Abstract
The endoplasmic reticulum (ER) localized unfolded protein response (UPR) sensors, IRE1α, PERK, and ATF6α, are activated by the accumulation of misfolded proteins in the ER. It is unclear how the endogenous UPR sensors are regulated by both ER stress and the ER luminal chaperone BiP, which is a negative regulator of UPR sensors. Here we simultaneously examined the changes in the endogenous complexes of UPR sensors by blue native PAGE immunoblotting in unstressed and stressed cells. We found that all three UPR sensors exist as preformed complexes even in unstressed cells. While PERK complexes shift to large complexes, ATF6α complexes are reduced to smaller complexes on ER stress. In contrast, IRE1α complexes were not significantly increased in size on ER stress, unless IRE1α is overexpressed. Surprisingly, depletion of BiP had little impact on the endogenous complexes of UPR sensors. In addition, overexpression of BiP did not significantly affect UPR complexes, but suppressed ER stress mediated activation of IRE1α, ATF6α and, to a lesser extent, PERK. Furthermore, we captured the interaction between IRE1α and misfolded secretory proteins in cells, which suggests that the binding of unfolded proteins to preformed complexes of UPR sensors may be crucial for activation.
INTRODUCTION
The endoplasmic reticulum (ER) is the major organelle for the synthesis of secretory and membrane proteins. These proteins enter the ER through the Sec61 translocon channel and mature with the help of a cascade of chaperones, folding enzymes, and posttranslocation modifications (van Anken and Braakman, 2005; Rapoport, 2007). Proteins that fail to achieve their native state are recognized and eliminated by the ER-associated degradation (ERAD) pathways (Brodsky, 2012; Christianson and Ye, 2014). Thus, only folded proteins are packaged into vesicles for their transport to the Golgi apparatus. However, environmental stress, nutrient overload, or expression of mutated proteins overwhelms ERAD machinery, resulting in accumulation of misfolded proteins in the ER. The excess of misfolded proteins in the ER activates the conserved unfolded protein response (UPR) pathway, which transmits the information of the folding status of the ER to the cytosol and nucleus (Walter and Ron, 2011). The UPR activates transcriptional and translational programs to increase the ER protein folding capacity (Lee et al., 2003; Shoulders et al., 2013). In case of failure to attenuate the UPR during prolonged stress, the cells commit suicide by initiating apoptotic pathways. The dysfunction of or overactive UPR signaling has been implicated in numerous human diseases including type 2 diabetes, neurodegenerative diseases, and cancer (Han et al., 2009; Hetz, 2012; Wang and Kaufman, 2016).
In metazoans, three UPR sensors, IRE1α, PERK, and ATF6α, are known to detect misfolded proteins in the ER lumen and transmit the signal across the ER membrane to the cytosol (Walter and Ron, 2011). IRE1α is a transmembrane kinase/endonuclease (RNase) that, on ER stress, initiates the unconventional splicing of XBP1 mRNA (Cox et al., 1993; Mori et al., 1993; Yoshida et al., 2001; Calfon et al., 2002). The spliced XBP1 mRNA encodes an active transcription factor that drives transcription of genes encoding chaperones, protein translocation, and ER degradation components (Lee et al., 2003; Shoulders et al., 2013). In addition, IRE1α can also reduce protein synthesis load at the ER through promiscuously cleaving ER-localized mRNAs encoding membrane and secretory proteins, a process known as IRE1α-dependent mRNA decay (RIDD) (Hollien and Weissman, 2006; Hollien et al., 2009). PERK is a transmembrane kinase, and its luminal domain shares a limited homology (∼12%) to the luminal domain of IRE1α (Zhou et al., 2006). On ER stress, PERK phosphorylates the α subunit of eukaryotic translation initiation factor 2 to shut down the overall protein synthesis, thus counteracting protein overload at the ER (Harding et al., 1999; Sood et al., 2000). However, some mRNAs that have small open reading frames in their 5′ untranslated regions are translated by phosphorylated eIF2α, and thereby production of transcription factors, such as ATF4 and CHOP (Ameri and Harris, 2008). ATF6α is an ER-localized transmembrane transcription factor (Haze et al., 1999). During ER stress conditions, ATF6α transported to the Golgi apparatus, where its cytoplasmic domain is released from membrane domain by S1P- and S2P-mediated proteolysis (Ye et al., 2000; Nadanaka et al., 2007; Schindler and Schekman, 2009). The cleaved ATF6α moves to the nucleus and drives transcription of genes encoding chaperones and ERAD machinery for restoring ER homeostasis (Lee et al., 2003; Shoulders et al., 2013).
While tremendous progress has been made in understanding the biology of UPR effectors, the mechanism of UPR sensors activation remains incompletely understood.
Two major models have been actively debated for the activation of UPR sensors (Walter and Ron, 2011; Snapp, 2012). The first model is similar to other stress sensing pathways such as the heat shock response, which is strongly regulated by the binding and availability of a chaperone (Arsene et al., 2000; Anckar and Sistonen, 2011). Similarly, for the UPR, binding immunoglobulin protein (BiP) is thought to bind to monomers of either IRE1α or PERK and prevent it from oligomerization and activation in unstressed cells. During ER stress, BiP is sequestered by misfolded proteins and displaced from IRE1α and PERK, leading to their oligomerization and activation (Walter and Ron, 2011). This model is supported by the evidence that BiP interacts with both IRE1α and PERK in unstressed cells and that the association is disrupted in the presence of ER stress (Bertolotti et al., 2000; Okamura et al., 2000; Oikawa et al., 2009; Carrara et al., 2015). The activation of ATF6α is slightly different from other two sensors since it appears to form large oligomers as well as associates with BiP under unstressed conditions (Nadanaka et al., 2007; Gallagher and Walter, 2016; Shen et al., 2002). On ER stress, ATF6α is dissociated from BiP and moves to the Golgi for the proteolytic processing (Shen et al., 2002).
In the second model, unfolded proteins may directly bind to the luminal sensor domains of UPR sensors. This binding may drive oligomerization and activation of UPR sensors (Walter and Ron, 2011). The first evidence supporting this model came from crystal structures of yeast Ire1p luminal domain, which resemble the peptide-binding groove of MHC-I (Credle et al., 2005). On the basis of this, Peter Walter’s group proposed the peptide-binding hypothesis. This idea is corroborated by the evidence that misfolded proteins directly bind to the luminal domain of Ire1p and drive oligomerization (Kimata et al., 2007; Gardner and Walter, 2011). Several studies have challenged the peptide-binding model. First, the human IRE1α luminal domain structure exhibits a narrow peptide-binding groove of MHC-1, which may not accommodate misfolded proteins. Nevertheless, a recent study suggests that a mutation in the groove of MHC-1 seems to interfere with the detection of misfolded proteins in the ER lumen (Kono et al., 2017). Second, unlike yeast Ire1p, human IRE1α does not seem to interact with misfolded proteins in cells (Oikawa et al., 2012). Finally, the fact that monomeric form of yeast Ire1p cannot bind to unfolded peptides in vitro raises the question of how monomers of IRE1α can efficiently bind to misfolded proteins in cells during ER stress conditions (Gardner and Walter, 2011).
It has been challenging to determine which of these models is occurring in mammalian cells. One of the key requirements to test these different models is to monitor the endogenous complexes of all three UPR sensors under homeostatic and ER stress conditions in cells. Although size fractionation assays to probe the complexes of UPR sensors were successful (Bertolotti et al., 2000), it was laborious to examine numerous fractionated samples derived from various time points of stress. This approach is further complicated by the fact that all three UPR sensors are relatively low abundance proteins in cells (Kulak et al., 2014). Thus, it is not feasible to detect these proteins in diluted size-fractionated samples. An imaging-based approach that monitors ER stress-dependent higher-order oligomers (or clusters) proves to be useful for probing IRE1α activation in human cells (Li et al., 2010). However, the ER stress-dependent cluster formation is not readily visible at the endogenous levels of IRE1α (Sundaram et al., 2017).
Blue native PAGE (BN–PAGE) immunoblotting has been successfully used to monitor the complex or oligomer formation of mitochondrial protein import machinery (Wittig et al., 2006). Recent studies have used BN–PAGE to follow the dynamics of Sec61 translocon complexes during protein translocation into the ER lumen (Conti et al., 2015) and ligand-dependent oligomerization of NLR family CARD domain-containing protein 4 (NLRC4) inflammasome (Kofoed and Vance, 2011). We have recently used this approach to specifically monitor the role of the Sec61 translocon in controlling IRE1α complexes (Sundaram et al., 2017). In the current study, we employed this technique to investigate dynamics of all three endogenous UPR complexes during ER stress. We found that all three UPR sensors exist as preformed complexes even in unstressed cells. While PERK shifts to large complexes on ER stress, there was no significant ER stress-dependent shift in the size of IRE1α complexes, unless IRE1α is overexpressed. ATF6α moved from large complexes to small complexes during ER stress. Neither depletion nor overexpression of BiP had significant effects on the complexes of UPR sensors both in unstressed and stressed cells. Furthermore, an in vivo cross-linking assay revealed a selective interaction between IRE1α and misfolded secretory proteins. Thus, our results support a model where misfolded proteins may bind and activate the preformed complexes of UPR sensors.
RESULTS
Probing the endogenous complexes of UPR sensors in unstressed and stressed cells
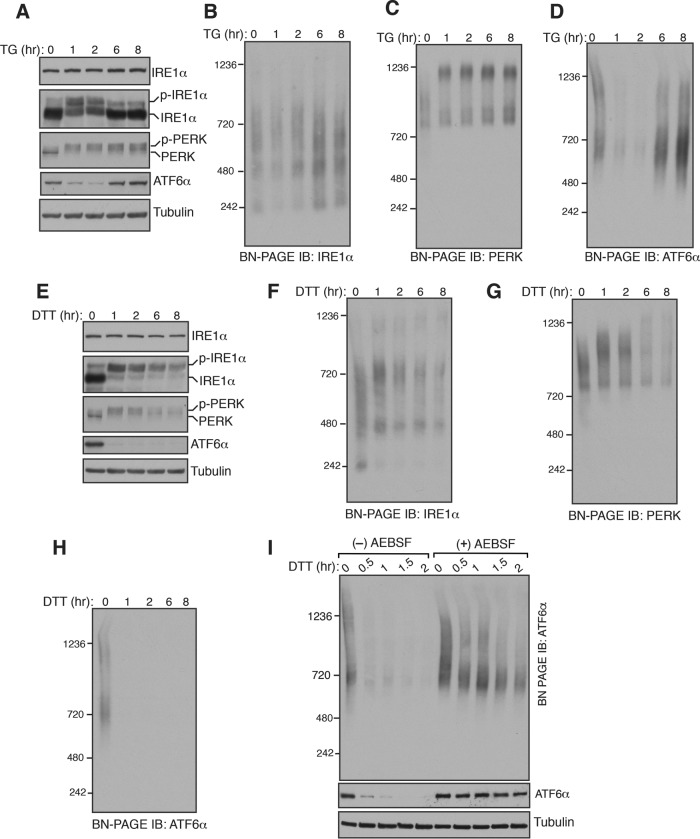
To monitor the changes in the endogenous complexes of IRE1α, PERK, or ATF6α during homeostatic and ER stress conditions, we used a BN–PAGE immunoblotting procedure (Wittig et al., 2006). HEK293 cells were treated with thapsigargin (TG), which induces ER stress by inhibiting calcium transport into the ER, and digitonin lysates were prepared for BN–PAGE immunoblotting. The activation of the endogenous IRE1α was monitored by probing its phosphorylation status using phos-tag–based immunoblotting (Yang et al., 2010; Sundaram et al., 2017). A significant proportion of IRE1α was activated after 1 h of ER stress and inactivated within 6 h of ER stress treatment (Figure 1A). In accordance with previous studies (Lin et al., 2007; Sundaram et al., 2017), PERK was activated throughout the stress period as evident by its phosphorylation (Figure 1A). ATF6α was activated on ER stress as shown by the loss of signal due to the proteolytic release of the N-terminal fragment after its migration to the Golgi apparatus (Figure 1A). The ATF6α antibody used in this study failed to detect the cleaved fragments of ATF6α. Similarly to IRE1α, ATF6α was robustly attenuated within 8 h of stress period, since the full-length ATF6α signal appeared back during the later hours of ER stress (Figure 1A).
FIGURE 1:
Changes in the endogenous complexes of IRE1α, PERK, and ATF6α during ER stress. (A) HEK293 cells were treated with 12 μM of thapsigargin (TG) for the indicated time points and lysed with digitonin. The lysates were separated by SDS–PAGE and immunoblotted for the indicated proteins. A phos-tag–based immunoblotting was performed for probing phosphorylated IRE1α. (B) The digitonin lysates from A were analyzed by BN–PAGE immunoblotting with IRE1α antibody, PERK antibody (C), or ATF6α antibody (D). (E–H) HEK293 cells were treated with 5 mM DTT for the indicated time points and analyzed as in A–D. (I) HEK293 cells were pretreated with a serine protease inhibitor AEBSF (250 μM) for 1 h and subsequently treated with DTT for the indicated time points to induce ER stress. The digitonin lysates were analyzed as in A and D. Experiments shown are representative of experiments repeated at least two times during different days.
Consistently with our previous findings (Sundaram et al., 2017), BN–PAGE immunoblotting revealed that IRE1α existed as predominantly two complexes: ∼480 and ∼720 kDa. An ER stress-dependent increase in the size of IRE1α complexes was not apparent, but the ∼240-kDa complex diminished on ER stress and appeared back in the later hours of ER stress (Figure 1B and Supplemental Figure S1A). In contrast, ER stress-dependent changes in PERK complexes were evident, since PERK moved from a ∼900-kDa complex to a ∼1200-kDa complex on ER stress (Figure 1C). BN–PAGE detected two large complexes of ATF6α, ∼720 and ∼1200 kDa, under homeostatic conditions. These bands nearly disappeared during the initial hours of ER stress and appeared back in the later hours of ER stress (Figure 1D). To rule out the possibility that some of the bands seen in BN–PAGE immunoblots are the result of spurious antibody cross-reactivity, we performed BN–PAGE with IRE1α- or ATF6α-depleted cells. Indeed, these antibodies are very specific since almost no background bands were seen in the depleted cells (Supplemental Figure S2, A and B). We next asked whether the changes in complexes of the UPR sensors on BN–PAGE are specific to TG treatment. We therefore performed BN–PAGE with cells treated with dithiothreitol (DTT), which induces protein misfolding in the ER by blocking protein disulfide bond formation. All three UPR sensors were robustly activated in cells treated with DTT (Figure 1E). In line with TG treatment, there was no significant increase in the size of IRE1α complexes, except the ∼240-kDa band disappeared upon DTT treatment (Figure 1F and Supplemental Figure S1B). Interestingly, changes in PERK complexes were less noticeable with DTT treatment compared with TG treatment since not all the 900-kDa complexes moved to the 1200-kDa complex (Figure 1G and Supplemental Figure S1C). ATF6α signal disappeared throughout DTT treatment, suggesting that the ER is continuously experiencing ER stress, and its homeostasis is not restored with DTT treatment (Figure 1H).
Since ATF6α is proteolytically cleaved during ER stress, we were not able to detect changes in ATF6α complexes. To determine the changes in ATF6α complexes during ER stress, we inhibited S1P and S2P proteases that are responsible for the cleavage of ATF6α using a previously described serine protease inhibitor, 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride (AEBSF) (Okada et al., 2003). In the presence of the inhibitor, ATF6α cleavage was nearly abolished as shown by immunoblotting (Figure 1I, bottom). Interestingly, the band signal of the 1200-kDa large complex was significantly reduced during ER stress, whereas little change occurred with the smaller complex of 720 kDa (Figure 1I), suggesting that ER stress-dependent dissociation of ATF6α complexes is necessary for its transport to the Golgi apparatus.
We next asked whether the architecture of UPR complexes is affected by choice of detergent used for preparing cell lysates. We therefore used Triton X-100 to solubilize the cells treated with either TG or DTT. BN–PAGE analysis with Triton X-100–solubilized cell lysates revealed that both PERK and ATF6α migrated similar to digitonin solubilized cell lysates in that PERK moved from a smaller complex to a larger complex, and ATF6 complexes disappeared on ER stress (Supplemental Figure S3, A, C, and D). In contrast, we observed two notable differences in the migration pattern of IRE1α with Triton X-100 compared with digitonin-based BN–PAGE. First, the 480-kDa complex of IRE1α seen in digitonin (Figure 1, B and F) was nearly absent in the Triton X-100–based BN–PAGE (Supplemental Figure S3B). The precise reason for the loss of the 480-kDa complex is not clear, but one possibility is that Triton X-100 might disrupt the interaction between IRE1α and the Sec61 translocon complex (Plumb et al., 2015). This interpretation is consistent with our previous findings that a Sec61 interaction defective IRE1α mutant also lacked the 480-kDa complex (Sundaram et al., 2017). The second difference is that ER stress-dependent appearance of a large complex (∼1000 kDa) of IRE1α on BN–PAGE was more prominent with the Triton X-100–solubilized samples (Supplemental Figure S3B).
Since all three UPR sensors appeared as large complexes on BN–PAGE, we wanted to exclude the possibility that the slow migration of UPR sensors on BN–PAGE was caused by their association with lipid or/and detergent micelles. We therefore tested whether the endogenous UPR sensors exist as preformed complexes by a chemical cross-linking approach. HEK293 cells were treated with a cysteine-reactive cross-linker and analyzed by a low percentage standard SDS–PAGE immunoblotting. Remarkably, consistent with BN–PAGE data, all three UPR sensors entirely shifted to high-molecular-weight cross-linked adducts both in unstressed and stressed cells (Supplemental Figure S4, A–C). Although BiP is a vastly more abundant chaperone than all three UPR sensors (Kulak et al., 2014), it showed significantly fewer cross-linked adducts compared with UPR sensors (Supplemental Figure S4D). Given that the total protein profile did not significantly change with all concentrations of the cross-linker, it is likely that only stable complexes, such as formed by the UPR sensors, can be efficiently cross-linked at these concentrations (Supplemental Figure S4E). For IRE1α and ATF6α, we do not expect to detect changes in cross-linked adducts between unstressed and stressed cells because the size of IRE1α complexes was not significantly increased with stress on BN–PAGE, and the signal for ATF6 was mostly disappeared with stress. Interestingly, PERK also did not show any noticeable change in cross-linked adducts between unstressed and stressed cells (Supplemental Figure S4B). This is presumably due to the limited resolution of SDS–PAGE to differentiate between ∼900-kDa complexes of PERK in unstressed cells and ∼1200-kDa complexes in stressed cells. Collectively, these results suggest that all three endogenous UPR sensors exist as preformed complexes and are activated on ER stress by changes in complex formation. PERK forms larger complexes on ER stress, ATF6α is reduced into smaller complexes, and IRE1α exhibits a less dramatic change in size, with only a diminishment of the 240-kDa complex.
Overexpressed IRE1α shows ER stress-dependent increases in the size of its complexes
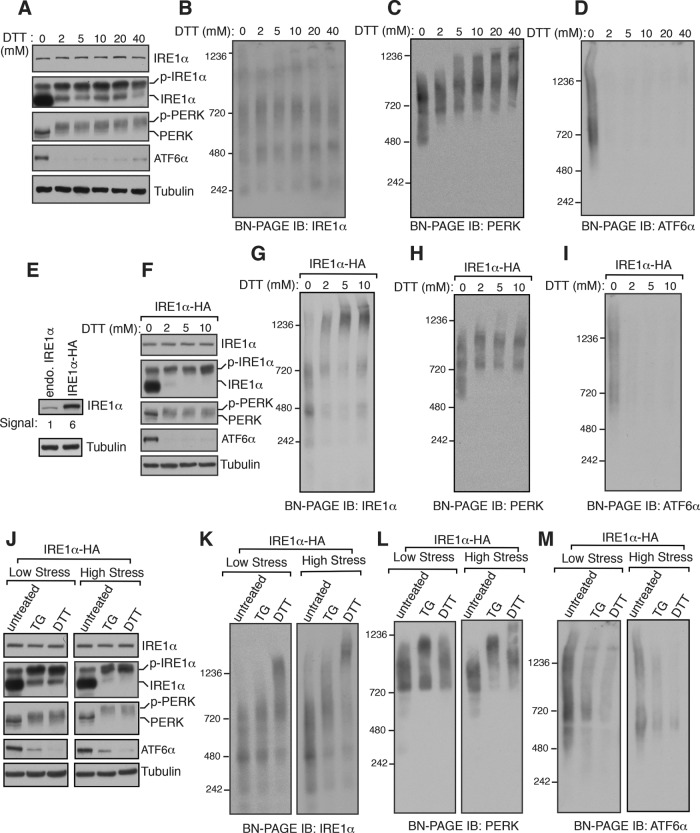
We were surprised by our observation that the size of the endogenous IRE1α complexes was not significantly increased on ER stress. This is inconsistent with the current model that IRE1α forms ER stress-dependent higher-order oligomerization (Walter and Ron, 2011). To rule out the possibility that the intensity of ER stress was not sufficient to induce IRE1α oligomerization, we treated cells from low to high concentrations of DTT. All three UPR sensors were robustly activated at these DTT concentrations (Figure 2A). Surprisingly, even at a high dosage of DTT treatment, we did not notice an appreciable increase in the size of IRE1α complexes (Figure 2B and Supplemental Figure S5A). In contrast, the size of PERK complexes increased with increasing concentration of the stress, whereas ATF6α signal disappeared at all concentrations of DTT treatment (Figure 2, C and D).
FIGURE 2:
The size of overexpressed IRE1α but not the endogenous IRE1α complexes increases on ER stress. (A) HEK293 cells were treated with the indicated concentrations of DTT for 2.5 h and analyzed for immunoblotting with the indicated antigens. (B–D) The samples from A were analyzed by BN–PAGE immunoblotting and probed for the indicated antigens. (E) An immunoblot comparing the expression levels between the endogenous IRE1α in HEK293 and IRE1α-HA complemented into HEK293 IRE1α–/– cells. (F) HEK293 IRE1α–/– complemented IRE1α-HA cells were treated with increasing concentration of DTT for 2.5 h and analyzed by immunoblotting for the indicated antigens. (G–I) The samples from F were analyzed by BN–PAGE immunoblotting and probed for the indicated antigens. (J) HEK293 IRE1α–/– complemented IRE1α-HA cells were induced with low stress using either 4 μM of TG or 2 mM DTT for 2.5 h. Alternatively, the cells were induced with high stress using either 39 μM of TG or 30 mM DTT for 2.5 h. The cell lysates were analyzed by standard immunoblotting for the indicated antigens. (K–M) The samples from J were analyzed by BN-PAGE immunoblotting for the indicated antigens.
We hypothesized that ER stress-dependent changes were not detected for IRE1α complexes since the concentration of the endogenous IRE1α (∼416 molecules/cell) is very low abundance compared with PERK (∼2299 molecules/cell) and ATF6α (15,598 molecules/cell) in human cells (Kulak et al., 2014). To test this, we used HEK293 IRE1α–/– cells complemented with recombinant IRE1α, expression of which is six times higher than the endogenous IRE1α, which shows little constitutive activation under homeostatic conditions (Figure 2, E and F). In support of our hypothesis, the overexpressed IRE1α cells showed an ER stress-dependent appearance of a large 1200-kDa complex (Figure 2G). While ER stress-dependent changes in the size of PERK complexes was less noticeable, ATF6α signal completely disappeared upon treating with DTT (Figure 2, H and I). We next simultaneously compared the effects of TG and DTT treatments on recombinant IRE1α complexes. Both low- and high-stress treatments with either TG or DTT resulted in efficient activation of all three UPR sensors (Figure 2J). Consistently with our previous studies (Sundaram et al., 2017), ER stress-dependent changes in recombinant IRE1α complexes were not obvious under a low-stress condition with TG treatment, but a modest increase in the size of IRE1α complexes occurred under a high-stress condition with TG treatment (Figure 2K and Supplemental Figure S5, B and C). However, the ER stress-dependent increase in the size of recombinant IRE1α complexes was conspicuous under both low and high-stress conditions with DTT treatment (Figure 2K and Supplemental Figure S5, B and C). The size of PERK complexes increased under both low and high-stress conditions with TG treatment, but their size increase was apparent only with a high-stress condition with DTT treatment (Figure 2L and Supplemental Figure S5, D and E). As expected, ATF6α complexes were responsive to both low- and high-stress conditions as ATF6α signal disappeared under both conditions (Figure 2M and Supplemental Figure S5, F and G). Together these results suggest that the endogenous IRE1α complexes are activated without a significant increase in their size on ER stress, unless IRE1α is overexpressed.
An IRE1α dimerization mutant disrupts IRE1α complexes in both unstressed and stressed cells
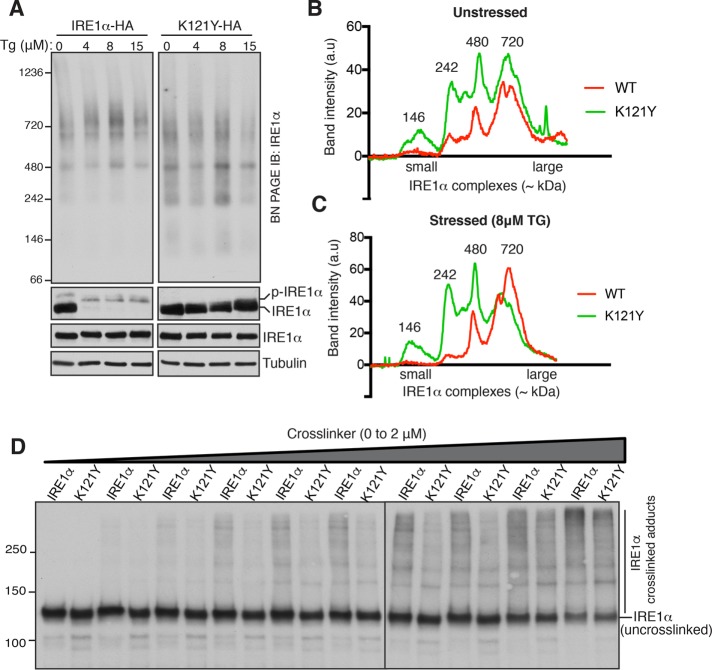
We hypothesized that larger complexes of UPR sensors might include both homo- and hetero-oligomers. To test this, we took advantage of the previously characterized IRE1α mutant K121Y that selectively disrupts homo-oligomerization of IRE1α (Li et al., 2010). As expected, we observed an ER stress-dependent phosphorylation of wild-type IRE1α, but K121Y showed significantly less phosphorylation (Figure 3A, bottom). BN–PAGE analysis of IRE1α K121Y showed more smaller size IRE1α species (∼146 and ∼242 kDa) compared with the wild type even under homeostatic conditions, indicating that a significant population of IRE1α exists as homo-oligomers in unstressed cells (Figure 3, A–C). In contrast to our prediction, we did not observe the total conversion of larger complexes of IRE1α into smaller species. It is likely that the dimerization mutant still forms complexes with other interacting proteins such as the Sec61 translocon complex, confounding the analysis of different IRE1α complexes on BN–PAGE. IRE1α is also known to form homo-oligomers through its cytosolic domain of IRE1α (Ghosh et al., 2014). Last, an in vivo chemical cross-linking assay further supported the idea that IRE1α exists as preassembled homo-oligomers even in unstressed cells, since the dimerization mutant of IRE1α K121Y showed markedly less cross-linked adducts compared with the wild type (Figure 3D). Taken together, these results suggest that IRE1α complexes already contain multiple copies of IRE1α in unstressed cells.
FIGURE 3:
An IRE1α dimerization mutant displays a reduction in the size of IRE1α complexes. (A) HEK293 IRE1α–/– cells were complemented either IRE1α-HA or IRE1α (K121Y)-HA and treated with increasing concentration of TG for 4 h. The cells were harvested and analyzed by either SDS–PAGE immunoblotting (bottom) with the indicated antibodies or BN-PAGE immunoblotting with IRE1α antibody (top). (B) The unstressed lanes (0 μM TG) from A were quantified and plotted. (C) The stressed lanes (8 μM TG) from A were quantified and plotted. (D) HEK293 IRE1α–/– cells expressing either IRE1α-HA or IRE1α (K121Y)-HA were permeabilized with a low concentration of digitonin. Subsequently, the permeabilized cells were treated with 0–2 μM BMH for 30 min on ice. The cells were directly harvested in SDS sample buffer and analyzed by immunoblotting.
Effects of BiP depletion on IRE1α, PERK, and ATF6α complexes and their activation
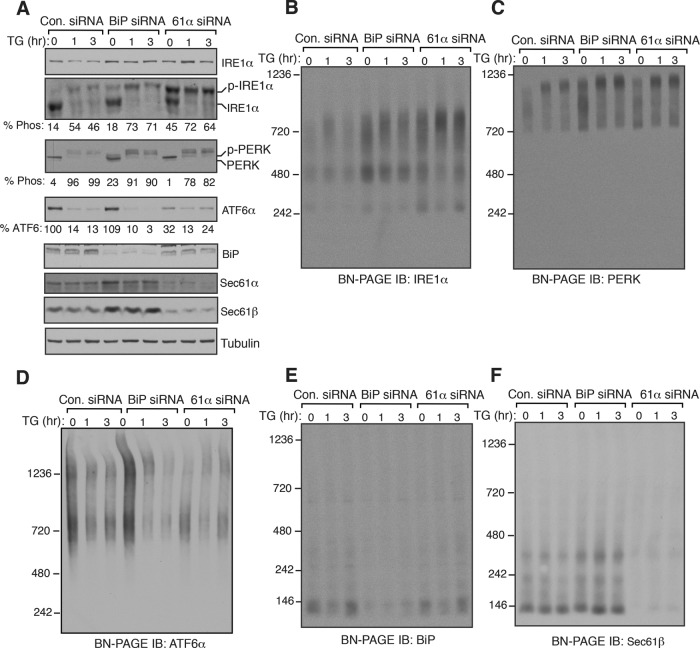
Since BiP has been suggested to be a negative regulator by inhibiting oligomerization and activation of UPR sensors (Bertolotti et al., 2000; Okamura et al., 2000), we expected that depletion of BiP might lead to an increase in the size of UPR complexes in both unstressed and stressed cells. On the other hand, the depletion of Sec61 translocon would serve as a control since its depletion selectively changes IRE1α complexes as well as activates IRE1α (Adamson et al., 2016; Sundaram et al., 2017). We therefore transiently depleted BiP or the Sec61 translocon in cells using small interfering RNA (siRNA)-mediated knockdown. Probing the phosphorylation status of IRE1α and PERK revealed that a small amount of IRE1α and PERK were activated in BiP-depleted cells during unstressed conditions, and they were fully activated on ER stress (Figure 3A). Depletion of BiP appeared to have little to no effect on the cleavage of ATF6α in unstressed as well as stressed cells relative to control siRNA-depleted cells (Figure 4A). Consistent with the previous studies (Adamson et al., 2016), depletion of the Sec61 translocon selectively activated ∼45% of IRE1α in unstressed cells, and IRE1α became fully activated on ER stress. While the depletion of the Sec61 translocon did not affect PERK, a significant loss of ATF6α signal occurred relative to the control (Figure 4A). Since our ATF6α antibody detects only the uncleaved form of ATF6α, we were not able to validate whether the loss of signal is due to the activation of ATF6α or due to the low expression ATF6α in the Sec61 translocon-depleted cells. ATF6α was also not efficiently activated in the Sec61 translocon-depleted cells on ER stress since it remains predominantly uncleaved on ER stress (Figure 4A).
FIGURE 4:
Effects of BiP depletion on IRE1α, PERK, and ATF6α complexes and their activation. (A) HEK293 cells were transfected with siRNA oligos directed against either BiP siRNA or Sec61α for two consecutive days. After 72 h of transfection, the cells were treated with 3.9 μM of TG for the indicated time points and analyzed by immunoblotting for the indicated antigens. (B–F) The samples from A were analyzed by BN–PAGE immunoblotting for the indicated antigens.
BN–PAGE analysis of BiP-depleted cells revealed that no significant changes occurred with complexes of all three UPR sensors under both unstressed cells and stressed conditions relative to the control siRNA-treated cells (Figure 4, B–D). Consistent with our recent studies (Sundaram et al., 2017), depletion of the Sec61 translocon led to an enrichment of 720-kDa complex of IRE1α compared with control or BiP siRNA-treated cells both under normal and stress conditions (Figure 4B). In contrast, the Sec61 translocon depletion did not affect either PERK or ATF6α complexes (Figure 4, C and D, and Supplemental Figure S6). Unlike all three UPR sensors, BiP migrated as predominantly a single species ∼140 kDa on BN–PAGE, whereas the Sec61 translocon ran predominantly as a ∼140-kDa form as well as a minor ∼350-kDa form, which is in agreement with previous studies (Figure 4, E and F) (Conti et al., 2015; Sundaram et al., 2017). Together these data suggest that the depletion of BiP had minor effects on the complexes and activation of all three UPR sensors both under unstressed and stressed conditions. However, it remains to be determined whether the residual amount of BiP in the depleted cells is sufficient to represses the activation of UPR sensors.
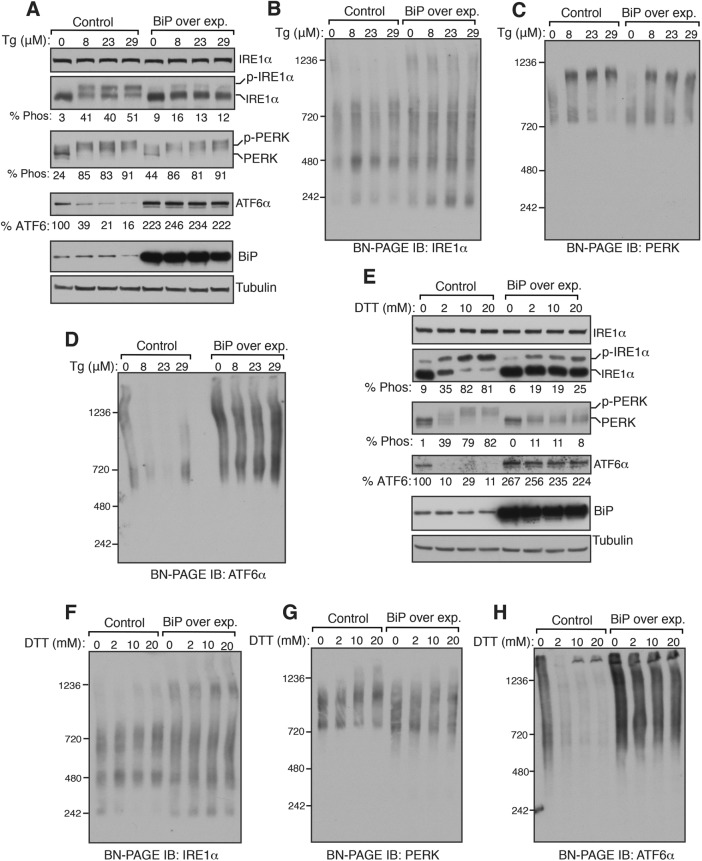
BiP overexpression does not significantly impact complexes of UPR sensors but suppresses activation of UPR sensors
We next reasoned that if BiP inhibits oligomerization of UPR sensors, its overexpression should reduce the size of UPR complexes on BN–PAGE as well as block the activation of UPR sensors. Consistent with the previous studies (Kohno et al., 1993; Bertolotti et al., 2000), overexpression of BiP above the endogenous level significantly suppressed the activation of IRE1α as reflected by significantly reduced phosphorylation of IRE1α on TG-induced ER stress (Figure 5A). To our surprise, PERK was activated as evident by its phosphorylation status even in BiP-overexpressing cells treated with TG, albeit slightly less efficient than the control. Interestingly, the ATF6α protein level was increased about twofold in BiP-overexpressing cells, and the ER stress-dependent cleavage of ATF6α was nearly blocked (Figure 5A). BN–PAGE analysis of IRE1α in BiP-overexpressing cells revealed that the overall pattern of IRE1α complexes was not significantly affected in the presence or absence of the TG treatment (Figure 5B). However, we noticed a large complex (∼1200 kDa) that specifically formed for IRE1α on BiP overexpression in a non-ER stress-dependent manner. BiP overexpression also did not affect the size of PERK complexes in unstressed cells. Consistent with the phosphorylation data (Figure 5A), PERK normally shifted from a smaller complex to a large complex on BN–PAGE in BiP-overexpressing cells upon treatment with TG (Figure 5C). BiP overexpression did not reduce the size of ATF6α complexes in unstressed cells, but it completely inhibited ER stress-dependent disappearance of ATF6α complexes (Figure 5D). These results were further corroborated by BiP-overexpressing cells treated with DTT (Figure 5, E–H). We observed two differences with UPR complexes between TG- and DTT-treated cells. First, the ER stress-dependent diminishment of ∼240 kDa of IRE1α complex was inhibited in BiP-overexpressing cells (Figure 5F). Second, while BiP overexpression had little effect on the activation of PERK when cells were treated with TG, it markedly blocked activation of PERK on DTT treatment (Figure 5, E and G). Collectively, our data suggest that the overexpression of BiP does not have a significant impact on the complexes of all three UPR sensors but interferes with the activation of UPR sensors on ER stress.
FIGURE 5:
Overexpression of BiP has little effect on complexes of IRE1α, PERK, and ATF6α but suppresses their activation. (A) HEK293 cells were transfected with either an empty plasmid (control) or BiP plasmid. After 24 h of transfection, the cells were treated with the indicated concentrations of TG for 2 h. The cells were harvested and analyzed by immunoblotting for the indicated antigens. (B–D) The samples from A were analyzed by BN–PAGE immunoblotting for the indicated proteins. (E) The cells were transfected as in A and treated with the indicated concentrations of DTT for 2 h. The cells were harvested and analyzed by immunoblotting. (F–H) The samples from E were analyzed by BN–PAGE immunoblotting for the indicated proteins. Experiments shown are representative of experiments repeated at least two times during different days.
IRE1α can interact with misfolded secretory proteins in cells
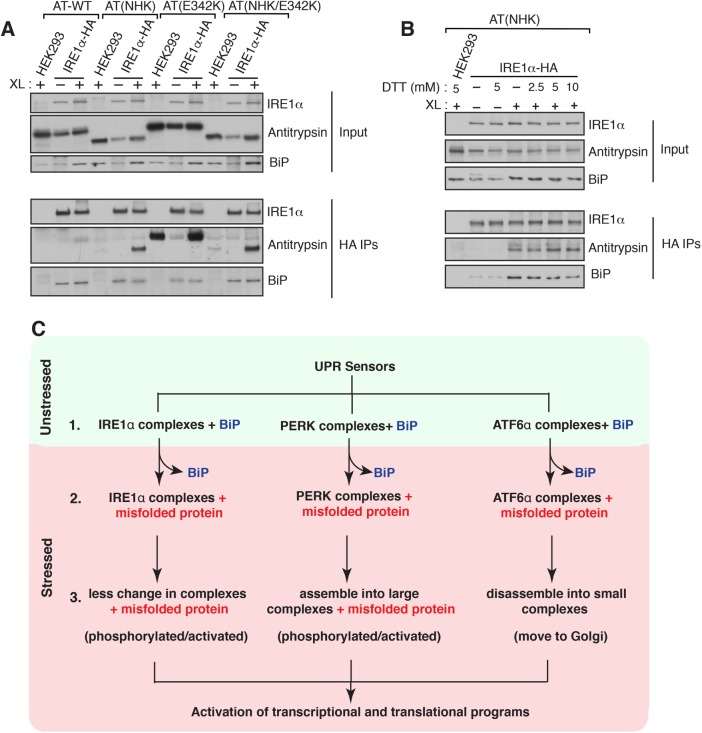
The above-mentioned observations suggest that BiP plays a minor role in regulating the endogenous complexes of UPR sensors. We therefore considered the model that misfolded proteins might bind and activate the preformed complexes of UPR sensors, which may have a higher affinity for misfolded proteins under ER stress conditions. To support this model, we attempted to capture the interaction between IRE1α and misfolded proteins in the ER lumen. We used three different mutants of α1-antitrypsin (α1-AT) as misfolded substrates: a highly misfolded nonpolymerogenic mutant of null Hong Kong (NHK) (Sifers et al., 1988), the polymerogenic mutant E342K (Lomas et al., 1992), and a combination of both. We first determined whether these α1-AT mutants could induce ER stress response when expressed in cells. Indeed, all three mutants of α1-AT, but not the wild-type α1-AT, can elicit IRE1-mediated UPR signaling, as measured by a luciferase reporter of IRE1α RNase activity (Supplemental Figure S7). As expected, α1-AT mutants produced only a background level of luciferase activities in HEK293 IRE1α-/- cells, validating that the reporter is specific to IRE1α branch (Supplemental Figure S7). To test the interaction between IRE1α and misfolded α1-AT, we expressed α1-AT variants into HEK293 IRE1α-/- expressing HA-tagged IRE1α. The cells were treated with a reversible chemical cross-linker to stabilize the interaction between IRE1α and misfolded proteins. The samples were immunoprecipitated for IRE1α using an HA antibody. Consistent with the misfolded protein binding model, we found an enhanced interaction between IRE1α and all three different α1-AT mutants compared with wild-type α1-AT (Figure 6A). This result is consistent with the ability of these α1-AT mutants to induce IRE1-mediated UPR signaling (Supplemental Figure S7). Interestingly, the polymerogenic α1-AT mutant (E342K) interacted with IRE1α, but it had a significant background binding to beads, possibly owing to its nature of forming larger polymers (Lomas et al., 1992). Interestingly, BiP interacted with IRE1α even in the absence of cross-linker. Furthermore, IRE1α interaction with the misfolded α1-AT was slightly increased in the presence of an external ER stress inducer (Figure 6B). In contrast, the interaction between IRE1α and BiP was reduced as the intensity of the stress increased (Figure 6B). Taken together, these results suggest that misfolded proteins may bind and activate preformed complexes of IRE1α.
FIGURE 6:
The UPR sensor IRE1α interacts with misfolded antitrypsin. (A) HEK293 IRE1α–/– cells expressing IRE1α-HA were transfected with wild-type (WT) α1 antitrypsin, null Hong Kong α1 antitrypsin (NHK), α1 antitrypsin (E342K), or NHK/E324K and induced with 10 ng of doxycycline to drive the expression of IRE1α-HA. After 24 h of transfection, the cells were either left untreated or treated with DSP cross-linker (XL). As a control, HEK293 cells were transfected antitrypsin variants and treated with the cross-linker. The cross-linked samples were immunoprecipitated with anti-HA antibody and analyzed by immunoblotting for the indicated antigens. (B) The procedure was identical to A, but the cells were transfected with α1 antitrypsin (NHK)-venus and treated with the indicated concentrations of DTT for 1 h before cross-linking. (C) Models for activation of UPR sensors. Step 1: all three endogenous UPR sensors exist as preformed complexes associated with BiP in cells. Step 2: on ER stress, complexes of UPR sensors bind to misfolded proteins with concomitant release of BiP from UPR sensors. Step 3: on binding to misfolded proteins, IRE1α may undergo conformational changes without dramatic changes in the size of the complexes, which in turn activate its kinase and RNase activities. PERK may be phosphorylated and activated through assembling into large complexes from small complexes on binding with misfolded proteins. Conversely, misfolded proteins binding to ATF6α complexes may induce disassembly of complexes into smaller complexes, thus migrating to Golgi for the proteolytic processing.
DISCUSSION
In mammalian cells, three UPR branches, IRE1α, PERK, and ATF6α, are activated on the accumulation of misfolded proteins in the ER (Walter and Ron, 2011). Once activated, these UPR sensors initiate transcriptional and translational programs to restore protein homeostasis in the ER. It is unclear how these UPR sensors detect the accumulation of misfolded proteins and how they are activated (Snapp, 2012). In the first model, the luminal sensor domains of all three UPR sensors bind to BiP under homeostatic conditions (Bertolotti et al., 2000; Okamura et al., 2000; Shen et al., 2002; Carrara et al., 2015). As misfolded proteins accumulate during ER stress, BiP is released from UPR sensors, allowing UPR sensors to oligomerize and activate. In the second model, misfolded proteins may directly bind and activate all three UPR sensors during ER stress (Gardner and Walter, 2011; Gardner et al., 2013). In both models, changes in the complexes of UPR sensors appear to play a crucial role in their activation. One critical barrier to test these different models is to reliably monitor the changes in the endogenous complexes of all three UPR sensors during ER stress conditions. In the present study, we have monitored all three UPR sensors side by side by employing a BN–PAGE immunoblotting technique.
Our BN–PAGE data suggest that all three UPR sensors exist as preformed complexes under homeostatic conditions (Figure 6C). At present, it is uncertain whether they are homo-oligomers or hetero-oligomers. Several of our observations suggest that these large complexes of UPR sensors include homo-oligomers. First, although IRE1α forms hetero-oligomeric complexes with the Sec61 translocon (Sundaram et al., 2017), IRE1α migrates as a large complex (∼720 kDa) even in the absence of the Sec61 translocon complex. This suggests that the interacting Sec61 complex proteins by themselves may not account for the preformed complexes of IRE1α. Second, BiP is a known interacting protein of all three UPR sensors, but its depletion does not result in any apparent changes in the size of UPR complexes, supporting the notion that the proteins interacting with UPR sensors may not significantly contribute to large complexes of UPR sensors. Third, the fact that IRE1α and, to a lesser extent, PERK can be activated by DTT with no major increase in the size of their complexes suggests that they already contain homo-oligomers, since the monomeric form of IRE1α has been shown to be inactive (Zhou et al., 2006; Li et al., 2010). Fourth, our BN–PAGE data with ATF6α is consistent with previous studies that ATF6α appears to be in higher-order oligomers even in unstressed cells (Nadanaka et al. 2007; Gallagher and Walter, 2016). Finally, IRE1α dimerization mutant K121Y exhibits an increased number of smaller species on BN–PAGE as well as reduced high-molecular-weight cross-linked adducts compared with the wild type. These results support the idea that IRE1α complexes already contain multiple copies of IRE1α in unstressed cells.
Unlike PERK and ATF6α, the endogenous IRE1α complexes do not exhibit wholesale rearrangement on ER stress, except that the 240-kDa complex of IRE1α diminishes on ER stress. It is unlikely that BN–PAGE is not suitable to detect an ER stress-dependent increase in the size of IRE1α complexes, because it can apparently detect an increased PERK complexes as well as a decreased ATF6α complexes. Moreover, an ER stress-dependent increase in the size of IRE1α complexes can be observed with a slight overexpression of IRE1α. It remains to be determined why the size of the endogenous IRE1α complexes does not completely change to larger complexes on ER stress. One possibility is that there are not sufficient numbers of IRE1α complexes (∼416 molecules/cell) in the ER membrane to form larger complexes on ER stress (Kulak et al., 2014). Alternatively, the Sec61 translocon may selectively interact and suppress the formation of large complexes of IRE1α, thereby attenuating IRE1α activity during ER stress (Plumb et al., 2015; Sundaram et al., 2017). Overexpression of IRE1α may lead to the formation of ER stress-dependent large complexes, since overexpression results in free IRE1α molecules that are not associated with the Sec61 translocon (Plumb et al., 2015). Interestingly, an ER stress-dependent increase in the size of recombinant IRE1α complexes is apparent with DTT treatment, but less noticeable with TG treatment, suggesting that IRE1α is more responsive to DTT treatment. Conversely, PERK is robustly activated by moving to a larger complex with TG-induced ER stress, but it is slightly less sensitive to DTT-induced oligomerization and activation, which is in agreement with the earlier studies (DuRose et al., 2006).
ATF6α signal quickly returns within 6 h of ER stress treatment, suggesting that the activation of ATF6α is attenuated despite the presence of ER stress. In contrast to previous findings (Lin et al., 2007), the rate of ATF6α inactivation is very similar to the attenuation of IRE1α signaling during ER stress. This discrepancy may be due to the use of heterologously expressed ATF6α, which may respond inefficiently to ER stress, in these studies, whereas the endogenous ATF6α in the current study and studies from the Mori group show a robust activation and inactivation to ER stress treatments (Okada et al., 2003). Unlike IRE1α and PERK, changes in ATF6α complexes cannot easily be monitored since the ATF6α signal is lost due to the proteolytic release of its cytosolic N-terminal domain during ER stress. However, the inhibition of ATF6α cleavage by AEBSF (Okada et al., 2003) proves to be a useful tool for monitoring ER stress-dependent conversion of two different ATF6α complexes to a predominantly single complex.
Our ability to monitor ER stress-dependent changes in all three UPR sensors by BN–PAGE motivated us to test the role of BiP in suppressing complexes of these sensor proteins. We predicted that the depletion of BiP should increase the size of UPR complexes, whereas overexpression of BiP should reduce the size of UPR complexes even under normal conditions. However, we found no significant changes in the complexes of all three UPR sensors on depletion of BiP in cells. This is also further supported by our observation that only a small proportion of UPR sensors is constitutively activated in BiP-depleted cells. This result is not consistent with a previous study, which found prominent IRE1α activation in cells exposed to a bacterial toxin that cleaves BiP (Hu et al., 2009). It is therefore possible that the knockdown of BiP is inadequate to deplete highly abundant BiP in our experiment. Thus, the residual amount of BiP is sufficient to bind and suppress the activation of UPR sensors under homeostatic conditions.
Consistent with the previous studies, overexpression of BiP effectively suppressed all three UPR sensors on DTT treatment (Kohno et al., 1993; Bertolotti et al., 2000). Surprisingly, PERK can still be activated by TG treatment, whereas the activation of IRE1α and ATF6α are suppressed. This result implies that PERK is the most sensitive arm of the UPR, which also may explain why it is not easily attenuated during ER stress conditions (Lin et al., 2007). Surprisingly, the overexpression of BiP has less impact on UPR complexes, but it inhibits the diminishment of ∼240-kDa IRE1α complex on ER stress. It is plausible that overexpression of BiP alone may not be sufficient to reduce IRE1α complexes since a recent study suggests that BiP requires a J-protein cochaperone to disassemble IRE1α homo-oligomers (Amin-Wetzel et al., 2017). Our observation of preformed UPR complexes in cells raises the question of how these complexes are activated under ER stress conditions. We hypothesize that preformed complexes of UPR sensors may be activated by directly binding to misfolded proteins. Importantly, preformed complexes of UPR sensors in unstressed cells may explain the robust nature of UPR response even at low levels of ER stress (Rutkowski et al., 2006), since oligomers of UPR sensors would have a higher affinity for misfolded proteins (Gardner et al., 2011). Our cross-linking data that specifically capture the interaction between IRE1α and misfolded proteins provide evidence for the misfolded protein binding model. Moreover, a recent study further supports this model as they can detect a direct interaction between unfolded peptides and the purified luminal domain of IRE1α (Karagoz et al., 2017). In our model, we hypothesize that the BiP interaction with preformed complexes of IRE1α might play an important role to shield the peptide-binding groove in IRE1α and prevent it from inappropriate activation in unstressed cells. This model is consistent with the observation that BiP not only binds to monomers but also binds to homodimers of IRE1α (Amin-Wetzel et al., 2017). Future studies focusing on the identification of mutations in UPR sensors that selectively either disrupt BiP or misfolded protein binding will be crucial for understanding the mechanism of UPR sensors activation.
MATERIALS AND METHODS
Antibodies and reagents
The sources of antibodies and dilutions used for immunoblotting are as follows: anti-IRE1α (1:1000 dilution; Cell Signaling #3294, RRID:AB_823545), anti-PERK (1:1000 dilution; Cell Signaling #3192, RRID:AB_2095847), anti-ATF6α (1:1000 dilution; Cell Signaling #65880), anti-tubulin (1:10,000 dilution; Abcam #65880, RRID:AB_2241126), and anti-BiP/GRP78 (1:1000; BD Biosciences #610979, RRID:AB_398292). Anti-Sec61α (1:4000), anti-GFP (1:5000), and antitrypsin (1:1000) were generously gifted by Ramanujan Hegde (MRC Laboratory of Molecular Biology, United Kingdom). Anti-mouse goat HRP (1:10,000; Jackson Immunoresearch #11-035-003), anti-rabbit goat HRP (1:10,000; Jackson Immunoresearch #111-035-003, RRID: AB_2313567), and anti-HA magnetic beads (Fisher scientific #88836).
Reagents were purchased: DMEM (10-013-CV; Corning), fetal bovine serum (FBS) (89510-186, VWR), penicillin/streptomycin (15140122; Life Technologies), lipofectamine 2000 (11668019; Invitrogen), doxycycline (631311; Clontech), AEBSF (A8456; Sigma), TG (BML-PE180-0005; Enzo Life Sciences), tunicamycin (T7765; Sigma), bismaleimidohexane (BMH) (22330; ThermoFisher), dithiobis(succinimidyl propionate (DSP) (22585; ThermoFisher), protease inhibitor cocktail (11873580001; Roche), digitonin (300410; EMD Millipore), Phos-tag (300-93523; Wako), 3–12% BN–PAGE Novex Bis–Tris Gel (BN1003BOX; Invitrogen), SuperSignal West Pico or Femto Substrate (34080 or 34095; Thermo Scientific).
Cell culture and ER stress treatment
HEK 293-Flp-In T-Rex cells were cultured in high-glucose DMEM (Corning) containing 10% FBS (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies) at 5% CO2. IRE1α−/− HEK293-Flp-In T-Rex cells and IRE1α complemented cells were previously described (Plumb et al., 2015). To create ATF6α knock-out cells using CRISPR/Cas9, ATF6α targeting sequence (5′ GACACTGATGAGCTGCAAT 3′) was cloned into the guide RNA (gRNA) expression vector (Mali et al., 2013) and transfected into HEK 293-Flp-In T-Rex cells as described previously (Plumb et al., 2015). ATF6α knock-out clones were examined by immunoblotting against endogenous ATF6α. For ER stress treatment, HEK293 cells ([1.5–1.7] × 106/well) were plated on polylysine-coated six-well plates and grown overnight. The confluent cells were then treated with either DTT or TG. Note that the high-density plating of cells is required for detecting the endogenous IRE1α complexes. Also, high concentrations of ER stress reagents are required to maximally induce ER stress in these confluent cells.
For the depletion experiments, 0.5 × 106 cells were plated and transfected the next day with either BiP siRNA (5′ GGAGCGCAUUGAUACUAGA 3′) or Sec61α siRNA (Plumb et al., 2015) using lipofectamine 2000. After 24 h of the first transfection, the cells were again transfected with siRNA oligos. After 72 h of the first transfection, the cells were treated with TG as indicated in the figure legends. For BiP overexpression experiment, 0.7 × 106 cells were plated and grown overnight. The cells were then transfected with rat BiP plasmid (a kind gift from Ramanujan Hegde) or empty vector. After 24 h of transfection, the cells were treated with ER stress inducers as mentioned in the figure legends. After the treatment, the cells were washed with 1x PBS, harvested in 1.2 ml 1x PBS. The cells were spun at 10,000 × g for 1 min, and the pellets were flash frozen and stored at –80°C.
BN–PAGE immunoblotting
The cell pellets were lysed using either 2% digitonin buffer (50 mM Bis–Tris, pH 7.2, 1x protease inhibitor cocktail [Roche], 100 mM NaCl, and 10% glycerol) for 30 min. In some cases, the cell pellets were lysed using 1% Triton X-100 buffer (50 mM Bis–Tris, pH 7.2, 1x protease inhibitor cocktail, 100 mM NaCl, and 10% glycerol) for 30 min. The cell lysates were then diluted to a final concentration of 1% digitonin and 50 mM NaCl and centrifuged at 18,500 × g for 20 min at 4°C. The supernatant was collected and mixed with BN–PAGE sample buffer (Invitrogen) and 5% G520 (Sigma).
The samples were run using 3–12% BN–PAGE Novex Bis–Tris (Invitrogen) gel at 150 V for 1 h with the dark blue buffer (50 mM Tricine, pH 7, 50 mM Bis–Tris, pH 7, and 0.02% G250) at room temperature. The dark blue buffer was then exchanged with the light blue buffer (50 mM Tricine, pH 7, 50 mM Bis–Tris, pH 7, and 0.002% G250) for 4 h in the cold room. To probe BiP, the gels were run for 1 h with the dark blue buffer at room temperature and 3 h with the light blue buffer in the cold room. After electrophoresis, the gel was gently shaken in 1x Tris-glycine–SDS transfer buffer for 20 min to remove the residual blue dye. The transfer was performed using polyvinylidene difluoride (PVDF) membrane (EMD Millipore) for 1 h and 30 min at 85 V. After transfer, the membrane was fixed with 4% acetic acid and followed with a standard immunoblotting procedure.
Chemical cross-linking
HEK293 cells or HEK293 IRE1α-/- cells expressing IRE1α variants were plated on 12-well plates (0.25 × 106 cells/well) and grown overnight. The cells were either left untreated or treated with 7 μM TG for 60 min. After the treatment, the cells were washed once with potassium, Hepes, and magnesium (KHM)-containing buffer (20 mM HEPES, pH 7.4, 110 mM KAc, 2 mM MgAc) and permeabilized with 0.005% digitonin for 5 min on ice. The digitonin buffer was removed and washed once with KHM buffer. Subsequently, the permeabilized cells on plates were treated with BMH (Thermo Scientific) in KHM buffer for 30 min on ice. The cells were directly harvested in 2X SDS sample buffer, boiled, separated on 6% Tris–glycine–based gels, and immunoblotted with the indicated antibodies in the Supplemental Figure S5.
For DSP cross-linking, HEK293 IRE1α–/– cells expressing IRE1α-HA were plated on six-well plates (0.75 × 106 cells/well). The cells were then transfected with wild-type α1-AT, α1-AT (NHK) (a kind gift from Ramanujan Hegde), α1-AT (E342K), or α1-AT (NHK/E342K) and induced with 10 ng of doxycycline to drive the expression of IRE1α-HA. After 24 h of transfection, the cells were washed with KHM and treated with 1 mM DSP for 30 min at room temperature. The cross-linking reaction was then quenched with 0.1 M Tris, pH 8.0, for 15 min and harvested in radioimmunoprecipitation assay (RIPA) buffer. The cell lysates were centrifuged at 18,500 × g for 15 min at 4°C. The supernatant was incubated with 15 μl anti-HA magnetic beads for 2 h at 4°C, washed, eluted with 2X SDS sample buffer, and processed for immunoblotting for the indicated antigens in the figure.
Immunoblotting
The cell lysates were electrophoresed under reducing conditions on 10% Tricine (RPI research products)-based SDS–PAGE gels and electroblotted onto nitrocellulose membranes (Bio-Rad). Blots were incubated with primary antibodies prepared in 1x PBS/Tween containing 5% bovine serum albumin/0.02% NaN3 for 1 h and 30 min at room temperature. The secondary antibodies were prepared in 5% milk with 1x PBS/Tween and incubated for 1 h at room temperature. Proteins were detected with either SuperSignal West Pico or Femto Substrate (Thermo Scientific), exposed to Kodak films (Dot Scientific), and developed. Immunoblots were quantified using ImageJ, and graphs were plotted by GraphPad Prism.
Phostag assay
IRE1α phosphorylation was detected by previously described method (Yang et al., 2010). Briefly, a 5% SDS–PAGE gel was made containing 25 μM Phos-tag (Wako). SDS–PAGE was run at 100 V for 2 h and 40 min. The gel was transferred to nitrocellulose (Bio-Rad) and followed with immunoblotting.
Luciferase assay
The luciferase assay was performed according to the manufacturer protocol (Roche) with the following modifications. Briefly, the luciferase reporter p5xUPRE-GL3 (formerly known as p5XATF6GL) (Wang et al., 2000), a kind gift from Ramanujan Hegde, was cotransfected with α1-AT variants. After 24 h of transfection, cells were harvested in luciferase lysis buffer and centrifuged to collect the supernatant for the assay. The samples were plated in a 96-well tissue culture plate and read using a BioTek Synergy plate reader.
Supplementary Material
Acknowledgments
We thank Mariappan lab members for useful discussions. We thank Sha Sun for providing comments on the manuscript. We thank Zairong Zhang for the help with quantification of native PAGE immunoblots. We thank Peter Cresswell for null Hong Kong α1-antitrypsin-venus plasmid. We are thankful to Ramanujan Hegde for providing various antibodies and plasmids. This work is supported by the Yale School of Medicine start-up package and National Institutes of Health Grant 1R01GM11738601.
Abbreviations used:
- ATF6α
activating transcription factor 6α
- ER
endoplasmic reticulum
- IRE1α
inositol-requiring enzyme-1α
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- UPR
unfolded protein response
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0594) on April 10, 2018.
REFERENCES
- Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. (2016). A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell , 1867–1882 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri K, Harris AL. (2008). Activating transcription factor 4. Int J Biochem Cell Biol , 14–21. [DOI] [PubMed] [Google Scholar]
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP, Ron D. (2017). A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell , 1625–1637 e1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. (2011). Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem , 1089–1115. [DOI] [PubMed] [Google Scholar]
- Arsene F, Tomoyasu T, Bukau B. (2000). The heat shock response of Escherichia coli. Int J Food Microbiol , 3–9. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol , 326–332. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. (2012). Cleaning up: ER-associated degradation to the rescue. Cell , 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature , 92–96. [DOI] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. (2015). Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife , e03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. (2014). Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol , 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti BJ, Devaraneni PK, Yang Z, David LL, Skach WR. (2015). Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell , 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell , 1197–1206. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. (2005). On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA , 18773–18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuRose JB, Tam AB, Niwa M. (2006). Intrinsic capacities of molecular sensors of the unfolded protein response to sense alternate forms of endoplasmic reticulum stress. Mol Biol Cell , 3095–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CM, Walter P. (2016). Ceapins inhibit ATF6alpha signaling by selectively preventing transport of ATF6alpha to the Golgi apparatus during ER stress. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. (2013). Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol , a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P. (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science , 1891–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, Prado K, Thamsen M, Caswell D, Macias H, et al. (2014). Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell , 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell , 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature , 271–274. [DOI] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell , 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. (2012). The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol , 89–102. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol , 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science , 104–107. [DOI] [PubMed] [Google Scholar]
- Hu CC, Dougan SK, Winter SV, Paton AW, Paton JC, Ploegh HL. (2009). Subtilase cytotoxin cleaves newly synthesized BiP and blocks antibody secretion in B lymphocytes. J Exp Med , 2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagoz GE, Acosta-Alvear D, Nguyen HT, Lee CP, Chu F, Walter P. (2017). An unfolded protein-induced conformational switch activates mammalian IRE1. Elife , e30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. (2007). Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol , 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. (2011). Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature , 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. (1993). The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol , 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Amin-Wetzel N, Ron D. (2017). Generic membrane-spanning features endow IRE1alpha with responsiveness to membrane aberrancy. Mol Biol Cell , 2318–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods , 319–324. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol , 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. (2010). Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA , 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. (2007). IRE1 signaling affects cell fate during the unfolded protein response. Science , 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas DA, Evans DL, Finch JT, Carrell RW. (1992). The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature , 605–607. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013). RNA-guided human genome engineering via Cas9. Science , 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell , 743–756. [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K. (2007). Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol , 1027–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. (2009). Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res , 2496–2504. [DOI] [PubMed] [Google Scholar]
- Oikawa D, Kitamura A, Kinjo M, Iwawaki T. (2012). Direct association of unfolded proteins with mammalian ER stress sensor, IRE1beta. PLoS One , e51290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Haze K, Nadanaka S, Yoshida H, Seidah NG, Hirano Y, Sato R, Negishi M, Mori K. (2003). A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J Biol Chem , 31024–31032. [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. (2000). Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun , 445–450. [DOI] [PubMed] [Google Scholar]
- Plumb R, Zhang ZR, Appathurai S, Mariappan M. (2015). A functional link between the co-translational protein translocation pathway and the UPR. Elife , e07426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature , 663–669. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. (2006). Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol , e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler AJ, Schekman R. (2009). In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc Natl Acad Sci USA , 17775–17780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell , 99–111. [DOI] [PubMed] [Google Scholar]
- Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, Yates JR, 3rd, Su AI, Kelly JW, Wiseman RL. (2013). Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep , 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. (1988). A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem , 7330–7335. [PubMed] [Google Scholar]
- Snapp EL. (2012). Unfolded protein responses with or without unfolded proteins? Cells , 926–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC. (2000). Pancreatic eukaryotic initiation factor-2alpha kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem J (Pt 2), 281–293. [PMC free article] [PubMed] [Google Scholar]
- Sundaram A, Plumb R, Appathurai S, Mariappan M. (2017). The Sec61 translocon limits IRE1alpha signaling during the unfolded protein response. Elife , e27187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Anken E, Braakman I. (2005). Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol , 191–228. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science , 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ. (2016). Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature , 326–335. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem , 27013–27020. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. (2006). Blue native PAGE. Nat Protoc , 418–428. [DOI] [PubMed] [Google Scholar]
- Yang L, Xue Z, He Y, Sun S, Chen H, Qi L. (2010). A Phos-tag-based approach reveals the extent of physiological endoplasmic reticulum stress. PLoS One , e11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell , 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell , 881–891. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. (2006). The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA , 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.