCC-endothelial cell axis and inhibit tumor growth. This has been achieved through antibodies that bind to VEGFA [10] or through small molecule inhibitors of its receptor VEGFR2 [8]. We therefore tested whether pharmacological angiogenesis blockade could be modeled in our microphysiological system. For these studies, we used a soluble recombinant VEGFR2-Fc chimera. Administration of this VEGF inhibitor into the media flowing through the central lumen effectively blocked ccRCC-induced sprouting (Figure 7). Thus, this microphysiological system provides a suitable platform to test the effects of novel antiangiogenesis agents on the complex 3D morphology of endothelial cell sprouting in response to tumor-derived growth factors.
Figure 7.
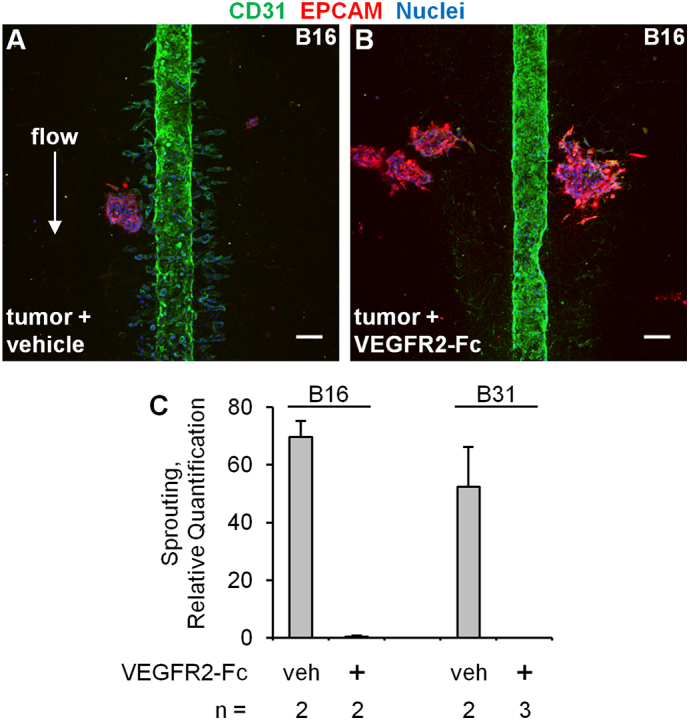
Pharmacological blockade of ccRCC-induced HUVEC sprouting. (A and B) Confocal z-stack volume projections (400 μm total thickness) of HUVEC vessels grown for 5 days in the presence of ccRCC tumor clusters with flow containing EGM media plus vehicle (0.0002% BSA) or 200 ng/ml recombinant VEGFR2-Fc chimera. Scale bar, 100 μm. (C) Quantification of sprouting. Error bars represent the average deviation. The P value is < .01 for the comparison of sprouting between vehicle versus VEGFR2-Fc across both patients.
Discussion
There is increasing interest in novel preclinical models of human tumors that more efficiently reconstitute the in vivo phenotype and key cell-cell interactions [32], [33], [34]. Recent advances in 3D microfluidic human tumor platforms have enabled the study of microenvironmental influences on properties such as tumor cell migration and T-cell homing and cytotoxicity [36], [37], while vascularized, perfusable 3D tumor models have permitted efficient examination of tumor cell chemosensitivity and extravasation [38], [39], [40], [41]. While 3D in vitro models have been developed to study tumor-induced angiogenesis, progress to date has been limited to cell lines [42], [43], [44], [45], [46], [47]. To our knowledge, we have developed the first vascularized, 3D, flow-directed culture system for primary human tumor cells that faithfully recapitulates tumor-induced angiogenesis. This tractable platform is easily adaptable to a standard tissue culture laboratory and provides a physiologically relevant alternative to artifact-prone tumor cell lines and more expensive and time-consuming transgenic mouse models and xenograft systems.
A key feature of our platform is tunability with respect to cellular components and extracellular matrix materials utilized. In these initial studies, we established the intrinsic potential of primary tumor cells to stimulate angiogenic sprouting of HUVECs. The use of primary tumor cells allows for parallel use of primary tubule cell cultures derived from the same patient as an isogenic control. Based on our own findings and those from the recent ccRCC-only organoid model [12], expression of key ccRCC biomarkers is enhanced in 3D cultures compared to 2D monolayers. Our system will also permit study of intratumoral heterogeneity [48] by generating primary tumor cultures from different anatomic regions of the tumor and from metastatic sites. We also plan to use this efficient platform to study mechanisms of transformation using genome engineering to introduce canonical ccRCC mutations (VHL, BAP1, SETD2, PBRM1) sequentially/simultaneously into primary tubule cells and assessing the angiogenic sprouting response of endothelial cells.
Perhaps most exciting is the suitability of this ccRCC microphysiological system to study mechanisms of action of novel pharmaceutics. In this study, we demonstrated the feasibility of blocking angiogenesis by delivering VEGFR2-Fc via the central lumen of the chip device. This platform is well suited to testing of additional antiangiogenesis agents but could also be utilized to test agents that block tumor cell proliferation and survival. Since constitutive HIF activation in ccRCC results in the production of numerous angiogenic factors, blockade of upstream HIF transcriptional activity itself may provide a more robust and durable blockade of angiogenesis. Encouragingly, novel HIF2 antagonists have been shown to disrupt ccRCC growth in a patient-derived xenograft model, but this effect was limited to only half of patient tumors tested [19]. Therefore, future studies are directed at testing whether the presently developed 3D culture model of ccRCC can be used to provide a more rapid and cost-efficient method of determining patient-specific responsiveness to emerging antiangiogenesis agents. As a fully in vitro system requiring relatively small numbers of input cells for reconstitution, this system presents an attractive alternative for personalized preclinical pharmaceutical testing.
Our system is not without limitations, chief of which is the absence of patient-derived microvascular endothelial cells derived from the kidney. Importantly, microvascular endothelial cells are known to exhibit tissue-specific molecular heterogeneity [49], [50]. While HUVEC vessels (which are derived from large vessels and exhibit some lot-specific variability) were sufficient to demonstrate that ccRCC cells produce functional angiogenic signals in the chip, it is more likely that organotypic endothelial cells will be needed to identify therapies that will be clinically efficacious and to accurately model the ccRCC microenvironment for mechanistic studies. Therefore, future iterations will investigate the use of primary microvascular endothelial cells isolated from tumor versus normal-adjacent kidney tissue. We recently reported the isolation and culture of normal human peritubular kidney microvascular endothelial cells (HKMEC) and demonstrated their nonangiogenic phenotype, a property which may contribute to the loss of capillaries that is universal to acute kidney injury [51]. We are presently optimizing methods for isolating HKMEC from primary tumors (t-HKMEC), and it will be of interest to compare the ability of patient-matched t-HKMEC versus HKMEC to sprout in the ccRCC-on-a-chip.
Another limitation is the current need for primary culture/limited expansion of primary tumor cells prior to seeding of the device. This may potentially bias our findings in favor of tumor cells that can temporarily adapt to 2D monolayer culture. One possible workaround we are exploring is isolation of sufficient tumor cells at the time of nephrectomy followed by direct seeding of the chips for study. A third limitation is the presence of only two cell types (tumor and vascular endothelial cells) in our model, which belies the true cellular complexity of the tumor environment in vivo. However, since the chip is flexible in terms of cellular and matrix components and even in terms of relative cell positioning (i.e., within the matrix or lining the central lumen), incorporation of additional cellular components can be explored. These might include stromal cells or perivascular endothelial cells incorporated into the matrix together with tumor cells. Another important cell type to incorporate will be immune cells such as receptor-engineered T cells specific for RCC antigens [52] to explore if they can traverse the vascular barrier to home to and destroy tumor clusters within the chip matrix. Future studies could also test an alternative extracellular matrix such as decellularized human kidney extracellular matrix [12]. Lastly, while we have initially focused our efforts on the clear cell variant of RCC, study of other RCC subtypes and other solid tumor types is possible.
Conclusions
In summary, we have developed the first vascularized, microphysiological biomimetic of ccRCC which retains the prominent angiogenic features of the tumor type. The system enables rapid morphological and pharmacologic studies using primary tumor samples, opening the door to personalization of drug selection for postoperative therapies. Its customizable design will permit mechanistic studies on tumorigenesis and incorporation of additional cell types. We envision that this efficient alternative to existing preclinical models will accelerate drug discovery and progress towards personalized therapies for ccRCC.
The following are the supplementary data related to this article.
Primary human ccRCC-induced HUVEC sprouting in a flow-directed 3D microphysiological device exposed to continuous flow for 7 days.
Histopathology of ccRCC Specimens
Real-Time PCR Primers
Acknowledgements
The authors would like to thank Sara Daniel, Daniel Chang, and Dr. Kim Muczynski for tissue procurement; Daniel Lih for assistance with isolating tumor and normal-adjacent cells, Dr. Elijah Weber, Dr. Kevin Lidbergh, Dr. Edward Kelly, and Dr. Henning Mann for instruction on the use of Nortis chips and insightful discussions; Dr. Dale Hailey for advice on imaging at The Lynn and Mike Garvey Imaging Core at the University of Washington; John Ruzinski for Luminex assays; and Dr. Theodore Gooley at the Fred Hutchinson Cancer Research Center for statistical advice. This work was supported by the National Institutes of Health (UH2/UH3 TR000504 to J.H. and Cancer Center Support Grant P30 CA015704 to the FHCRC/UW Cancer Consortium); The Damon Runyon Cancer Research Fellowship (DRG114-13 to S.A.); The Mary Gates Endowment for Students Research Scholarship and The Washington Research Foundation Fellowship (to C.T.); and an unrestricted gift from the Northwest Kidney Centers to the Kidney Research Institute.
Footnotes
Conflicts of interest: none.
Contributor Information
Chris P. Miller, Email: cpmiller@uw.edu.
Connor Tsuchida, Email: ctsuchida51@yahoo.com.
Ying Zheng, Email: yingzy@uw.edu.
Jonathan Himmelfarb, Email: Jhimmelfarb@Nephrology.washington.edu.
Shreeram Akilesh, Email: shreeram@uw.edu.
References
- 1.National Cancer Institute Surveillance, epidemiology, and end results program. https://seer.cancer.gov/statfacts/html/kidrp.html Available from:
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 5.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L. Identification of the von Hippel–Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 6.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wettersten HI, Aboud OA, Lara PN, Jr., Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–419. doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- 8.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 9.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang JC, Haworth L, Sherry RM, Hwu P, Schwartzentruber DJ, Topalian SL, Steinberg SM, Chen HX, Rosenberg SA. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med. 2003;349:427–434. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter SN, Baker LC, Mittelman D, Porteus MH. Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 2014;15:R75. doi: 10.1186/gb-2014-15-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batchelder CA, Martinez ML, Duru N, Meyers FJ, Tarantal AF. Three dimensional culture of human renal cell carcinoma organoids. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 14.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel–Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleymenova E, Everitt JI, Pluta L, Portis M, Gnarra JR, Walker CL. Susceptibility to vascular neoplasms but no increased susceptibility to renal carcinogenesis in Vhl knockout mice. Carcinogenesis. 2004;25:309–315. doi: 10.1093/carcin/bgh017. [DOI] [PubMed] [Google Scholar]
- 16.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel–Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. Generation of a mouse model of Von Hippel–Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1alpha. Cancer Res. 2011;71:6848–6856. doi: 10.1158/0008-5472.CAN-11-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlander S, Schonenberger D, Toussaint NC, Prummer M, Catalano A, Brandt L, Moch H, Wild PJ, Frew IJ. Combined mutation in Vhl, Trp53 and Rb1 causes clear cell renal cell carcinoma in mice. Nat Med. 2017;23:869–877. doi: 10.1038/nm.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavia-Jimenez A, Tcheuyap VT, Brugarolas J. Establishing a human renal cell carcinoma tumorgraft platform for preclinical drug testing. Nat Protoc. 2014;9:1848–1859. doi: 10.1038/nprot.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivanand S, Pena-Llopis S, Zhao H, Kucejova B, Spence P, Pavia-Jimenez A, Yamasaki T, McBride DJ, Gillen J, Wolff NC. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran TA, Leong HS, Pavia-Jimenez A, Fedyshyn S, Yang J, Kucejova B, Sivanand S, Spence P, Xie XJ, Pena-Llopis S. Fibroblast growth factor receptor-dependent and -independent paracrine signaling by sunitinib-resistant renal cell carcinoma. Mol Cell Biol. 2016;36:1836–1855. doi: 10.1128/MCB.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90:627–637. doi: 10.1016/j.kint.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Network TCGAR Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjerregaard H, Pedersen S, Kristensen SR, Marcussen N. Reference genes for gene expression analysis by real-time reverse transcription polymerase chain reaction of renal cell carcinoma. Diagn Mol Pathol. 2011;20:212–217. doi: 10.1097/PDM.0b013e318212e0a9. [DOI] [PubMed] [Google Scholar]
- 26.Cross VL, Zheng Y, Won Choi N, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials. 2010;31:8596–8607. doi: 10.1016/j.biomaterials.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagao RJ, Xu J, Luo P, Xue J, Wang Y, Kotha S, Zeng W, Fu X, Himmelfarb J, Zheng Y. Decellularized human kidney cortex hydrogels enhance kidney microvascular endothelial cell maturation and quiescence. Tissue Eng Part A. 2016;22:1140–1150. doi: 10.1089/ten.tea.2016.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.COMSOL Multiphysics Modeling Software. https://www.comsol.com/ Available from:
- 30.Cho CH, Park J, Nagrath D, Tilles AW, Berthiaume F, Toner M, Yarmush ML. Oxygen uptake rates and liver-specific functions of hepatocyte and 3T3 fibroblast co-cultures. Biotechnol Bioeng. 2007;97:188–199. doi: 10.1002/bit.21225. [DOI] [PubMed] [Google Scholar]
- 31.Rudge JS, Holash J, Hylton D, Russell M, Jiang S, Leidich R, Papadopoulos N, Pyles EA, Torri A, Wiegand SJ. VEGF Trap complex formation measures production rates of VEGF, providing a biomarker for predicting efficacious angiogenic blockade. Proc Natl Acad Sci U S A. 2007;104:18363–18370. doi: 10.1073/pnas.0708865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn J, Sei YJ, Jeon NL, Kim Y. Tumor microenvironment on a chip: the progress and future perspective. Bioengineering (Basel) 2017;4 doi: 10.3390/bioengineering4030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol. 2017;24:1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp Biol Med (Maywood) 2014;239:1264–1271. doi: 10.1177/1535370214539228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Hebert JD, Lee TA, Xing H, Boussommier-Calleja A, Hynes RO, Lauffenburger DA, Kamm RD. Macrophage-secreted TNFalpha and TGFbeta1 influence migration speed and persistence of cancer cells in 3D tissue culture via independent pathways. Cancer Res. 2017;77:279–290. doi: 10.1158/0008-5472.CAN-16-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen MB, Whisler JA, Frose J, Yu C, Shin Y, Kamm RD. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat Protoc. 2017;12:865–880. doi: 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moya M, Tran D, George SC. An integrated in vitro model of perfused tumor and cardiac tissue. Stem Cell Res Ther. 2013;4(Suppl 1):S15. doi: 10.1186/scrt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobrino A, Phan DT, Datta R, Wang X, Hachey SJ, Romero-Lopez M, Gratton E, Lee AP, George SC, Hughes CC. 3D microtumors in vitro supported by perfused vascular networks. Sci Rep. 2016;6 doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bray LJ, Werner C. Evaluation of three-dimensional in vitro models to study tumor angiogenesis. ACS Biomater Sci Eng. 2017 doi: 10.1021/acsbiomaterials.7b00139. [DOI] [PubMed] [Google Scholar]
- 43.Correa de Sampaio P, Auslaender D, Krubasik D, Failla AV, Skepper JN, Murphy G, English WR. A heterogeneous in vitro three dimensional model of tumour-stroma interactions regulating sprouting angiogenesis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janvier R, Sourla A, Koutsilieris M, Doillon CJ. Stromal fibroblasts are required for PC-3 human prostate cancer cells to produce capillary-like formation of endothelial cells in a three-dimensional co-culture system. Anticancer Res. 1997;17:1551–1557. [PubMed] [Google Scholar]
- 45.Kim S, Lee H, Chung M, Jeon NL. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13:1489–1500. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 46.Szot CS, Buchanan CF, Freeman JW, Rylander MN. In vitro angiogenesis induced by tumor-endothelial cell co-culture in bilayered, collagen I hydrogel bioengineered tumors. Tissue Eng Part C Methods. 2013;19:864–874. doi: 10.1089/ten.tec.2012.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y, Sun Y, Yu X, Shao Y, Zhang P, Dai G, Fu J. Angiogenesis in liquid tumors: an in vitro assay for leukemic-cell-induced bone marrow angiogenesis. Adv Healthc Mater. 2016;5:1014–1024. doi: 10.1002/adhm.201501007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357 doi: 10.1126/science.aal2379. [DOI] [PubMed] [Google Scholar]
- 50.Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding BS, Schachterle W, Liu Y, Rosenwaks Z. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ligresti G, Nagao RJ, Xue J, Choi YJ, Xu J, Ren S, Aburatani T, Anderson SK, MacDonald JW, Bammler TK. A novel three-dimensional human peritubular microvascular system. J Am Soc Nephrol. 2016;27:2370–2381. doi: 10.1681/ASN.2015070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tykodi SS, Satoh S, Deming JD, Chou J, Harrop R, Warren EH. CD8+ T-cell clones specific for the 5T4 antigen target renal cell carcinoma tumor-initiating cells in a murine xenograft model. J Immunother. 2012;35:523–533. doi: 10.1097/CJI.0b013e318261d630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary human ccRCC-induced HUVEC sprouting in a flow-directed 3D microphysiological device exposed to continuous flow for 7 days.
Histopathology of ccRCC Specimens
Real-Time PCR Primers