Abstract
Metabolic pathway assembly typically involves the expression of enzymes from multiple organisms in a single heterologous host. Ensuring that each enzyme functions effectively can be challenging, since many potential factors can disrupt proper pathway flux. Here, we compared the performance of two enzyme homologs in a pathway engineered to allow Escherichia coli to grow on 4-hydroxybenzoate (4-HB), a byproduct of lignocellulosic biomass deconstruction. Single chromosomal copies of the 4-HB 3-monooxygenase genes pobA and praI, from Pseudomonas putida KT2440 and Paenibacillus sp. JJ-1B, respectively, were introduced into a strain able to metabolize protocatechuate (PCA), the oxidation product of 4-HB. Neither enzyme initially supported consistent growth on 4-HB. Experimental evolution was used to identify mutations that improved pathway activity. For both enzymes, silent mRNA mutations were identified that increased enzyme expression. With pobA, duplication of the genes for PCA metabolism allowed growth on 4-HB. However, with praI, growth required a mutation in the 4-HB/PCA transporter pcaK that increased intracellular concentrations of 4-HB, suggesting that flux through PraI was limiting. These findings demonstrate the value of directed evolution strategies to rapidly identify and overcome diverse factors limiting enzyme activity.
Keywords: Lignin, Protocatechuate, Experimental evolution
Highlights
-
•
Experimental evolution identified beneficial mutations for heterologous enzymes.
-
•
Codon optimization can confound bioprospecting by introducing shared features.
-
•
Beneficial mutations can be general, or homolog-specific.
1. Introduction
Synthetic biologists frequently transfer metabolic pathways from native hosts into more-tractable production strains (Nielsen and Keasling, 2016). By combining enzymes from different organisms, they can construct novel pathways to produce valuable biochemicals (Yim et al., 2011, Galanie et al., 2015). However, combining diverse enzymes into complex pathways is challenging, since the interactions between enzymes within a pathway, or between a pathway and its host, can limit productivity (Michener et al., 2012, Kim and Copley, 2012). A common solution is to screen multiple enzyme homologs, with the goal of identifying a variant that lacks deleterious interactions (Bayer et al., 2009, Narcross et al., 2016). A deeper understanding of the reasons that enzyme homologs function poorly will increase the success rate of screening homolog libraries, and thereby allow faster and cheaper pathway assembly.
The catabolism of lignin-derived aromatic chemicals provides a representative example of this engineering challenge. The thermochemical depolymerization of lignin produces a complex mixture of aromatic compounds (Rodriguez et al., 2017). Though a process described as biological funneling, microbes can convert this low-value mixture of substrates into a small set of core intermediates and then into valuable products (Linger et al., 2014). However, no single microbe has yet been isolated that is capable of catabolizing and valorizing all the constituents of a typical mixture (Bugg et al., 2011). Instead, researchers seek to augment the catabolic capabilities of foundation organisms by introducing new metabolic pathways for conversion of recalcitrant substrates (Strachan et al., 2014). Changes in the feedstock composition and pretreatment strategy will alter the composition of the depolymerized mixture (Ragauskas et al., 2014), necessitating the construction and optimization of an entire suite of microbes, each specific to a particular substrate mixture. Consequently, the facile valorization of lignin-derived aromatics will require the ability to rapidly engineer metabolic networks using pathways from diverse microbes.
Heterologous metabolic pathways are frequently expressed from plasmids. These vectors are easy to construct, and the resulting high DNA copy numbers can compensate for low specific enzyme activity. However, plasmid-based pathways are unstable and difficult to scale for larger pathways, such as those that will be required for catabolism of a mixture of lignin-derived aromatic compounds (Jiang et al., 2013). Moving heterologous enzymes from plasmids to the chromosome increases stability and scaling, and due to new techniques is increasingly practical (Jiang et al., 2015). However, the reduced copy number increases the challenge of providing sufficient enzyme activity (Alonso-Gutierrez, 2018, Wang and Pfeifer, 2008).
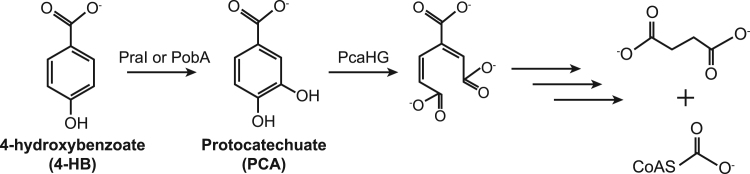
In this work, we explored the challenges involved in extending a metabolic pathway using chromosomally-expressed enzyme homologs, focusing specifically on pathways for the catabolism of 4-hydroxybenzoate (4-HB) and protocatechuate (PCA) (Fig. 1). These compounds are the core metabolites for catabolism of hydroxyphenyl (H) and guaiacyl (G) lignans, respectively, and therefore are important targets for engineering biological conversion of lignin components. We previously constructed and optimized a pathway in E. coli that converts PCA to succinate and acetyl-CoA using the nine-gene pca pathway from Pseudomonas putida KT2440 (Supplementary Fig. 1). After optimization, the pathway enabled growth with PCA as the sole source of carbon and energy (Clarkson et al., 2017). We then extended the pathway to convert 4-HB into PCA using a 4-hydroxybenzoate monooxygenase, praI, from Paenibacillus sp. JJ-1B (Kasai et al., 2009), allowing E. coli to grow with 4-HB as the carbon source, with limitations that we have now discovered. In the present work, we introduced an alternative 4-hydroxybenzoate monooxygenase homolog, pobA, from Pseudomonas putida KT2440 (Harwood and Parales, 1996, Jimenez et al., 2002). Using either of the monooxygenases, we selected mutant strains that grew efficiently with 4-HB as the sole source of carbon and energy, at the same rate as with PCA, and without the previous limitations.
Fig. 1.
Catabolism of 4-HB. The 4-HB monooxygenases PraI and PobA convert 4-HB into PCA. The first step in PCA degradation is ring cleavage by the PCA 3,4-dioxygenase PcaHG, ultimately yielding succinate and acetyl-CoA.
Characterizing the resulting evolutionary solutions identified multiple interacting factors that limited 4-HB catabolism and revealed unexpected differences between enzyme homologs. Similar RNA secondary structures, unintentionally introduced during codon optimization, initially decreased expression of both 4-HB monooxygenases. After resolving this issue, we found that the monooxygenases are not fully interchangeable, as different optimization solutions produced varied results with the two homologs. Ultimately, we identified pathway modifications for both homologs that allowed similar levels of growth with either PCA or 4-HB. These modifications included duplications of the core PCA degradation pathway and point mutations in the PCA/4-HB transporter, PcaK. Unexpectedly, these modifications had very different impacts on praI and pobA strains. Either allowed pobA strains to grow on 4-HB, but only the PcaK mutations allowed praI strains to grow. The results provide examples of the modifications required to optimize the function of chromosomally-expressed enzyme homologs, facilitating the rapid, predictable construction and debugging of novel metabolic pathways.
2. Materials and methods
2.1. Media and chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Fairlawn, NJ) and were molecular grade. All oligonucleotides were ordered from IDT (Coralville, IA). E. coli strains were routinely cultivated at 37 °C in LB broth containing the necessary antibiotics (50 mg/L kanamycin or 50 mg/L spectinomycin). Growth assays with PCA and 4-HB were performed in M9 salts medium containing 300 mg/L thiamine and 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). PCA and 4-HB were dissolved in water at 5 g/L, filter sterilized, and added at a final concentration of 1 g/L. The pH of the substrate stock solutions was not adjusted, as PCA oxidation in air occurred more rapidly at neutral pH.
2.2. Plasmid construction
Expression for sgRNA plasmids targeting chromosomal loci were constructed as described previously (Clarkson et al., 2017). Briefly, an inverse PCR was used to amplify the vector backbone. Overlapping oligonucleotides containing the new 20-nt targeting sequence were inserted by Gibson assembly and transformed into 10-β E. coli (NEB, Waltham, MA). Correct assembly was verified by Sanger sequencing. The plasmids used are listed in Supplementary Table 2.
2.3. Strain construction
Strain JME17 was previously optimized for growth with PCA. For growth with 4-HB, the appropriate 4-HB monooxygenase was introduced into the chromosome of JME17. Construction of JME50, expressing the 4-HB monooxygenase PraI, was described previously (Clarkson et al., 2017). Strain JME38 was constructed in a similar fashion by introducing a commercially-synthesized pobA expression cassette (Gen9, Cambridge, MA) into JME17. The promoter and terminator for pobI expression were chosen from previously characterized genetic parts (Supplementary Fig. 1B; Chen et al., 2013; Kosuri et al., 2013). The expression cassette was integrated into the gfcAB locus using plasmid pJM168 (Jiang et al., 2015). To reconstruct evolved mutations, the mutant locus was amplified from the appropriate genomic DNA and introduced into the selected recipient strain in the same fashion. To introduce a second copy of pcaHGBDC, the expression cassette was amplified from JME17, combined with homology arms for yiaU by overlap-extension PCR, and transformed into JME17 together with plasmid pJM193. All modifications were verified by colony PCR and Sanger sequencing, or by whole-genome resequencing. The strains used are listed in Supplementary Table 1.
2.4. Experimental evolution
Parental strains were streaked to single colonies. Three colonies were grown to saturation in LB + 1 mM IPTG, then diluted 128-fold into M9 + 1 mM IPTG + 1 g/L 4-HB + 50 mg/L PCA and grown at 37 °C. When the cultures reached saturation, they were diluted 128-fold into fresh medium. Initially, the saturated cultures had a low optical density (~ 0.2). Over time, the final density increased. When the final density passed 0.8, the PCA concentration was reduced, first to 20 mg/L, then to 10 mg/L, 5 mg/L, and finally 0 mg/L. After a reduction in the PCA concentration, a culture often required one to two additional days to reach saturation. After 300 generations, each population was streaked to single colonies. Eight replicate colonies were picked for further characterization.
2.5. Growth rate measurements
Cultures were grown overnight to saturation in M9 + 1 mM IPTG + 2 g/L glucose. They were then diluted 100-fold into fresh M9 + IPTG containing the appropriate carbon source and grown as triplicate 100 µL cultures in a Bioscreen C plate reader (Oy Growth Curves Ab Ltd, Helsinki, Finland). Growth rates were calculated using CurveFitter software based on readings of optical density at 600 nm (Delaney et al., 2013).
2.6. Genome resequencing
Genomic DNA (gDNA) was prepared using a Blood and Tissue kit (Qiagen, Valencia, CA) according to the manufacturer's directions. The gDNA was quantified using a Qubit fluorimeter (Thermo Fisher, Waltham, MA) and resequenced by the Joint Genome Institute on a MiSeq (Illumina, San Diego, CA) to approximately 100 × coverage.
2.7. Proteomics
Parent, evolved and engineered E. coli strains were grown in triplicate and processed for LC-MS/MS analysis. Whole-cell lysates were prepared by bead beating in sodium deoxycholate lysis buffer (4% SDC, 100 mM ammonium bicarbonate, pH 7.8) using 0.15 mM zirconium oxide beads followed by centrifugation to clear debris. After measuring protein concentration via bicinchoninic acid (BCA) assay, samples were adjusted to 10 mM dithiothreitol and incubated at 95 °C for 10 min to denature and reduce proteins. Cysteines were alkylated/blocked with 30 mM iodoacetamide during 20 min incubation at room temperature in the dark. Protein samples (250 µg) were then transferred to a 10-kDa MWCO spin filter (Vivaspin 500, Sartorius) and digested in situ with proteomics-grade trypsin as previously described (Clarkson et al., 2017). The tryptic peptide solution was then filtered through the MWCO membrane, adjusted to 1% formic acid to precipitate residual SDC, and SDC precipitate removed from the peptide solution with water-saturated ethyl acetate. Peptide samples were then concentrated via SpeedVac and measured by BCA assay.
Peptide samples were analyzed by automated 2D LC-MS/MS analysis using a Vanquish UHPLC plumbed directly in-line with a Q Exactive Plus mass spectrometer (Thermo Scientific) outfitted with a triphasic MudPIT back column (RP-SCX-RP) coupled to an in-house pulled nanospray emitter packed with 30 cm of 5 µm Kinetex C18 RP resin (Phenomenex). For each sample, 5 µg of peptides were loaded, desalted, separated and analyzed across two successive salt cuts of ammonium acetate (50 mM and 500 mM), each followed by 105 min organic gradient as previously described (Clarkson et al., 2017). Eluting peptides were measured and sequenced by data-dependent acquisition on the Q Exactive MS.
MS/MS spectra were searched against the E. coli K-12 proteome concatenated with relevant exogenous protein sequences and common protein contaminants using MyriMatch v.2.2 (Tabb et al., 2007). Peptide spectrum matches (PSM) were required to be fully tryptic with any number of missed cleavages; a static modification of 57.0214 Da on cysteine (carbamidomethylated) and a dynamic modification of 15.9949 Da on methionine (oxidized) residues. PSMs were filtered using IDPicker v.3.0 (Ma et al., 2009) with an experiment-wide false-discovery rate (assessed by PSM matches to decoy sequences) initially controlled at< 1% at the peptide-level. Peptide intensities were assessed by chromatographic area-under-the-curve using IDPicker's embed spectra/label-free quantification option, and unique peptide intensities summed to estimate protein-level abundance. Protein abundance distributions were then normalized across samples and missing values imputed to simulate the MS instrument's limit of detection. Significant differences in protein abundance were assessed by one-way ANOVA and pairwise T-test. Statistical significance was evaluated using a Benjamini-Hochberg false discovery rate of 5%.
2.8. qPCR
Gene copy numbers were determined by quantitative PCR using Phusion DNA Polymerase (NEB, Ipswich, MA) and EvaGreen (Biotium, Fremont, CA) on a CFX96 Touch thermocycler (Bio-Rad, Hercules, CA). Whole cells from triplicate cultures were used as the DNA templates. Copy number was normalized to 16S rRNA gene copies.
2.9. Intracellular metabolite measurements
Strains were grown overnight in M9 + glucose. They were then diluted 100 × into fresh M9 + glucose and regrown to mid-log phase. The cells were centrifuged and resuspended in fresh M9 + glucose containing the appropriate substrate at an optical density of ~ 3, 1 mL per replicate. Samples were incubated at 37 °C for 15 min, washed twice in M9 + glucose, transferred to a fresh tube, and stored at − 80 °C until analysis.
Metabolites (4-HB and PCA) were analyzed by gas chromatography/mass spectrometry (GC/MS) as follows. Frozen cell pellets were thawed and resuspended in 100 µL of water by vortex mixing, and lysozyme was added to a final concentration of 100 µg/mL by addition of 5 µL of a 2 mg/mL stock solution. Cells were incubated at room temperature for 30 min and then acidified with 10 µL of 0.1 M HCl containing 10 µg/mL of 4-HB-d4 as an internal standard. Metabolites were extracted by adding 1.0 mL of ethyl acetate, vortex mixing for 2 × 10 s, centrifuging (16,000 × g for 2 min), transferring 0.8 mL of the ethyl acetate layer to a 2-mL glass autosampler vial, and evaporating the solvent under a stream of argon. Metabolites were derivatized by adding 100 µL of N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% chlorotrimethylsilane, sealing the vials, heating to 60 °C for 30 min, and diluting with 900 µL of n-hexane.
GC/MS analysis was performed using an Agilent 7890A gas chromatograph equipped with a 7693A automatic liquid sampler, an HP-5 ms capillary column (30 m long × 0.25 mm inside diameter with a 0.25-µm capillary film of 5% phenyl methylsilicone) and a 5975 C mass-sensitive detector. Splitless injections of 1 µL were made at an inlet temperature of 270 °C with a 15-s dwell time (needle left in the inlet after injection), an initial column temperature of 50 °C and helium carrier gas at a constant flow of 1 mL/min. After 2 min at 50 °C, the temperature was ramped at 25 °C/min to 300 °C and held for 2 min. The detector was operated with a transfer line temperature of 300 °C, source temperature of 230 °C and quadrupole temperature of 150 °C. After a 5-min solvent delay, electron-impact mass spectra from 50 to 400 amu were collected continuously (~ 4/s) for the duration of the run. Relative amounts of metabolites were determined by integration of selected ion chromatograms for PCA(TMS)3, 4-HB(TMS)2 and 4-HB-d4(TMS)2 at m/z 193, 267 and 271, respectively, and then dividing by the area of the 271 peak (internal standard).
3. Results and discussion
3.1. Pathway construction and characterization
We previously described the construction of E. coli strain JME50 (Clarkson et al., 2017), containing praI from Paenibacillus sp. JJ-1B (Kasai et al., 2009). To explore the different optimization challenges of enzyme homologs, we used a similar process in this work to construct JME38, introducing pobA from Pseudomonas putida KT2440 (Harwood and Parales, 1996, Jimenez et al., 2002). These two genes are 60% identical at the nucleotide level, and the associated enzymes have 54% amino acid identity.
We reported that JME50 could grow in minimal medium containing 4-HB as the sole source of carbon and energy, in contrast to the parental strain JME17 lacking praI (Clarkson et al., 2017). Upon further investigation, however, we discovered that this strain was unable to grow in minimal medium with 4-HB if it had previously been cultured in minimal medium with glucose or 4-HB as the carbon source (Supplementary Fig. 2). The same was true of JME38: expression of a 4-hydroxybenzoate monooxygenase was necessary for growth with 4-HB, but was not sufficient for repeated propagation. We were unable to identify a trace mineral or vitamin that could replace the LB preculture and allow repeated growth in minimal medium. However, supplementation with 0.1 g/L of PCA, in addition to 1 g/L of 4-HB, did allow moderate growth. We expect that the nutrient-replete conditions of an LB preculture provided a similar supplementation. Rather than screening more enzyme homologs, in the hopes of finding an enzyme with sufficient activity, we decided to understand why these enzymes failed to function effectively and to modify both the enzymes and host to enable efficient catabolism of these challenging carbon sources.
3.2. Experimental evolution identifies improved variants
To uncover the factors limiting growth with 4-HB, we serially passaged three replicate cultures each of strains JME38 and JME50 for 300 generations in minimal medium containing 1 g/L 4-HB. Initially, cultures were supplemented with 50 mg/L of PCA to allow slight growth of the parental strains. The PCA concentration was reduced over time and eliminated after generation 100. After 300 generations, we isolated eight mutant strains from each replicate culture and measured their ability to grow with PCA and 4-HB. We then selected one representative isolate from each replicate population for further characterization.
All six isolates grew at similar rates with 4-HB (Fig. 2). Two of the isolates showed reduced growth with PCA, as compared to the parental strain. Changes in growth rate with glucose were minor. We then resequenced the genome of each isolate. All six isolates had mutations at the 5′ end of the praI or pobA coding sequence, five of which were silent mutations (Supplementary Table 4). Based on sequence coverage, all three pobA mutants were predicted to have duplications of the pcaHGBDC operon. Conversely, all three praI mutants had unique, non-synonymous mutations in the gene encoding the PCA/4-HB transporter, pcaK. Each isolate had one or two additional mutations in the genome that were unique to that isolate.
Fig. 2.
Evolved strains grow efficiently with 4-HB. Isolates from each of three replicate populations were assayed for growth in minimal medium with 2 g/L glucose, 1 g/L PCA, or 1 g/L 4-HB. Error bars show one standard deviation, calculated from three biological replicates.
For verification, we used quantitative PCR to measure the copy number of pcaH in the parental and evolved mutants of JME38, since this is the first enzyme in the operon and PcaH and PcaG together catalyze the first step in PCA degradation (Fig. 1). The relative copy number of pcaH was two- to three-fold higher in the evolved isolates relative to the parental strain (Supplementary Fig. 3A). To understand the effects of the pcaHGBDC duplication, we measured expression levels of the entire proteome in both the parental and evolved isolates (Supplementary Fig. 3B). Consistent with our previous observations (Clarkson et al., 2017) and the qPCR results, we saw increased expression of PcaH and PcaG in the evolved strain (p < 0.002). Although all five genes form a single operon, duplication of pcaHGBDC predominantly increased abundance of only PcaH and PcaG. We hypothesize that another factor, such as aggregation-dependent proteolysis, is limiting expression of the remaining genes in the operon.
3.3. Reconstruction identifies lineage-specific causal mutations
We introduced the silent praI and T388A pcaK mutations from JME73, individually and in combination, into JME50 to verify their effects. Neither mutation alone was sufficient for growth with 4-HB, while the double mutant was able to grow efficiently (Fig. 3A). Similarly, we tested the silent pobA mutation from JME70 and an engineered duplication of the pcaHGBDC operon, alone or in combination. Only the double mutant was able to grow with 4-HB (Fig. 3B). To understand why we saw different evolutionary outcomes in the two lineages, we then swapped the pcaK and pcaHGBDC mutations. The pcaK mutation supported rapid growth of a strain with pobA, while the pcaHGBDC duplication had minimal effect on a strain with praI (Fig. 3C). Since only two mutations were necessary to recover growth with 4-HB, we conclude that the additional mutations observed in the evolved isolates had relatively minor fitness effects.
Fig. 3.
Two mutations are necessary for efficient growth with 4-HB. (A) Strains with either the wild-type or mutant alleles of praI and pcaK were grown with PCA or 4-HB as the sole source of carbon and energy. Mutation numbers for praI and pobA correspond to the strain from which those mutations were identified, as shown in Fig. 4. (B) As in A, except using strains with either the wild-type or mutant allele of pobA and one or two copies of pcaHGBDC. (C) As in A, except using strains with the mutant alleles of praI or pobA and either the mutant allele of pcaK or a second copy of pcaHGBDC. Error bars show one standard deviation, calculated from three biological replicates.
To understand the effect of the pcaHGBDC duplication, we measured the growth rate of strains JME17, with one copy of pcaHGBDC, and JME146, with a second copy, across a range of PCA concentrations (Supplementary Fig. 4). While the growth rate of the two strains was similar at high PCA concentrations, JME17 grew more slowly at low PCA concentrations.
3.4. Silent mutations increase expression of heterologous proteins
Mutations to pobA and praI that do not affect the primary structure of the protein were necessary for growth with 4-HB. To understand the effects of these mutations, we measured the expression of PobA and PraI in strains with wild-type and mutant alleles. In both cases, the mutations increased expression of the monooxygenase by more than three-fold (Fig. 4, p < 0.01). We expect that the remaining silent mutations have similar effects.
Fig. 4.
Silent mutations increase protein expression. All six evolved isolates had mutations to the 5′ end of the coding region of the monooxygenase gene, five of which were silent. The silent mutations present in JME70 and JME73 were introduced individually into the appropriate parental strains, and enzyme expression was measured by global proteomics. Each silent mutation significantly increased the abundance of the associated enzyme. Error bars show one standard deviation, calculated from three biological replicates.
Secondary structures at the 5′ end of an mRNA can decrease protein abundance (Kudla et al., 2009, Boël et al., 2016). We calculated the predicted folding energy of the 5′ UTR and first 54 coding nucleotides for each of the wild-type and mutant expression constructs. Each of the mutations increased the predicted local mRNA folding energy, i.e. destabilized the mRNA folding, by 1.4–4.1 kcal/mol. Changes in folding energy at this scale have been shown to be capable of producing changes in protein abundance of approximately three-fold (Kudla et al., 2009). The parallelism in mutations between pobA and praI likely results from a combination of high local amino acid identity combined with codon optimization. While the enzymes are 54% identical across their entire length, the first 18 amino acids are 78% identical. Codon optimization for E. coli eliminated any pre-existing nucleotide variation and yielded nucleotide sequences that were 83% identical across this region.
3.5. Mutations to pcaK affect transport of 4-HB
To understand how mutations to pcaK affect growth with 4-HB, we studied one particular mutation, T388A, in more detail. Expression of membrane proteins in a heterologous host can be difficult, and mutations to the protein may improve expression. Therefore, we measured changes in the expression of PcaK in otherwise-isogenic strains containing either the wild-type or mutant alleles of pcaK. Expression of PcaK was roughly two-fold higher for the mutant allele (Fig. 5A, p < 0.001).
Fig. 5.
A mutation topcaKaffects transport of 4-HB. (A) PcaK abundance was measured in two strains that differ only in a point mutation (T388A) to pcaK. (B) The intracellular concentrations of PCA and 4-HB were measured by GC/MS in strains with no transporter, the wild-type transporter, or the mutant transporter. None of the strains have a 4-HB monooxygenase or the pcaHGBDC operon, and therefore are unable to catabolize PCA or 4-HB. (C) Strains JME17 and JME87 were grown with PCA as the sole source of carbon and energy in media containing various concentrations of 4-HB. The two strains differ by a single point mutation to pcaK in JME87. Neither strain contains a 4-HB monooxygenase, preventing catabolism of 4-HB. Error bars in A-C show one standard deviation, calculated from three biological replicates. Lines in C are a guide for the eye.
We next sought to understand the effect of this mutation on transport. We first measured the intracellular concentration of PCA and 4-HB in strains that can import both compounds but cannot metabolize either. Relative concentrations were measured using GC/MS in cells exposed for 5 min to 1 g/L of PCA or 4-HB, then quickly harvested by centrifugation and washed. We found that the mutant transporter led to increased accumulation of both PCA and 4-HB, relative to the wild-type transporter, and that the increase was greater for 4-HB (Fig. 5B). No significant accumulation was seen in cells lacking PcaK. PCA and 4-HB compete for transport by PcaK, so the addition of 4-HB can prevent the import of PCA (Nichols and Harwood, 1997). Using strains that can import both PCA and 4-HB but can metabolize only PCA, we measured the inhibitory effect of 4-HB (Fig. 5C). In the strain with a wild-type copy of pcaK, the addition of 4-HB has no effect. However, 4-HB prevents growth with PCA in the pcaK mutant. No inhibitory effect was seen for either strain during growth with glucose (Supplementary Fig. 5). In combination, these results are consistent with the hypothesis that growth inhibition with PCA in the pcaK mutant was due to competition for transport rather than inherent toxicity of 4-HB. Two other mutations to pcaK were observed, L276Q and S207N, and the evolved strains containing these mutations grew more slowly with PCA than the parent. We hypothesize that these mutant transporters similarly increase import of 4-HB, but at the expense of decreasing import of PCA. Consistent with this hypothesis, the appearance of the pcaK mutations in populations B and C coincided with a decrease in the growth rate with PCA (Supplementary Table 5).
3.6. Evolutionary trajectories match physiological effects
Mutations to the pcaK transporter are beneficial in strains containing pobA, yet were not observed in our isolates. In order to estimate the order of occurrence of the mutations, we measured the growth rates, mutation frequencies by Sanger amplicon sequencing, and pcaH copy number by qPCR of mixed populations saved at generations 50, 100, 200, and 300 (Supplementary Table 5 and Supplementary Fig. 6). The mutations to pobA and praI are difficult to track by Sanger sequencing of mixed amplicons, since we expect that multiple mutations could produce similar effects, leading to soft selective sweeps (Lee et al., 2013).
In replicate populations of strain JME38, the pcaH duplication fixed before any single mutation in pobA rose to an observable frequency. In replicate population A of this strain, the pcaH duplication fixed even before the population was able to grow with 4-HB, likely due to increased growth with PCA during the initial selection with 0.1 g/L PCA and 1.0 g/L 4-HB. Similarly, in replicate populations of JME50, mutations to pcaK are observed earlier than mutations to praI. However, in contrast to duplication of pcaHGBDC, mutations to pcaK represent a tradeoff between growth with PCA and growth with 4-HB. Consequently, we saw mutations to pcaK occur only in populations that had also acquired the ability to grow with 4-HB.
Additionally, insertion-sequence-mediated duplications, such as those seen with pcaHGBDC, occur at much higher frequencies than specific point mutations (Andersson and Hughes, 2009). While strains with pobA can benefit from either mutations in pcaK or duplication of pcaHGBDC, we would expect the duplication to occur more often and therefore rise to fixation before a point mutation to pcaK would likely arise. Conversely, strains with praI were unable to benefit from the frequent duplications and instead waited for the less common mutations to pcaK. The observance of independent parallel mutations suggests that potentially-beneficial mutations or duplications were common, and therefore that a combination of pcaHGBDC duplication and pcaK mutation occurred in the same strain. Since the pobA/pcaK/pcaHGBDC triple mutant did not enrich in our populations, we conclude that it likely has a small fitness benefit under the experimental conditions.
3.7. Functional accommodation to enzyme homologs can be homolog-specific
Two different genetic solutions can rescue growth with 4-HB in the original engineered strains, either mutations in pcaK or duplication of pcaHGBDC. We have shown that the mutations to pcaK increase both the expression of PcaK and its affinity for 4-HB. Conversely, duplication of pcaHGBDC increased expression of PcaHG and sustained rapid growth at lower concentrations of PCA. In combination, these results suggest that PcaHG has low catalytic activity in E. coli. When the internal PCA concentration is high, such as during growth with 1 g/L of PCA, the level of PcaHG activity present in JME17 is sufficient. However, at low external PCA concentrations or when the PCA is produced intracellularly by oxidation of 4-HB, the intracellular concentration will be much lower. The two genetic solutions, therefore, represent two separate biochemical solutions to increase the flux from 4-HB to PCA, either increasing transport of 4-HB or increasing expression of PcaHG (Fig. 6).
Fig. 6.
Homologous enzymes require different solutions to enable rapid growth with 4-HB. A model is shown in which mutations to the PcaK transporter increase the internal 4-HB concentration (vertical dashed lines), while modifications to the core PCA catabolism pathway allow growth at lower PCA flux (horizontal dashed lines indicate minimal flux to sustain growth). The mutant transporter thus allows growth with any combination of monooxygenase and pcaHGBDC copy number (green circles). The wild-type transporter cannot supply enough 4-HB to allow PraI to support growth (red circle). However, the wild-type transporter, in combination with PobA, can support growth only if the pcaHGBDC duplication accommodates a reduced flux to PCA (yellow circle). Semi-log Michaelis-Menten curves for PobA and PraI are drawn for illustrative purposes.
Increased transport of 4-HB is sufficient for strains with either pobA or praI. Increased transport would act to raise the intracellular 4-HB concentration and these results suggest that, at a higher 4-HB concentration, both enzymes are capable of generating sufficient flux to PCA to support rapid growth. Conversely, increased expression of PcaHG lowers the threshold for flux to PCA. Increased expression of PcaHG rescues only pobA strains, suggesting that PraI might have a higher KM for 4-HB and be unable to meet even this reduced threshold using the wild-type PcaK.
4. Conclusions
We have successfully engineered strains of E. coli that grow efficiently with both 4-HB and PCA, using different homologous 4-HB monooxygenases. These strains provide the foundation for effective processes to valorize lignin through initial catabolism of complex mixtures of phenolics. Our results demonstrate that standard engineering strategies can confound bioprospecting attempts by introducing shared features such as similar RNA structures. Accordingly, we showed that silent mutations to the 5′ end of the codon-optimized genes encoding 4-HB monooxygenases increased protein expression by approximately three-fold. However, increasing enzyme expression alone was not sufficient to support growth. After making this change, we demonstrated that the effects on enzyme activity of mutations to the host can differ between homologs. Specifically, we identified mutations to the PcaK transporter that support growth using either the PobA or PraI enzymes, while duplication of the core PCA pathway only rescued growth with PobA. Using these methods to rapidly identify the best enzyme homolog for a given pathway, or the best genetic context for a given homolog, will greatly speed the process of metabolic pathway construction.
Acknowledgments
Acknowledgements
Genome resequencing and analysis was performed by Christa Pennacchio, Natasha Brown, Anna Lipzen, and Wendy Schackwitz at the Joint Genome Institute.
Acknowledgments
Funding sources
This work was supported by the BioEnergy Science Center and Center for Bioenergy Innovation, both U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the DOE under Contract No. DE-AC05-00OR22725. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2018.04.002.
Appendix A. Supplementary material
Supplementary material
References
- Alonso-Gutierrez J. Towards industrial production of isoprenoids in Escherichia coli: lessons learned from CRISPR-Cas9 based optimization of a chromosomally integrated mevalonate pathway. Biotechnol. Bioeng. 2018;115(4):1000–1013. doi: 10.1002/bit.26530. [DOI] [PubMed] [Google Scholar]
- Andersson D.I., Hughes D. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 2009;43(1):167–195. doi: 10.1146/annurev-genet-102108-134805. [DOI] [PubMed] [Google Scholar]
- Bayer T.S. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009;131(18):6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- Boël G. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529(7586):358–363. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg T.D.H., Ahmad M., Hardiman E.M., Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011;28(12):1883. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- Chen Y.-J. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013;10(7):659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- Clarkson S.M., Kridelbaugh D.M., Elkins J.G., Guss A.M., Michener J. Construction and optimization of a heterologous pathway for protocatechuate catabolism in Escherichia coli enables rapid bioconversion of model lignin monomers. Appl. Environ. Microbiol. 2017;83(18):e01313–e01317. doi: 10.1128/AEM.01313-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney N.F. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One. 2013;8(4):e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349(6252):1095–1100. doi: 10.1126/science.aac9373. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C.S., Parales R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996;50(1):553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- Jiang M., Fang L., Pfeifer B.A. Improved heterologous erythromycin A production through expression plasmid re-design. Biotechnol. Prog. 2013;29(4):862–869. doi: 10.1002/btpr.1759. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81(7):2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J.I., Minambres B., Garcia J.L., Diaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 2002;4(12):824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- Kasai D. Uncovering the protocatechuate 2,3-cleavage pathway genes. J. Bacteriol. 2009;191(21):6758–6768. doi: 10.1128/JB.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Copley S.D. Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. Proc. Natl. Acad. Sci. USA. 2012;109(42):E2856–E2864. doi: 10.1073/pnas.1208509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2013;110(34):14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924) doi: 10.1126/science.1170160. 〈http://science.sciencemag.org/content/324/5924/255.full〉 (80-, Accessed 10 August 2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-C., Marx C.J., Treves D.S., Adams J. Synchronous waves of failed soft sweeps in the laboratory: remarkably rampant clonal interference of alleles at a single locus. Genetics. 2013;193(3):943–952. doi: 10.1534/genetics.112.148502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger J.G. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. USA. 2014;111(33):12013–12018. doi: 10.1073/pnas.1410657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z.-Q. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009;8(8):3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener J.K., Nielsen J., Smolke C.D. Identification and treatment of heme depletion attributed to overexpression of a lineage of evolved P450 monooxygenases. Proc. Natl. Acad. Sci. USA. 2012;109(47):19504–19509. doi: 10.1073/pnas.1212287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narcross L., Bourgeois L., Fossati E., Burton E., Martin V.J.J. Mining enzyme diversity of transcriptome libraries through DNA synthesis for benzylisoquinoline alkaloid pathway optimization in yeast. ACS Synth. Biol. 2016;5(12):1505–1518. doi: 10.1021/acssynbio.6b00119. [DOI] [PubMed] [Google Scholar]
- Nichols N.N., Harwood C.S. PcaK, a high-affinity permease for the aromatic Compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 1997;179(16):5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164(6):1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Ragauskas A.J. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;344(6185):1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Rodriguez A. Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustain. Chem. Eng. 2017;5(9):8171–8180. [Google Scholar]
- Strachan C.R. Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. USA. 2014;111(28):10143–10148. doi: 10.1073/pnas.1401631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D.L., Fernando C.G., Chambers M.C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pfeifer B.A. 6-Deoxyerythronolide B production through chromosomal localization of the deoxyerythronolide B synthase genes in E. coli. Metab. Eng. 2008;10(1):33–38. doi: 10.1016/j.ymben.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Yim H. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011;7(7):445–452. doi: 10.1038/nchembio.580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material