Abstract
The unicellular heterotrophic thraustochytrids are attractive candidates for commercial polyunsaturated fatty acids (PUFA) production. However, the reactive oxygen species (ROS) generated in their aerobic fermentation process often limits their PUFA titer. Yet, the specific mechanisms of ROS involvement in the crosstalk between oxidative stress and intracellular lipid synthesis remain poorly described. Metabolic engineering to improve the PUFA yield in thraustochytrids without compromising growth is an important aspect of economic feasibility. To fill this gap, we overexpressed the antioxidative gene superoxide dismutase (SOD1) by integrating it into the genome of thraustochytrid Schizochytrium sp. PKU#Mn4 using a novel genetic transformation system. This study reports the ROS alleviation, enhanced PUFA production and transcriptome changes resulting from the SOD1 overexpression. SOD1 activity in the recombinant improved by 5.2–71.6% along with 7.8–38.5% decline in ROS during the fermentation process. Interestingly, the total antioxidant capacity in the recombinant remained higher than wild-type and above zero in the entire process. Although lipid profile was similar to that of wild-type, the concentrations of major fatty acids in the recombinant were significantly (p ≤ 0.05) higher. The PUFA titer increased up to 1232 ± 41 mg/L, which was 32.9% higher (p ≤ 0.001) than the wild type. Transcriptome analysis revealed strong downregulation of genes potentially involved in β-oxidation of fatty acids in peroxisome and upregulation of genes catalyzing lipid biosynthesis. Our results enrich the knowledge on stress-induced PUFA biosynthesis and the putative role of ROS in the regulation of lipid metabolism in oleaginous thraustochytrids. This study provides a new and alternate strategy for cost-effective industrial fermentation of PUFA.
Keywords: Polyunsaturated fatty acids, Schizochytrium sp., Superoxide dismutase, Transgene, Transcriptomic analysis
Highlights
-
•
SOD1 gene overexpressed in Schizochytrium sp. PKU#Mn4 for enhanced PUFA production.
-
•
Recombinant strain yielded 1.37-fold higher PUFA production than wild-type strain.
-
•
ROS alleviation and increased T-AOC in recombinant strain without affecting growth.
-
•
Probing transcriptome revealed strong downregulation of fatty acids β-oxidation.
-
•
Most downregulated genes were peroxisomal suggesting reduced β-oxidation of C22:6.
1. Introduction
As a significant component of cellular membranes, polyunsaturated fatty acids (PUFA) are known to play vital structural and functional roles in humans including regulation of immunity (Armenta and Valentine, 2013) and coronary heart disease (Lopez-Huertas, 2010). A typical ω–3 PUFA – docosahexaenoic acid (DHA, C22:6) – has been recommended to prevent coronary heart disease, hypertension, diabetes, renal disease, rheumatoid arthritis and other diseases (Simopoulos, 1997). With an increasingly better understanding of the health benefits of PUFA, their demand is rising enormously. Although fish oils have long been the main source of PUFA (Pike and Jackson, 2010), their supply is largely limited by fish stocks (Lahsen and Iddya, 2014). The risks of potential contamination, unpleasant odors, and multifarious purification procedures further impact the fish oils market (de Oliveira Finco et al., 2017). Therefore, microbial sources of PUFA are currently being developed as an alternative source (Shi et al., 2017). Of these, thraustochytrids, a unicellular heterotrophic fungus-like marine protist (Leyland et al., 2017), are becoming an increasingly promising source of PUFA on account of their fast growth rate and high content of DHA (Aasen et al., 2016).
Much work focused on the thraustochytrid culture optimization demonstrated significant improvements in their biomass and DHA yields (Chen et al., 2016, Jiang et al., 2017, Ludevese-Pascual et al., 2016, Lung et al., 2016, Patil and Gogate, 2015, Zhao et al., 2017, Zhu et al., 2008). The biomass can reach up to 171.5 g/L under the optimal cultivation condition (Bailey et al., 2003), and with modified medium the total fatty acids (TFA) could account for 83.84% of biomass (Li et al., 2015) with PUFA content as high as 76.5% (Marchan et al., 2017). Interestingly, from these studies, it was evident that cell proliferation and fatty acids (FA) yield are enhanced under an oxygen-rich environment (Jakobsen et al., 2008, Ren et al., 2010); however, PUFA gets easily oxidized due to their high degree of unsaturation (Else and Kraffe, 2015, Guichardant et al., 2011). Thus, lipid peroxidation not only reduces the content of PUFA but also activates the accumulation of high levels of reactive oxygen species (ROS) (Johansson et al., 2016, Ruenwai et al., 2011). Besides lipids, these ROS cause severe oxidative damage to proteins and DNA resulting in the loss of protein function and even cell death (Shi et al., 2017). However, they could be scavenged by different antioxidative defense components under a steady physiological state (Finkel and Holbrook, 2000). Both non-enzymatic and enzymatic mechanisms have been implicated in the detoxication process of ROS (Apel and Hirt, 2004, Gupta et al., 2013, Sun et al., 2016). Based on these mechanisms, several strategies have been adopted to enhance removal of ROS (Apel and Hirt, 2004).
Apart from the external addition of antioxidants (Ren et al., 2017), overexpression of antioxidative enzymes also seems to be an alternative way of alleviating ROS. Of these enzymes, superoxide dismutase (SOD) acts as the first line of defense against ROS (Yanase et al., 2009) and converts superoxide radical (O2-) into hydrogen peroxide (H2O2). Various transgenic plants that expressed increased amounts of SOD have been developed in past to better tolerate the oxidative stress (Tang et al., 2006, Zambounis et al., 2002). Overexpression of sod1 gene and sod2 gene in Streptomyces peucetius enhanced secondary metabolites as a result of the increased transcriptional level of regulatory genes and biomass increment (Bashistha et al., 2011). Although metabolic engineering seemed an effective and promising way to alleviate ROS, no attempts have yet been made, to our knowledge, which demonstrates the effect of oxidative stress in thraustochytrids at the molecular level. Despite the knowledge of a versatile transformation system for thraustochytrids (Hong et al., 2013, Sakaguchi et al., 2012, Sun et al., 2015), only a few studies were focused on the improvement of FA productivity through gene manipulation (Table S1). Up to now, there are no comprehensive studies focused on ROS quenching to enhance PUFA productivity in thraustochytrids.
To elucidate the effect of oxidative stress alleviation on PUFA production, we first designed a genetic transformation system for a previously isolated high PUFA yielding thraustochytrid strain – Schizochytrium sp. PKU#Mn4 (Liu et al., 2014). In an attempt to alleviate oxidative stress, we integrated the SOD1 gene overexpression cassette driven by a newly isolated promoter into the PKU#Mn4 genome at the 18S rDNA loci. The recombinant strain exhibited increased PUFA production resulting from ROS alleviation and increased total antioxidant capacity (T-AOC). Further transcriptomic analysis revealed strong regulation of key genes implicated in the FA metabolism. This study highlights an alternative direction for enhanced PUFA productivity in thraustochytrids and provides the framework for a cost-effective industrial fermentation process.
2. Materials and methods
2.1. Strains and culture conditions
Schizochytrium sp. PKU#Mn4 (GenBank accession number JX847360), previously isolated from mangrove leaves from Pearl River Delta region of China (Liu et al., 2014), was used in this study. The strain was maintained on 2% modified Vishniac's (MV) agar plates (Damare and Raghukumar, 2006) prepared with artificial seawater (ASW) (Nagano et al., 2009) at room temperature and subcultured every 4 weeks. The seed culture was prepared by cultivating a single colony from the agar plate in M4 medium (20 g/L glucose, 0.25 g/L KH2PO4, 1.5 g/L peptone, 1 g/L yeast extract, dissolved in ASW, pH = 7) (Jain et al., 2005) at 28 °C on an orbital shaker at 170 rpm for 36 h. The seed culture was subsequently transferred to 100 mL shake flask containing 40 mL fresh M4 medium at a final concentration of 5%, which was then cultivated on an orbital shaker for 4 days under the same conditions until harvest.
2.2. Construction of EGFP and superoxide dismutase (SOD) expression cassettes
We identified a polyubiquitin promoter region with the length of 1484 bp in the whole genome dataset of Schizochytrium sp. PKU#Mn4. The egfp gene was expressed under this promoter region. Polyubiquitin promoter was amplified from PKU#Mn4 genomic DNA using primers pup-F2 and pup-gfp-R, egfp gene was amplified from PX458 (plasmid #48138, Addgene, USA) with primers pup-gfp-F and gfp-cyc-R, and the CYC1 terminator was amplified from pGAPZαA (Invitrogen, USA) with gfp-cyc-F and cyc-sc-R (Table S2). The three fragments (polyubiquitin promoter, egfp gene, and CYC1 terminator) were then ligated seamlessly by Overlap Extension PCR with the outer primer pair pup-FU-BamHI/cyc-FU-SalI and designated PGC. Vector pMG201M (Dubeau et al., 2009), containing the neomycin-resistant gene (neor), was digested with BamHI to remove the codA gene and designated pNeoR. The PGC segment was subcloned into pNeoR at the BamHI and SalI site and designated pNeoR-PGC, which was used for transformation and EGFP gene expression (Fig. S1a). To confirm the fragments obtained by fusion PCR, they were ligated to pEASY-Blunt vector (TransGen Biotech Inc., Beijing) and transformed into Escherichia coli DH5α (Tiangen Inc, Beijing). The plasmid was extracted using TIANprep Mini Plasmid Kit (TIANGEN Inc., Beijing) and sequenced with primers M13F and M13R at AuGCT Inc., Beijing.
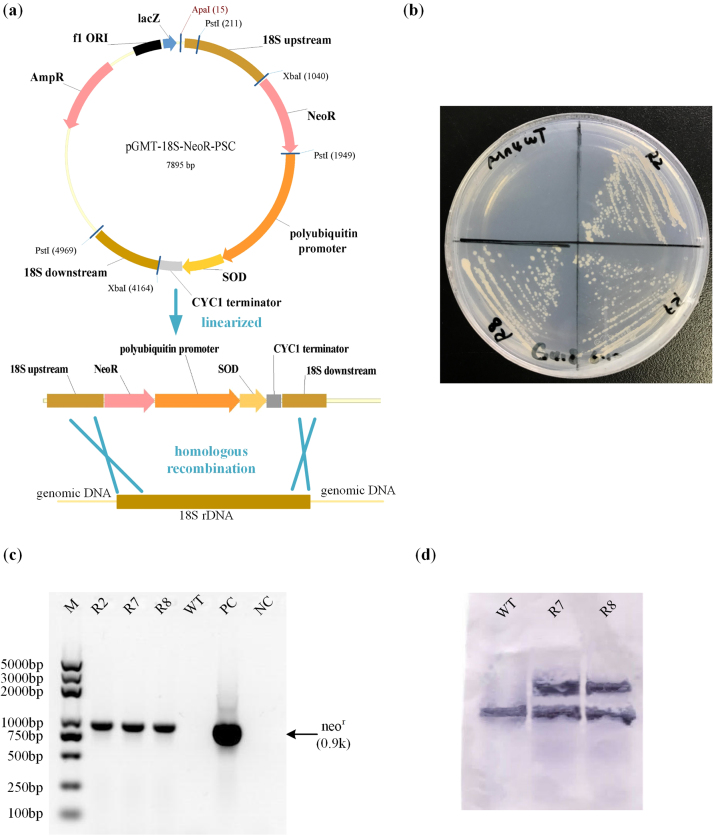
The SOD overexpression cassette was integrated into the genome by 18S rDNA homologous recombination. The sod1 gene was amplified from PKU#Mn4 genome with pup-sod-F and sod-cyc-R and was ligated seamlessly with polyubiquitin promoter and CYC1 terminator by Overlap Extension PCR. The 18S rDNA fragment of PKU#Mn4 was amplified using primers 18S001 and 18S13 (Honda et al., 1999) and cloned into pGM-T vector (Tiangen, China). The plasmid was designated pGMT-18S. The insert DNA was confirmed by sequencing (AuGCT Inc., Beijing) pGMT-18S with primers M13F and M13R. The neomycin resistant gene (neor) was amplified from pNeoR with primers NeoR-F-XbaI and NeoR-R-PstI. The neor gene and sod1 expression cassette were cloned into pGMT-18S and designated pGMT-18S-NeoR-PSC (Fig. 2b). The resulting PCR fragments were confirmed by sequencing as described above. The vector pGMT-18S-NeoR-PSC was linearized with ApaI REase digestion before transformation. The PCR protocols used in this study are provided in Table S3.
Fig. 2.
Overexpression in Schizochytrium sp. PKU#Mn4. (a) Scheme of integration of sod1 expression cassette into the genome of Mn4, (b) G418 - resistant phenotype of recombinants generated by linearized 18S rDNA-targeted homologous recombination structure pGMT-18S-NeoR-PSC, (c) Genomic PCR for detection of integration in the recombinants. Lanes: M, Trans2K plus marker; R2, R7, R8, Mn4-SOD recombinant R2, R7 and R8, respectively; WT, Mn4 wild-type; PC, positive control (pNeoR); NC, no template control, (d) Southern blot detecting sod1 in wild-type strain and recombinants with a gene-specific DIG-labeled probe. Wild-type shows the original one copy of sod1 on the genome, while two hybridized bands are seen in the two recombinants.
2.3. Preparation of competent cells, transformation, and screening
The seed culture, prepared as above, was transferred to fresh M4 medium and grown to the exponential phase (OD660 = 7–8). One mL culture was centrifuged (4000× g, 4 °C, 10 min) and the resulting cell pellet was washed twice with the ice-cold sterile water, then treated with 25 mM DTT in 10 mM PBS for 10 min and washed with 1 M ice-cold sterile sorbitol to remove the remaining DTT. The cells were then placed into 2 mL enzyme medium and incubated at 28 °C for 5 h to further weaken the cell wall. The enzyme medium (Cheng et al., 2012) with modification, containing 0.7 M KCl, 20 g/L pectinase, and 20 g/L snailase, was prepared with 10 mM PBS and sterilized by filtration. Cells were then collected from enzyme medium by centrifugation and washed twice with 1 M ice-cold sterile sorbitol to remove the remaining medium. A 250 μL aliquot of ice-cold sterile sorbitol was added to the resuspended competent cells. Approximate 5 × 106 competent cells were mixed with around 2 μg circular or linearized plasmids in a 0.1 cm cuvette for electroporation. Electroporator (Bio-Rad, Gene Pulser Xcell, USA) were set to exponential decay, 1000 V, 50 μF and 500 Ω.
One mL fresh M4 medium was added immediately after electroporation and the resulting cells were incubated at 28 °C for 20 h before plating on agar plates containing antibiotics. An aliquot of the culture was spread on MV agar plates containing 0.8 mg/mL G418, which was then incubated at 28 °C for 3–5 days to obtain visible colonies. The colonies resistant to G418 were isolated and cultured for subsequent analysis. The putative transformants carrying EGFP and SOD1 expression cassette were identified by fluorescence assay and genomic PCR analysis, respectively. For fluorescence assay, putative transformants were directly observed under the LSM880 confocal laser microscope (Zeiss, Germany). For genomic PCR analysis, putative transformants cultivated in M4 medium containing 0.8 mg/mL G418 were harvested by centrifugation and the genomic DNA (gDNA) was extracted with the CTAB method using a gDNA extraction kit (Generay, China). The integrity of gDNA was checked by electrophoresis and PCR was performed to amplify neor with primers NeoR-F and NeoR-R (Table S2) using gDNA as the template.
2.4. Southern blot analysis
The gDNA (~2 μg) extracted from wild-type or recombinant cells was digested with the HindIII REase, separated on 0.8% agarose gel, and then transferred onto Hybond N+ nylon membranes (Amersham, United Kingdom). A DIG-labeled DNA probe was synthesized at AuGCT Co. Ltd, China. Hybridization and detection were performed using DIG Easy Hyb Granules and DIG Nucleic Acid Detection Kits (Roche, Germany) following instructions of the manufacturer.
2.5. Fatty acid analysis
Cells were collected by centrifugation (4000× g, 4 °C, 10 min) and washed twice with distilled water followed by lyophilization for 24 h. The dry cell weight was determined by the gravimetric method. Fatty acid methyl esters (FAME) were prepared as described previously (Liu et al., 2014) with minor modification. Briefly, approximately 50 mg lyophilized cells were weighed, mixed with 2 mL of 4% sulfuric acid in methanol (v/v), 10 μL BHT (100 μg/mL hexane) and 100 μL nonadecanoic acid (1 mg/mL hexane), vortexed for 20 s and incubated at 80 °C for 1 h. After transesterification, 1 mL of hexane was added when the mixture was cooled to room temperature. The upper hexane layer with FAME was then washed with 1 mL 5% NaCl (w/v) and 1 mL 2% KHCO3 (w/v) sequentially. The hexane layer was centrifuged, collected and dried by passing nitrogen gas. The FAME samples were resolved in 1 mL hexane and analyzed with a 7890 Gas Chromatograph (Agilent Technologies, USA) system equipped with a flame ionization detector and DB-23 capillary column (60 m × 0.25 µm × 0.32 µm) (Agilent Technologies, USA) with nitrogen as the carrier gas. One μL of FAME samples were injected under split mode at the ratio of 50:1 keeping the inlet at 250 °C. An internal standard nonadecanoic acid (C19:0) was used for quantification of FA. All the analyses were performed in triplicates.
2.6. Quantitative real-time PCR (RT-qPCR)
The total RNA of culture sample was isolated using the RNAsimple total RNA kit following the manufacturer's protocol (Tiangen, China) and quantified by NanoDrop 2000c (Thermo Fisher Scientific, USA). Genomic DNA was removed with gDNase and cDNA was synthesized using random hexamers with FastKing RT kit (Tiangen, China). Quantitative PCR assays were performed in triplicate with the CFX Connect Real-Time PCR system (Bio-Rad, USA) using the CharmQ SYBR qPCR master mix (Vazyme, China). Gene-specific primers were designed for superoxide dismutase [Cu-Zn] (sod1) and reference gene β-actin (actb) (Table S2). PCR amplification was carried out in a 10 μL reaction volume containing 5 μL qPCR master mix, 0.25 μL each primer (10 μM), 1 μL cDNA and 3.5 μL nuclease-free water. The PCR program was set to 95 °C for 3 min for activation followed by 40 cycles of amplification step at 95 °C for 10 s, 55 °C for 20 s and then 72 °C for 10 s. The expression level of sod1 mRNA relative to β-actin was calculated following the 2-ΔΔCt method (Livak and Schmittgen, 2001). The amplification efficiencies for sod1 and reference gene were 102.25% and 105.32%, respectively, and melt curve analysis showed a single peak for each of these genes (Fig. S2).
2.7. Analysis of superoxide dismutase activity
For SOD activity, an aliquot of the culture was collected and the cells were centrifuged as described above. After washing twice with distilled 10 mM PBS, the cells were crushed by liquid nitrogen grinding. The powdered cells were resuspended in 1 mL of 10 mM PBS and centrifuged (13,400 × g, 4 °C, 30 min). According to the manufacturer's instruction, the inhibition rate of WST-1 formazan formation should range from 40% to 60% to ensure an accurate measurement. Thus, we diluted the supernatant appropriately to ensure the accuracy. The diluted supernatant (20 μL) containing crude cytosolic extracts was used for total SOD activity analysis. The activity was examined by a WST-1 method using a superoxide dismutase assay kit (Nanjing Jiancheng Bioengineering Institute, China) following the manufacturer's instructions. The 50% inhibition rate of WST-1 formazan formation in the reaction was defined as one unit. The SOD activity was calculated as follows:
2.8. Determination of total antioxidant capacity (T-AOC)
Cells were collected and crushed as described above for SOD activity assay. The cell powders were collected and resuspended in the extraction buffer provided in the T-AOC assay kit (Solarbio, China) followed by centrifugation (10,000× g, 4 °C, 10 min) to obtain the supernatant for intracellular T-AOC measurement. The fluorescence recovery after photobleaching (FRAP) method was used to measure the T-AOC. The FRAP assay was based on the formation of ferrous tripyridyltriazine (Fe(II)-TPTZ) by a reductant at low pH and absorbance measurement at 593 nm (Benzie and Strain, 1996). The T-AOC was calculated with the standard curve provided in the manufacturer's protocol.
2.9. Determination of intracellular reactive oxygen species
The ROS level in the cell samples was determined using a cell-permeable probe – 2′,7′-Dichloro-dihydro-fluorescein diacetate (DCFH-DA) (Bass et al., 1983). The cells were first treated with DTT and snailase to weaken the rigid cell wall and then incubated with DCFH-DA. The probe DCFH-DA was diluted to 10 μM with 10 mM PBS before adding into the cell aliquot. The cells (ca. 1 × 106 to 2 × 107 cells/mL diluted DCFH-DA) were fully exposed to the probe by incubation at 37 °C for 40 min. The excess probe was washed thrice with 10 mM PBS to ensure only the intracellular ROS was measured. Fluorescence intensity was measured at an excitation wavelength of 488 nm, an emission wavelength of 525 nm, and 450 V gain on a fluorescence Spectrophotometer F97 Pro (Lengguang, China).
2.10. RNA library preparation, sequencing, and RNA-seq data analysis
Messenger RNA was purified from total RNA with poly-T oligo-attached magnetic beads. First-strand cDNA was synthesized using random hexamer primers after fragmentation and second-strand cDNA synthesis was subsequently carried out using RNase H and DNA polymerase I. The overhangs were converted into blunt ends, 3′ ends of the cDNA fragments were adenylated and adaptors were ligated to the fragments. The cDNA fragments with a specific length were separated by agarose gel electrophoresis and purified. The size-selected cDNA fragments were PCR amplified to prepare the library which was then sequenced on an Illumina HiSeq. 2500 platform by Gene Denovo Biotechnology Co. (Guangzhou, China).
Raw reads were filtered by removing the reads with adapter and those containing more than 10% unknown bases or had low quality. Clean reads were then mapped to Ribosomal Database Project with Bowtie 2 (Langmead and Salzberg, 2012) and the reads that belonged to rRNA were removed. The remaining clean reads were mapped to the reference genome – the built-in PKU#Mn4 genome – by TopHat2 (Kim et al., 2013). The transcriptome was assembled with Cufflinks (Trapnell et al., 2012) and Cuffmerge was used to merge assembly of two replicate samples. Differential expression analysis was conducted using the edgeR package. The gene expression level was normalized by using FPKM (fragments per kilobases of transcript per million mapped reads) method. This method is able to eliminate the influence of different gene lengths and sequencing data amount on the calculation of gene expression. The false discovery rate (FDR) was used to determine the p-value threshold in multiple tests. The genes with FDR ≤ 0.05 and the absolute value of log2change fold ≥1 were regarded as differential expression genes (DEGs). Sequence analysis was performed by Gene Denovo Biotechnology Co. (Guangzhou, China).
3. Results
3.1. Validation of promoter function
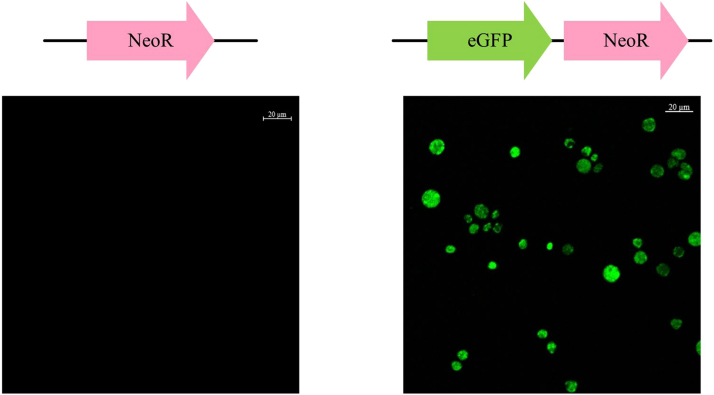
To validate the function of the newly isolated polyubiquitin promoter, the EGFP gene was expressed in the strain PKU#Mn4 with the neor conjugated egfp expression cassette (Fig. S1b). Circular plasmid pNeoR and pNeoR-PGC were introduced into PKU#Mn4 separately and the resulting transformants were observed under a confocal laser microscope. As shown in Fig. 1, only the transformants harboring pNeoR-PGC containing both neor and egfp construct showed green fluorescence. These results suggest that the polyubiquitin promoter was functional in driving heterologous gene expression systems in the strain PKU#Mn4. However, loss of plasmid upon subsequent subcultures into antibiotic-free medium instigated integration of the neor conjugated sod1 expression cassette into the genome of the PKU#Mn4 by 18S rDNA homologous recombination (Fig. 2a).
Fig. 1.
Expression of eGFP in Schizochytrium sp. PKU#Mn4 (bars = 20 µm). neor conjugated egfp expression cassette was introduced to express the eGFP by electroporation and the neomycin resistant gene was introduced as a negative control. Transformants were selected and subsequently observed under the confocal laser microscope. Only the transformants that harbored both neor and egfp show green fluorescence.
3.2. Genome integration and overexpression of SOD1 gene
Genomic PCR amplifying neor was carried out on the G418-resistant colonies (Fig. 2b) to verify the DNA insertion. As shown in Fig. 2c, a PCR product of 900 bp corresponding to the size of neor was amplified in each of the three different recombinants of PKU#Mn4 (R1, R7, R8). Southern blot was carried out with a DIG-labeled sod1 specific probe. Only one hybridized band was detected in the wild type of PKU#Mn4 while two hybridized bands were detected in the recombinants, indicating the successful integration of a copy of SOD1 gene by 18S rDNA homologous recombination (Fig. 2d).
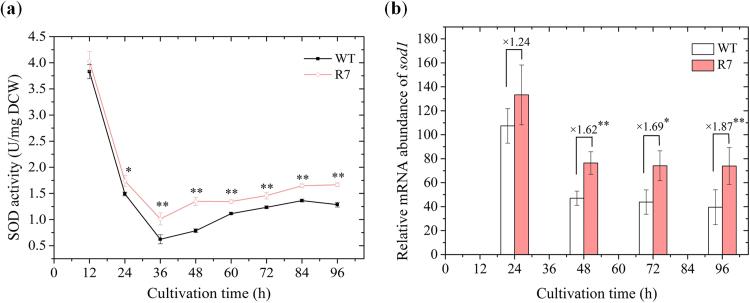
To evaluate overexpression of SOD1 gene in the Schizochytrium sp., SOD activity (Fig. 3a) and relative sod1 mRNA abundance (Fig. 3b) were determined and compared with the wild-type strain (WT). With the peak values at 12 h, the SOD activity in both strains decreased sharply during the exponential phase and increased slightly to the late stationary phase. The SOD activity of recombinant R7 was 5.16–71.65% higher than the wild type (Fig. 3a) during the stationary growth phase. Similarly, the relative abundance of sod1 mRNA in recombinant R7 was significantly greater (1.62- to 1.87- fold) than in wild-type strain during the same phase. These results clearly demonstrated high relative levels of SOD activity and sod1 mRNA abundance in the recombinant strain, further validating the successful integration and function of the sod1 expression system developed in the study.
Fig. 3.
Comparisons of antioxidative parameters. (a) SOD activity and (b) sod1 expression dynamics. * indicates the data with statistical significance at p < 0.05; ** indicates the data with statistical significance at p < 0.01.
3.3. Growth and fatty acid profiles of recombinant strain
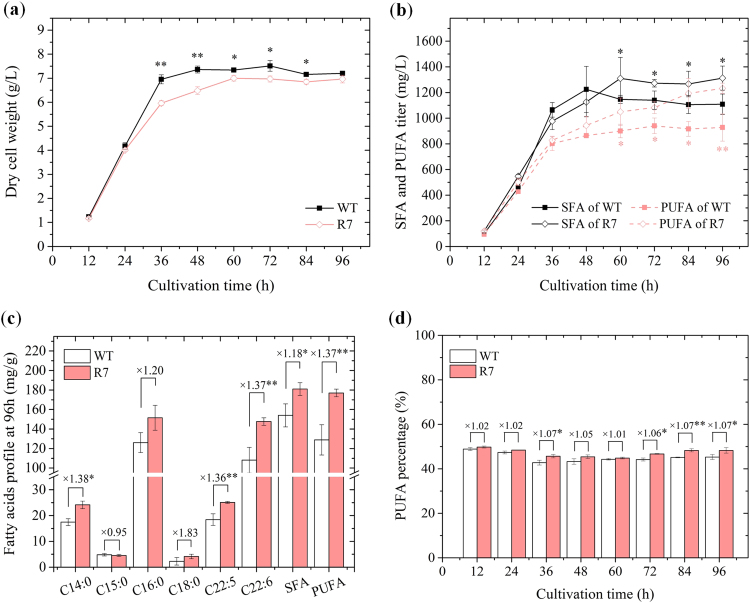
The typical cell growth, FA profile and titer of recombinant strain (R7) were analyzed and compared with those of the wild-type strain (WT) (Fig. 4). The cell biomass of R7 remained slightly lower than that of WT during the entire stationary phase, possibly due to increased metabolic load resulting from sod1 overexpression. Yet, R7 achieved its peak biomass of 6.96 ± 0.16 g/L at 96 h that was only 3.4% lower than the value of WT (Fig. 4a). Saturated fatty acids (SFA) and PUFA titers were significantly higher in R7 relative to WT during the late stationary phase (60–96 h) (Fig. 4b). Their titers at 96 h reached up to 1312 ± 94 mg/L and 1232 ± 41 mg/L, higher by 18.3% and 32.9% than that of WT, respectively. Although the FA composition of R7 and WT were similar, the percent titers of major FA increased significantly (p ≤ 0.05) in R7 culture compared to WT (Fig. 4c). Myristic acid (C14:0) and palmitic acid (C16:0) were the main constituents of SFA, while docosapentaenoic acid (DPA, C22:5) and DHA (C22:6) were the major fatty acids among PUFA. DPA and DHA titers were 25 ± 0.37 mg/g DCW and 147 ± 3.8 mg/g DCW, higher by 36.3% and 36.6% to that of WT, respectively. The average PUFA percentage varied within the range of 44.8 – 49.8% and 42.7 – 48.9% during the entire growth period of R7 and WT, respectively (Fig. 4d). Taken together, the results demonstrated a significant effect of sod1 overexpression on the growth and FA titer of R7, suggesting potential regulation of lipid metabolism.
Fig. 4.
Growth and fatty acid accumulation of SOD overexpression strain R7. (a) Cell growth profile, (b) Titers of SFA and PUFA, (c) Fatty acids profile of Mn4 wild-type and R7 strain after 96 h cultivation, (d) Changes in PUFA percentage. * indicates the data with statistical significance at p < 0.05; ** indicates the data with statistical significance at p < 0.01.
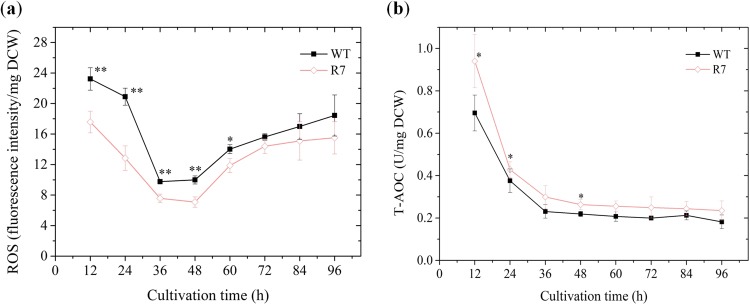
3.4. ROS and T-AOC profiles
We speculated oxidative stress alleviation for the evidence of higher FA production in R7 compared to WT. To validate our hypothesis, we measured the cellular oxidative stress parameters – ROS and T-AOC – at 12 h intervals during the fermentation process (Fig. 5). ROS consistently remained lower in R7 culture compared to WT and decreased by 24.3% at 12 h (start of exponential phase) (Fig. 5a). The dynamics of ROS equated to that of SOD activity, which suggests the dependence of these two parameters on the pattern of intracellular oxygen concentration tightly associated with growth phases. The excess intracellular oxygen when cell growth is minimal or static leads to the generation of ROS, which in turn induces SOD activity that alleviates cell damage. Thus, cell growth profile strongly determines the evolution of these two parameters. In addition to ROS alleviation, the T-AOC of R7 was relatively higher than that of WT and maintained above zero during the entire fermentation process (Fig. 5b). A significantly alleviated ROS in conjunction with increased T-AOC clearly supports our hypothesis and explains the enhanced production of FA in the recombinant strain.
Fig. 5.
Effect of SOD1 overexpression on (a) reactive oxygen species (ROS) and (b) total antioxidant capacity (T-AOC).
3.5. Relative changes in the transcriptome of recombinant strain
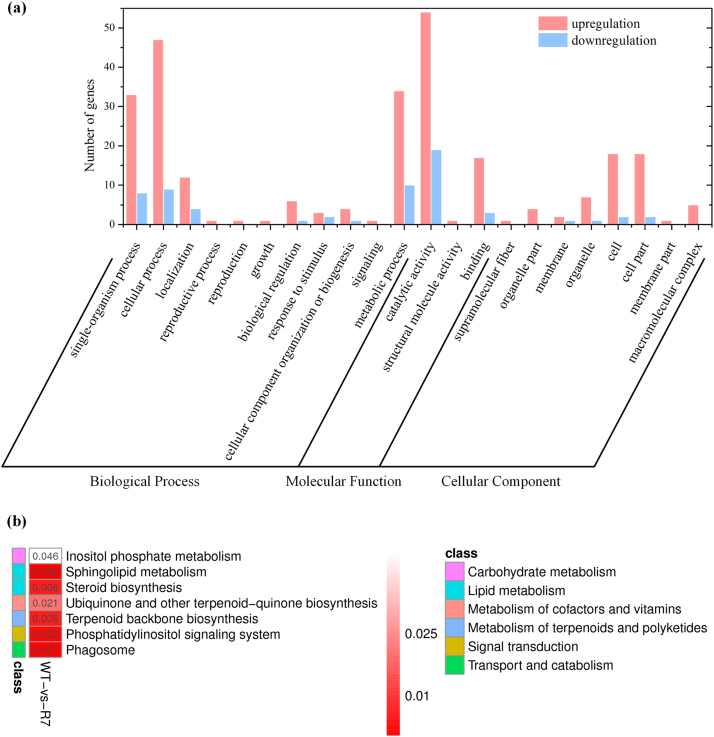
To understand the molecular mechanisms of the enhanced production of FA, we elucidated the transcriptional changes involved in the regulation of FA biosynthesis under SOD overexpression by sequencing a cDNA library prepared from total mRNA of WT and R7 strains using Illumina HiSeq platform. After removing low-quality and rRNA sequences, we obtained 36,193,947 (WT) and 41,451,821 (R7) clean reads, which were further assembled into 13,791 (WT) and 14,286 (R7) unigenes (Table S4). We identified 1962 differentially expressed genes (DEGs) comprising 1347 upregulated and 615 downregulated when q-value ≤ 0.05 and log2 fold change ≥ 1 was applied as the filter condition. A majority of these DEGs were significantly enriched in the biological process, followed by molecular function and cellular component (Fig. 6A). On closer inspection of the transcriptome analysis results, we found several key DEGs directly or indirectly involved in the lipid metabolism (Table 1, Fig. 6B). The putative DEGs (alpha-methylacyl-CoA racemase, 1-acyl-sn-glycerol-3-phosphate acyltransferase, hydroxyacyl-thioester dehydratase, 2-hydroxyacyl-CoA lyase, acetyl-CoA acyltransferase, acyl-CoA oxidase) indicated in the β-oxidation of FA were significantly downregulated while those (acyl-CoA dehydrogenase, 3-ketoacyl-acp reductase, acyl-protein thioesterase 1, acyl-CoA synthetase) involved in the FA biosynthesis were strongly upregulated. In addition, many (alpha-methylacyl-CoA racemase, acyl-CoA oxidase, acetyl-CoA acyltransferase) of these key DEGs were involved in peroxisome pathway (KEGG map04146) (Fig. S3), which indicates their role in FA oxidation and free radical detoxification. These results provide clear evidence of the mechanism underlying the regulation of the FA metabolism that resulted in the higher FA accumulation under SOD1 overexpression.
Fig. 6.
Functional classification of DEGs in the transcriptome of recombinant Schizochytrium sp. PKU#Mn4. (A) Histogram presentation of Gene Ontology (GO) classification (p ≤ 0.001), (B) Significant KEGG orthology (KO) enrichment (p ≤ 0.05).
Table 1.
Differential expression of putative genes associated with fatty acid metabolism in the recombinant strain.
| Gene name | Gene ID | |log2(FC)| |
|---|---|---|
| alpha-methylacyl-CoA racemase | Mn4_18183 | −2.02 |
| 1-acyl-sn-glycerol-3-phosphate acyltransferase | Mn4_02144 | −1.99 |
| Hydroxyacyl-thioester dehydratase | Mn4_07562 | −1.91 |
| 2-hydroxyacyl-CoA lyase 1 | Mn4_10486 | −1.25 |
| Acetyl-CoA acyltransferase | Mn4_07147 | −1.07 |
| Acyl-CoA oxidase | Mn4_05075 | −1.01 |
| Acyl-CoA dehydrogenase | Mn4_14961 | 10.34 |
| 3-ketoacyl-acp reductase | Mn4_15767 | 1.05 |
| Acyl-protein thioesterase 1 | Mn4_05203 | 1.12 |
| Acyl-CoA synthetase | Mn4_05938 | 1.16 |
4. Discussion
4.1. Genetic transformation system and overexpression of SOD1 gene
Schizochytrium sp. is a potential thraustochytrid known for its fast growth rate and high DHA yield (Chang et al., 2013a, Patil and Gogate, 2015, Qu et al., 2013a, Qu et al., 2013b, Sun et al., 2016). Much of the previous work has demonstrated dual roles of oxygen on fermentation process which significantly affects its growth and FA accumulation (Chang et al., 2013a, Chang et al., 2013b, Chang et al., 2014, Chi et al., 2009, Ren et al., 2010, Xie and Wang, 2015). Stepwise oxygen supply strategies have been proposed in the past (Chi et al., 2009, Ren et al., 2010); however, the accurate determination of time point for the shift appeared challenging, which makes the fermentation process difficult to control. Since the ROS mediated oxidative damage under the oxygen-rich fermentation was one of the reasons that cause deterioration of PUFA, manipulation of the T-AOC was an effective way to achieve high PUFA titer. Therefore, in an attempt to improve the DHA titer, we genetically modified Schizochytium sp. PKU#Mn4 by insertion of sod1 gene into the 18S rDNA region of its genome and generated a recombinant R7 (Fig. 2a).
Construction of tailor-made strains depends on efficient genetic methods, and in order to design genetically-stable strains, chromosomal integration is often desirable (Englaender et al., 2017, Solem et al., 2008, Wu et al., 2016). Multiple copies of 18S rDNA provide more integration chances and ensure non-lethal loci, which make this region a suitable recombination site in thraustochytrids (Hong et al., 2013, Yan et al., 2013). As expected, the growth of SOD overexpression strain R7 was unaffected by the genome integration of SOD1 gene at the 18S rDNA loci. The sod1 overexpression cassette was successfully integrated and efficiently expressed under the newly isolated promoter. However, since the introduced cassette contained a long polyubiquitin promoter besides 18S rDNA gene (Fig. S1a), the integration could occur at several sites theoretically. We found single-crossover homologous recombination (integrated at polyubiquitin promoter region) occurred with much lower frequency than double-crossover homologous recombination (integrated at 18S rDNA loci) and was in agreement with the previous study on thraustochytrid transformation (Sakaguchi et al., 2012). Moreover, double cross-over is more stable and the vector backbone can be removed to get only the desired gene integrated without the rest of the vector body. The hybridized bands with the same size detected in two recombinants R7 and R8 confirmed the double-crossover homologous structure that was favored in the integration (Fig. 2d).
4.2. Overproduction of FA and transcriptome changes
The R7 strain exhibited higher PUFA titer than WT and reduced ROS by improved T-AOC without much decrease in its biomass. Although titers of both SFA and PUFA were significantly higher than those of WT, the moderated intracellular oxidative stress affected their temporal evolution differentially. In general, the PUFA titer was more sensitive to the overexpression of SOD than the SFA and continued to increase reasonably until the late stationary phase (Fig. 4b). Moreover, the PUFA titer increment was relatively more (1.37-fold) than that of SFA (1.18-fold) (Fig. 4c). Our results clearly demonstrated the antioxidant capacity manipulation as an effective control strategy in PUFA production. The advantage of this control strategy was also evident when ascorbic acid was added to the culture (Lujing et al., 2017). However, the addition of ascorbic acid to culture medium would elevate the operational cost of industrial fermentation. Thus, the recombinant R7 provide a favorable Schizochytrium sp. strain for stable and cost-effective PUFA production. Thus, this is an efficient metabolic engineering strategy for oleaginous thraustochytrids.
The transcriptome changes in the recombinant R7 elucidate the gene regulation underlying the SOD1 overexpression and ROS alleviation. Work focusing on lipid biosynthesis has suggested fatty acid synthase (FAS) pathway in thraustochytrids, but the complete pathway is still not fully characterized (Lippmeier et al., 2009, Metz et al., 2001, Nagano et al., 2011). Nevertheless, we identified several unigenes related to FA metabolism that were strongly regulated by SOD1 overexpression. Interestingly, genes related to FA oxidation and biosynthesis were strongly downregulated and upregulated, respectively (Table 1), and many of those genes were peroxisomal (Fig. S3). The main PUFA in WT and R7 strains were very long chain fatty acids containing more than 20 carbon atoms. These fatty acids are almost exclusively β-oxidized in peroxisomes due to the lack of certain enzymes in mitochondria (Kalish et al., 1995, Poulos, 1995). The downregulated genes related to β-oxidation of FA might be another interpretation for higher sensitivity of PUFA titer. We also identified three unigenes (Mn_02824, Mn_04576, and Mn_14213) that encoded fatty acids desaturase, but failed to characterize them well. On the other hand, two unigenes (Mn_10535 and Mn_12937) encoding PKS enzyme were identified and one (Mn_12937) of them was significantly upregulated. These results perhaps indicate the presence of both FAS and PKS pathways for FA production in our strain; however, they are yet to be fully assembled and characterized. Taken together, the enhanced PUFA titer might be a comprehensive result caused by ROS alleviation, enhanced T-AOC, and strongly regulated genes of FA metabolism.
5. Conclusions
In this study, we developed an efficient and stable modified strain of Schizochytrium sp. PKU#Mn4 by insertion of SOD1 gene into the 18S rDNA loci of its genome for overproduction of PUFA. The recombinant strain (R7) successfully yielded higher production of PUFA (1.37-fold) and SFA (1.18-fold) than the WT strain. Overexpression of SOD1 gene resulted in the ROS alleviation and increased T-AOC in R7 with similar biomass yield to that of WT. Transcriptome level changes clearly demonstrated the downregulation of key DEGs in FA β-oxidation, and upregulation of few DEGs related to FA synthesis. Moreover, most of the downregulated DEGs were associated with peroxisome suggesting reduced β-oxidation of DHA. Our results provide the first report of a successful genome integration of SOD1 gene into a thraustochytrid strain and enrich the current understanding of the effect of oxidative damage to the cell at the transcriptome level.
Acknowledgements
This work was partially supported by National Key R&D Program of China (2016YFA0601400) and National Science Foundation of China (31670044, 91751115,and 31602185). The views expressed herein are those of the authors and do not necessarily reflect the views of the funding agencies or any of its subagencies.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2018.03.002.
Appendix A. Supplementary material
Supplementary material
References
- Aasen I.M., Ertesvåg H., Heggeset T.M.B., Liu B., Brautaset T., Vadstein O., Ellingsen T.E. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl. Microbiol. Biotechnol. 2016;100:4309–4321. doi: 10.1007/s00253-016-7498-4. [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Armenta R.E., Valentine M.C. Single-cell oils as a source of omega-3 fatty acids: an overview of recent advances. J. Am. Oil Chem. Soc. 2013;90:167–182. [Google Scholar]
- Bailey, R.B., Don, D., Hansen, J.M., Mirrasoul, P.J., Ruecker, C.M., Veeder, G.T.I., Kaneko, T., Barclay, W.R., 2003. Enhanced Production of Lipids Containing Polyenoic Fatty Acid by Very High Density Cultures of Eukaryotic Microbes in Fermentors. US Patent No. 6607900, Vol. 6607900, US.
- Bashistha K.K., Jnawali H.N., Narayan Prasad N., Sohng J.K. Superoxide dismutase (SOD) genes in Streptomyces peucetius: effects of SODs on secondary metabolites production. Microbiol. Res. 2011;166:391–402. doi: 10.1016/j.micres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Bass D.A., Parce J.W., Dechatelet L.R., Szejda P., Seeds M.C., Thomas M. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J. Immunol. 1983;130:1910. [PubMed] [Google Scholar]
- Benzie I.F., Strain J.J. The ferric reducing ability of plasma (FRAP) as a Measure of "Antioxidant Power": the FRAP assay. Anal. Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- Chang G., Gao N., Tian G., Wu Q., Chang M., Wang X. Improvement of docosahexaenoic acid production on glycerol by Schizochytrium sp. S31 with constantly high oxygen transfer coefficient. Bioresour. Technol. 2013;142:400–406. doi: 10.1016/j.biortech.2013.04.107. [DOI] [PubMed] [Google Scholar]
- Chang G., Luo Z., Gu S., Wu Q., Chang M., Wang X. Fatty acid shifts and metabolic activity changes of Schizochytrium sp. S31 cultured on glycerol. Bioresour. Technol. 2013;142:255–260. doi: 10.1016/j.biortech.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Chang G., Wu J., Jiang C., Tian G., Wu Q., Chang M., Wang X. The relationship of oxygen uptake rate and kLa with rheological properties in high cell density cultivation of docosahexaenoic acid by Schizochytrium sp. S31. Bioresour. Technol. 2014;152:234–240. doi: 10.1016/j.biortech.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Chen W., Zhou P., Zhu Y., Xie C., Ma L., Wang X., Bao Z., Yu L. Improvement in the docosahexaenoic acid production of Schizochytrium sp. S056 by replacement of sea salt. Bioprocess Biosyst. Eng. 2016;39:315–321. doi: 10.1007/s00449-015-1517-1. [DOI] [PubMed] [Google Scholar]
- Cheng R., Ma R., Li K., Rong H., Lin X., Wang Z., Yang S., Ma Y. Agrobacterium tumefaciens Mediated Transformation of Marine Microalgae Schizochytrium. Microbiol. Res. 2012;167:179–186. doi: 10.1016/j.micres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Chi Z., Liu Y., Frear C., Chen S. Study of a two-stage growth of DHA-producing marine algae Schizochytrium limacinum SR21 with shifting dissolved oxygen level. Appl. Microbiol. Biotechnol. 2009;81:1141–1148. doi: 10.1007/s00253-008-1740-7. [DOI] [PubMed] [Google Scholar]
- Damare V., Raghukumar S. Morphology and physiology of the marine straminipilan fungi, the aplanochytrids isolated from the equatorial Indian Ocean. Indian J. Mar. Sci. 2006;35:326–340. [Google Scholar]
- de Oliveira Finco A.M., Goyzueta Mamani L.D., de Carvalho J.C., de Melo Pereira G.V., Thomaz-Soccol V., Soccol C.R. Technological trends and market perspectives for production of microbial oils rich in Omega-3. Crit. Rev. Biotechnol. 2017;37:656–671. doi: 10.1080/07388551.2016.1213221. [DOI] [PubMed] [Google Scholar]
- Dubeau M.P., Ghinet M.G., Jacques P.É., Clermont N., Beaulieu C., Brzezinski R. Cytosine deaminase as a negative selection marker for gene disruption and replacement in the genus Streptomyces and other actinobacteria. Appl. Environ. Microbiol. 2009;75:1211–1214. doi: 10.1128/AEM.02139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else P.L., Kraffe E. Docosahexaenoic and arachidonic acid peroxidation: it's a within molecule cascade. Biochim. Biophys. Acta (BBA) - Biomembr. 2015;1848:417–421. doi: 10.1016/j.bbamem.2014.10.039. [DOI] [PubMed] [Google Scholar]
- Englaender J.A., Jones J.A., Cress B.F., Kuhlman T.E., Linhardt R.J., Koffas M.A. Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli. ACS Synth. Biol. 2017;6(4):710–720. doi: 10.1021/acssynbio.6b00350. [DOI] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Guichardant M., Chen P., Liu M., Calzada C., Colas R., Véricel E., Lagarde M. Functional lipidomics of oxidized products from polyunsaturated fatty acids. Chem. Phys. Lipids. 2011;164:544–548. doi: 10.1016/j.chemphyslip.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Gupta A., Singh D., Colin J.B., Puri M. Exploring potential use of Australian thraustochytrids for the bioconversion of glycerol to omega-3 and carotenoids production. Biochem. Eng. J. 2013;78:11–17. [Google Scholar]
- Honda D., Yokochi T., Nakahara T., Raghukumar S., Nakagiri A., Schaumann K., Higashihara T. Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18s ribosomal RNA gene. J. Eukaryot. Microbiol. 1999;46:637–647. doi: 10.1111/j.1550-7408.1999.tb05141.x. [DOI] [PubMed] [Google Scholar]
- Hong W.-K., Heo S.-Y., Oh B.-R., Kim C.H., Sohn J.-H., Yang J.-W., Kondo A., Seo J.-W. A transgene expression system for the marine microalgae aurantiochytrium sp. KRS101 using a mutant allele of the gene encoding ribosomal protein L44 as a selectable transformation marker for cycloheximide resistance. Bioprocess Biosyst. Eng. 2013;36:1191–1197. doi: 10.1007/s00449-012-0846-6. [DOI] [PubMed] [Google Scholar]
- Jain R., Raghukumar S., Tharanathan R., Bhosle N.B. Extracellular polysaccharide production by thraustochytrid protists. Mar. Biotechnol. 2005;7:184–192. doi: 10.1007/s10126-004-4025-x. [DOI] [PubMed] [Google Scholar]
- Jakobsen A.N., Aasen I.M., Josefsen K.D., Strøm A.R. Accumulation of docosahexaenoic acid-rich lipid in thraustochytrid Aurantiochytrium sp. strain T66: effects of N and P starvation and O2 limitation. Appl. Microbiol. Biotechnol. 2008;80:297–306. doi: 10.1007/s00253-008-1537-8. [DOI] [PubMed] [Google Scholar]
- Jiang X., Zhang J., Zhao J., Gao Z., Zhang C., Chen M. Regulation of lipid accumulation in Schizochytrium sp. ATCC 20888 in response to different nitrogen sources. Eur. J. Lipid Sci. Technol. 2017:119. [Google Scholar]
- Johansson M., Chen X., Milanova S., Santos C., Petranovic D. PUFA-induced cell death is mediated by Yca1p-dependent and -independent pathways, and is reduced by vitamin C in yeast. FEMS Yeast Res. 2016;16:fow007. doi: 10.1093/femsyr/fow007. [DOI] [PubMed] [Google Scholar]
- Kalish J.E., Chen C.I., Gould S.J., Watkins P.A. Peroxisomal activation of long- and very long-chain fatty acids in the yeast Pichia pastoris. Biochem. Biophys. Res. Commun. 1995;206:335–340. doi: 10.1006/bbrc.1995.1046. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahsen A., Iddya K. The state of world fisheries and aquaculture: opportunities and challenges. State World Fish. Aquac. 2014;4:40–41. [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyland B., Leu S., Boussiba S. Are thraustochytrids algae? Fungal Biol. 2017;121:835–840. doi: 10.1016/j.funbio.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Li J., Liu R., Chang G., Li X., Chang M., Liu Y., Jin Q., Wang X. A strategy for the highly efficient production of docosahexaenoic acid by Aurantiochytrium limacinum SR21 using glucose and glycerol as the mixed carbon sources. Bioresour. Technol. 2015;177:51–57. doi: 10.1016/j.biortech.2014.11.046. [DOI] [PubMed] [Google Scholar]
- Lippmeier J.C., Crawford K.S., Owen C.B., Rivas A.A., Metz J.G., Apt K.E. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids. 2009;44:621–630. doi: 10.1007/s11745-009-3311-9. [DOI] [PubMed] [Google Scholar]
- Liu Y., Singh P., Sun Y., Luan S., Wang G. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl. Microbiol. Biotechnol. 2014;98:3241–3255. doi: 10.1007/s00253-013-5391-y. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas E. Health effects of oleic acid and long chain omega-3 fatty acids (EPA and DHA) enriched milks. A review of intervention studies. Pharmacol. Res. 2010;61:200–207. doi: 10.1016/j.phrs.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Ludevese-Pascual G., Dela Peña M., Tornalejo J. Biomass production, proximate composition and fatty acid profile of the local marine thraustochytrid isolate, Schizochytrium sp. LEY7 using low-cost substrates at optimum culture conditions. Aquac. Res. 2016;47:318–328. [Google Scholar]
- Lujing R., Xiaoman S., Xiaojun J., Shenglan C., Dongsheng G., He H. Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour. Technol. 2017;223:141–148. doi: 10.1016/j.biortech.2016.10.040. [DOI] [PubMed] [Google Scholar]
- Lung Y.-T., Tan C.H., Show P.L., Ling T.C., Lan J.C.-W., Lam H.L., Chang J.-S. Docosahexaenoic acid production from crude glycerol by Schizochytrium limacinum SR21. Clean. Technol. Environ. Policy. 2016;18:2209–2216. [Google Scholar]
- Marchan L.F., Lee Chang K.J., Nichols P.D., Polglase J.L., Mitchell W.J., Gutierrez T. Screening of new British thraustochytrids isolates for docosahexaenoic acid (DHA) production. J. Appl. Phycol. 2017:1–13. doi: 10.1007/s10811-017-1149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J.G., Roessler P., Facciotti D., Levering C., Dittrich F., Lassner M., Valentine R., Lardizabal K., Domergue F., Yamada A., Yazawa K., Knauf V., Browse J. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- Nagano N., Sakaguchi K., Taoka Y., Okita Y., Honda D., Ito M., Hayashi M. Detection of genes involved in fatty acid elongation and desaturation in thraustochytrid marine eukaryotes. J. Oleo Sci. 2011;60:475–481. doi: 10.5650/jos.60.475. [DOI] [PubMed] [Google Scholar]
- Nagano N., Taoka Y., Honda D., Hayashi M. Optimization of culture conditions for growth and docosahexaenoic acid production by a marine thraustochytrid, Aurantiochytrium limacinum mh0186. J. Oleo Sci. 2009;58:623–628. doi: 10.5650/jos.58.623. [DOI] [PubMed] [Google Scholar]
- Patil K.P., Gogate P.R. Improved synthesis of docosahexaenoic acid (DHA) using Schizochytrium limacinum SR21 and sustainable media. Chem. Eng. J. 2015;268:187–196. [Google Scholar]
- Pike I.H., Jackson A. Fish oil: production and use now and in the future. Lipid Technol. 2010;22:59–61. [Google Scholar]
- Poulos A. Very long chain fatty acids in higher animals—a review. Lipids. 1995;30:1–14. doi: 10.1007/BF02537036. [DOI] [PubMed] [Google Scholar]
- Qu L., Ren L.-J., Huang H. Scale-up of docosahexaenoic acid production in fed-batch fermentation by Schizochytrium sp. based on volumetric oxygen-transfer coefficient. Biochem. Eng. J. 2013;77:82–87. [Google Scholar]
- Qu L., Ren L.J., Sun G.N., Ji X.J., Nie Z.K., Huang H. Batch, fed-batch and repeated fed-batch fermentation processes of the marine thraustochytrid Schizochytrium sp. For producing docosahexaenoic acid. Bioprocess Biosyst. Eng. 2013;36:1905–1912. doi: 10.1007/s00449-013-0966-7. [DOI] [PubMed] [Google Scholar]
- Ren L., Ji X., Huang H., Qu L., Feng Y., Tong Q., Ouyang P. Development of a stepwise aeration control strategy for efficient docosahexaenoic acid production by Schizochytrium sp. Appl. Microbiol. Biotechnol. 2010;87:1649–1656. doi: 10.1007/s00253-010-2639-7. [DOI] [PubMed] [Google Scholar]
- Ren L., Sun X., Ji X., Chen S., Guo D., Huang H. Enhancement of docosahexaenoic acid synthesis by manipulation of antioxidant capacity and prevention of oxidative damage in Schizochytrium sp. Bioresour. Technol. 2017;223:141–148. doi: 10.1016/j.biortech.2016.10.040. [DOI] [PubMed] [Google Scholar]
- Ruenwai R., Neiss A., Laoteng K., Vongsangnak W., Dalfard A.B., Cheevadhanarak S., Petranovic D., Nielsen J. Heterologous production of polyunsaturated fatty acids in Saccharomyces cerevisiae causes a global transcriptional response resulting in reduced proteasomal activity and increased oxidative stress. Biotechnol. J. 2011;6:343–356. doi: 10.1002/biot.201000316. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Matsuda T., Kobayashi T., Ohara J.-i., Hamaguchi R., Abe E., Nagano N., Hayashi M., Ueda M., Honda D., Okita Y., Taoka Y., Sugimoto S., Okino N., Itoa M. Versatile transformation system that is applicable to both multiple transgene expression and gene targeting for thraustochytrids. Appl. Environ. Microbiol. 2012;78:3193–3202. doi: 10.1128/AEM.07129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K., Gao Z., Shi T.-Q., Song P., Ren L.-J., Huang H., Ji X.-J. Reactive oxygen species-mediated cellular stress response and lipid accumulation in oleaginous microorganisms: the state of the art and future perspectives. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simopoulos A.P. Essential fatty acids in health and chronic disease. Am. J. Clin. Nutr. 1997;70:560S–569S. doi: 10.1093/ajcn/70.3.560s. [DOI] [PubMed] [Google Scholar]
- Solem C., Defoor E., Jensen P.R., Martinussen J. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl. Environ. Microbiol. 2008;74:4772–4775. doi: 10.1128/AEM.00134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Chen H., Zang X., Hou P., Zhou B., Liu Y., Wu F., Cao X., Zhang X. Application of the Cre/loxP site-specific recombination system for gene transformation in Aurantiochytrium limacinum. Molecules. 2015;20:10110–10121. doi: 10.3390/molecules200610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Ren L., Ji X., Chen S., Guo D., Huang H. Adaptive evolution of Schizochytrium sp. by continuous high oxygen stimulations to enhance docosahexaenoic acid synthesis. Bioresour. Technol. 2016;211:374–381. doi: 10.1016/j.biortech.2016.03.093. [DOI] [PubMed] [Google Scholar]
- Tang L., Kwon S.-Y., Kim S.-H., Kim J.-S., Choi J.S., Cho K.Y., Sung C.K., Kwak S.-S., Lee H.-S. Enhanced tolerance of transgenic potato plants expressing both superoxide dismutase and ascorbate peroxidase in chloroplasts against oxidative stress and high temperature. Plant Cell Rep. 2006;25:1380–1386. doi: 10.1007/s00299-006-0199-1. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Yan Q., Jones J.A., Tang Y.J., Fong S.S., Koffas M.A. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016;34(8):652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Xie Y., Wang G. Mechanisms of fatty acid synthesis in marine fungus-like protists. Appl. Microbiol. Biotechnol. 2015;99:8363–8375. doi: 10.1007/s00253-015-6920-7. [DOI] [PubMed] [Google Scholar]
- Yan J., Cheng R., Lin X., You S., Li K., Rong H., Ma Y. Overexpression of Acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl. Microbiol. Biotechnol. 2013;97:1933–1939. doi: 10.1007/s00253-012-4481-6. [DOI] [PubMed] [Google Scholar]
- Yanase S., Onodera A., Tedesco P., Johnson T.E., Ishii N. SOD-1 deletions in Caenorhabditis elegans alter the localization of intracellular reactive oxygen species and show molecular compensation. J. Gerontol. Ser. A-Biol. Sci. Med. Sci. 2009;64A:530–539. doi: 10.1093/gerona/glp020. [DOI] [PubMed] [Google Scholar]
- Zambounis A., Obeidant I.N., Tsaftaris A. Cloning of superoxide dismutase (Cu/Zn Sod) gene in peppers for stress tolerance. Acta Hortic. 2002;579:101–106. [Google Scholar]
- Zhao B., Li Y., Mbifile M.D., Li C., Yang H., Wang W. Improvement of docosahexaenoic acid fermentation from Schizochytrium sp. AB-610 by staged pH control based on cell morphological changes. Eng. Life Sci. 2017;17:981–988. doi: 10.1002/elsc.201600249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Zhang X., Ren X., Zhu Q. Effects of culture conditions on growth and docosahexaenoic acid production from Schizochytrium limacinum. J. Ocean Univ. China. 2008;7:83–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material