Abstract
In many organisms, early embryonic development is characterized by a series of reductive cell divisions that result in rapid increases in cell number and concomitant decreases in cell size. Intracellular organelles, such as the nucleus and mitotic spindle, also become progressively smaller during this developmental window, but the molecular and mechanistic underpinnings of these scaling relationships are not fully understood. For the mitotic spindle, changes in cytoplasmic volume are sufficient to account for size scaling during early development in certain organisms. This observation is consistent with models that evoke a limiting component, whereby the smaller absolute number of spindle components in smaller cells limits spindle size. Here we investigate the role of a candidate factor for developmental spindle scaling, the microtubule polymerase XMAP215. Microinjection of additional XMAP215 protein into Xenopus laevis embryos was sufficient to induce the assembly of larger spindles during developmental stages 6.5, 7, and 8, whereas addition of a polymerase-incompetent XMAP215 mutant resulted in a downward shift in the in vivo spindle scaling curve. In sum, these results indicate that even small cells are able to produce larger spindles if microtubule growth rates are increased and suggest that structural components are not limiting.
INTRODUCTION
In eukaryotic cells, accurate segregation of chromosomes during cell division requires the assembly of a spindle of the appropriate size and shape. For a given cell type, the constancy of the spindle form is remarkable, but spindle size can vary considerably from one cell type to the next. This is also evident during early embryonic development when rapid cycles of cell division, coupled with little or no cell growth, quickly produce large numbers of smaller and smaller cells (Wuhr et al., 2008; Wilbur and Heald, 2013; Crowder et al., 2015). Concomitant with these changes, the sizes of many internal components of the cell, including the nucleus (Levy and Heald, 2010; Jevtic and Levy, 2015) and mitotic spindle (Wuhr et al., 2008; Wilbur and Heald, 2013), also decrease.
How changes in cell size affect changes in spindle size remains an open question. Spindles assembled in discrete droplets of cytoplasmic egg extract scale with droplet volume, suggesting that spindle size is regulated by cell size even in a developmentally and compositionally static system (Good et al., 2013; Hazel et al., 2013). This process likely involves passive regulation of spindle size through component limitation, a model that posits that as cells become smaller, so does the absolute number of components from which the spindle can be made, with the association rates of these components decreasing more quickly, ultimately resulting in smaller steady-state spindles (Goehring and Hyman, 2012; Reber and Goehring, 2015). Although the extract droplets used in those experiments represent a reasonable proxy for blastomeres during the reductive divisions that characterize early Xenopus development, it is clear that the underlying mechanisms in vivo are likely more complicated. For example, developmental changes in the localization and activity of effector proteins can affect spindle size without gross changes in the expression of those proteins (Wilbur and Heald, 2013).
Even if a limiting-component model is evoked to explain spindle scaling in vivo, it is unclear which component (or components) becomes limiting. For clarity, it should be noted that we have adopted a broad definition of what is meant by “component” and one that is in alignment with a recent review on scaling mechanisms (Reber and Goehring, 2015). In contrast to a narrower definition that limits “component” to structural elements, we define a component as being any element that associates with the assembly/organelle being considered. Thus, a limiting component need not be a structural building block, and the effects of its limitation on size can be exerted indirectly by affecting the dynamic balance of building block incorporation. In the context of spindle scaling, this might manifest itself as negative regulation of tubulin association with growing spindle microtubules (MTs). Several putative spindle scaling factors have been identified. For example, in Caenorhabditis elegans the relationship between cell and spindle size during early development depends on centrosome size (Greenan et al., 2010) and, more specifically, on the amount of SPD-2 (Decker et al., 2011), a centrosomal protein that becomes limiting as the available pool is divided among a growing number of total centrosomes in the embryo. In vertebrates, the MT severing enzyme katanin exhibits different levels of inherent activity in two closely related, but disparately sized, frog species, X. laevis and X. tropicalis, and contributes to the observed differences in interspecies spindle length (Loughlin et al., 2011). In these two species, the activity of katanin is inversely proportional to spindle size (Loughlin et al., 2011), and, therefore, a reduction in the number of katanin molecules through reductive cell divisions would be expected to result in larger, not smaller, spindles. Thus, though it is possible that katanin regulates spindle size during early development, it is difficult to envision katanin playing the role of a limiting component in this context.
Another putative scaling factor is the processive MT polymerase XMAP215 (Brouhard et al., 2008; Reber et al., 2013), which has long been known to affect MT plus-end dynamics (Gard and Kirschner, 1987; Vasquez et al., 1994). Orthologues include Mini-spindles in Drosophila melanogaster, TOGp in humans, Dis1 in Schizosaccharomyces pombe, and ZYG-9 in C. elegans. XMAP215 is a multidomain protein containing multiple NH2-terminal tumor overexpressed gene (TOG) domains that facilitate binding to GTP-tubulin heterodimers (Popov et al., 2001; Al-Bassam et al., 2007; Ayaz et al., 2012). The NH2-terminal region also contains a MT lattice binding domain that is thought to work in conjunction with the TOG domains to confer MT polymerase activity to the protein (Brouhard et al., 2008; Widlund et al., 2011; Fox et al., 2014; Byrnes and Slep, 2017). A mutant form of the protein, XMAP215TOG1-5AA, in which all five of the protein’s TOG domains have been mutated such that they are unable to bind tubulin heterodimers yet retains an intact lattice binding domain, is able to bind to MTs and growing MT ends but is unable to facilitate incorporation of tubulin dimers into those ends (Widlund et al., 2011). Interestingly, and in contrast to the inverse relationship between katanin activity and spindle size, spindle size is directly proportional to XMAP215 concentration/activity in X. laevis egg extracts (Reber et al., 2013), making it a plausible candidate for an in vivo spindle scaling factor. To test this hypothesis, we modulated the levels and activity of XMAP215 by injection of recombinant wtXMAP215 or the XMAP215TOG1-5AA mutant (herein referred to as “mutXMAP215”) protein into X. laevis embryos and characterized the effects of these manipulations on spindle scaling during development.
RESULTS AND DISCUSSION
Addition of wtXMAP215 increases spindle length in early X. laevis embryos
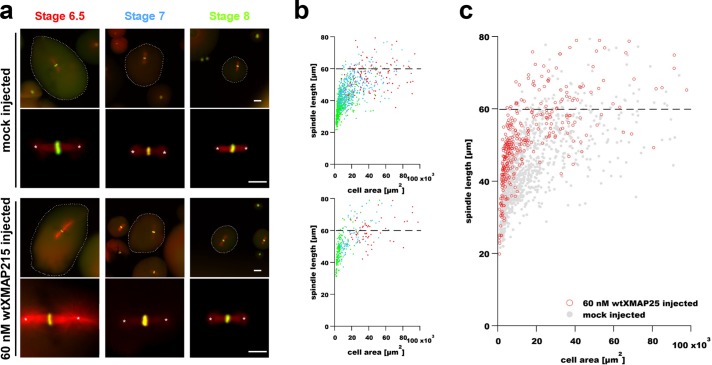
An increase in the amount of a putative spindle scaling factor is expected to result in a concomitant shift in the scaling relationship between cell and spindle size, provided other components are not limiting. Therefore, we hypothesized that the addition of exogenous XMAP215 to developing embryos would shift the cell–spindle scaling curve upward, with larger-than-normal spindles being assembled for a given cell size. To test this, different concentrations of purified wild-type XMAP215 protein (wtXMAP215; Brouhard et al., 2008) were microinjected into X. laevis embryos at the one-cell stage. The concentration of endogenous XMAP215 is ∼120 nM in early embryos (Reber et al., 2013; Peshkin et al., 2015) and is thought to remain relatively constant until transcriptional activation at the mid-blastula transition (MBT) at stage 8.5 (Peshkin et al., 2015). Embryos were allowed to progress through pre-MBT developmental stages before being processed for immunofluorescence with subsequent measurement of their spindle lengths. Embryos at developmental stages 6.5, 7, and 8 (Nieuwkoop and Faber, 1967) were dissociated and fixed, and spindle MTs and DNA were labeled (Nieuwkoop and Faber, 1967; Jevtic and Levy, 2015). Figure 1a shows representative images of spindles in blastomeres isolated from embryos that were either mock injected with a buffer-only control or injected with 60 nM recombinant wtXMAP215 (to a final concentration of ∼180 nM; top two and bottom two rows, respectively; see also Supplemental Figure S2, a and b). Spindle length was measured as the distance between centrosomal aster centers (Figure 1a) and plotted as a function of cell cross-sectional area (Figure 1, b and c, and Supplemental Figure S1). In mock-injected controls, scatter plots of spindle size versus cell cross-sectional area revealed a scaling relationship between spindle and cell size that is consistent with previously reported in vivo results (Wuhr et al., 2008). This plot exhibits a characteristic scaling regime in smaller cells in which spindle size is dependent on cell size and a second regime that is cell-size independent and indicates an intrinsic upper limit to spindle size in larger cells (Figure 1b). Admittedly, the source of the large variation seen in spindle length measurements at a specific cell size is not fully understood. We interpret this variation as a being the result of different isovolumetric geometries that cell shape can take and that longer cells might produce longer spindles compared with shorter cells of the same volume (Li and Jiang, 2017). However, we cannot entirely rule out some sort of measurement artifact. Interestingly, the addition of 60 nM excess wtXMAP215 caused a statistically significant upward shift in spindle size (Figure 1, a–c, and Supplemental Figure S1) even in large early-stage blastomeres (Supplemental Figure S2, c and d, “stage 6.5″), which are purportedly well within the size-independent regime of the X. laevis cell–spindle scaling curve (Wuhr et al., 2008). We interpret this result to mean that each of the growing MT ends in stage 6.5 blastomeres could bind more polymerase molecules provided the concentration of XMAP215 was raised. This result is consistent with other published data, indicating that spindle length can indeed be increased above the apparent upper limit in vivo (Novakova et al., 2016; Kyogoku and Kitajima, 2017). Regardless of the nature of the upper limit to spindle size, the upward shift in the scaling curve caused by wtXMAP215 addition implicitly suggests that X. laevis blastomeres, even small ones, have an ample supply of spindle components and are capable of building larger spindles but might be limited in their ability to efficiently incorporate the components, that is, these other putative components are not limiting.
FIGURE 1:
Microinjection of recombinant XMAP215 results in increased spindle size in X. laevis early development. (a) Images show representative spindles in blastomeres isolated from embryos and fixed at the indicated stages. The two top rows show spindles in blastomeres from control (mock-injected) embryos, whereas the two bottom rows show spindles from blastomeres isolated from embryos injected with 60 nM wtXMAP215. Spindle MTs were imaged using indirect immunofluorescence and are shown pseudo-colored in red, and DNA was labeled using Sytox Green nucleic acid stain and is shown pseudo-colored in green. Scale bar = 50 µm (images in first and third rows). Scale bar = 30 µm (higher-magnification spindle images in second and fourth rows). Asterisks mark aster centers, and the distance between each pair of asterisks was used for measurements of spindle lengths contained in subsequent panels. Cell boundaries, indicated by dashed outlines shown in lower-magnification spindle images, were manually traced and used to determine cell cross-sectional area for scaling plots. (b) Spindle length measurements are shown plotted as a function of blastomere cross-sectional area in mock-injected (top plot) and in embryos injected with 60 nM wtXMAP215 (bottom plot). Data point colors correspond to stage-specific text colors shown in panel a. (c) Data from both graphs in b were plotted in a single graph and show a general upward shift in spindle length with the injection of 60 nM wtXMAP215. Dashed horizontal lines indicate the putative upper limit to spindle size in X. laevis embryos (Wuhr et al., 2008).
Assuming there is not a proportional increase in the number of growing MT ends with cell size, a limited number of XMAP215 binding sites at each growing MT end (Brouhard et al., 2008) suggests that the effect of excess XMAP215 addition on spindle length changes should be saturable (Reber et al., 2013). To test this prediction, we injected additional wtXMAP215 (300 nM) into single-celled embryos and measured spindle lengths in early-stage blastomeres (Supplemental Figure S2). Western blot analyses were used to determine the amount of total wtXMAP215 present within injected embryos at the indicated stages (Supplemental Figure S2). The monotonic decrease in total protein levels through developmental stage 8 suggests that the exogenous protein is most likely partially unstable and is degraded during developmental progression. This instability made interpretation of the data somewhat difficult, however, with the exception of stage 6.5 embryos, stage-specific trends in spindle length typically exhibited a biphasic response to excess wtXMAP215, with higher levels (300 nM of injected wtXMAP215) having a less-pronounced effect on spindle length than intermediate levels (60 nM of injected wtXMAP215; Supplemental Figure S2, c and d). Because XMAP215 binds tubulin as well as MTs, it is possible that at these high concentrations, XMAP215 might actually deplete tubulin from the cytoplasm. Taken as a whole, these results suggest that the assumption of a relatively constant number of growing MT ends might be invalid at higher wtXMAP215 concentrations, which is consistent with recent observations implicating wtXMAP215 as playing a role in MT nucleation (unpublished data).
Addition of a polymerase-deficient mutant form of XMAP215 decreases spindle length in early X. laevis embryos
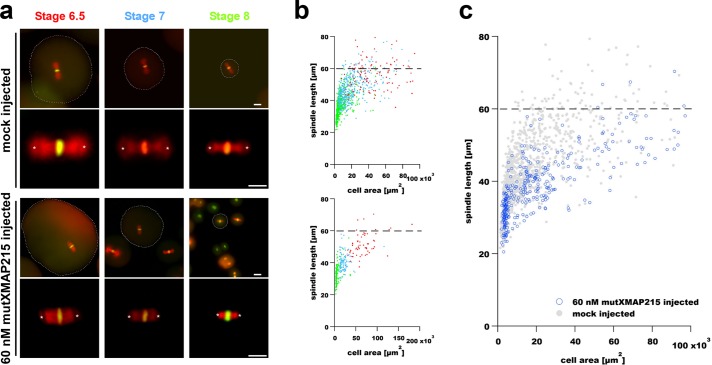
On the basis of the results of wtXMAP215 addition experiments, we reasoned that addition of the XMAP215TOG1-5AA mutant (mutXMAP215) should result in a titratable and downward shift in the spindle length scaling curve. For this analysis, isolated blastomeres were mock injected (buffer-only control) or were injected with mutXMAP215 (at 60 nM excess), and spindle length was again analyzed relative to the cross-sectional area of the cells at different developmental time points (Figure 2). In contrast to injection of wtXMAP215, the same concentration of injected mutXMAP215 resulted in smaller-than-expected spindles in early-stage blastomeres (Figure 2, b and c). Stage-specific estimations of protein levels via Western blot analyses indicated that, like wtXMAP215, the mutant form of the polymerase was also unstable, with amounts of total XMAP215 protein decreasing as a function of developmental time (Supplemental Figure 3). For unknown reasons, the effects of mutXMAP215 addition were most pronounced in stage 7 embryos (Supplemental Figure S3, c and d); however, the downward shift in the scaling curve was statistically significant over the total range of measurements (Figure 2c and Supplemental Figure 1). In summary, these results add credence to an in vivo model of spindle scaling whereby XMAP215 levels directly impact spindle length via binding at growing MT ends.
FIGURE 2:
Microinjection of a polymerase-deficient mutant XMAP215 decreases spindle length in early X.laevis embryos. (a) Images show representative spindles in blastomeres isolated from embryos at the indicated stages. The two top rows show spindles in blastomeres from control (mock-injected) embryos, whereas the two bottom rows show spindles from blastomeres isolated from embryos injected with 60 nM mutXMAP215. Spindle MTs and DNA were labeled as in Figure 1. Scale bar = 50 µm (images in first and third rows). Scale bar = 30 µm (higher-magnification spindle images in second and fourth rows). Spindle length was again measured as the aster center–to–aster center distance (centers are marked with white asterisks). Cell boundaries, indicated by dashed outlines shown in lower-magnification spindle images, were manually traced and used to determine cell cross-sectional area for scaling plots. (b) Spindle length measurements are shown plotted as a function of blastomere cross-sectional area in mock-injected embryos (top plot) and in embryos injected with 60 nM mutXMAP215 (bottom plot). Data point colors correspond to stage-specific text colors shown above images in panel a. (c) Data from both graphs in b were plotted in a single graph and show a general downward shift in spindle length with the injection of 60 nM mutXMAP215. Dashed horizontal lines indicate the putative upper limit to spindle size in X. laevis embryos (Wuhr et al., 2008).
High levels of wtXMAP215 induce the formation of supernumerary MT-organizing centers in early X. laevis embryos
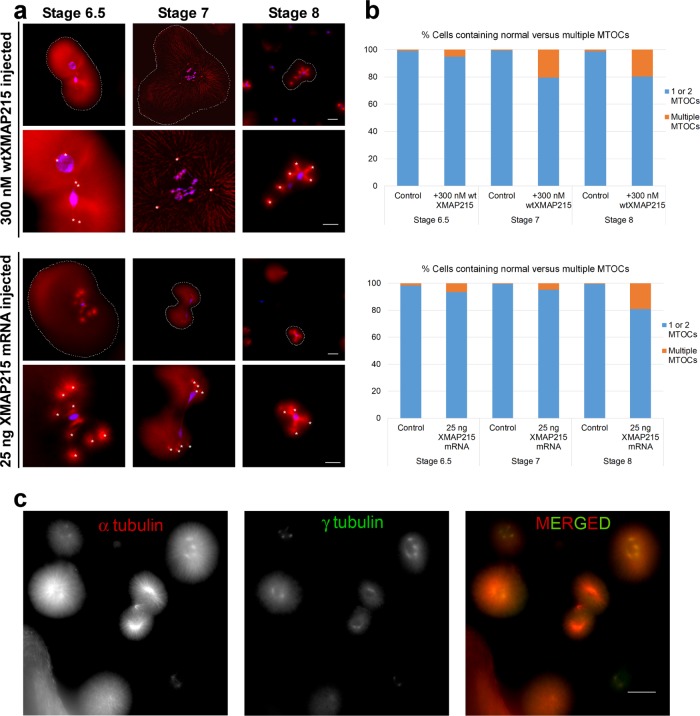
The biphasic response of spindle size to increasing XMAP215 levels might be caused by increased MT nucleation throughout the cytoplasm of early-stage blastomeres, resulting in an increased absolute number of growing MT ends and more competition for a limited supply of XMAP215 (and perhaps tubulin or other microtubule associated proteins). Indeed, when we counted the detectable MT foci per cell in embryos injected with 300 nM wtXMAP215, we observed an increase in the proportion of cells containing supernumerary MT foci, that is, >2 MT-organizing centers (MTOCs)/cell (Figure 3, a and b). The proportion of cells with supernumerary MT foci increased with developmental time. We speculate that this might be an indirect effect of increases in surface area to volume ratio during reductive divisions or perhaps of changes in cytoplasmic composition.
FIGURE 3:
High concentrations of wtXMAP215 induce the formation of supernumerary MTOCs in early X. laevis embryos. (a) Embryos were injected at the one-cell stage with either 300 nM wtXMAP215 (top two rows) or 25 ng wtXMAP215 mRNA (bottom two rows), fixed, and then processed for immunofluorescence. MTs were imaged using indirect immunofluorescence and are shown pseudo-colored in red, whereas DNA was labeled using Sytox Green nucleic acid stain (shown pseudo-colored in blue). Images show examples of cells with supernumerary MT foci displays in blastomeres isolated at the indicated stages. Cell boundaries are indicated by dashed outlines (first and third rows). For higher-magnification images, supernumerary MTOCs are overlaid with white asterisks. Scale bar = 50 µm (images in first and third rows). Scale bar = 30 µm (images in second and fourth rows). (b) The MT foci per cell were counted and plotted at the indicated stages as the percentage of cells containing a normal number of MTOCs (i.e., 1 or 2) or supernumerary MTOC numbers (i.e., >2). The top graph shows data from embryos injected with 300 nM wtXMAP215; the bottom graph shows data taken from single-celled blastomeres injected with 25 ng XMAP215 mRNA. (c) Embryos were injected at the one-cell stage with 25 ng XMAP215 mRNA and processed for immunofluorescence for α-tubulin (pseudo-colored in red) and γ-tubulin (pseudo-colored in green). Representative images of blastomeres from stage 8 embryos containing multiple MTOCs are shown.
On the basis of these findings, we reasoned that the addition of even higher levels of XMAP215 should result in a more severe phenotype. Because of concerns about the stability of our exogenous protein in embryos stemming from our Western analyses, instead of simply injecting higher amounts of wtXMAP215 protein, we injected mRNA encoding XMAP215 into one-cell-stage embryos. Although this treatment led to notable increases in wtXMAP215 levels at all developmental stages quantified (Supplemental Figure S4), the proportion of cells containing supernumerary MT foci showed either no differences or modest increases over embryos injected with 300 nM wtXMAP215 (Figure 3, a and b). Interestingly, MT foci that formed in the presence of excess wtXMAP215 were positive for γ tubulin (Figure 3c), suggesting that these foci are capable of nucleating MTs (Petry and Vale, 2015), thus potentially increasing the effective density of growing MT ends in the affected blastomeres.
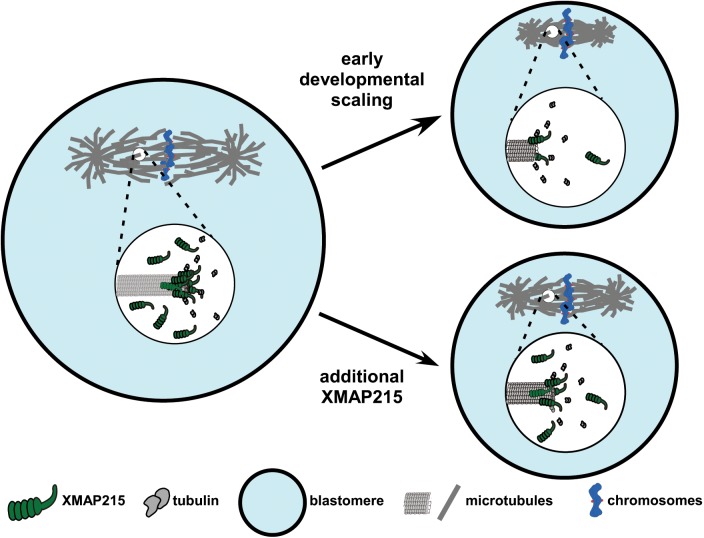
In sum, our data support a scaling model in which relative numbers of XMAP215 molecules and growing MT ends, regulate spindle length during development (Figure 4).
FIGURE 4:
Model of XMAP215 regulation of MT end dynamics and spindle scaling during X. laevis development. The cartoon depicts the mechanism of XMAP215-dependent spindle scaling during development. As cell size decreases during early development, the number of XMAP215 molecules in each blastomere is concomitantly reduced, resulting in fewer XMAP215 molecules per MT plus-end, slower MT growth rates and, by mass action, smaller spindles. Additional XMAP215 can compensate for this loss and, because other components are not limiting, allow for the generation of larger-than-expected spindles for a blastomere of a given size.
MATERIALS AND METHODS
Embryos
Embryos were the product of in vitro fertilization of freshly laid X. laevis eggs using crushed X. laevis testes. Eggs were obtained from X. laevis frogs (Nasco). After being dejellied in 2.5% cysteine (pH 7.8) dissolved in 1/3 × Marc’s modified Ringer’s (MMR; 20 × MMR = 2 mM EDTA, 2 M NaCl, 40 mM KCl, 20 mM MgCl2, 40 mM CaCl2, 100 mM HEPES, pH 7.8), the embryos were cultured at 16°C or 23°C in 1/3 × MMR using standard methods (Sive et al., 2010). Embryos were staged according to Nieuwkoop and Faber (1967). All the experiments involving X. laevis were in compliance with the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and were performed in accordance with national regulatory standards.
Recombinant XMAP215 proteins
His-tagged recombinant wild-type and mutant XMAP215 proteins were kind gifts from Simone Reber (Humboldt-Universität zu Berlin). One-cell-stage embryos were injected with recombinant XMAP215 protein to a final concentration of 180 and 420 nM (endogenous concentration is ∼120 nM). Control embryos were mock injected with Tris-buffered saline (TBS).
RNA
For ectopic expression, capped mRNA was transcribed from constructs in vitro using SP6 or T7 mMessage mMachine Kit (Ambion). RNA was purified with LiCl precipitation and resuspended in nuclease-free double-distilled H2O.
Microinjections
Embryos at the one-cell stage were transferred to 1/3x MMR plus 2.5% Ficoll, and the dorsal region (i.e., the animal cap) was injected using a PicoSpritzer III (Parker). Microinjection was conducted as described (Jevtic and Levy, 2015). After 45 min, the buffer was changed to either 1/3 × MMR or 1 × Ca++- and Mg++-free medium (5 M NaCl, 1 M KCl, 1 M NaHCO3, 1 M Tris-Cl, pH 7.6). and embryos were allowed to develop to the indicated stages.
Western blot analyses
Embryo samples were prepared as described (Edens and Levy, 2014). Proteins were separated on SDS–PAGE gels and transferred to polyvinylidene difluoride membranes using a wet blotting apparatus. Membranes were blocked in Odyssey PBS Blocking Buffer (Li-Cor). The primary antibody rabbit anti-XMAP215 was used at 1:1000 (Novus; NBP1-06513) and DM1A mouse anti–α-tubulin was used at 1:1000 (Santa Cruz Biotechnology; sc-32293). Secondary antibodies IRDye 800CW–conjugated anti-rabbit (Li-Cor) and IRDye 680RD–conjugated anti-mouse (Li-Co) were used at 1:20,000. Blots were scanned on a Li-Cor Odyssey CLx instrument, and band quantification was performed with ImageStudio. For a given stage, XMAP215 band intensity was normalized to the α-tubulin signal in the sample.
Whole-mount fluorescence immunocytochemistry
Whole-mount fluorescence immunocytochemistry was conducted as described (Jevtic and Levy, 2015). Briefly, after removing the vitelline membrane, embryos were moved to a Ca++- and Mg++-free medium, which promotes cell disassociation. At the desired stages, isolated blastomeres were fixed and processed for immunocytochemistry. After bleaching of their pigment granules, blastomeres were incubated overnight at 4°C with primary mouse anti–α-tubulin DM1A (1:200 dilution in BBT, 1x PBS with 0.1% bovine serum albumin and 0.1% Triton X-100, plus 5% boiled donkey serum) and rabbit anti–γ-tubulin (Abcam, ab11321; 1:200 dilution in BBT plus 5% boiled donkey serum). After a few washes in BBT and blocking for 2 h in BBT plus 5% boiled donkey serum at room temperature, blastomeres were incubated at 4°C overnight with Alexa Fluor 568–conjugated donkey anti-mouse secondary antibody and Alexa Fluor 488–conjugated goat anti-rabbit secondary antibody (Invitrogen; A10037 and A32731, respectively; 1:200 dilution in BBT plus 5% donkey serum). To visualize DNA, isolated blastomeres were washed a few times in TBST (0.1% Tween 20 in 1x TBS) and incubated overnight at 4°C with Sytox Green nucleic acid stain (Life Technologies; 1:1000 dilution in TBST). Finally, embryonic cells were dehydrated in methanol and cleared in 2:1 benzyl benzoate/benzyl alcohol before imaging.
Microscopy and imaging
All the images for spindle measurements were acquired using either a QIClick Digital charged-coupled device camera, Mono, 12-bit (model QIClick-F-M-12) mounted on an Olympus BX51 fluorescence microscope or a scientific complementary metal oxide semiconductor camera (Flash 4.0; Hamamatsu) mounted on an IX81 stand equipped with a spinning-disk confocal head (CSU-X1; Yokogawa). Confocal illumination was provided by an LMM5 laser launch (Spectral Applied Research). Integration of all imaging system components was provided by Biovision Technologies. Image acquisition was performed using either Metamorph 7.7 software (Molecular Devices) or Olympus cellSens software. The following objectives were used in these studies: 10× (0.25 NA, air) and 20× (0.50 NA, air).
Statistics
Indicated statistical tests were conducted using Igor Pro 7 software (ver 7.0.4.1; Wavemetrics). For t tests, two-tailed comparisons of means were used with α = 0.5. Linear regressions of log-log plots of spindle length versus cell area were also performed using Igor Pro 7 with dashed lines indicating the boundaries of 95% confidence intervals for the fits.
Supplementary Material
Acknowledgments
We thank current members of the Gatlin and Levy labs for their invaluable comments and discussions related to the work. Additionally, we thank T. Mitchison, C. Field, J. Oakey, other members of the Marine Biological Laboratory Cell Division Group, and Tony Hyman for helpful discussions; M. Tomschik and J. Bisht for help with handling the frogs; Simone Reber for providing the XMAP215 constructs and purified XMAP215 proteins used in these studies; and Amy Fluet for her editing of manuscript drafts. This work was supported by grants R01GM102428 (to J.C.G.) and R01GM113028 to (D.L.L.) from the National Institute of General Medical Sciences (NIGMS) as well as fellowships from the Marine Biological Laboratory Whitman Center (to J.C.G.). J.C.G.’s research is also funded by the Pew Scholars Program in the Biomedical Sciences. This project was also supported in part by a grant from the NIGMS (2P20GM103432) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- MT
microtubule
- MTOC
microtubule-organizing center
- TOG
tumor overexpressed gene
- XMAP215
Xenopus microtubule-associated protein 215
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-01-0011) on April 5, 2018.
REFERENCES
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. (2007). Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure , 355–362. [DOI] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. (2012). A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science , 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. (2008). XMAP215 is a processive microtubule polymerase. Cell , 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes AE, Slep KC. (2017). TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J Cell Biol , 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder ME, Strzelecka M, Wilbur JD, Good MC, von Dassow G, Heald R. (2015). A comparative analysis of spindle morphometrics across metazoans. Curr Biol , 1542–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, Hyman AA. (2011). Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol , 1259–1267. [DOI] [PubMed] [Google Scholar]
- Edens LJ, Levy DL. (2014). cPKC regulates interphase nuclear size during Xenopus development. J Cell Biol , 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JC, Howard AE, Currie JD, Rogers SL, Slep KC. (2014). The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol Biol Cell , 2375–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. (1987). A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol , 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Hyman AA. (2012). Organelle growth control through limiting pools of cytoplasmic components. Curr Biol , R330–R339. [DOI] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. (2013). Cytoplasmic volume modulates spindle size during embryogenesis. Science , 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Jaensch S, Gharakhani J, Julicher F, Hyman AA. (2010). Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr Biol , 353–358. [DOI] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. (2013). Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science , 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtic P, Levy DL. (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Curr Biol , 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku H, Kitajima TS. (2017). Large cytoplasm is linked to the error-prone nature of oocytes. Dev Cell , 287–298. [DOI] [PubMed] [Google Scholar]
- Levy DL, Heald R. (2010). Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell , 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang H. (2017). Geometric asymmetry induces upper limit of mitotic spindle size. Biophys J , 1503–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell , 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. (eds.) (1967). Normal Table of Xenopus laevis [Daudin]—A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis, Amsterdam: North-Holland. [Google Scholar]
- Novakova L, Kovacovicova K, Dang-Nguyen TQ, Sodek M, Skultety M, Anger M. (2016). A balance between nuclear and cytoplasmic volumes controls spindle length. PLoS ONE e0149535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wuhr M, Pearl E, Haas W, Freeman RM, Jr, Gerhart JC, Klein AM, Horb M, Gygi SP, Kirschner MW. (2015). On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev Cell , 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Vale R. (2015). Microtubule nucleation at the centrosome and beyond. Nat Cell Biol , 1089–1093. [DOI] [PubMed] [Google Scholar]
- Popov AV, Pozniakovsky A, Arnal I, Antony C, Ashford AJ, Kinoshita K, Tournebize R, Hyman AA, Karsenti E. (2001). XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J , 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Julicher F. (2013). XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol , 1116–1122. [DOI] [PubMed] [Google Scholar]
- Reber SB, Goehring NW. (2015). Intracellular scaling mechanisms. Cold Spring Harbor Perspect Biol , a019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. (2010). Early Development of Xenopus laevis: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Vasquez RJ, Gard DL, Cassimeris L. (1994). XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol , 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA , 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. (2013). Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. Elife , e00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. (2008). Evidence for an upper limit to mitotic spindle length. Curr Biol , 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.