Abstract
The Florida red tide dinoflagellate, Karenia brevis, is the major harmful algal bloom dinoflagellate of the Gulf of Mexico and plays a destructive role in the region. Blooms of K. brevis can produce brevetoxins: ladder-shaped polyether (LSP) compounds, which can lead to adverse human health effects, such as reduced respiratory function through inhalation exposure, or neurotoxic shellfish poisoning through consumption of contaminated shellfish. The endogenous role of the brevetoxins remains uncertain. Recent work has shown that some forms of NADPH dependent thioredoxin reductase (NTR) are inhibited by brevetoxin-2 (PbTx-2). The study presented herein reveals that high toxin and low toxin K. brevis, which have a ten-fold difference in toxin content, also show a significant difference in their ability, not only to produce brevetoxin, but also in their cellular redox status and distribution of xanthophyll cycle pigments. These differences are likely due to the inhibition of NTR by brevetoxin. The work could shed light on the physiological role that brevetoxin fills for K. brevis.
Keywords: Karenia brevis, brevetoxin, thioredoxin, thioredoxin reductase, NPQ, xanthophyll cycle
Graphical abstract
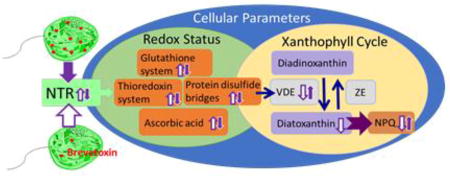
1 Introduction
The brevetoxins are a suite of structurally related polyether ladder type neurotoxins produced by the Florida red tide organism Karenia brevis. This dinoflagellate blooms almost annually in the Gulf of Mexico, resulting in massive fish kills and deaths of marine mammals, sea turtles, and aquatic birds (Backer et al., 2005; Fire et al., 2015; Flewelling et al., 2005). Brevetoxins have been associated with numerous manatee epizootic events (Flewelling et al., 2005; Walsh et al., 2010) and dolphin strandings (Flewelling et al., 2005; Hoagland et al., 2009).
Exposure to brevetoxins as aerosols during red tide events can result in respiratory distress (Cheng et al., 2005a; Cheng et al., 2005b; Fleming et al., 2005a; Fleming et al., 2007; Milian et al., 2007). The consumption of tainted shellfish induces a syndrome known as neurotoxic shellfish poisoning (NSP) whose symptoms include severe gastrointestinal distress, reversal of temperature sensation and parasthesias (Errera et al., 2010; Fleming et al., 2005a; Fleming et al., 2005b; Kirkpatrick et al., 2004; Trainer et al., 1994). The economic effects of the Florida red tide resulting from the closure of fisheries, loss of tourism and public health costs have been estimated between $2 and $24 million (Hoagland et al., 2014).
The cellular toxin content in field samples of K. brevis can vary significantly. One study reports toxin content in field samples ranging from 1 – 68 pg/cell (Hardison et al., 2014). The reasons for this variation remain unclear, and could be a result of genetic variation among strains or response to environmental conditions. Several studies report a negative correlation between cellular toxin content and the availability of nutrients (Hardison et al., 2013; Ransom Hardison et al., 2012; Waggett et al., 2012) or a positive correlation with osmotic stress (Errera and Campbell, 2011). Although, others have disputed the later claim (Sunda et al., 2013). Laboratory strains of K. brevis can also exhibit a wide range of cellular toxin content, even under identical culture conditions, differing by as much as 10-fold (Errera et al., 2010). On at least one occasion, different cultures of the Wilson strain of K. brevis have unexpectedly demonstrated sustained differences in toxin production, by as much as 10-fold. The availability of cultures of the same strain with different cellular toxin loads offers a unique opportunity to study both the biosynthetic pathway of the brevetoxins as well as their endogenous function.
Recent efforts have focused on the application of brevetoxin probes to uncover the endogenous role of the brevetoxins. Using fluorescent and photoaffinity derivatives of brevetoxin, it was demonstrated that exogenously applied brevetoxin localizes to the chloroplast of K. brevis where it bound to two cholorplast proteins: the light harvesting complex II (LHCII) and thioredoxin (Trx) (Cassell et al., 2015). The light harvesting complex II is an integral part of the photosynthetic apparatus where it harvests and distributes light energy using fourteen bound chlorophyll and four carotenoid molecules. It is also central to the process of non-photochemical quenching (NPQ) or the dissipation of light energy as heat under conditions of high light. Thioredoxin is the parent of a family of enzymes, which mediate the redox state within a cell via thiol disulfide exchange (Hanschmann et al., 2013; Holmgren, 1995; Jeng et al., 1998). Thioredoxins are present in almost all living organisms. In photosynthetic organisms, Trxs are located within the chloroplast stroma and numerous thioredoxin-dependent regulatory networks have been identified within the chloroplasts with over 400 confirmed or potential Trx targets (Nikkanen and Rintamäki, 2014).
Alterations in the normal function of either LHCII or Trx have the potential to disrupt the oxidative status of the cell, redox signaling and photosynthetic parameters. Indeed, high toxin and low toxin K. brevis (Wilson) have been found to have differences in their ability, not only to produce brevetoxin, but to perform NPQ and in the production of ROS (Cassell et al., 2015).
In terrestrial plants and green algae, LHCII binds to four carotenoids: two luteins, neoxanthin and zeaxanthin (Zx) (under high light conditions) or violaxanthin (Vx) (under low light conditions) (Standfuss et al., 2005). The interconversion of Zx↔Vx modulates light energy dissipation through the xanthoyphyll cycle (Fuciman et al., 2012). Under high light conditions, the xanthoyphyll cycle (Fig. 1) is initiated in which Vx (a di-epoxide) is converted to Zx in a two-step process catalyzed by violaxanthin de-epoxidase (VDE). The light harvesting complex II then undergoes a conformational change moving Zx into close proximity to the Chl molecule permitting the quenching of Chl fluorescence. Under low-light conditions Zx is converted back to Vx by the enzyme zeaxanthin epoxidase (ZE). Dinoflagellates fall into six unique chloroplast pigment types, with the genus Karenia defining a unique group having principally fucoxanthins and gyroxanthins (Zapata et al., 2012). Dinoflagellate LHCII has not been as well characterized as those of terrestrial plants and the specific binding site for each of these carotenoids is not known, nor is it known if there is a one to one correlation between plant and dinoflagellate carotenoids. Like diatoms and other dinoflagellates, in K. brevis Zx/Vx are replaced by diatoxanthin (Dt) and diadinoxanthin (Dd) respectively (Fig. 1). Furthermore, the xanthophyll cycle has been demonstrated to be functional in K. brevis through the interconversions of the monoepoxide Dd and the non-epoxide Dt (Evens et al., 2001; Schaeffer et al., 2009).
Fig.1.
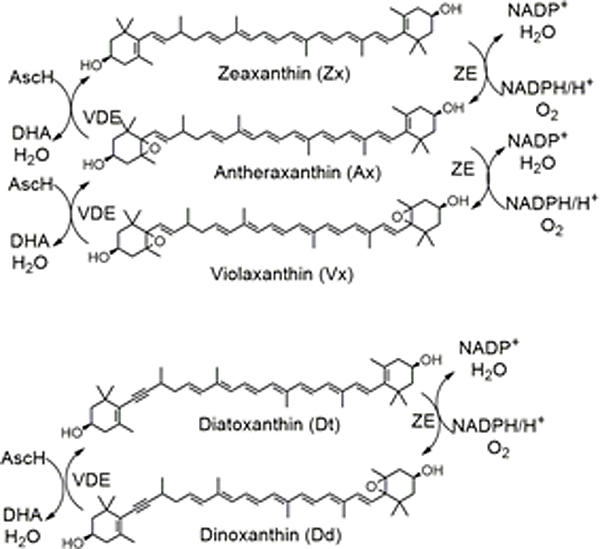
The xanthoyphyll cycle.
Trx is converted from its inactive disulfide form to its active dithiol through reduction by thioredoxin reductases (TR). In plants and algae, two types of thioredoxin reductase are typically present: ferredoxin-thioredoxin reductase (FTR) and NADPH dependent thioredoxin reductase (NTR). Ferredoxin-thioredoxin reductase is exclusive to the chloroplast, has an iron-sulfur center and a disulfide bridge, with electrons provided by ferredoxin. NADPH dependent thioredoxin reductases are pyridine nucleotide disulfide oxidoreductases, which transfer electrons from NADPH via FAD, to a cysteine residue by reduction of a disulfide bridge. Both classes reduce Trx by thiol-disulfide exchange. It is believed that the two reductases are selective for different Trx targets. Ferredoxin-thioredoxin reductase is active only in the light and is believed to be responsible for regulation of light reactions. NADPH dependent thioredoxin reductases are present in both photosynthetic and non-photosynthetic organisms. When found in the chloroplast, NTR is often referred to as NADPH dependent thioredoxin reductase of the chloroplast (NTRC) to distinguish it from non-chloroplastic NTR and FTR. In nature, numerous forms of NTR have been identified (Jacquot et al., 2009) falling into two broad categories referred to as small and large NTR. These are distinguished principally by their size and the redox active disulfide motifs. Small NTR will have one or more active site CxxC motifs. These are found in terrestrial plants and bacteria. Whereas large NTR contains one N-terminal CxxxxC motif and a C-terminal catalytic CC or CU (cysteine-selenocysteine) motif. In other words, one cysteine residue may be replaced by a selenocysteine to form an active site selenosulfide at the C-terminal redox center. Large NTR having a CU redox center was first identified in mammals and is sometimes referred to generally as “mammalian” NTR, but has since been found to be widespread in the animal kingdom, including in eukaryotic algae. It has been recently established that PbTx-2 is an inhibitor of selenocysteine-containing large NTR (Chen et al., 2017). Evidence suggests that the mechanism of inhibition is by alkylation of the nucleophilic Sec residue via a Michael addition to the K-ring side chain of PbTx-2, an α, β-unsaturated aldehyde. PbTx-3, which has an allylic alcohol side chain, does not inhibit TrxR, underscoring the importance of the side chain functionality. The presence of a Sec containing large NTRC in K. brevis, would indicate that PbTx-2 may regulate the redox status of a subset of proteins that are NTRC targets and that cellular redox status of K. brevis may be modulated by PbTx-2. These observations prompted an assessment of the diversity of NTR in K. brevis and a comparative examination of the redox status and carotenoid content of high toxin and low toxin K. brevis.
2 Material and Methods
2.1 General
Enzyme activity kits (thioredoxin, thioredoxin reductase, glutaredoxin) were purchased from Cayman Chemical. All other reagents were purchased from Thermo Fisher Scientific Co. or Sigma-Aldrich unless otherwise specified. Ultraviolet/Visible measurements were performed in 96 or 384 well microplates using a Synergy® 2 (Biotek Instrument, Inc.) microplate reader. Fluorescence measurements were performed in 96 or 384 well microplates using an Infinite® M1000 PRO (Tecan Group Ltd.) microplate reader.
2.2 Query of K. brevis transcriptome library
Analysis of a K. brevis (Wilson strain) transcriptome library consisting of 86,580 predicted transcripts (Ryan et al., 2014) by tBLASTn (Altschul et al., 1990) used Arabidopsis thaliana small NTRC and Emiliania huxleii large NTRC (NCBI Accession numbers: AEE86518.1, BAH20464.1, respectively), Chlamydomonas reinhardtii and Emiliania huxleyi selenocysteine synthase (NCBI Accession numbers: EDP02915.1 and XP_005760065.1 respectively), Spironucleus salmonicida and Dictyostelium discoideum selenophosphate synthase (NCBI Accession numbers: EST43726.1 and Q94497.3 respectively) and Cryptosporidium parvum, Chrysochromulina sp. and Ectocarpus siliculosus SECIS binding protein (NCBI Accession numbers: EAK89005.1, KOO29690.1 and CBN78485.1 respectively) as queries. Each of the identified NTRs were analyzed for the presence of N-terminal organelle targeting sequences using the following programs: SignalP (http://www.cbs.dtu.dk/services/SignalP/), (Petersen et al., 2011) TargetP (plant version) (http://www.cbs.dtu.dk/services/TargetP), (Emanuelsson et al., 2000); iPSORT (http://ipsort.hgc.jp/), (Bannai et al., 2002); PATS (http://gecco.org.chemie.unifrankfurt.de/pats/pats-index.php), (Zuegge et al., 2001) and PredictProtein (https://www.predictprotein.org/), (Waller et al., 1998; Waller et al., 2000; Yachdav et al., 2014). Each of the identified NTRs were analyzed for the presence of a Sec insertion element (SECIS) using the program SECISSearch3 (http://gladyshevlab.org/SelenoproteinPredictionServer/), (Mariotti et al., 2013).
2.3 Culture methods
Cultures of K. brevis (Wilson strains) were obtained from Mote Marine Laboratory (Sarasota, Florida) and maintained in L1-Si medium, with the exception that the NH 15 vitamin supplement (Gates and Wilson, 1960) replaced the L-1 supplement. Cultures were maintained in a growth chamber at ~20°C under under artificial light (40 W, 3.4 × 103 Lux; PFD ca. 46 µmol phota•m−2•sec−1). Growth was monitored by counting a 1:10 dilution of culture in Z pak reagent using a Beckman Z-series Coulter Counter with aperture size between 10~30 µm, according to the manufacturer’s instructions.
2.4 Preparation of K. brevis homogenate
K. brevis cells (200–500 mL) were concentrated by centrifugation of cultures (5 min at 466 × g) and the supernatant was discarded. The cells were resuspended in phosphate-buffered saline (PBS) and vortexed for 1 min. This suspension was centrifuged (10 min at 14,000 × g) to remove cellular debris and the supernatant was transferred to a clean tube. An aliquot (50 µL) was reserved for a protein assay. Samples were quantitated against BSA standards (2, 1, 0.5, 0.25, 0.125, 0.0625 and 0 mg /mL) prepared in PBS using the Coomassie Protein Assay Reagent (BioRad) according to the manufacturer’s instructions. Final protein concentrations is adjusted to 0.2–1.5 mg / mL.
2.5 Total antioxidant capacity
Total antioxidant capacity of K. brevis homogenate was determined by 2,2-azinobis (3-ethenelbenzothiazoline-6-sulfonic acid) radical cation (ABTS•+) decolorization and expressed as trolox ((6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid)) equivalents (Re et al., 1999). ABTS•+ was produced by oxidation of aqueous ABTS (1.3 mL, 7 mM) with ammonium persulfate (21.2 µL, 2.45 mM). The mixture was allowed to stand at room temperature in the dark for 12–16 h before use. The ABTS•+ solution was diluted with PBS to an absorbance of 0.7. Either 50 uL K. brevis homogenate or trolox standard (0 µM, 25 µM, 50 µM, 100µM, 200 µM) was added to the ABTS•+ solution (100 µL). The absorbance was read at 734 nM after incubation for 5 min. Ascorbic acid concentration (as trolox equivalents) of K. brevis homogenate was estimated by incubating homogenate with ascorbic acid oxidase (2 µL, 0.01 U/ µL) for 30 min prior to the assay and taking the difference in absorbance of parallel samples.
2.6 Modified papain assay for high and low molecular weight thiols
Preparation of papain-S-SCH3
Papain S-methyl sulfide (~1.2 mg / mL) was prepared as previously described (Singh et al., 1995). Briefly, 1.5 mL of papain stock solution (26.4 mg / mL in 0.05 M acetate buffer, pH 4.5, containing 0.01% thymol) was added to a solution of cysteine (0.6 mM, 28.5 mL of 20 mM sodium phosphate, 1 mM EDTA buffer). The solution was kept at room temperature for 30 min. Methyl methanethiosulfonate (117 µmol, 1.337 g/ml in chloroform) was added in mixture. The reaction mixture was kept on ice for 4 hr. The mixture was dialyzed (Mr cutoff 6000–8000) against 5 mM sodium acetate buffer, containing 50 mM NaCl, pH 4.7 at 4 °C 12 hr.
Modified papain assay
The assay was performed as described previously (Singh et al., 1995). Briefly, Papain-S-SCH3 solution (0.5 mL, 0.6 mg / mL), N-benzoyl-L-arginine-p-nitroanilide (0.7 mL of a 3.4 mM solution in Bis/EDTA buffer, pH 6.3) and 15 µL cystamine (4 mM in 0.05 M acetate buffer, pH 4.5, containing 0.01% thymol) were added to K. brevis homogenate (200 µL, 0.8 mg protein/ml in PBS buffer). Absorbance was measured at 410 nm every 10 min for 2 hr, then every 60 min for up to 15 hr. Samples were analyzed in triplicate. Samples with acetate buffer (0.05 M, pH 4.5) instead of homogenate were used to determine background absorbance.
2.7 Determination of reduced glutathione
Homogenate of K. brevis (300 µL, 0.3–0.5 mg / mL protein) was mixed with reaction buffer (700 µL, 0.1 M sodium phosphate, 1 mM EDTA, pH 8.0) and Ellman’s (Ellman, 1959) reagent (25 µL, 4 mg / mL). After incubation for 15 min at room temperature, absorbance was measured at 410 nm. Background absorbance of homogenate alone was subtracted from each sample. Ellman’s reagent in reaction buffer served as a control. Samples were quantitated against cysteine standards (0, 0.3, 0.6 and 1.2 mM) prepared in reaction buffer. Samples and standards were prepared in triplicate. Thiol content was normalized to protein concentration in the homogenate.
2.8 Determination of total glutathione
Homogenate of K. brevis (275 µL, 0.3–0.5 mg / mL) was mixed with NADPH (100 µL, 1 mg / mL), glutathione reductase (3 µL, 10 µM, Cayman Chemical) and Ellman’s reagent (25 µL, 4 mg / mL). Absorbance was measured immediately every 10 min at 410 nm at room temperature for up to 30 min. Background absorbance of homogenate alone was subtracted from each sample. Ellman’s reagent in reaction buffer served as a control. Samples were quantitated against cysteine standards (0, 0.3, 0.6 and 1.2 mM) prepared in reaction buffer. Samples and cysteine standards were analyzed in triplicate. Thiol content was normalized to total protein in the homogenate.
2.9 Determination of free selenol
The selenol selective probe, Sel-green, was synthesized according to published methods (Zhang et al., 2015). Homogenate of K. brevis was adjusted to a protein concentration of 0.8 mg / mL. Sel-green (20 µL, 20 µM) was added to K. brevis homogenate (80 µL). Fluorescence was monitored (λex = 370 nm/ λem 502 nm) at 0, 2.5 and 5 min then every 10 min for 120 mins. Samples were prepared in triplicate. A control sample was prepared by the addition of an equal volume of PBS buffer instead of homogenate. A blank sample was prepared with homogenate and PBS.
2.10 Thioredoxin, thioredoxin reductase and glutaredoxin activities of K. brevis
The enzymatic activities of Trx, TrxR and Grx were determined using commercial kits (Cayman Chemical) according to the manufacturer’s instructions. For each assay, homogenate containing 30 µg of protein was used. Assays were performed in triplicate. Fluorescence was normalized to protein concentration.
2.11 Extraction and analysis of carotenoids
Log phase cultures of K. brevis were both light or dark adapted and extracted for carotenoids and chlorophyll on each of five different days. Cultures were light-adapted by placing the culture under 1.2 × 105 Lux (full sun: PFD ca. 2,220 µmol phota•m−2•sec−1) for 25 minutes. Cultures were dark-adapted by placing culture in the dark for 25 minutes. On two of the 5 days, cultures were filtered through a pre-conditioned (according to the manufacturer’s instructions, 100 mL of methanol followed by 50 mL of water) Varian C-18 disc (47 mm). Disks were washed with water (10 mL) and eluted with methanol (10 mL). The methanol was evaporated in vacuo and resuspended in aqueous methanol (10% NH4OAc, 50 mM, pH 7.0, 1.5 mL). Samples were immediately stored at −80°C. On three of the five days, cultures were vacuum filtered through Whatman GF/F #30 and stored at −80°C.
Frozen filters were added to a 10 mL Potter-Elvehjem tissue grinder and 3.0 mL of extraction solvent (methanol:acetone:dimethylformamide:water, 30:03:30:10, v/v/v/v) containing a known amount of an internal standard (IS = copper-mesoporphyrin-IX dimethyl ester) was added. The pestle of the tissue grinder was modified by carving off bits of the PFTE tip to make it somewhat pointed rather than round in order to aid disruption of the glass fiber filter. The Pestle was inserted into the chuck of a motorized mixer and the filter in the mortar (aka tube) was ground at moderate rpm while maintaining the glass mortar in an ice filled Teflon beaker. Following grinding, the extract within the mortar was sonicated in a water bath several times in short (< 3 sec) spurts. The tube (mortar) was then covered with aluminum foil and placed in the refrigerator (~ 4°C) to steep for 1–2 hours (Hagerthey et al., 2006). All procedures were performed at ice-bath temperatures under dim yellow light to avoid photooxidative and/or thermal alterations of the pigments. The tube was then centrifuged for 2–3 minutes, the extract decanted and filtered using a syringe filter (0.45 µm). The UV-Vis spectrum of the crude extract was recorded between 350–800nm for estimating total chlorophyll at ca. 662-4 nm (e.g. a = 87.7 L•g−1•cm−1) (Egeland et al., 2011) and to make sure that the extract was not too concentrated (highest peak over < 1.2 AU) for injection into the liquid chromatograph. Prior to injection, 0.125 mL of an aqueous ion pairing (aka ion suppression) solution consisting of 15.0 g of tetrabutylammonium acetate (TBAA) and77.0 g ammonium acetate per liter final volume (Louda et al., 1998; Mantoura and Llewellyn, 1983) was added to 1.0 mL of the filtered extract.
High performance liquid chromatography (HPLC) –photodiode array detection (PAD) involved the injection of 100 or 500µL of the prepared injectate onto a 3.9 × 300mm Waters NovaPak C18 column using a ternary gradient (Louda et al., 1998; Louda et al., 2002) provided by Thermo-Separations-Products (TSP) P4000 quaternary pump. Pigments were detected and PDA generated UV-Vis spectra recorded by a Waters 990 or 996 photodiode array detector with Waters software. Pigment identifications by retention times and UV-Vis spectra (Louda et al., 2002) were verified versus known mixed and pure (e.g. fucoxanthin, 19’-hexanoyloxyfucoxanthin, 19’-butanoyloxyfucoxanthin etc.) standards purchased from DHI Lab Products (Agern Allé 5, 2970 Hørsholm, Denmark). Quantitation of separated pigments involved integration of the peak area and application of millimolar or specific extinction coefficients given in the literature (Egeland et al., 2011).
3 Results
3.1 Analysis of NTR in K. brevis
Analysis of a K. brevis (Wilson strain) transcriptome library (Ryan et al., 2014) consisting of 86,580 transcripts by tBLASTn (Altschul et al., 1990) using Arabidopsis thaliana NTRC and Emiliania huxleyi large NTR (NCBI Accession numbers: AEE86518.1 and BAH20464.1, respectively) as queries each returned two transcripts with significant homologies to the query. These results are summarized in Tables 1 and 2.
Table 1.
Results of tBLASTn Search Using Various NTR as Queries
| Query | Locus | E-value | NTR |
|---|---|---|---|
| Emiliania huxleyi TrxR BAH20464.1 (large NTRC) | 29890 | 4e−141 | NTR 1–3 |
| 38966 | 1e−132 | NTR4 | |
| Arabidopsis thaliana TrxR AEE86518.1 (small NTRC) | 21223 | 3e−65 | NTR5 |
| 67462 | 5e−70 | NTR6 |
Table 2.
Characteristics of NTR from K. brevis
| NTR | Redox center N-terminal |
Redox center C-terminal |
Small /large? |
3' UTR SECIS |
Signal peptide |
|---|---|---|---|---|---|
| NTR1 | CVNVGC | CC | large | yes | no |
| NTR2 | CVNVGC | CU | large | yes | no |
| NTR3 | absent | CU | unknown | yes | no |
| NTR4 | CVGQAC, CVGNAC, CIPC, CVNVGC, CGGGKC | unknown | no | yes | |
| NTR5 | CGPC | CATC | small | no | no |
| NTR6 | CAAC | absent | small | no | no |
Inspection of the nucleotide sequence of locus 29890, reveals a polycistronic mRNA encoding 5 thioredoxin reductases (ORF1-5). Open Reading Frames 1 and 3 are identical 657 nucleotide ORFs, each coding for a 218 amino acid large NTR (NTR1) having an N-terminal CVNVGC motif and a C-terminal CC. Open Reading Frames 2 and 4 are identical 1461 nucleotide open reading frames, each coding for a 486 amino large NTR (NTR2) having an N-terminal CVNVGC motif. The nucleotide sequence of NTR2 ends with UGC UGA AGC UAA (Cys-Sec-Ser-Stop). A UGA codon is found at position 1453 of ORF2 and ORF4. The UGA codon typically represents a stop codon, but the Sec residue of selenoproteins is also encoded by UGA with UAA as the true stop codon. In large NTR, the Sec is followed by a single amino acid. In E. huxleyi NTR, the last amino acid is a glycine. For the K. brevis NTR2, this position appears to be occupied by a serine. Finally, ORF5 is an 828 nucleotide open reading frame coding for a 275 amino acid NTR (NTR3). Like NTR2, the nucleotide sequence of NTR3 ends with UGC UGA AGC UAA. The UGA codon is found at position 820 of ORF5. It is difficult to classify NTR3. While it does appear to incorporate a C-terminal CU motif, an N-terminal CxxxxC motif was not present.
Inspection of the nucleotide sequence of locus 38966, reveals a 2907 nucleotide open reading frame coding for a 968 amino acid protein (NTR4) with high homology to other large NTR. This NTR appears to be atypical in that five potential redox centers were identified: four having the CxxxxC motif typical of a large NTR and one CxxC motif typical of a small NTR. Furthermore, it lacks a C-terminal CC or CU motif. Large NTRs, having multiple CxxxxC motifs have been identified in other Apicomplexa (Plasmodium falciparum, Toxoplasma gondii, Cryptosporidium spp., and Theilera spp.) (McCarty et al., 2015). In fact, the C-terminal CGGGKC sequence is identical to that of Plasmodium falciparum Large NTR (Hirt et al., 2002).
Inspection of the nucleotide sequence of locus 21223, reveals an open reading frame coding for a 711 amino acid protein with high homology to other small NTRC (NTR5) found in terrestrial plants and green algae. Finally, locus 67462 encodes a 354 amino acid small NTR (NTR6) with a single disulfide redox center CAAC. This type of TR is also found in terrestrial plants, green algae, red algae, diatoms and cyanobacteria (Jacquot et al., 2009). Each of the NTRs was analyzed for the presence of N-terminal organelle targeting sequences using the following programs: SignalP, TargetP (plant version), chloroP, iPSORT and PredictProtein. SignalP identified a signal peptide sequence in NTR4 (probability 0.809 or 0.985 using the hidden Markov model). TargetP predicted chloroplast localization for NTR4, although with a low probability (0.341). Among the other NTRs, SignalP identified no other signal peptides. ChloroP did not indicate chloroplast localization for any of the NTRs. iPSORT identified a chloroplast transit peptide for NTR4 and predicted localization to the mitochondria for NTR6. While PredictProtein localized one to the chloroplast (NTR5), three to mitochondria (NTR1, 2 and 4) and two to the cytosol (NTR3 and 6).
The recoding of the UGA from a termination signal to a Sec residue requires the presence of a Sec Insertion Sequence (SECIS) in the 3’ UTR of the selenoprotein mRNA. The SECIS element forms a stem loop structure, which is recognized by a SECIS binding protein. The SECIS binding protein positions a Sec-tRNA for incorporation of Sec at the UGA codon. Analysis of the nucleotide sequence of K. brevis locus 29890 using the program SECISearch3 (Mariotti et al., 2013) identified a 73 nucleotide SECIS immediately following the TAA stop codon of ORF5. Additionally, a search of the K. brevis, Wilson transcriptome library revealed a SECIS binding protein (locus 14693) which recruits translation and assembly factors and positions the Sec-tRNA. Other enzymes required for selenoprotein biosynthesis, which were identified in the K. brevis transcriptome library, include seleneocysteine tRNA synthase (locus 48280) and seleophosphate synthetase (loci 41392, 52141, 18903). Seleneocysteine tRNA synthase is required to convert serine, which is pre-loaded on to a Sec tRNA, into Sec, using selenophosphate and seleophosphate synthetase is necessary to synthesize selenophosphate from selenide and ATP.
3.2 Trolox equivalent antioxidant capacity (TEAC) of K. brevis homogenates
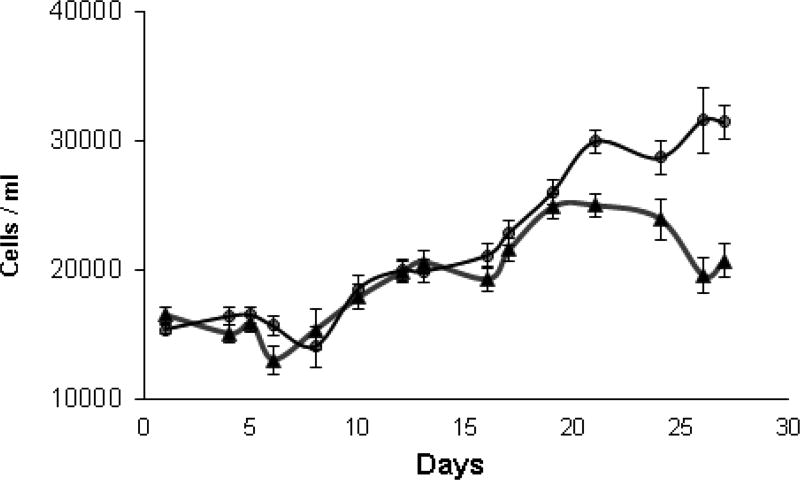
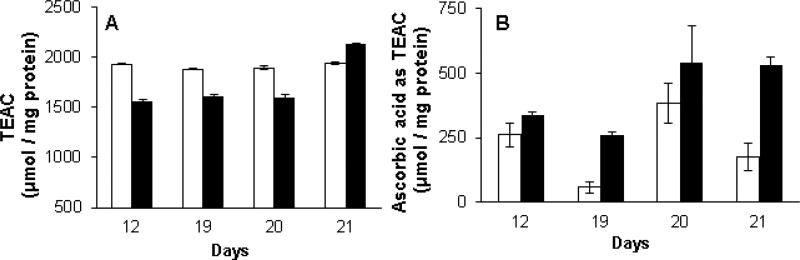
Over a period of ten days, total antioxidant capacity of high and low toxin K. brevis homogenates was determined by ABTS radical cation (ABTS•+) decolorization (Re et al., 1999) and expressed as trolox equivalent antioxidant capacity (TEAC). Cell counts over this period are shown in Fig. 2. Fig. 3A shows that the TEAC for high toxin K. brevis was fairly constant during this period, ranging from 1,880–1,936 nmol TEAC/mg protein: A coefficient of variation of only 1.40 % (n=12). The TEAC for low toxin K. brevis was fairly constant on three of the four days tested, ranging from 1,559–1,598 nmol TEAC/mg protein: A coefficient of variation of only 1.98 % (n=9). On three of the four days tested, high toxin K. brevis had a higher TEAC (16–23 %) than low toxin K. brevis. On day 21 the TEAC for low toxin K. brevis was 2,135 TEAC/mg protein, which was 9.3% higher than that of the high toxin K. brevis. A clear trend for low/high was not apparent.
Fig.2.
Cell density of K. brevis cultures over the lifetime of high toxin (△) and low toxin (●) strains.
Fig.3.
A) The antioxidant capacity of the cell homogenates of K. brevis expressed as trolox equivalents/mg of protein. ABTS•+ was produced by oxidation of aqueous ABTS and decolorization of ABTS•+ was determined by measuring absorbance at 734 nM and quantitated against trolox standards. B) Ascorbic acid as trolox equivalents, was determined by incubation of homogenate with AAO for 30 minutes prior to the addition of ABTS•+ and taking the difference in absorbance of parallel samples. High toxin (white bar) and low toxin (black bar) strains.
Ascorbic acid concentration (also as trolox equivalents) was estimated by pre-incubation of parallel samples with ascorbic acid oxidase (AAO) prior to the addition of ABTS•+. Ascorbic acid oxidase catalyzes the oxidation of ascorbic acid to dehydroascorbic acid. Thus, the difference in trolox equivalents in the presence and absence of AAO provides an estimate of ascorbic acid in the homogenate. In contrast to the TEAC results, low toxin K. brevis consistently showed higher levels of ascorbic acid (Fig. 3B) with a maximum 4.5-fold difference on day 19. The ascorbic acid content of low toxin K. brevis consistently represented a larger percentage of the TEAC.
3.3 Low molecular weight thiols
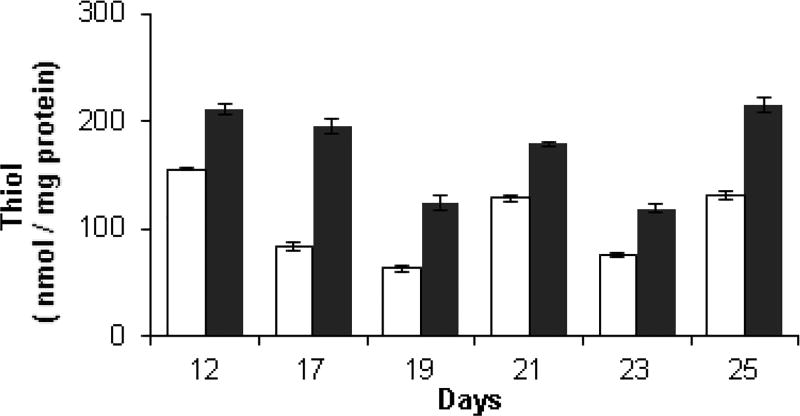
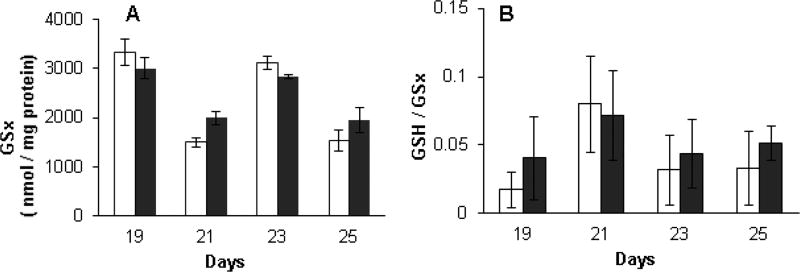
Low molecular weight thiols in high and low toxin K. brevis homogenates were quantified against cysteine standards, on selected days over a 13-day period using Ellman’s reagent, 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB) and normalized to protein concentration. The thiols detected by Ellman’s assay are believed to be composed principally of reduced glutathione (GSH) as cellular glutathione levels are typically in the mM range. Other low molecular weight thiols and some higher molecular weight thiols, which are accessible to DTNB may also be detected. Additionally, on selected days, glutathione reductase (GR) was added to parallel samples to reduce any glutathione which is present as the disulfide GSSG to the thiol GSH. In this way, the total glutathione can be quantified and the ratio of GSH to total glutathione (GSx) can be calculated. Inspection of Fig. 4 reveals that low toxin K. brevis consistently had a higher thiol concentration (from 1.5 to 2.3-fold) than high toxin K. brevis. Total glutathione content, measured after the addition of GR (Fig. 5A) did not reveal a consisted pattern. Indeed on three of the four days that this analysis was performed (19, 23 and 25) total glutathione content was quite similar. Low toxin K. brevis consistently had a higher fraction of reduced to total glutathione than high toxin K. brevis, ranging from 1.5 to 2-fold higher (Fig. 5B).
Fig. 4.
The content of thiol (GSH) in high toxic and low toxic K. brevis homogenates (90–150 µg protein /ml) by DTNB reduction (0.2 mM). Samples were quantified against cysteine standards and normalized to protein concentration. High toxin (white bar) and low toxin (black bar) strains.
Fig. 5.
A) Total glutathione in high and low toxin K. brevis homogenates (90–150 µg protein /ml) by DTNB reduction (0.2 mM) reduction after treatment of homogenate with GR. Samples were quantified against cysteine standards and normalized to protein concentration. (B) The ratio of GSH to total glutathione (GSx) was calculated based on the normalized results from Figs. 4 and 5A. High toxin (white bar) and low toxin (black bar) strains.
3.4 Total low and high molecular weight thiols
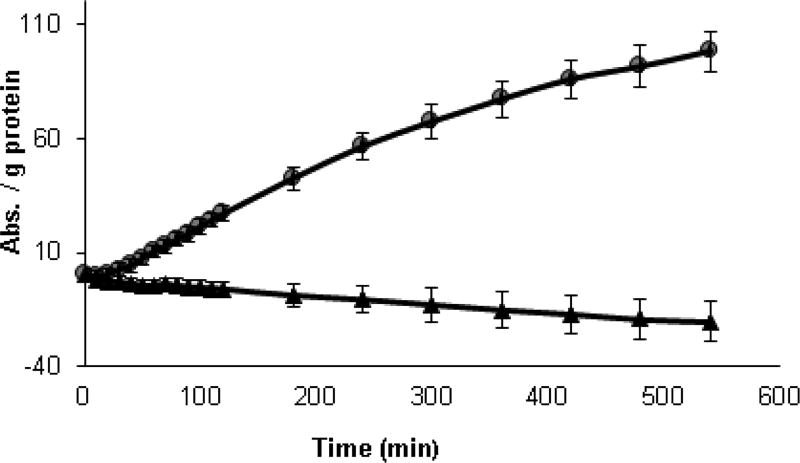
While Ellman’s reagent may detect some accessible protein thiols, it cannot detect those which are inaccessible or buried in the interior of proteins. The redox state of both protein and non-protein thiols in K. brevis homogenates can be determined with an indirect method using a modified papain assay (Singh et al., 1995). This assay is more sensitive than the Ellman’s assay as the signal is amplified by the enzymatic activity of papain. The active site cysteine of papain is converted to an inactive mixed disulfide which may be reduced by low MW thiols to the active form. The active papain then catalyzes the hydrolysis of N-benzoyl-L-arginine-p-nitroanilide to yield p-nitroanilide. The addition of the disulfide cystamine to the assay allows for the detection of inaccessible protein thiols. Cystamine is reduced to cysteamine by protein thiols and the resulting cysteamine acts as a shuttle between high MW inaccessible protein thiols and modified papain. Thus the difference in papain activity with and without cystamine is a reflection of high MW inaccessible thiols. Results of the modified papain assay using homogenates from high and low toxin K. brevis are shown in Fig. 6. Because the results shown represent the difference in absorbance with and without cystamine, the values for the high toxin K. brevis became negative over time. The rate for the low toxin K. brevis was two-fold higher that of the high toxin before subtracting the background (Fig. 6).
Fig.6.
Determination of high MW thiols in K. brevis homogenate by the modified papain assay. Papain-S-SCH3 (0.21 mg/mL), N-benzoyl-L-arginine-p-nitroanilide (1.7 mM), cystamine (68 mM), K. brevis homogenate (0.11 mg protein/ml). High toxin (△) and low toxin (●) strains
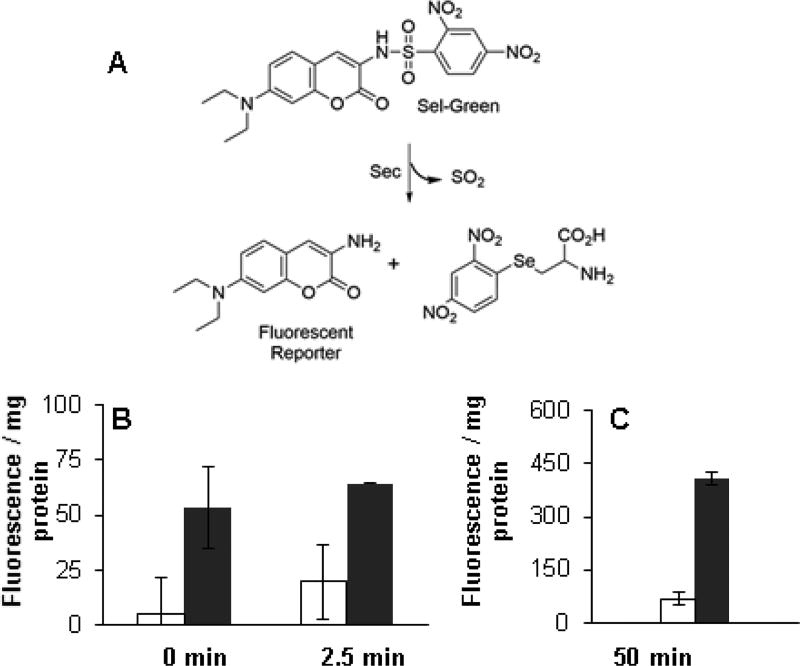
3.5 Free thiols using Sel-green probe
The selenol selective probe, Sel-green (Fig. 7A) reacts with selenols (including the Sec residue of large NTR) to release a fluorescent reporter and may be used for the determination of free selenols. Brevetoxin (PbTx-2) was shown to inhibit this reaction (Chen et al., 2017) by the alkylation of Sec. Sel-green was added to the K. brevis homogenates and fluorescence was monitored (λex/em = 370/502 nm) for a period of two hours. The 0, 2.5 and 50 min time points are shown in Figs. 7B and 7C. Fluorescence is clearly reduced in low high toxin K. brevis homogenates.
Fig.7.
A) The reaction of sel-green probe with selenols. The fluorescence is released by the reaction of sel-green probe with selenol groups in K. brevis homogenate. B) Sel-green (4 µM) was incubated with K. brevis homogenate (0.6 mg protein/ml) and fluorescence was monitored (λex/em = 370/502 nm) at 0, 2.5 and C) 50 min. The fluorescence was normalized to protein concentration in the homogenate. High toxin (white bar) and low toxin (black bar) strains.
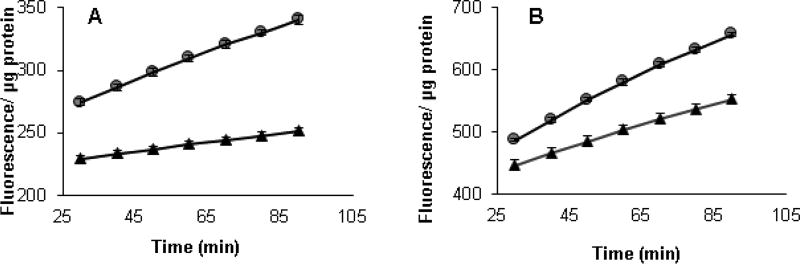
3.6 The activity of Trx and Grx
The activities of TrxR, Trx and Grx in K. brevis homogenates were determined using fluorescent activity assay kits (Fig. 8A and 8B). Both Trx and Grx activities are higher in the low toxin K. brevis (3-fold and 1.6-fold respectively) by comparison of the rates between 25 and 85 min. Even though Trx and Grx activity was detectable in the homogenates, the activity of NTR was not significantly higher than background. This is not to say that there was no NTR activity. The kits use human NTR or Trx and cell homogenates replace the missing enzyme in the NTR/Trx system. Apparently, at least one K. brevis Trx may reduce insulin (photosynthetic organisms typically express multiple Trxs) and may be reduced by human NTR hence activity was detectable in the Trx assay, but it appears that K. brevis NTR may not reduce human Trx.
Fig.8.
A) Trx activity in K. brevis homogenate (30 µg total protein). B) Grx activity in K. brevis homogenate (30 µg total protein). high toxin (△) and low toxin (●) strains
3.7 Carotenoid Analysis
Log phase cultures of high and low toxin K. brevis were light or dark adapted on each of five days and analyzed for seventeen known and three unidentified carotenoids as well as chlorophyll-a, -c3 and -c2. When normalized to chlorophyll, no significant difference in total carotenoid content between high and low toxin K. brevis either in the light (n = 5, p = 0.25) or dark (n = 7, p = 0.86) was observed. Nonetheless, some differences in carotenoid distribution were observed. The mole % of individual carotenoids in the high toxin and low toxin K. brevis in the light and dark-adapted samples are shown in Table 3. For this analysis, samples from cultures which were split, then light or dark adapted were used. Samples which were taken from light-adapted then to dark-adapted were not included in this analysis because the starting carotenoid distribution would be different from the former. In the high toxin K. brevis, when comparing the light and dark adapted samples, significant differences were observed only between the two xanthophyll cycle pigments: diatoxanthin (n = 5, p = 0.04) and diadinoxanthin (n = 5, p = 0.05). This difference between Dd and Dt in the high toxin light vs. high toxin dark samples translated to a significant difference (n = 5, p = 0.04) in epoxidation state (EPS = Dd/(Dd+Dt)). No significant difference was observed for these two xanthophylls or for any other carotenoids in the low toxin K. brevis.
Table 3.
Carotenoid content of high and low toxin K. brevis. Carotenoids having significant differences (p ≤ 0.05) in the light vs dark are shown in bold.
| High mole% (SD) (n = 5) | Low mole% (SD) (n = 5) | |||
|---|---|---|---|---|
| Pigment* | light | dark | light | dark |
| 19'-but-Fucoxanthin | 12.21 (1.91) | 12.96 (1.81) | 8.39 (3.55) | 7.43 (2.58) |
| 19'-but- Fucoxanthin | 12.56 (1.50) | 11.68 (1.63) | 7.66 (2.86) | 6.70 (2.30) |
| 19'-but-Fucoxanthin | 5.723 (1.10) | 4.99 (1.46) | 6.89 (3.83) | 6.54 (1.02) |
| Fucoxanthin | 2.564 (1.04) | 2.26 (1.17) | 5.47 (3.13) | 5.92 (1.93) |
| 19'-hex- Fucoxanthin | 13.44 (2.94) | 14.33 (2.22) | 12.21 (6.42) | 15.83 (0.61) |
| 19'-hex- Fucoxanthin | 15.82 (2.46) | 15.11 (1.64) | 13.20 (7.04) | 17.10 (2.55) |
| 19'-hex- Fucoxanthin | 2.666 (1.23) | 2.34 (1.10) | 5.67 (3.56) | 6.26 (2.99) |
| DiaDinoChrome | 0.12 (0.27) | ND | 0.32 (0.72) | 0.26 (0.59) |
| Diadinoxanthin (Dd) | 15.43 (5.72) | 21.63 (5.41) | 19.54 (6.42) | 22.68 (5.19) |
| Diatoxanthin (Dt) | 6.18 (3.68) | 1.22 (0.35) | 5.56 (5.29) | 2.32 (1.50) |
| Gyroxanthin | 9.59 (1.21) | 9.49 (1.34) | 8.21 (1.75) | 6.81 (0.74) |
| alpha-carotene | 0.19 (0.43) | 0.87 (1.07) | ND | 0.40 (0.63) |
| Tns-β-Carotene | 1.66 (1.74) | 1.38 (0.79) | 2.16 (1.83) | 0.78 (0.75) |
| Cis- β-Carotene | ND | 0.48 (1.06) | ND | ND |
| UNKN#1 | 1.04 (1.20) | 0.67 (0.91) | 1.26 (1.21) | 0.49 (1.10) |
| UNKN#2 | 0.43 (0.77) | 0.30 (0.67) | 1.37 (2.50) | ND |
| UNKN#3 | 0.57 (0.83) | ND | 0.92 (1.28) | ND |
| Zeaxanthin | 0.33 (0.75) | 0.28 (0.63) | 1.14 (1.22) | 0.46 (1.03) |
| DD+DT | 21.61 (5.46) | 22.86 (5.56) | 25.11 (7.84) | 25.01 (5.14) |
| EPS | 0.71 (0.16) | 0.94 (0.018) | 0.80 (0.19) | 0.90 (0.06) |
| Total Fucoxanthin | 64.44 (6.56) | 63.67 (5.78) | 59.49 (10.55) | 65.78 (6.48) |
| But Fucoxanthin | 30.48 (2.00) | 29.63 (3.43) | 22.95 (7.82) | 20.66 (5.38) |
| Hex Fucoxanthin | 31.39 (5.49) | 31.78 (4.05) | 31.08 (15.98) | 39.19 (1.38) |
cis fucoxanthin and fucoxanthinol were not present.
ND = not detected.
Carotenoids showing significant differences between the high and low toxin K. brevis are shown in Table 4. Comparison of the light and dark-adapted samples within the high or the low toxin K. brevis indicated that the majority of carotenoids (with the exception of Dd and Dt in the high toxin K. brevis, Table 3) do not vary in the light vs dark. For this reason, all samples were used in this analysis (n = 7) for comparison of high vs low toxin, unless otherwise noted. Significant differences were observed for the fucoxanthins, with more 19’-butanoyl fucoxanthin A and B and less fucoxanthin and 19’-hexanoyl fucoxanthin C in the in the high toxin K. brevis. These variations in individual fucoxanthins seem to offset one another, as there was not a significant difference observed in total fucoxanthins. A significant difference in Dd in the light-adapted samples was observed, which corresponds to a significant difference in EPS with the low toxin K. brevis having a higher epoxidation state (more Dd) in the light-adapted samples than the high toxin K. brevis. There was also a significant difference in the total xanthophyll cycle pigments (Dd+Dt) with the low toxin K. brevis having more.
Table 4.
Carotenoids with significant (p ≤ 0.05) differences between high and low toxin K. brevis. Unless otherwise noted n = 7.
| mole% (SD) | ||||
|---|---|---|---|---|
| Pigment | High Tox | Low Tox | P = | Avg. High-Low |
| 19'-but- Fucoxanthin | 12.58 (1.73) | 8.11 (2.81) | 1.17 E−5 | 4.68 |
| 19'-but- Fucoxanthin | 12.31 (1.47) | 7.24 (2.31) | 3.37 E−5 | 5.07 |
| Fucoxanthin | 5.53 (1.03) | 5.73 (2.54) | 1.17 E-−3 | −3.48 |
| 19'-hex- Fucoxanthin | 2.25 (1.03) | 5.93 (2.88) | 1.45 E−3 | −3.39 |
| Diadinoxanthin (Dd) light (n = 5) | 15.43 (5.72) | 19.54 (6.42) | 2.75 E−2 | −4.11 |
| Gyroxanthin | 9.50 (1.13) | 7.48 (1.35) | 1.36 E−3 | 2.02 |
| But Fucoxanthin Total | 30.62 (2.76) | 22.00 (6.00) | 6.08 E−4 | 8.62 |
| Dd+Dt | 21.28 (5.51) | 24.73 (6.47) | 1.04 E−2 | −3.45 |
| EPS light (n = 5) | 0.71 (0.16) | 0.80 (0.18) | 2.80 E−2 | −0.09 |
4 Discussion
Recent studies have demonstrated that PbTx-2 is in inhibitor of the selenoprotein, large NTR (Chen et al., 2017). This observation has implications for the endogenous function of PbTx-2 only if K. brevis also produces a large NTR, which incorporates a Sec residue in one of its redox active sites. The requisite biosynthetic machinery for selenoprotein biosynthesis (SECIS binding protein, seleneocysteine tRNA synthase and seleophosphate synthetase) was found in the K. brevis transcriptome library. A search of the library also uncovered six unique NTRs, both large and small including two that could not be classified (NTR3 and 4) with two of them incorporating a Sec residue at the C-terminal redox center (NTR2 and 3). Using multiple search programs, a signal peptide, which would target enzymes to the chloroplast, was identified only in NTR4 and not in either of the NTRs which include a C-terminal Sec residue. These algorithms can fail in the case of dinoflagellates due to the complex evolutionary origins of the dinoflagellate plastid (Minge et al., 2010). Dinoflagellate plastids have arisen from secondary and tertiary endosymbiotic events, with intervening endosymbiotic gene transfer. These events have produced plastids with multiple membranes and chimeric plastid proteomes (Nosenko et al., 2006). The K. brevis plastid arose from a tertiary endosymbiotic event in which a chromalveolate acquired a fucoxanthin-containing plastid of haptophyte origin and lost its peridinin-containing plastid. It may be noteworthy that plastid targeting sequences characteristic of peridinin-containing plastids were present in NTR2. Specifically these transit peptide sequences have a Phe-Val at or near the N-terminus followed by two uncharged residues (typically FVAP). This is followed by mostly hydrophobic, non-polar residues (~25% Ala), and in order of decreasing composition, polar uncharged residues, polar basic residues and very few polar acidic residues within the following 22 amino acids (Patron et al., 2005). In NTR2, the N-terminal sequence F10VIGGGSGGLAAAKGAQALGAKVAVA fits these criteria having 84.6% hydrophobic non-polar residues, of which 30.7% are Ala, 7.7% polar uncharged, 7.7% polar basic and no polar acidic residues. While NTR2 is likely to have originated from the chromalveolate peridinin type chloroplast, the trafficking of this protein is uncertain. The related dinoflagellate Karlodinium micrum, which also has a tertiary plastid of haptophyte origin, has at least one chloroplast protein having an FVAP motif (Patron et al., 2006).
Having identified a large NTR selenoprotein in the K. brevis transcriptome library, it was reasoned that the presence of brevetoxin may influence the activity of NTR in K. brevis. A comparative analysis of the biochemical parameters of low and high toxin producing strains of K. brevis, which might be effected by the inhibition of NTR was undertaken. The NTR/Trx and GSH systems are the major systems maintaining redox homeostasis within a cell. Other antioxidants play an important role in protecting cells from oxidative stress. Reduced glutathione, ascorbic acid, Trx, carotenoids and vitamin E are central to the antioxidant defense in most of species. Specifically, trolox equivalent antioxidant capacity (TEAC), total glutathione, reduced glutathione, ascorbic acid, total free thiols, total free selenols, Trx, Grx and NTR activities were examined. These results are summarized in relative terms in Table 5.
Table 5.
Comparison of various biochemical parameters in high and low toxin K. brevis.
| Low/ High range |
Low/ High Average |
|
|---|---|---|
| TEAC | 0.81–1.10 | 0.904 (n=4) |
| Ascorbic acid | 1.29–4.49 | 1.90 (n=4) |
| GSH | 1.36–2.34 | 1.63 (n=6) |
| GSH/(GSx) | 1.29–2.1 | 1.58 (n=4) |
| High MW thiols | 2.2 | - |
| Sel-green 0 min | 11.0 | - |
| Sel-green 2.5 min | 3.3 | - |
| Sel-green 50 min | 5.6 | - |
| Trx activity | 3.0 | - |
| Grx activity | 1.6 | - |
Trolox equivalent antioxidant capacity (TEAC) was determined by the ability of homogenates to decolorize ABTS•+. On three of four days, the total antioxidant capacity was 20% higher in the high toxin K. brevis (p=0.01, n=3). The difference in TEAC between the low and high toxin K. brevis including all four of the days tested was not statistically significant (p=0.25, n=4).
While TEAC was not significantly different between high and low toxin K. brevis, when individual antioxidants were assessed, significant differences became evident. Antioxidants which can decolorize ABTS•+ include ascorbic acid, GSH, carotenoids, and others. Ascorbic acid, defined as the difference in TEAC in parallel samples with and without ascorbic acid oxidase, was consistently and significantly higher in the low toxin homogenates (p=0.04, n=4). Free thiol was monitored via Ellman’s assay over several days of growth. The reduction of Ellman’s reagent is taken as an indicator of GSH because it can reach millimolar concentrations and typically comprises the majority of low molecular weight thiols in cells (Armstrong, 1997; Van Bladeren, 2000). Similarly, the reduction of Ellman’s reagent after incubation with GR is taken as an indicator of total glutathione (GSx = GSH+GSSG). This assay revealed that GSH was consistently higher (p=0.001, n=6) in the low toxin K. brevis with the ratio of low/high ranging from 1.36 to 2.34 with an average ratio of 1.63. Total glutathione was less variable between the two cultures (p=0.41, n=4) with an average low/high ratio of 1.11. As a result, the ratio of GSH/GSx was consistently higher in the low toxin K. brevis, ranging from 1.29 – 2.1 with an average of 1.58 (p=0.02, n=4). Ellman’s reagent can detect low MW and some accessible high MW thiols. On the other hand, the modified papain assay can detect these and inaccessible high MW thiols. The addition of cystamine acts as a shuttle between less accessible high MW thiols and papain methyl sulfide. The rate of hydrolysis of N-benzoyl-L-arginine-p-nitroanilide is indicative of the activity of papain, which in turn is indicative of total thiols. Consistent with Ellman’s assay, modified papain activity in the low toxin homogenate was twice that of the high toxin homogenate. The selenol selective probe reacts rapidly with selenols and very slowly with thiols (Zhang et al., 2015). Therefore, the 0 and 2.5 min readings likely represent the reaction with Sec while the fluorescence reading at 50 minutes represent the reaction with both Cys and Sec. Fluorescence was elevated in the low toxin samples at all time points. Both Trx and Grx activities were elevated in the low toxin K. brevis. Thioredoxin activity is dependent on NTR activity.
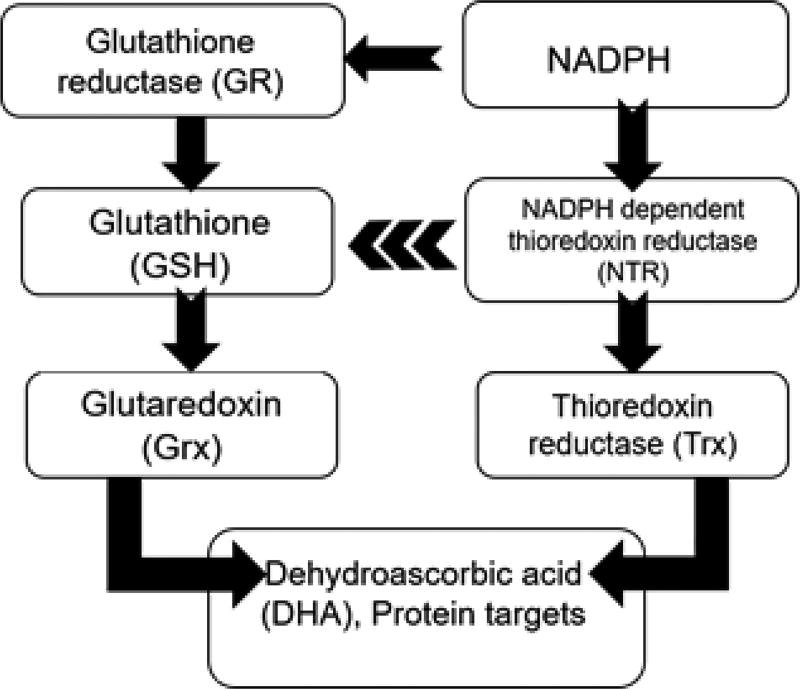
Taking the results from these assays under consideration, there is clearly a difference in redox status between high and low toxin K. brevis. Is it possible that NTR inhibition is responsible? Fig. 9 shows the major redox regulatory systems, NTR/Trx and glutathione, and the relationship between the two. The accepted concept is that the glutathione system and the NTR/Trx systems work independently of each other in maintaining redox homeostasis. Nonetheless, crosstalk between these two systems has been found in yeast (Grant, 2001), plants (Meyer et al., 2012) and in Apicomplexan parasites (Kanzok et al., 2000). In fact, the NTR/Trx system in Plasmodium falciparum has been found to efficiently reduce GSSG (Kanzok et al., 2000). Dehydroascorbate can be reduced to ascorbate both by NTR and Grx. Several types of NTR are present in K. brevis and the specific role and subcellular localization of each NTR remains undetermined. Nonetheless, antioxidants such as thiols and ascorbic acid as well as Trx and Grx activities are all negatively correlated with brevetoxin content in K. brevis. These data provide evidence to support the hypothesis that brevetoxin impacts the redox system in cells through reaction with TrxR. While the effects of PbTx-2 on Sec containing large NTR have been demonstrated, the possibility of inhibition of other enzymes with dithiol active sites has not been ruled out. Indeed, PbTx-2 has been shown to react with GSH and cysteine residues in proteins (Wang and Ramsdell, 2010). This will be the subject of further studies.
Fig. 9.
The major redox regulatory systems
The recent finding that the low toxin K. brevis is deficient in NPQ (Cassell et al., 2015) prompted us to examine the carotenoid content and distribution in low and high toxin K. brevis. When normalized to chlorophyll, there appears to be no difference in total carotenoids between the two cultures. A comparison of the carotenoid distribution in the light vs. dark adapted samples of the high toxin K. brevis, revealed differences only in the mole% Dd and Dt in with Dd higher in the dark-adapted and Dt higher in the light-adapted samples (Table 3). This behavior would be expected when NPQ is functioning normally. Conversely, the differences in Dd and Dt in the light vs. dark adapted samples of the low toxin K. brevis were not statistically significant. This behavior would result in NPQ deficiency. A comparison of carotenoid content of the high and low toxin K. brevis indicates a statistically significant difference for Dd in the light (Table 4), with the low toxin K. brevis having more of the epoxide form (Dd). As a result, there was a statistically significant difference in EPS between the high and low toxin K. brevis in the light, with the low toxin having a higher EPS. There was not a significant difference in Dt between the low and high toxin cells. Higher levels of Dd in the light would suggest that the de-epoxidation of Dd functions less efficiently in low toxin K. brevis. As previously mentioned, VDE catalyzes the de-epoxidation of Vx to Zx in a two-step process, using ascorbic acid as a co-factor. Results presented earlier in this work establish that more ascorbic acid is available in the low toxin K. brevis. Again, these results suggest that VDE (the dinoflagellate version of VDE) is not functioning properly. VDE is an oxidoreductase, which has six disulfide bridges which are critical to its activity. Reduction of these disulfide bridges, or mutation of the cysteines renders the enzyme inactive (Simionato et al., 2015). The modified papain assay indicates that the low toxin K. brevis does have more high MW thiols (Fig. 6). The reduction of the disulfide bonds in VDE by Trx has been demonstrated in vitro (Hall et al., 2010) suggesting that higher Trx activity could suppress VDE. It seems unlikely that VDE is a direct target for Trx. In plants, VDE is located in the chloroplast lumen while Trx is located in the stroma. Another possibility is simply the reducing environment of the low toxin K. brevis suppresses VDE activity. In fact, it has long been known that treatment of chloroplasts with DTT shuts down NPQ by virtue of reduction of VDE’s disulfide bonds. The more likely scenario is the transfer of reducing equivalents across the thylakoid membrane from the stroma to the lumen. In plants and cyanobacteria, reducing equivalents are transferred across the thylakoid membrane by a “Trx-like” transmembrane enzyme called HCF164 (Motohashi and Hisabori, 2006; Nikkanen and Rintamäki, 2014). A tBLASTn search of the K. brevis Wilson transcriptome library using HCF164 from Arabidopsis lyrata (Accession EHF452731.1) returned a single transcript (locus 3952) with high homology to the query (E value = 5 e10−13). Therefore, the effects of inhibition of NTRC in the chloroplast stroma could ultimately be transferred to the lumen. Significant differences in the distribution of other carotenoids of the high and low toxin K. brevis were observed, specifically the butanoyl and hexanoyl-fucoxanthins, but the significance of these differences remains unclear.
In summary, differences in the redox status of low and high toxin K. brevis have been detected. The inhibition by PbTx-2 of NTR having a C-terminal Sec residue has been previously demonstrated and herein the existence of such NTRs in K. brevis has been established. Additionally, the xanthophyll cycle has been shown to be less efficient in low toxin K. brevis, and this work suggest the link between the presence of PbTx-2 and NPQ in K. brevis is via NTR inhibition.
Highlights.
Redox status is significantly different in high and low-toxic K. brevis.
Thioredoxin and glutaredoxin activities are higher in low toxic K. brevis.
Reduced diadinoxanthin in low toxic K. brevis results in NPQ insufficiency.
Inhibition of NTR by brevetoxin results in differences between K. brevis strains.
Acknowledgments
Research described in this publication was supported, in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number R25 GM061347.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Armstrong RN. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10(1):2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- Backer LC, Kirkpatrick B, Fleming LE, Cheng YS, Pierce R, Bean JA, Clark R, Johnson D, Wanner A, Tamer R, Zhou Y, Baden DG. Occupational exposure to aerosolized brevetoxins during Florida red tide events: effects on a healthy worker population. Environ. Health Perspect. 2005;113(5):644–649. doi: 10.1289/ehp.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18(2):298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Cassell RT, Chen W, Thomas S, Liu L, Rein KS. Brevetoxin, the dinoflagellate neurotoxin, localizes to thylakoid membranes and interacts with the light-harvesting complex II (LHCII) of Photosystem II. ChemBioChem. 2015;16(7):1060–1067. doi: 10.1002/cbic.201402669. [DOI] [PubMed] [Google Scholar]
- Chen W, Tuladhar A, Rolle S, Lai Y, del Rey FR, Zavala CE, Liu Y, Rein KS. Brevetoxin-2, is a unique inhibitor of the C-terminal redox center of mammalian thioredoxin reductase-1. Toxicol. Appl. Pharmacol. 2017 doi: 10.1016/j.taap.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YS, Villareal TA, Zhou Y, Gao J, Pierce RH, Wetzel D, Naar J, Baden DG. Characterization of red tide aerosol on the Texas coast. Harmful Algae. 2005a;4(1):87–94. doi: 10.1016/j.hal.2003.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YS, Zhou Y, Irvin CM, Pierce RH, Naar J, Backer LC, Fleming LE, Kirkpatrick B, Baden DG. Characterization of marine aerosol for assessment of human exposure to brevetoxins. Environ. Health Perspect. 2005b:638–643. doi: 10.1289/ehp.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeland E, Garrido J, Clementson L, Andresen K, Thomas C, Zapata M, Airs R, Llewellyn C, Newman G, Rodríguez F. Data sheets aiding identification of phytoplankton carotenoids and chlorophylls. Phytoplankton pigments: characterization, chemotaxonomy and applications in oceanography. 2011:665–822. [Google Scholar]
- Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82(1):70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Errera RM, Bourdelais A, Drennan M, Dodd E, Henrichs D, Campbell L. Variation in brevetoxin and brevenal content among clonal cultures of Karenia brevis may influence bloom toxicity. Toxicon. 2010;55(2):195–203. doi: 10.1016/j.toxicon.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Errera RM, Campbell L. Osmotic stress triggers toxin production by the dinoflagellate Karenia brevis. Proc. Natl. Acad. Sci. U. S. A. 2011;108(26):10597–10601. doi: 10.1073/pnas.1104247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evens TJ, Kirkpatrick GJ, Millie DF, Chapman DJ, Schofield OM. Photophysiological responses of the toxic red-tide dinoflagellate Gymnodinium breve (Dinophyceae) under natural sunlight. J. Plankton Res. 2001;23(11):1177–1194. [Google Scholar]
- Fire SE, Flewelling LJ, Stolen M, Noke Durden W, de Wit M, Spellman AC, Wang Z. Brevetoxin-associated mass mortality event of bottlenose dolphins and manatees along the east coast of Florida, USA. Mar. Ecol. Prog. Ser. 2015;526:241–251. [Google Scholar]
- Fleming LE, Backer LC, Baden DG. Overview of aerosolized Florida red tide toxins: exposures and effects. Environ. Health Perspect. 2005a;113(5):618–620. doi: 10.1289/ehp.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming LE, Jerez E, Stephan WBB, Cassedy A, Bean JA, Reich A, Kirkpatrick B, Backer L, Nierenberg K, Watkins S. Evaluation of harmful algal bloom outreach activities. Mar. Drugs. 2007;5(4):208–219. doi: 10.3390/md504208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming LE, Kirkpatrick B, Backer LC, Bean JA, Wanner A, Dalpra D, Tamer R, Zaias J, Cheng YS, Pierce R. Initial evaluation of the effects of aerosolized Florida red tide toxins (brevetoxins) in persons with asthma. Environ. Health Perspect. 2005b:650–657. doi: 10.1289/ehp.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flewelling LJ, Naar JP, Abbott JP, Baden DG, Barros NB, Bossart GD, Bottein M-YD, Hammond DG, Haubold EM, Heil CA, Henry MS, Jacocks HM, Leighfield TA, Pierce RH, Pitchford TD, Rommel SA, Scott PS, Steidinger KA, Truby EW, Van Dolah FM, Landsberg JH. Brevetoxicosis: Red tides and marine mammal mortalities. Nature. 2005;435(7043):755–756. doi: 10.1038/nature435755a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuciman M, Enriquez MM, Polívka Ts, Dall’Osto L, Bassi R, Frank HA. Role of xanthophylls in light harvesting in green plants: a spectroscopic investigation of mutant LHCII and Lhcb pigment–protein complexes. J. Phys. Chem. B. 2012;116(12):3834–3849. doi: 10.1021/jp210042z. [DOI] [PubMed] [Google Scholar]
- Gates JA, Wilson WB. The toxicity of Gonyaulax monilata Howell to Mugil cephalus. Limnol. Oceanogr. 1960;5(2):171–174. [Google Scholar]
- Grant CM. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001;39(3):533–541. doi: 10.1046/j.1365-2958.2001.02283.x. [DOI] [PubMed] [Google Scholar]
- Hagerthey SE, William Louda J, Mongkronsri P. Evaluation of pigment extraction methods and a recommended protocol for periphyton chlorolhyll a determination and chemotaxonomic assessment. J. Phycol. 2006;42(5):1125–1136. [Google Scholar]
- Hall M, Mata-Cabana A, Åkerlund HE, Florencio FJ, Schröder WP, Lindahl M, Kieselbach T. Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics. 2010;10(5):987–1001. doi: 10.1002/pmic.200900654. [DOI] [PubMed] [Google Scholar]
- Hanschmann E-M, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signaling. 2013;19(13):1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison DR, Sunda WG, Shea D, Litaker RW. Increased toxicity of Karenia brevis during phosphate limited growth: ecological and evolutionary implications. PLoS One. 2013;8(3):e58545. doi: 10.1371/journal.pone.0058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison DR, Sunda WG, Tester PA, Shea D, Litaker WR. Increased cellular brevetoxins in the red tide dinoflagellate Karenia brevis under CO2 limitation of growth rate: Evolutionary implications and potential effects on bloom toxicity. Limnol. Oceanogr. 2014;59(2):560–577. [Google Scholar]
- Hirt RP, Müller S, Embley TM, Coombs GH. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 2002;18(7):302–308. doi: 10.1016/s1471-4922(02)02293-6. [DOI] [PubMed] [Google Scholar]
- Hoagland P, Jin D, Beet A, Kirkpatrick B, Reich A, Ullmann S, Fleming LE, Kirkpatrick G. The human health effects of Florida Red Tide (FRT) blooms: An expanded analysis. Environ. Int. 2014;68:144–153. doi: 10.1016/j.envint.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Hoagland P, Jin D, Polansky LY, Kirkpatrick B, Kirkpatrick G, Fleming LE, Reich A, Watkins SM, Ullmann SG, Backer LC. The costs of respiratory illnesses arising from Florida Gulf Coast Karenia brevis blooms. Environ. Health Perspect. 2009;117(8):1239. doi: 10.1289/ehp.0900645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3(3):239–243. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- Jacquot J-P, Eklund H, Rouhier N, Schürmann P. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009;14(6):336–343. doi: 10.1016/j.tplants.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Jeng MF, Reymond MT, Tennant LL, Holmgren A, Dyson HJ. NMR characterization of a single-cysteine mutant of Escherichia coli thioredoxin and a covalent thioredoxin-peptide complex. Eur. J. Biochem. 1998;257(2):299–308. doi: 10.1046/j.1432-1327.1998.2570299.x. [DOI] [PubMed] [Google Scholar]
- Kanzok SM, Schirmer RH, Türbachova I, Iozef R, Becker K. The thioredoxin system of the malaria parasite Plasmodium falciparum glutathione reduction revisited. J. Biol. Chem. 2000;275(51):40180–40186. doi: 10.1074/jbc.M007633200. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick B, Fleming LE, Squicciarini D, Backer LC, Clark R, Abraham W, Benson J, Cheng YS, Johnson D, Pierce R. Literature review of Florida red tide: implications for human health effects. Harmful algae. 2004;3(2):99–115. doi: 10.1016/j.hal.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louda JW, Li J, Liu L, Winfree MN, Baker EW. Chlorophyll-a degradation during cellular senescence and death. Org. Geochem. 1998;29(5):1233–1251. [Google Scholar]
- Louda JW, Liu L, Baker EW. Senescence-and death-related alteration of chlorophylls and carotenoids in marine phytoplankton. Org. Geochem. 2002;33(12):1635–1653. [Google Scholar]
- Mantoura R, Llewellyn C. The rapid determination of algal chlorophyll and carotenoid pigments and their breakdown products in natural waters by reverse-phase high-performance liquid chromatography. Anal. Chim. Acta. 1983;151:297–314. [Google Scholar]
- Mariotti M, Lobanov AV, Guigo R, Gladyshev VN. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013;41(15):e149–e149. doi: 10.1093/nar/gkt550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty SE, Schellenberger A, Goodwin DC, Fuanta NR, Tekwani BL, Calderón AI. Plasmodium falciparum thioredoxin reductase (PfTrxR) and its role as a target for new antimalarial discovery. Molecules. 2015;20(6):11459–11473. doi: 10.3390/molecules200611459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld J-P, Riondet C. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signaling. 2012;17(8):1124–1160. doi: 10.1089/ars.2011.4327. [DOI] [PubMed] [Google Scholar]
- Milian A, Nierenberg K, Fleming LE, Bean JA, Wanner A, Reich A, Backer LC, Jayroe D, Kirkpatrick B. Reported respiratory symptom intensity in asthmatics during exposure to aerosolized Florida red tide toxins. J. Asthma. 2007;44(7):583–587. doi: 10.1080/02770900701539251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minge MA, Shalchian-Tabrizi K, Tørresen OK, Takishita K, Probert I, Inagaki Y, Klaveness D, Jakobsen KS. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 2010;10(1):191. doi: 10.1186/1471-2148-10-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T. HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 2006;281(46):35039–35047. doi: 10.1074/jbc.M605938200. [DOI] [PubMed] [Google Scholar]
- Nikkanen L, Rintamäki E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Phil. Trans. R. Soc. B. 2014;369(1640):20130224. doi: 10.1098/rstb.2013.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T, Lidie KL, Van Dolah FM, Lindquist E, Cheng J-F, Bhattacharya D. Chimeric plastid proteome in the Florida “red tide” dinoflagellate Karenia brevis. Mol. Biol. Evol. 2006;23(11):2026–2038. doi: 10.1093/molbev/msl074. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Archibald JM, Keeling PJ. Complex protein targeting to dinoflagellate plastids. J. Mol. Biol. 2005;348(4):1015–1024. doi: 10.1016/j.jmb.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Keeling PJ. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 2006;357(5):1373–1382. doi: 10.1016/j.jmb.2006.01.084. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ransom Hardison D, Sunda WG, Wayne Litaker R, Shea D, Tester PA. Nitrogen limitation increases brevetoxins in Karenia brevis (Dinophyceae): Implications for bloom toxicity. J. Phycol. 2012;48(4):844–858. doi: 10.1111/j.1529-8817.2012.01186.x. [DOI] [PubMed] [Google Scholar]
- Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radical Biol. Med. 1999;26(9–10):1231–1237. doi: 10.1016/s0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Ryan DE, Pepper AE, Campbell L. De novo assembly and characterization of the transcriptome of the toxic dinoflagellate Karenia brevis. BMC genomics. 2014;15(1):888. doi: 10.1186/1471-2164-15-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer BA, Kamykowski D, McKay L, Sinclair G, Milligan E. Lipid class, carotenoid, and toxin dynamics of Karenia brevis (dinophyceae) during diel vertical migration. J. Phycol. 2009;45(1):154–163. doi: 10.1111/j.1529-8817.2008.00627.x. [DOI] [PubMed] [Google Scholar]
- Simionato D, Basso S, Zaffagnini M, Lana T, Marzotto F, Trost P, Morosinotto T. Protein redox regulation in the thylakoid lumen: The importance of disulfide bonds for violaxanthin deepoxidase. FEBS Lett. 2015;589(8):919–923. doi: 10.1016/j.febslet.2015.02.033. [DOI] [PubMed] [Google Scholar]
- Singh R, Blättler WA, Collinson AR. Assay for thiols based on reactivation of papain. Methods Enzymol. 1995;251:229–237. doi: 10.1016/0076-6879(95)51125-3. [DOI] [PubMed] [Google Scholar]
- Standfuss J, van Scheltinga ACT, Lamborghini M, Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 2005;24(5):919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunda WG, Burleson C, Hardison DR, Morey JS, Wang Z, Wolny J, Corcoran AA, Flewelling LJ, Van Dolah FM. Osmotic stress does not trigger brevetoxin production in the dinoflagellate Karenia brevis. Proc. Natl. Acad. Sci. U. S. A. 2013;110(25):10223–10228. doi: 10.1073/pnas.1217716110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer VL, Baden DG, Catterall WA. Identification of peptide components of the brevetoxin receptor site of rat brain sodium channels. J. Biol. Chem. 1994;269(31):19904–19909. [PubMed] [Google Scholar]
- Van Bladeren PJ. Glutathione conjugation as a bioactivation reaction. Chem.-Biol. Interact. 2000;129(1):61–76. doi: 10.1016/s0009-2797(00)00214-3. [DOI] [PubMed] [Google Scholar]
- Waggett RJ, Hardison DR, Tester PA. Toxicity and nutritional inadequacy of Karenia brevis: synergistic mechanisms disrupt top-down grazer control. Mar. Ecol. Prog. Ser. 2012;444:15–30. [Google Scholar]
- Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 1998;95(21):12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19(8):1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CJ, Leggett SR, Carter BJ, Colle C. Effects of brevetoxin exposure on the immune system of loggerhead sea turtles. Aquat. Toxicol. 2010;97(4):293–303. doi: 10.1016/j.aquatox.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ramsdell JS. Analysis of interactions of brevetoxin-B and human serum albumin by liquid chromatography/mass spectrometry. Chem. Res. Toxicol. 2010;24(1):54–64. doi: 10.1021/tx1002854. [DOI] [PubMed] [Google Scholar]
- Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Hönigschmid P, Schafferhans A, Roos M, Bernhofer M. PredictProtein—an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014:gku366. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Fraga S, Rodríguez F, Garrido JL. Pigment-based chloroplast types in dinoflagellates. Mar. Ecol. Prog. Ser. 2012;465:33–52. [Google Scholar]
- Zhang B, Ge C, Yao J, Liu Y, Xie H, Fang J. Selective selenol fluorescent probes: design, synthesis, structural determinants, and biological applications. J. Am. Chem. Soc. 2015;137(2):757–769. doi: 10.1021/ja5099676. [DOI] [PubMed] [Google Scholar]
- Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. Deciphering apicoplast targeting signals–feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280(1):19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]