Abstract
ASH2L encodes a trithorax group protein that is a core component of all characterized mammalian histone H3K4 methyltransferase complexes, including Mixed Lineage Leukemia (MLL) complexes. ASH2L protein levels in primary leukemia patient samples have not yet been defined. We analyzed ASH2L protein expression in 511 primary AML patient samples using reverse phase protein array technology. We discovered that ASH2L expression is significantly increased in a subset of patients carrying FLT3 mutations. Furthermore, we observed that low levels of ASH2L are associated with increased overall survival. We also compared ASH2L levels to the expression of 230 proteins previously analyzed on this array. ASH2L expression was inversely correlated with 32 proteins, mostly involved in cell adhesion and cell cycle inhibition, while a positive correlation was observed for 50 proteins, many of which promote cell proliferation. Together, these results indicate that a lower level of ASH2L protein is beneficial to AML patients.
Keywords: ASH2L, leukemia, epigenetics, histone modifications, chromatin, methyltransferase
INTRODUCTION
Chromatin modifying enzymes play essential roles during hematopoiesis, and a number of these enzymes are aberrantly expressed and/or exhibit altered catalytic activity in hematological malignancies.[1] Modifications added to DNA and histone proteins are often associated with transcriptional competency. Histone H3 lysine 4 trimethylation (H3K4me3) is frequently found near transcription start sites of active genes and is added by several SET domain-containing enzymes, including those belonging to the mixed lineage leukemia (MLL) family.[2] Translocations involving MLL (11q23) are observed in AML and ALL and correlate with poor prognosis.[3–5]
Absent, small, or homeotic discs-like protein (ASH2L) is a trithorax group family member that functions within mammalian histone H3K4 methyltransferase complexes to maintain global levels of H3K4me3.[6–8] Emerging evidence points toward a role for ASH2L in regulating cell proliferation and cell fate decisions. ASH2L is mechanistically involved in regulating the transition between pluripotent and differentiated states by coordinating the activities of histone H3K4 methyltransferase complexes and transcription factors that drive the expression of pluripotency genes through its direct interactions with Sox2 and c-Myc.[9] Ash2L is widely expressed in the developing mouse embryo[10–12], with highest expression observed in the fetal lung and liver.[11] Mice lacking Ash2L die prior to implantation (E3.5–8.5), indicating that Ash2L is essential for viability.[12] However, the in vivo function of Ash2L during early embryogenesis has not been defined because embryonic stem (ES) cells lacking Ash2L were not attainable [12], further emphasizing the functional significance of this epigenetic regulator during this crucial window for cell fate decisions.
Regulation of ASH2L expression appears to be required for differentiation decisions post-developmentally as well. ASH2L mRNA is highly expressed in leukemia cell lines with erythroleukemia and megakaryoblastic features. ASH2L expression remains high if cells are induced toward erythroid differentiation but is downregulated during megakaryocytic differentiation.[13] Furthermore, depletion of Ash2L in murine erythroid leukemia cells inhibits their terminal differentiation [14], which is attributed to simultaneous loss of H3K4me3 at the β-globin locus control region and β-globin gene transcription.[15] It remains to be determined whether downregulation of ASH2L expression is a cause or consequence of megakaryocyte differentiation. Nevertheless, these results suggest that regulation of ASH2L expression and function is important during hematopoietic cell differentiation.
In addition to an apparent role in differentiation, ASH2L has intrinsic oncogenic properties.[16] ASH2L interacts with MYC and cooperates with activated Ha-RAS to transform rat embryo fibroblasts, which readily form highly proliferative, disorganized, and poorly differentiated tumors in vivo.[16],[17] Furthermore, ASH2L protein expression is increased in solid tumors and leukemia cell lines compared to matched controls, while no difference in mRNA expression is detected[16], suggesting that the increase in ASH2L protein expression observed in cancer cells is governed by a post-transcriptional mechanism. Indeed, at the post-translational level, ASH2L is methylated by protein arginine methyltransferases, PRMT1 and PRMT5 [18], enzymes with established ties to hematological malignancies.[19–21]
Taken together these data strongly suggest that ASH2L function is required for appropriate cell proliferation and differentiation in leukemia model systems. To our knowledge, ASH2L expression has not been evaluated in primary leukemia patient samples. Therefore, we set out to determine if ASH2L protein expression is altered in AML, and whether ASH2L can be used as a prognostic marker. We used reverse phase protein array (RPPA) technology to assess ASH2L protein expression in 511 AML patients. We observed that low expression of ASH2L protein correlates with a favorable outcome for patients.
METHODS
Patient Population
Peripheral blood (PB) and bone marrow (BM) samples were collected from 511 patients with newly diagnosed AML and from 21 patients with newly diagnosed APL evaluated at the University of Texas MD Anderson Cancer Center (MDACC) between September 1999 and March 2007. Informed consent was acquired from patients in accordance with the Declaration of Helsinki and samples were obtained during routine diagnostic assessments. All samples were analyzed in accordance with regulations and protocols approved by the MDACC Institutional Review Board. The composition of this sample set and the treatments received have been previously reported.[22,23]
Sample Preparation, RPPA Methodology, and Western Blot Analysis
Samples were enriched for mononuclear cells by performing Ficoll-Hypaque (Sigma-Aldrich) density gradient separation, and contaminating B and T cells were removed by CD3/CD19 depletion if they were calculated to be > 5% based on the post-ficoll differential. Whole cell lysates were prepared, and the proteomic profiles of the samples from patients with AML were analyzed by RPPA as previously described [23–25] using a rabbit monoclonal antibody against ASH2L (Cell Signaling Technology, cat. # 5019). Whole cell extracts were prepared in lysis buffer (250 mM NaCl, 5 mM EDTA, 50 mM HEPES [pH 7.5], 0.1% NP-40, 0.1% [v/v] protease inhibitor cocktail, 0.5 mM DTT), and western blot analysis was carried out as previously described [18] using the same antibody as for RPPA.
FLT3 and NPM1 Mutational Analysis
Mutational analysis of FLT3 and NPM1 was performed by RT-PCR amplification, followed by sequencing using standard primers. For most patients, the mutation status was determined as part of routine care by the Clinical Laboratory Improvement Amendments (CLIA)-approved molecular hematology laboratory at MDACC. The same primers and conditions were used by the S.M.K. laboratory to determine the FLT3 and NPM1 mutational status of older patients where FLT3 status was not assessed as part of routine care.
Statistical Analysis
Statistical analyses were carried out similarly to those previously described.[22,23] To correlate ASH2L expression with clinical characteristics, as shown in Table I, F-tests were performed. ASH2L expression was compared to that of normal bone marrow derived CD34+ cells to divide the cohort into those with above (15.74%), equal to (73.15%), or below (11.11%) normal expression.
Table I.
Clinical characteristics of AML patient cohort
| Expression relative to Normal CD34+ | ||||||
|---|---|---|---|---|---|---|
| All Cases |
Equal | Above | Below | P | ||
| Number | 216 | 158 | 34 | 24 | ||
| Gender | M | 111 | 79 | 19 | 13 | 0.79 |
| F | 105 | 79 | 15 | 11 | ||
| Age | Min | 60 | 19.47 | 17.39 | 22.71 | 0.85 |
| Max | 87 | 87.23 | 84.31 | 84.01 | ||
| Median | 62 | 64.7 | 60.37 | 63.01 | ||
| WHO | Abnormal Cytogenetics | 27 | 20 | 4 | 3 | 0.046 |
| Therapy Related | 27 | 20 | 4 | 3 | ||
| Multilineage Dysplasia | 35 | 23 | 3 | 9 | ||
| Not in Others | 127 | 95 | 23 | 9 | ||
| Zubrod PS | 3 or 4 | 8 | 8 | 0 | 0 | 0.218 |
| AHD | >=2 mo | 71 | 54 | 4 | 13 | 0.003 |
| Infection | Yes | 60 | 48 | 11 | 1 | 0.023 |
| WBC | Median | 20.80 | 25.65 | 23.85 | 11.85 | 0.039 |
| Absolute Blast Count | Median | 5.28 | 6.145 | 16.875 | 1.2675 | 0.0047 |
| Platelet | Median | 50.00 | 52 | 43.5 | 75 | 0.0003 |
| Hemoglobin | Median | 9.55 | 9.5 | 9.55 | 10.05 | 0.89 |
| % Marrow Blasts | Median | 59.00 | 62 | 75 | 37.5 | 0.0002 |
| % Blood Blasts | Median | 35.00 | 34 | 75 | 10.5 | <0.00001 |
| Treated | 185 | 135 | 31 | 19 | ||
| Response | CR | 109 | 76 | 17 | 16 | 0.06 |
| Resistant | 57 | 46 | 10 | 1 | ||
| Fail | 19 | 13 | 4 | 2 | ||
| Relapse | Yes | 69 | 49 | 9 | 11 | 0.596 |
| Alive | Yes | 43 | 27 | 9 | 7 | 0.079 |
The table shows clinical characteristics of patients with ASH2L protein levels equal to, above, or below that of normal CD34+ cells, along with data for the total patient population.
RESULTS
Characterization of ASH2L Protein Expression in AML Patient Samples
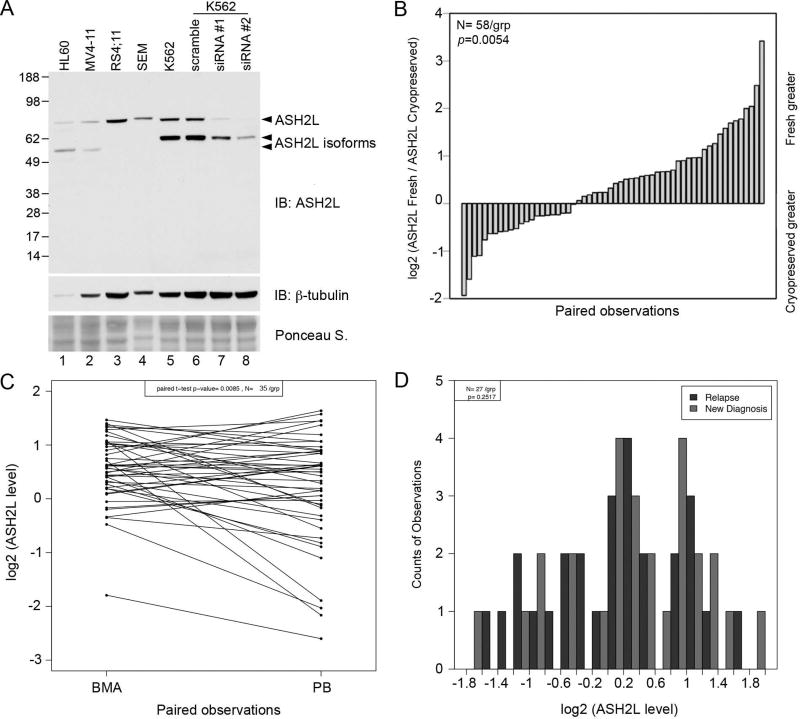
RPPA was used to determine ASH2L protein expression in 511 newly diagnosed AML and 21 newly diagnosed APL patients. We first characterized the specificity of the ASH2L antibody used for RPPA by western blot. Two human ASH2L RefSeq transcripts, NM_004674 and NM_001105214.1 exist, which encode the full-length 628 aa protein (ASH2L or ASH2L1; 70 kDa) and a 534 aa protein (ASH2L2; 59 kDa) that lacks amino acids encoded by the first exon. A search of the Ensembl database revealed that the human ASH2L gene encodes six protein-coding transcripts. In addition to ASH2L1 and ASH2L2, one transcript, ASH2L-201, lacking exons 1, 2, and 14, has been described in the literature [13], and is predicted to produce a 501 amino acid protein (55 kDa). The antibody used to probe the RPPA recognizes an internal epitope, and therefore detects full length ASH2L as well as these shorter isoforms (Figure 1A). Western blot analysis revealed that ASH2L is expressed in a variety of leukemic cell lines, although the expression of the shorter isoforms appears to be dependent on lineage (lanes 1 and 2; HL60, MV4;11 – myeloblastic/myelomonocytic), (lanes 3 and 4; RS4;11, SEM – lymphoblastic), (lanes 5–8; K562 – erythroleukemia) (Figure 1A). Following transient knock-down using ASH2L-specific siRNAs in K562 cells, full-length ASH2L protein was nearly undetectable, while expression of the shorter isoforms was reduced to a lesser extent (Figure 1A, lanes 5–8). No additional proteins were detected, indicating that the antibody used to probe the RPPA recognizes only ASH2L protein products.
Figure 1. Analysis of ASH2L protein expression in leukemia cell lines and patient samples.
(A) Western blot analysis of ASH2L protein expression in leukemia cell lines of varying lineages. (B) RPPA was used to determine ASH2L levels in 58 paired cryopreserved and freshly prepared samples, which were compared by paired t test (P = 0.0054). (C) ASH2L levels were determined by RPPA in 35 paired peripheral blood (PB) and bone marrow (BMA) samples, and compared by paired t test (P = 0.0085). (D) ASH2L expression was measured by RPPA, and a comparison was made using 27 paired samples that were collected from patients when newly diagnosed and upon relapse (P = 0.2517).
Analysis of paired samples indicated that ASH2L protein levels were significantly higher in samples prepared fresh on the day of collection compared to those prepared from cells that were cryopreserved (P = 0.0054, t = 6.05; Figure 1B). This result suggests that the cryopreservation process affected ASH2L protein levels. To ensure that ASH2L protein levels were not affected by the duration of cryopreservation of the prepared lysates, we analyzed the entire collection date range and found that no correlation exists between the duration of cryopreservation and ASH2L expression. To avoid a confounding factor, we limited our analysis to the 216 AML cases with protein prepared from fresh cells on the day of collection. In addition, ASH2L expression analysis in APL samples is reported here as part of the complete study, however, these samples were excluded from most of the outcomes analyses.
All normal CD34+ bone marrow cell samples analyzed express ASH2L (Figure S1A). The patient samples were divided into 3 cohorts depending on whether ASH2L protein expression was above (15.74%), equal to (73.15%), or below (11.11%) expression observed in normal CD34+ cells (Figure S1B). We began by analyzing the relationship between ASH2L protein expression as defined by RPPA and various demographic and clinical features of the patient samples included in this array, as shown in Table I. Patients with infections at the time of diagnosis tended to have higher levels of ASH2L protein (P = 0.023), while patients who had an antecedent hematologic disorder (AHD) tended to have lower ASH2L protein expression (P = 0.003).
Equal to or above normal levels of ASH2L protein expression correlate with the percentage of blasts found in the bone marrow and peripheral blood (P = 0.0002, P < 0.00001). In addition, ASH2L protein expression is positively correlated with increased white blood cell count (P = 0.0009; Figure S2A), absolute blast count (P = 0.0003; Figure S2B), and negatively correlates with platelet count (P < 0.00001; Figure S2C). ASH2L protein expression was significantly different in paired samples that were obtained from bone marrow and peripheral blood (P = 0.009; Figure 1C). However, further analyses revealed that despite slightly higher levels of ASH2L in bone marrow samples, the range, mean, and distribution of ASH2L levels are similar between the sample types and overall clinical evaluation is not affected by the sample type. ASH2L protein levels did not differ significantly between paired samples obtained from patients at the time of diagnosis and at relapse (P = 0.252; Figure 1D).
ASH2L Protein Expression Differs Amongst Subtypes of AML
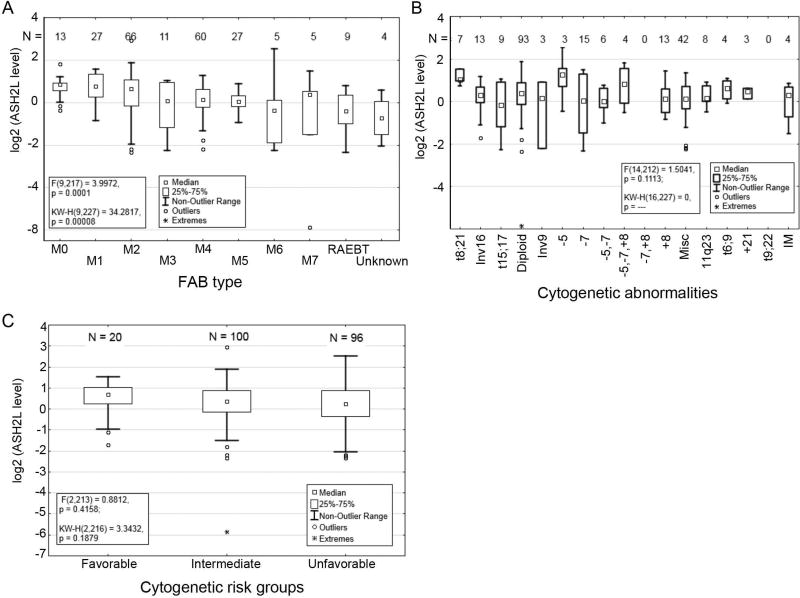
ASH2L protein expression varied significantly across the French-American-British (FAB) classification system of AML (P = 0.0001; Figure 2A). Higher expression of ASH2L was observed in the M0, M1, and M2 classes, which represent myeloblastic leukemias that are characterized by an increased presence of minimally differentiated cells, while lower than average expression was observed in M6 and RAEBT classes. Variable ASH2L protein expression was also observed across AML groups divided by common cytogenetic abnormalities (P = 0.111; Figure 2B). However, levels did not significantly differ if patients were divided into broader groups with favorable, intermediate or unfavorable cytogenetic profiles (P = 0.416; Figure 2C).
Figure 2. ASH2L protein expression and clinical characteristics of AML patient samples.
(A) Distribution of ASH2L protein levels among FAB subtypes of AML (P = 0.0001). (B) ASH2L protein levels were determined in AML patient samples by RPPA and categorized by cytogenetic abnormalities (P = 0.111). (C) ASH2L protein levels in cytogenetic risk groups (P = 0.416).
ASH2L Protein Expression Correlates with Specific Molecular Mutations
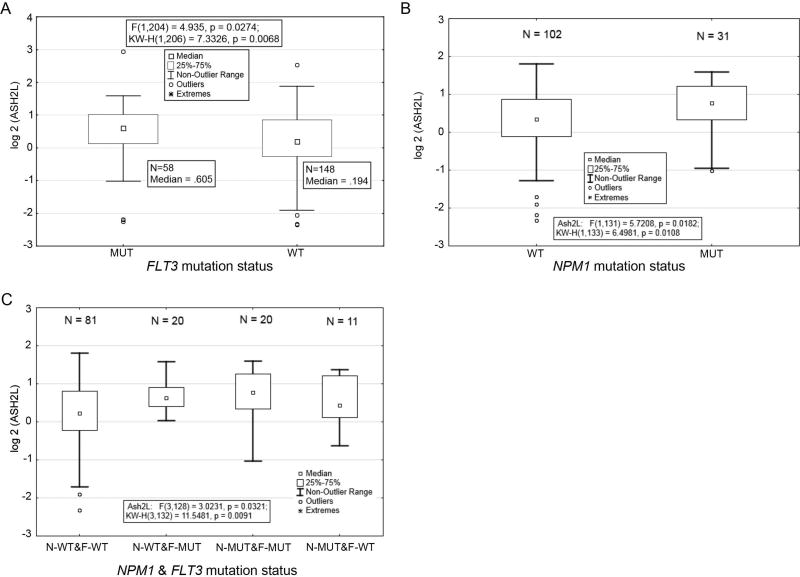
We next analyzed ASH2L protein expression in the context of molecular mutations commonly found in the AML patient population. ASH2L protein expression significantly correlated with mutations in fms-related tyrosine kinase 3 (FLT3) and nucleophosmin (NPM1) (Figure 3). ASH2L expression is significantly higher in patients with FLT3 mutations, including both internal tandem duplication (ITD) and D835Y activating mutations (P = 0.027; Figure 3A). Increased ASH2L expression was also observed in patients with NPM1 mutations (P = 0.018; Figure 3B). The increase in ASH2L protein level is more pronounced in patients with combined FLT3 and NPM1 mutations than those with individual mutations (compare boxplot 3 with 2 and 4; Figure 3C). ASH2L expression did not correlate with the mutation status of TP53, RAS, or IDH1 (Figure S3A–C), suggesting a biological relevance for the association between ASH2L expression level and FLT3 and NPM1 mutations.
Figure 3. ASH2L protein levels in AML patients with FLT3 and NPM1 mutations.
(A) Comparison of ASH2L protein expression in AML patient samples with wild-type (WT) or mutant (MUT; ITD and D835Y) FLT3 (P = 0.0274). (B) Comparison of ASH2L protein levels in patient samples with WT or mutant (4 BP Ins) NPM1 (P = 0.0182). (C) Comparison of ASH2L protein expression in AML patient samples with combined FLT3 and NPM1 mutations (P = 0.0321).
Low Expression of ASH2L Correlates with a Favorable Outcome for AML Patients
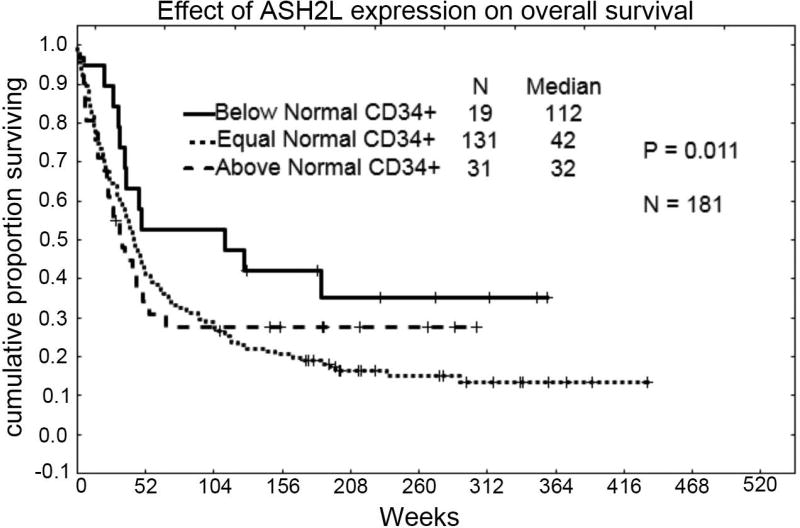
We analyzed the effect of ASH2L protein expression on overall survival using samples that were collected from newly diagnosed patients and were freshly prepared on the day of collection. The trends observed following Kaplan Meier analysis suggest that patients with the lowest level of ASH2L expression survive the longest compared to those with medium or high levels (median, 112 weeks compared to 42 or 32 weeks, P = 0.011; Figure 4). While multivariate analysis indicates that low expression of ASH2L is nearly an independent favorable prognostic factor, the limited number of cases demonstrating a “below normal” level in this dataset preclude statistical significance (n = 24; P = 0.067).
Figure 4. ASH2L protein levels and clinical outcome of AML patients.
Kaplan-Meier analysis of AML patient samples grouped as high, middle, or low expression of ASH2L protein (P = 0.011).
ASH2L Expression Correlates with the Expression of Proteins Involved in Cell Survival and Proliferation
To identify potential molecular mechanisms whereby ASH2L might contribute to AML pathology, ASH2L expression was compared to the expression of 230 other proteins simultaneously analyzed on the array. ASH2L protein expression was strongly and significantly correlated (R > 0.3, P <0.00001) with the expression of 82 other proteins, in a negative (n = 32) or positive (n = 50) manner (Figure 5A). This represents an unusually high number of strongly correlated proteins, which suggests a prominent role for ASH2L in AML pathogenesis. The strongest correlation observed is between ASH2L and the lysine demethylase LSD1 (KDM1A) (R = 0.792). Of the observed positive correlations, many proteins are involved in cell survival and proliferation, such as the previously identified ASH2L interacting partner MYC (R = 0.3823) [16],[17], as well as signaling kinases that have not previously been associated with ASH2L, such as MTOR (R = 0.4568), KIT (R = 0.4627), and AKT1 (R = 0.326). Given the role for ASH2L-containing methyltransferase complexes in transcriptional activation, it is not surprising that positive correlations were observed with several transcription factors, including ETS family members Fli1 (R = 0.6169), ERG (R = 0.4736), ELK1 pS383 (R = 0.5174), and SPI1 (R = 0.3786). In addition, several SMAD family transcription factors positively correlated with ASH2L protein expression, including SMAD2 (R = 0.3097), SMAD4 (R = 0.5959), and SMAD5 (R = 0.3493).
Figure 5. ASH2L protein expression significantly correlates with the expression of proteins involved in various AML pathways.
(A) Levels of ASH2L and 230 other proteins were measured by RPPA in the sample set containing 216 freshly prepared samples from AML patients. Positive and negative correlations with an R > |0.3| and P < 0.00001 are depicted in the waterfall plot. Asterisks denote ASH2L-correlated proteins involved in different cellular functions; red - transcription factors; green – positive regulators of cell proliferation; blue - RNA processing. (B) A heatmap was generated in which patient samples were organized in a hierarchical arrangement based on ASH2L protein expression levels (low, mid, high), cytogenetic group (favorable, intermediate, unfavorable), and FLT3 mutation status (negative or ITD).
ASH2L protein expression highly correlated with WTAP (R = 0.7667) and hnRNPK (R = 0.3908), both of which are involved in pre-mRNA splicing.[26],[27] Remarkably, ASH2L protein expression correlated not only with the mutational status of NPM1 (Figure 3B), but also with the total protein level (R = 0.6731). In addition, correlations between ASH2L and factors involved in ribosome biogenesis and protein synthesis were revealed, and include ribosomal assembly factors nucleolin (NCL) (R = 0.7522) and EBP1 (PA2G4) (R = 0.4704), and translation factors EIF2 (R = 0.3675) and EIF4E (R = 0.4001). Taken together, these results suggest a previously unrecognized association between ASH2L and RNA processing.
Inverse correlations were demonstrated between ASH2L protein expression and proteins that promote cell adhesion, such as CD49b (ITGA2), (R = −0.4872), Integrin B3 (ITGB3) (R = −0.3388), FAK (PTK2) (R = −0.3187), and VEGFR2 (KDR) (R = −0.3071). Surprisingly, the expression of SRC family kinases, including SRC (R = −0.3448) and LCK (R = −0.3536), were inversely correlated with ASH2L expression. Perhaps this relationship reflects the roles of these kinases in promoting cell adhesion rather than cell proliferation.
In order to determine whether any of the observed correlations clustered together in particular patient samples, the top 25 positively correlated and top 25 negatively correlated proteins were chosen to generate a heatmap in which the patient samples were organized in a hierarchical arrangement based on ASH2L protein expression levels (low, mid, high), cytogenetic group (favorable, intermediate, unfavorable), and FLT3 mutation status (negative or ITD) (Figure 5B). Following stringent statistical filtering to remove outliers at +/− 2.5%, 32 significantly associated proteins remained (P = 0.00001). The three proteins most highly correlated with ASH2L protein expression (Figure 5A), LSD1, WTAP, NCL, are clustered together irrespective of FLT3 mutation status or cytogenetic group, further suggesting a functional association with ASH2L.
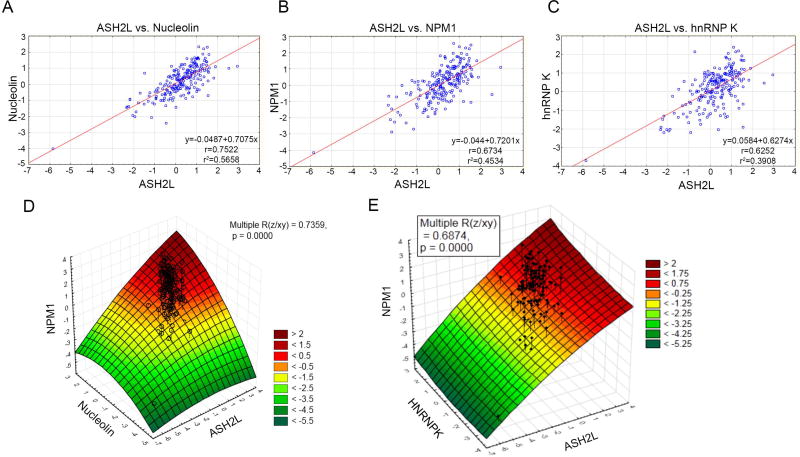
Because the expression levels of several RNA-associated proteins were highly correlated with ASH2L protein levels (Figure 5A), we analyzed these correlations with two-way comparisons visualized in individual patient samples (Figure 6A, B, C). The strictest correlation is observed between ASH2L and NCL (R2 = 0.5658) (Figure 6A), although ASH2L is also highly correlated with NPM1 (R2 = 0.4534) (Figure 6B) and hnRNPK (R2 = 0.3908) (Figure 6C) in individual patient samples. Three-dimensional surface plots were generated in order to determine whether relationships exist between multiple components in individual patient samples (Figure 6D, E). A clear interaction is observed between ASH2L, NPM1, and NCL, with similar expression levels of each protein observed in each individual sample. A similar, yet less robust interaction is also observed between ASH2L, NPM1 and hnRNPK, suggesting that the association with NCL may be of greater functional significance.
Figure 6. ASH2L protein expression is highly correlated with proteins involved in RNA associated processes.
(A) Following RPPA analysis, ASH2L protein levels in AML patient samples were compared to the total levels of (A) hnRNPK, (B) NPM1, and (C) NCL. (D) A three-dimensional surface plot was generated using Statistica software and depicts a statistically significant relationship between three components: ASH2L, NCL, and NPM1 (P < 0.0001). The color scale indicates the expression of the z-axis component (NPM1), and the scale ranges from < −5.25 to > + 2. (E) The plot was generated as in (D), and shows the relationship between ASH2L, hnRNPK, and NPM1 (P < 0.0001).
DISCUSSION
ASH2L is a component of all characterized mammalian histone H3K4 methyltransferase complexes, and expression of ASH2L affects global activity of these complexes.[7] ASH2L protein expression is elevated in leukemia cell lines [16], and we demonstrated lineage-specific expression of ASH2L isoforms. There are currently six human ASH2L protein-coding transcripts annotated in the Ensembl database, however, only two of these ASH2L transcripts were previously reported in leukemia cell lines.[13] Characterization of ASH2L isoforms expressed in different hematopoietic lineages will be required to determine the functional significance of these proteins.
We used RPPA to define ASH2L protein expression in primary AML samples and to correlate this data with clinical features. Our results indicate that ASH2L protein level correlates with select FAB classification subgroups and cytogenetic abnormalities. ASH2L expression was significantly higher in classes defined by the presence of minimally differentiated cells, suggesting that ASH2L promotes survival and/or proliferation of undifferentiated or progenitor cells, in agreement with the unviability of Ash2l−/− murine ES cells.[12]
ASH2L protein levels were significantly elevated in patients that carried D835Y and ITD mutations in FLT3. Because ITD and D835Y mutations result in constitutive activation of FLT3 kinase activity in about 30% of AML cases [28], it will be interesting to determine whether FLT3 signaling directly influences ASH2L protein levels. For example, ASH2L could be a substrate of FLT3 kinase. Or, perhaps transcription of ASH2L is upregulated by downstream effectors of FLT3 activity, such as STAT proteins. Although FLT3-ITD remains a therapeutic target in AML [29], evidence suggests that in many instances additional lesions are necessary for leukemogenesis. Given the significant increase in protein level in this subset, our results reveal ASH2L as a potential alternative target in patients with FLT3 mutations.
Increased ASH2L protein expression was observed in patient samples carrying mutations in NPM1. NPM1 normally shuttles between the nuclear, nucleolar, and cytoplasmic compartments to participate in various cellular functions, including ribosome biogenesis and centrosome duplication.[30] In addition, NPM1 has been shown to bind histone H3 and function in chromatin remodeling and transcriptional activation of rRNA genes in the nucleolus.[31,32] In AML, mutations in NPM1 are considered an early initiating event and result in aberrant localization and accumulation of the protein in the cytoplasm.[33] ASH2L is a nuclear protein making it presently unclear why mutations that cause cytosolic localization of NPM1 would correlate with increased ASH2L protein levels. However, our data also indicate that ASH2L protein expression highly correlates with total NPM1 protein levels, further implying a functional relationship between these proteins.
Our results indicate that patients expressing lower levels of ASH2L have increased overall survival. Remarkably, ASH2 deficiency extends the lifespan in C. elegans.[34,35] Furthermore, deficiency of ASH2 in the parental generation confers the extended lifespan in the descendants, suggesting that ASH2 is a key factor mediating epigenetic inheritance. Taken together, these results suggest that the level of ASH2L protein expression may serve as an overall predictor for longevity across species.
We compared ASH2L protein expression with that of 230 additional proteins analyzed on this RPPA. Positive correlations were demonstrated between ASH2L protein level and a number of proteins that promote cell proliferation, including MYC, MTOR, KIT, and AKT1. These correlations fit well with previously published work demonstrating that knock-down of ASH2L inhibits cell proliferation, specifically, S-phase progression.[16],[36] It is unclear whether these relationships are a cause or consequence of ASH2L overexpression, or whether they exist at the transcriptional or post-transcriptional level. ASH2L was previously implicated in the regulation of cell growth pathways as a downstream target of mTOR signaling.[37] Conversely, because ASH2L-containing H3K4 methyltransferase complexes activate transcription, it is likely that increased ASH2L expression contributes to the upregulation of some of these genes. Moreover, ASH2L protein level was highly correlated with transcription factors that might enhance the targeting and activity of H3K4 methyltransferase complexes. Defining transcriptional targets of ASH2L and associated H3K4 methyltransferase activity will aid in defining these relationships.
ASH2L protein expression positively correlates with proteins that function in pre-mRNA splicing in the nucleus, such as WTAP [26] and hnRNP K.[38] WTAP was recently implicated as a regulator of differentiation and proliferation in AML.[39] The N-terminal region of ASH2L harbors a DNA binding domain [40,41] but RNA binding activity has not been reported. It is possible that a physical association between ASH2L and these RNA-associated proteins is mediated by post-translational modifications. PRMT1-mediated methylation of hnRNP K affects its distribution between the nucleus and cytoplasm and also inhibits its association with Src kinase.[42,43] We previously identified ASH2L as a substrate of PRMT1 [18], and even though methylation does not affect the nuclear localization of ASH2L, it will be interesting to determine whether methylation of this site mediates physical interactions with these nuclear mRNA binding proteins.
We also observed that ASH2L protein expression is highly correlated with proteins involved in ribosomal biogenesis, including NPM1, NCL, and EBP1. In lower eukaryotes, the activity of ASH2L-containing H3K4 methyltransferase complexes, specifically H3K4me3 activity, is required for transcriptional silencing of rDNA genes.[44–46] Our data indicates a novel correlation between human ASH2L and nucleolar rRNA processing proteins. Heterozygous mutations in NPM1 are observed in 30% of all AML patients, and therapeutic targeting of the mutant protein has not been straightforward.[47,48] It has been proposed that targeting the wild-type NPM1 molecules remaining in AML cells might be a better approach because they likely confer a survival advantage.[33] Therefore defining functional relationships between wild-type NPM1 and candidates such as ASH2L might provide alternative therapeutic targeting strategies.
In conclusion, our results indicate that a lower level of ASH2L protein is beneficial to AML patients. Furthermore, we discovered novel correlations between ASH2L protein expression and proteins involved in promoting cell proliferation and survival. Even though mutations in ASH2L have not been described in AML, our results suggest that ASH2L contributes to leukemogenesis by cooperating with other proteins that aberrantly upregulate cellular growth and proliferation pathways.
Supplementary Material
Acknowledgments
The authors would like to thank the following for funding support of this work: the Cancer Prevention Research Institute of Texas, grant RP100429 to S.Y.R.D., and the Leukemia and Lymphoma Society, research grant to S.M.K.
Footnotes
Contribution:
J.S.B. conceptualized the study, analyzed the data, and wrote the manuscript
Y.H.Q. performed experiments
N.Z., S.-Y. Y., and K.R.C. performed statistical analysis and analyzed the data
S.Y.R.D. analyzed the data and edited the manuscript
S.M.K. performed RPPA, analyzed the data, and wrote and edited the manuscript
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Butler JS, Dent SY. The role of chromatin modifiers in normal and malignant hematopoiesis. Blood. 2013;121:3076–3084. doi: 10.1182/blood-2012-10-451237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 3.Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci U S A. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila Trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen CS, Sorensen PH, Domer PH, et al. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81:2386–2393. [PubMed] [Google Scholar]
- 6.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural and Molecular Biology. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 7.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 8.Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem. 2009;284:24242–24256. doi: 10.1074/jbc.M109.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Augustin J, Hu J, Jiang H. Physical Interactions and Functional Coordination between the Core Subunits of Set1/Mll Complexes and the Reprogramming Factors. PLoS One. 2015;10:e0145336. doi: 10.1371/journal.pone.0145336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan CC, Sindhu KV, Li S, et al. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc Natl Acad Sci U S A. 2008;105:7472–7477. doi: 10.1073/pnas.0711896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet Cell Genet. 1999;84:167–172. doi: 10.1159/000015248. [DOI] [PubMed] [Google Scholar]
- 12.Stoller JZ, Huang L, Tan CC, et al. Ash2l interacts with Tbx1 and is required during early embryogenesis. Exp Biol Med (Maywood) 2010;235:569–576. doi: 10.1258/ebm.2010.009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Zhou Y, Yin B, et al. ASH2L: alternative splicing and downregulation during induced megakaryocytic differentiation of multipotential leukemia cell lines. J Mol Med (Berl) 2001;79:399–405. doi: 10.1007/s001090100222. [DOI] [PubMed] [Google Scholar]
- 14.Demers C, Chaturvedi CP, Ranish JA, et al. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Chaturvedi CP, Tremblay V, et al. A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation. Genes Dev. 2015;29:123–128. doi: 10.1101/gad.254870.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luscher-Firzlaff J, Gawlista I, Vervoorts J, et al. The human trithorax protein hASH2 functions as an oncoprotein. Cancer Res. 2008;68:749–758. doi: 10.1158/0008-5472.CAN-07-3158. [DOI] [PubMed] [Google Scholar]
- 17.Ullius A, Luscher-Firzlaff J, Costa IG, et al. The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification. Nucleic Acids Res. 2014;42:6901–6920. doi: 10.1093/nar/gku312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler JS, Zurita-Lopez CI, Clarke SG, Bedford MT, Dent SY. Protein-arginine methyltransferase 1 (PRMT1) methylates Ash2L, a shared component of mammalian histone H3K4 methyltransferase complexes. J Biol Chem. 2011;286:12234–12244. doi: 10.1074/jbc.M110.202416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YI, Hua WK, Yao CL, et al. Protein-arginine methyltransferase 1 suppresses megakaryocytic differentiation via modulation of the p38 MAPK pathway in K562 cells. J Biol Chem. 2010;285:20595–20606. doi: 10.1074/jbc.M109.092411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28:6262–6277. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:1865–1874. doi: 10.1158/1078-0432.CCR-09-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornblau SM, Qiu YH, Zhang N, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornblau SM, Coombes KR. Use of reverse phase protein microarrays to study protein expression in leukemia: technical and methodological lessons learned. Methods Mol Biol. 2011;785:141–155. doi: 10.1007/978-1-61779-286-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 26.Ortega A, Niksic M, Bachi A, et al. Biochemical function of female-lethal (2) D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 27.Venables JP, Koh CS, Froehlich U, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3:650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- 29.Smith CC, Wang Q, Chin CS, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstrom MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 2001;506:272–276. doi: 10.1016/s0014-5793(01)02939-8. [DOI] [PubMed] [Google Scholar]
- 32.Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol Cell Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falini B, Gionfriddo I, Cecchetti F, Ballanti S, Pettirossi V, Martelli MP. Acute myeloid leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy? Blood Rev. 2011;25:247–254. doi: 10.1016/j.blre.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Greer EL, Maures TJ, Hauswirth AG, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greer EL, Maures TJ, Ucar D, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali A, Veeranki SN, Tyagi S. A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia. Nucleic Acids Res. 2014;42:7611–7624. doi: 10.1093/nar/gku458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guertin DA, Guntur KV, Bell GW, Thoreen CC, Sabatini DM. Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol. 2006;16:958–970. doi: 10.1016/j.cub.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 38.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 39.Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–1174. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Wan B, Wang KC, et al. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011;12:797–803. doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarvan S, Avdic V, Tremblay V, et al. Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nat Struct Mol Biol. 2011;18:857–859. doi: 10.1038/nsmb.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang YI, Hsu SC, Chau GY, et al. Identification of the methylation preference region in heterogeneous nuclear ribonucleoprotein K by protein arginine methyltransferase 1 and its implication in regulating nuclear/cytoplasmic distribution. Biochem Biophys Res Commun. 2011;404:865–869. doi: 10.1016/j.bbrc.2010.12.076. [DOI] [PubMed] [Google Scholar]
- 43.Ostareck-Lederer A, Ostareck DH, Rucknagel KP, et al. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J Biol Chem. 2006;281:11115–11125. doi: 10.1074/jbc.M513053200. [DOI] [PubMed] [Google Scholar]
- 44.Briggs SD, Bryk M, Strahl BD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- 46.Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28761–28765. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falini B, Martelli MP, Bolli N, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117:1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 48.Falini B, Bolli N, Liso A, et al. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23:1731–1743. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.