Abstract
Introduction:
Reducing misdiagnosis has long been a goal of medical informatics. Current thinking has focused on achieving this goal by integrating diagnostic decision support into electronic health records.
Methods:
A diagnostic decision support system already in clinical use was integrated into electronic health record systems at two large health systems, after clinician input on desired capabilities. The decision support provided three outputs: editable text for use in a clinical note, a summary including the suggested differential diagnosis with a graphical representation of probability, and a list of pertinent positive and pertinent negative findings (with onsets).
Results:
Structured interviews showed widespread agreement that the tool was useful and that the integration improved workflow. There was disagreement among various specialties over the risks versus benefits of documenting intermediate diagnostic thinking. Benefits were most valued by specialists involved in diagnostic testing, who were able to use the additional clinical context for richer interpretation of test results. Risks were most cited by physicians making clinical diagnoses, who expressed concern that a process that generated diagnostic possibilities exposed them to legal liability.
Discussion and Conclusion:
Reconciling the preferences of the various groups could include saving only the finding list as a patient-wide resource, saving intermediate diagnostic thinking only temporarily, or adoption of professional guidelines to clarify the role of decision support in diagnosis.
Introduction
Reducing misdiagnosis and costly “diagnostic odysseys” by using diagnostic decision support software (DDSS) has been a key goal from the outset of medical informatics [1,2]. However it has long been appreciated that standalone systems that are not integrated into the clinical workflow are a barrier to adoption, particularly for tools that require significant data entry [3]. Since physicians are increasingly using Electronic Health Records (EHRs), bringing DDSS into the clinical workflow has focused on integration into EHRs [4].
In integrating DDSS into EHRs, there are important questions, including: (1) how to avoid double entry of information, while still capturing the data with sufficient richness and consistency to improve diagnostic accuracy; (2) how much “intermediate thinking” (before a diagnosis is reached) should be saved in a legally discoverable record; and (3) how much information can be stored in a way that is accessible to multiple decision support tools.
To approach these questions we integrated into 2 EHRs a DDSS already shown to improve accuracy in clinical diagnosis and in genome analysis [5,6]. We describe the process for design of the workflow, and the evaluations by users as to advantages and disadvantages of EHR integration.
Some of this data has been presented in preliminary form [7].
Methods
Design of the integration
The workflow integration was planned with input from semi-structured interviews with 4 physicians who deal with genetic disorders; thematic saturation appeared to be achieved with this number The questions addressed in these interviews included how to do the following: incorporate the DDSS into the clinic workflow; present the diagnostic advice; organize and display information to be saved in the EHR; choose what to include from each DDSS session in the reports and EHR; and deal with diagnostic codes. In addition to report mockups, participants were given the opportunity to review the DDSS as it functions on a standalone basis. The results of the interviews and observations led to an understanding of the clinician workflow that informed the subsequent implementation.
The resulting workflow (Figure 1) was implemented in two EHRs: Epic (a vendor supplied system at Geisinger Health System), and HELP2 (a locally designed EHR solution at Intermountain Healthcare). At both sites, at the request of the institutions, the DDSS was installed locally such that the software did not communicate outside the institution except if a user clicked a link to an external reference such as an article about a disease. All patient information was saved to a server in the institution’s data center Other decisions about the interfaces with the EHRs were dictated by institutional preferences for button location, displays of existing records and location of their storage, and need for encryption messaging.
Figure 1.
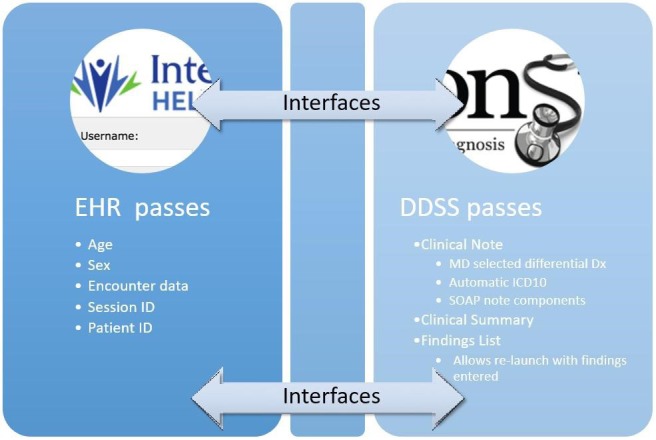
Integration Diagram
The integrations required significant local customization to implement the desired functionality as emerging standards for EHR resource interoperability e.g., SMART-on-FHIR, [8] were as yet unable to support the complexity of the workflow.
Training and interviews
Once the DDSS-EHR integrations were implemented, semi-structured interviews were conducted with 10 physicians to assess their reaction, including 4 from the original design group and 6 additional participants. A single interviewer (AKR) met with all participants, who were asked to review the implementation and practice using the tool in a training environment. The participants were a convenience sample of physicians at Geisinger and Intermountain. Medical specialties represented were radiology (3), pediatric neurology (1), neurodevelopmental pediatrics (2) and genetics (4), a mix chosen to assess the flow of clinical information throughout the diagnostic workup.
A semi-structured interview guide was created to walk individual physicians through the workflow processes for a typical patient being seen for an initial diagnostic workup of a rare genetic disease. This workflow included how the EHR is used during diagnostic encounters.
Physicians were then given a demonstration of the new integrated DDSS using a mock case in a training environment within the EHR and shown its use in the diagnostic process and the EHR-integrated features. Feedback was gathered on how those features fit with charting workflow and if the physicians felt that these features would save them time during their diagnostic workflow. All training was done by two authors (MMS and AKR). Physicians were later asked a series of questions about their current process of diagnosis (by AKR), including the time spent on various tasks before adopting the DDSS, their thoughts about the DDSS and their likely use of it. Interviews were audio recorded, transcribed, reviewed, summarized, and coded for themes related to utility and perceptions. Results were continually reviewed for consensus with team members. The coded responses were analyzed in the framework of qualitative description, in which the participant’s perceptions about an object or event are reported [9].
Part way through the study, at the point of testing the full integration, Intermountain began implementing a switch from the HELP2 EHR to that from Cerner, and as a result the post-implementation interviews and use are primarily from Geisinger.
Functionality in the DDSS
The DDSS and its outputs have been described previously [5,6,10,11,12]. Briefly a user enters patient findings (signs, symptoms and test results), which can include onset age of individual findings and pertinent negative findings. The patient information is compared to the detailed DDSS database that has quantitative and temporal information on >8,000 findings in >6,000 diseases, with comprehensive coverage of Mendelian genetics as well as deep coverage in pediatric neurology and rheumatology. After each input, a differential diagnosis with graphical display of disease probabilities is presented to the user In keeping with the iterative nature of diagnosis, [13] findings likely to be useful in distinguishing among diseases in the differential diagnosis are offered in rank order of their likelihood of changing the differential diagnosis in a cost effective way that takes into account treatability of each disease, with separate displays for clinical and lab findings [14]. Findings in the patient profile are tagged with “pertinence”, computed based on how much the differential diagnosis would be different if that information were not available [10]. In studies using vignettes of clinical cases, the DDSS reduces diagnostic error and workup costs significantly and requires minutes to use [5].
The genome-phenome version of the DDSS can import a genomic variant table and identify pertinent genes, likely to account for the pathogenesis of observed findings [6,10]. The genomic analysis includes an export of information about the genomic diagnosis, including detailed prognosis tables showing how each disease unfolds over time, generated using information from the DDSS database [11,12].
As requested by the institutions, the DDSS loads from an internal server, and runs on the client computer as a Java applet (and later as a browser-independent Java Web Start application) with database bundled into the applet’s JAR file. Patient and physician IDs are received from the EHR, as well as sex, age and contact session numbers, and passed back to the EHR as identifiers (Figure 1). No patient data is made available to external sources under any circumstances, consistent with the Health Insurance Portability and Accountability Act (HIPAA). One institution with multiple networks requested encryption for communication between the EHR and DDSS.
The physician could choose desired diseases from the differential diagnosis, and the DDSS then provided outputs only for those diseases, together with ICD-10 coding. This was made possible by adding Intelligent Medical Objects (IMO) codes to the database, which necessitated creation of thousands of IMO codes to support clinical genetics.
Functionality added to EHRs
The EHRs were configured to receive 3 outputs, as described in the results section:
Note: text for a chart note or selected sub-sections
Summary: findings, differential diagnosis and useful tests
Finding List with additional parameters: patient findings represented by codes plus analysis parameter values such as whether disease incidence was used. The diagnostic session can be recreated by importing this information into the software, appearing identical to the original session if the DDSS diagnostic database hasn’t changed.
The ability to launch the software from within a patient record was added to the EHRs, with tables of summaries and finding lists from previous sessions available for each patient. Patient identifiers were passed securely and received securely to tag outputs. From these, one could relaunch the DDSS with the patient’s findings and settings already entered.
Results
Specialists spend considerable time in diagnosis
In specialties where diagnosis is difficult, such as genetics and neurology, physicians reported an average of 3.4 hours per new patient evaluation (Table 1; range of 2.3 – 6.1), not including the time spent by non-physician clinicians such as genetic counselors and nurses. Of that 3.4 hours, 10 percent is spent in preparation – reading the notes from the referring physician. The process of taking a history and physical, including a family history, takes approximately half of the time. Synthesis and test selection usually take 6 percent. Documentation, whether via dictation or typed directly into the chart, typically takes about one-third of the time, averaging 59 minutes.
Table 1.
Time Spent by 4 Specialists in Initial Diagnosis
| NEURO DEVELOPMENTAL PEDIATRICIAN | PEDIATRIC NEUROLOGIST | PEDIATRIC GENETICIST | NEURO DEVELOPMENTAL PEDIATRICIAN | AVERAGE | PORTION OF TOTAL (%) | |
|---|---|---|---|---|---|---|
| Years in practice | 19 | 25 | 22 | 22 | 22 | |
| Chart review or reading referring notes | 17.5 | 17.5 | 45 | 20 | 10 | |
| History & physical & family history | 25 | 105 | 77.5 | 180 | 97 | 48 |
| Developing differential diagnosis | 17.5 | 15 | 8 | 4 | ||
| Research and test selection | 5 | 15 | 5 | 2 | ||
| Discussion with family ordering tests, getting family consents | 20 | 30 | 7.5 | 14 | 7 | |
| Encounter Note in EHR | 70 | 30 | 30 | 105 | 59 | 29 |
| TOTAL minutes | 155 | 135 | 155 | 367.5 | 203 | 100 |
| Hours | 2.6 | 2.3 | 2.6 | 6.1 | 3.4 | |
Physician opinions on diagnostic decision support
Physicians valued the diagnostic decision support, and in particular they liked suggestions on useful findings and a differential diagnosis ranked by likelihood, with the relative probabilities displayed. They felt these features could be helpful in their practice to improve diagnostic accuracy (Table 2).
Table 2.
Reactions to the Diagnostic DDSS
| Pediatric neurologist | “I see the tool as very useful, and it covers my specialty very well.” |
| Pediatric geneticist | “I like the way it gives key features for making sure not to miss a finding and suggestions about what other diseases to think about when considering a specific disease.” |
| Pediatric geneticist | “I love the probability graph for the differential diagnosis. I wish the probabilities were pulled into the EHR, not just the diseases in rank order. You don't get this with London Dysmorphology or OMIM [Online Mendelian Inheritance in Man]” |
| Pediatric geneticist | “I like seeing how the differential shifts real time as the findings are entered.” |
| Neurodevelopmental pediatrician | “The system is very useful to guide and standardize the etiology, and especially for users needing help with the etiology, getting suggestions on other diseases and useful tests to consider is helpful.” |
Design of the EHR integration based on physician interviews
During the design phase of the DDSS-EHR integration, physicians told us that they value not only the advice from using the DDSS, but also outputs from the DDSS to the EHR. They wanted to ensure they had complied with the requirements necessary to achieve the highest appropriate level of billing and they liked that they had choices of how to accomplish it. Many were comfortable with having the pertinent positive and negative findings in a list format, rather than the more traditional prose style used in most patient notes, as this allowed the text be more easily editable. In the initial part of the study in which the integration functionality was designed, 3 key types of output from the DDSS to the EHR emerged, which we refer to as the (1) Note, (2) Summary and (3) Finding List.
(1) Note:
There was universal feedback that one output from the DDSS to the EHR should take the form of an editable block of text that provides the basis for a standard chart note. The chart note is a legal record representing the assessment of the physician. Providing such output minimizes the need for double entry of information.
Accordingly, a note was constructed as plain text and transmitted to the EHR, where it could be edited by the physician while preparing a chart note. The SOAP note (Subjective Objective Assessment Plan) format was used. The note was offered as a whole or as separable sections (e.g., physical exam) that could be imported individually to accommodate physicians who use pre-existing note templates into which content could be pasted. The note was designed to minimize unnecessary prose, and made use of lists, where appropriate. Because the note was editable, it could be changed by the user (Figure 2).
Figure 2.

Patient Note
Assignment of data to relevant sections of the note was made possible by tagging all findings in the DDSS with the type of information (e.g., history exam, lab). The Assessment section includes a recapitulation of the subset of information about the patient that was most pertinent, something made possible because the DDSS assigns pertinence to each finding, [6] in contrast to systems with low awareness of diagnosis. For the differential diagnosis, many diseases are listed, ranked by probability The physician is offered a screen on which to select diagnoses to save, and only those chosen by the physician are included in the note, together with the appropriate ICD-10 codes for those diagnoses based on the Intelligent Medical Objects (IMO) codes added to the database as part of this study The Plan section suggested not only specific tests, but specific findings to check using tests, (e.g., not just head MRI, but checking for hydrocephalus and agenesis of the corpus callosum). Since neither EHR had a computerized order entry module for genetics, there was no effort made to connect the recommended tests to an ordering system.
(2) Summary:
Physicians also requested snapshots of the information in the DDSS, i.e., patient findings (pertinent positives with onsets and pertinent negatives), a differential diagnosis with a display of the relative likelihood, and recommendation of useful test findings (Figure 3). They envisioned this as an “informatics lab report” documenting the DDSS session. We refer to this HTML format output with findings, differential diagnosis and useful tests as the summary.
Figure 3.

Patient Summary
Physicians asked for the ability to see information as it developed over time and over different specialties involved in the diagnosis process, and to use previous summaries as a starting point for their own analysis. Some physicians told us that they wanted to begin with their own previous snapshot and add new information, and radiologists told us that they wanted to begin with the snapshot from a referring physician and add results from radiological testing. Accordingly, the ability was created for multiple summaries to be saved in the EHR, and displayed as a table with timestamp, physician name and physician department, and a link to display each summary and a link to relaunch the DDSS with the previous set of findings already entered using the patient’s Findings List (Figure 4).
Figure 4.

Schema for Table in EHR of Saved Versions
To accommodate the flow of information to and from specialists for interpreting laboratory tests, the display of test-related findings suggested by the usefulness calculation [14] could be filtered in the DDSS database by the relevant laboratory specialty. This enabled radiologists, geneticists and pathologists to use the filtered display of tests to focus on the findings in their area of specialty most relevant to the differential diagnosis. For example, a radiologist was presented useful findings determinable from an MRI scan for the patient, facilitating comment on the most pertinent positive and negative findings for that patient.
In contrast to the note, physicians asked for the summary to have all patient findings shown together ranked by pertinence, [6,10] instead of by type of information (history exam, lab), and they asked for diseases to be ranked by probability. Diseases were hyperlinked to web-based articles cited in the DDSS database and tagged with the IMO codes in the DDSS database. In addition, Human Phenotype Ontology (HPO) codes were added to the database for the findings, allowing such tagging as well.
(3) Findings List:
The fundamental input of computable information for this DDSS is the patient findings, including not just presence, but also onsets for pertinent positive findings, and including pertinent negative findings, together with the patient age and sex. The patient findings are tagged with internal codes, and HPO codes were added to the database to allow greater interoperability with other resources, and new HPO codes are being assigned to remaining terms. Other settings when using the DDSS were also included with the finding list (e.g., whether disease incidence was considered), resulting in a description of the inputs so complete so that this could be imported to regenerate the DDSS session. The information technology experts at both institutions had a strong preference for storing the findings list separately from the note and summary using tabular displays of sessions by various physicians for each patient (Figure 4).
Building on previous sessions
Most physicians liked the convenience of a link to launch the DDSS within the EHR and have findings previously saved populate the DDSS session, thereby achieving the ability to share the information across the team, while avoiding the need to re-enter the findings (Table 3).
Table 3.
Ability to Return to Previously Entered Patients
| Neurodevelopmental pediatrician | “Being able to save and relaunch with the previously saved findings is very helpful, especially to be able to continue to add and refine findings as new information is gathered.” |
| Pediatric neurologist | “I like the ability to reopen the saved findings and to continue to add new information findings as more information is known, such as after lab results are returned.” |
| Radiologist | “I like being able to sort the tests by specialty and see the top 3-5 radiological findings that would be most useful to narrow the differential diagnosis. I can focus on those when reading the scans, and it will allow easy commenting on a referring physician’s thinking for diagnosis to help them rule in or out a particular diagnosis.” |
Radiologists valued the ability to get more clinical information than they usually get from referring physicians. Radiologists also valued the ability of the DDSS to identify useful findings, and filter them to see only radiology findings. When radiologists receive a request for imaging together with more detailed information about the differential diagnosis, they thought this made it easier to comment on the most pertinent negatives to help them rule in or out a particular diagnosis.
Opinions varied among different specialties
The most positive reception to the DDSS integration was among geneticists, where diagnosis is particularly difficult, testing is particularly comprehensive, and choice and interpretation of tests involves considerable information. Geneticists derived additional value from the DDSS because they could use its genome-phenome analysis features to analyze results from whole exome or whole genome testing in the clinical context. The DDSS analysis is particularly successful when the patient is characterized robustly using a set of pertinent positives and pertinent negatives. One of the institutions extended the DDSS integration by adding interoperability for “return of results” to patients, passing variant and diagnosis information specified in the DDSS in XML format to the EHR, which passed it to a module for reporting the genetic testing results. The DDSS output was also notable for using the detailed quantitative information about frequency and age of onset of findings in a disease to generate a prognosis table showing how a diagnosis changes over time, (Figure 5), much valued by referring physicians and families [11,12].
Figure 5.
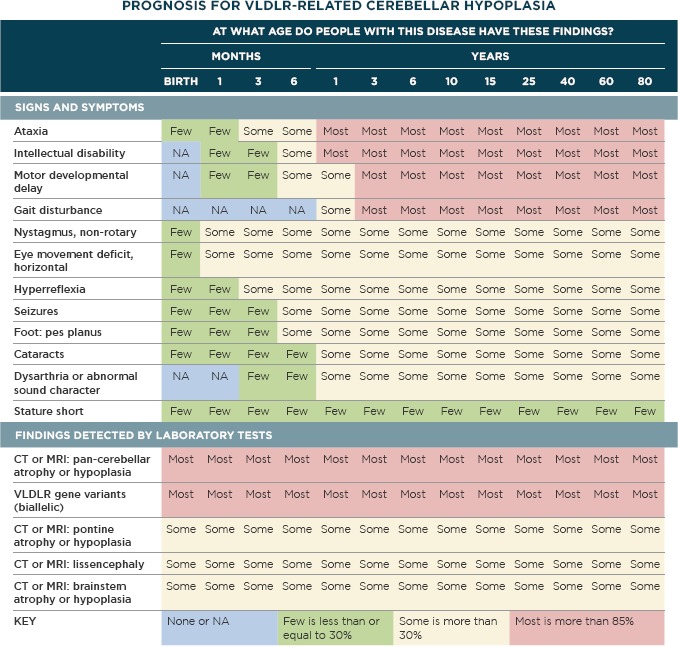
Prognosis Table
Generated using patented SimulConsult® software, database © 1998–2016 and prognosis tables © 2015–2016. All rights reserved. Generated on 13 December 2016 11:24 using software of 6 December 2016 17:34 and database of 8 December 2016 13:12.
Geneticists focusing on cardiology initially found the granularity for specifying cardiac ultrasound findings to be insufficient. Expansion of that granularity and curation of those findings in the DDSS database resulted in satisfaction by those geneticists without concerns from other physicians that the granularity of findings was excessive. There were also concerns that prenatal ultrasound findings needed to be more complete and granular, which were addressed with curation of additional information, resulting in more choices of prenatal ultrasound findings to enter.
Some physicians with highly specialized practices initially found that entry of findings was slow because of the thousands of findings in the database, while they use a much smaller set of findings for their subspecialty To address this issue, checklist screens were created in the DDSS, tailored for clinical areas such as metabolism. Up to 46 findings fit on such screens, representing the core findings typically seen in a referral. The findings were assembled using lists provided by specialists and using findings that occurred frequently in vignettes of clinical cases [5]. These findings are actively assessed in curation of diseases for the database, adding to the value in using these lists. These lists allow a user to quickly enter several findings before jumping to the differential diagnosis, and then being offered a much wider range of findings in the displays of useful clinical and lab findings.
Liability issues
Physicians could quit the DDSS without saving, having benefited from the advice, as they would after reading a textbook or article. Alternatively they could save output to the EHR, with the goal of reducing the duplication of entry when the DDSS is launched again to add further clinical or laboratory findings. Some specialists expressed concern that the use of the DDSS and storing of DDSS output in the EHR by those outside their specialty resulted in a need for formally documented discussion and exclusion of the diagnoses present in summaries that were generated by referring physicians. Furthermore, there was even a concern among some physicians about storing of their own intermediate thinking prior to making a decision on diagnosis. This concern was expressed both in terms of increased malpractice risk and increased testing costs. The reluctance was not only if the DDSS output was stored in the official EHR; the concern was that the DDSS being an option in the EHR made the material legally discoverable, even if stored on a server outside the EHR system.
One physician stated that even if the hospital system indemnified physicians from the liability associated with recording intermediate thinking, the concern would remain, because the loss of time to testimony would overwhelm any efficiencies (Table 4).
Table 4.
Reactions to Saving Diagnostic Thinking in the EHR
| Neurodevelopmental pediatrician | “I wouldn’t want to save the differential diagnosis to the chart until I have all the findings, I don’t want to save my intermediate thinking to a chart.” |
| Pediatric geneticist | “I am really concerned about the malpractice implications. Having to defend even one lawsuit stemming from use of the DDSS would negate any time saved.” |
| Pediatric geneticist | “The whole exome or genome tests coverage trumps concerns about legal liability because the provider cannot be faulted for failing to consider and order the appropriate test.” |
Such concerns varied by institution, perhaps reflecting different experiences with issues such as malpractice, as well as different institutional and individual views about the plusses and minuses of systematization in medicine. Such concerns also varied strongly by role, with those conducting testing such as MRI scans or whole exome analysis being least concerned about the issue of “if you think of it, you have to test for it” because of the comprehensiveness of the diagnostic testing being done (Table 4).
Genetic laboratories and radiologists complain that they get very little information from the referring physician, and in particular, little or no information about the differential diagnosis behind the referral, typically just descriptions such as “short stature” or “disproportionate” [15]. Physicians even suggested that the reason for such sparse communication is concern about saving intermediate thinking; sometimes there is even “informal feedback” to referring physicians from the specialists instructing them to avoid recording too much in the record that would later have to be explained or corrected.
Neither the Geisinger Epic EHR nor the Intermountain HELP2 system have a formal mechanism for temporarily saving intermediate thinking. In contrast, some other EHRs have implemented what is often termed a “scratch pad” that persists until the chart note is finalized in the EHR and then is wiped out.
As a result, most users chose not to save to the EHR, thereby reducing the convenience of being able to avoid duplicate entry and the ability to have communication of patient findings in a computable form from one physician to another. That meant that use of the summary and finding list was realized primarily for cases “where they know they are stuck”, or where previous testing had not produced a diagnosis and the physician was planning on comprehensive testing such as exome analysis.
Discussion
The paucity of good diagnostic documentation and communication is often blamed on the physician being busy It is well recognized that poor communication is a problem and a contributor to diagnostic errors, which affect 5 percent of the population per year [16]. The data from this study provide insights into other reasons for sparse documentation. We focus the discussion on such issues related to EHR integration, not discussing here general issues of stand-alone diagnostic software discussed in earlier reports about the software in clinical and genomic diagnosis [5,6].
Streamlining diagnosis and its documentation are important in some specialties
The time spent diagnosing patients in difficult specialties is long (3.4 hours) – well beyond the compensation typically received. Of that, fully one third is spent doing documentation – suggesting streamlining the process of diagnosis and its documentation are important.
Benefits of information sharing
This study and related work focusing on genomic analysis [6,15] show that there are major benefits from treating a detailed finding list as a collaborative tool to be shared among the various physicians involved in care of a particular patient. The benefits are also well exemplified by a radiologist receiving a computable patient summary. The radiologist gets a detailed understanding of the thinking of the referring physician and receives guidance from the DDSS on which radiological findings are most useful in the clinical diagnosis. This allows the radiologist to be maximally helpful by commenting explicitly not only on abnormalities noticed and on a standard set of findings routinely commented upon, but also on “pertinent negative” findings that are important for the differential diagnosis of the individual patient.
More generally this study highlights the benefits of creating a finding list with patient-wide status in the EHR similar to other resources such as a problem list, medication list, and allergy list. As implemented here, the finding list is represented using finding codes, and is thus a computable resource that can be shared by multiple tools operating in an EHR. EHRs having a standard format for the finding list would represent a major simplification of the task of scanning an EHR for findings, without the compromises inherent in Natural Language Processing, [17] and without the need for physicians to be involved in looking up codes, other than selecting terms in the DDSS. Such capabilities could be used as done here, with DDSS sessions that build one upon another in an EHR, or in other ways, such as feeding into ordering systems, reporting to databases such as ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), or sharing information among many decision support tools. Ideally, the finding list would support both positive findings and negative findings. There are of course many issues of granularity, terminology systems and formats to be addressed to make such interoperability commonplace.
Risks of information sharing
Such information sharing also has perceived risks, best exemplified by the concerns among some physicians that failure to test for any diagnosis mentioned in the DDSS analysis exposed the physician to legal risk. Some physicians are concerned that mentioning a diagnosis anywhere in an EHR or associated materials such as a summary could expose them to malpractice risk for not performing tests associated with that condition, and they doubt the ability of their institution to protect them from such risk, and in particular, the time lost to preparing and testifying in a case.
However, there is legal risk with either approach. There is a countervailing risk to not using a DDSS. Plaintiffs might point out that entering the findings from a narrative chart note into a widely available DDSS demonstrates that the correct diagnosis should have been considered, and not using decision support was a failure to meet the prevailing standard of care. At present there is no case law or malpractice experience that allows comparison of these two types of risk. However a recent systematic review of malpractice in primary care physicians identifies delayed or missed diagnosis as the most common reason for a suit being filed [18].
Conclusion
It is important to resolve the tradeoff between risks and benefits of information sharing. Many physicians perceive an incentive for sharing as little information as possible, yet the benefit to care of each patient comes from having a culture of robust information sharing among physicians, labs and insurers [16,19].
One established approach to managing such tradeoffs is by implementing professional guidelines that weigh the benefits and risks, and set forth a policy that is legally defensible as a result of a professional consensus. A key issue to be addressed is storage of intermediate thinking before a diagnosis is reached. In the era of paper charts, many physicians maintained “shadow charts” in addition to the official chart, in part to represent this intermediate thinking [20]. There are many important questions to answer: Should there be a separate section of an EHR that is understood to hold intermediate thinking, and be used for collaboration but not “discoverable” for use in court for “if you think of it, you have to test for it” scrutiny? Should intermediate thinking be retained for specific time periods (such as until the note is finalized) or the duration of a hospital admission? Should the EHR contain only the finding list and not the summary (the “informatics lab report” of the DDSS session)? Should storage of information for decision support be in the EHR or at an external site, maintained by the DDSS provider? Is the existence of a computable finding list different legally from similar information in a narrative note? Resolving these issues is essential to prevent domination of the incentives to the physician to share as little information as possible in the EHR.
The surfacing of these issues is an opportunity for convening the various parties involved, including patients, and developing a consensus that can optimize the balance of information sharing to improve care versus secrecy to avoid litigation. While such issues are being resolved, it may be that decision support will be documented in charts mostly as text-format findings, with or without finding codes, and natural language processing or finding code processing could offer this information to many different decision support systems. The exception may be for comprehensive testing such as MRI or genome analysis, where the risks of not testing have been minimized because of the extensiveness of the testing and because the need to document the intermediate thinking in order to get the testing reimbursed.
Specific next steps should include the following: (1) Making use of the shift to EHR plug-in approaches such as SMART-on-FHIR [8] to use the lessons learned in this study and define the relationships between EHRs and DDSS, (2) Making use of the plug-in nature of SMART-on-FHIR [8] to adapt the functionality in a continuous way once in use, both in functions, and in technology such as the porting being done of this DDSS from client-side Java to a non-Java architecture that runs on all computers and smartphones without needing special permissioning or installation of additional software, (3) Testing in a less specialized physician group, for example by using the recently added rheumatology support in the DDSS to help general pediatricians deal with the critical shortage of pediatric rheumatologists, [21] and (4) Testing in a randomized controlled way in which the impact on diagnostic accuracy and clinician time can be assessed.
Acknowledgements
This research was funded through the National Library of Medicine (NLM) under Grant Number: 2R44LM011585. Project title: Interoperable decision support to improve diagnostic workflow across multiple EHRs.
List of abbreviations
- DDSS –
Diagnostic Decision Support Software
- EHR –
Electronic Health Record
- FHIR –
Fast Healthcare Interoperability Resources
- HELP2 –
Health Evaluation through Logical Programming version 2, an electronic health record
- HIPAA –
Health Insurance Portability and Accountability Act
- HPO –
Human Phenotype Ontology
- ICD10 –
International Classification of Diseases (10th edition)
- IMO –
Intelligent Medical Objects
- JAR –
Java Application Resource
- MRI –
Magnetic Resonance Imaging
- OMIM –
Online Mendelian Inheritance in Man
- PHI –
Protected Health Information
- SMART –
Substitutable Medical Applications and Reusable Technology
- SOAP –
Subjective Objective Assessment Plan
- XML –
Extensible Markup Language
References
- 1.Schwartz WB. Medicine and the Computer: The Promise and Problems of Change. NEJM 1970; 283:1257-64. [DOI] [PubMed] [Google Scholar]
- 2.Kassirer JP. A report card on computer-assisted diagnosis--the grade: C. N Engl J Med. 1994; 330:1824-5. [DOI] [PubMed] [Google Scholar]
- 3.Clayton PD, Hripcsak G. Decision support in healthcare. Int J Biomed Comput. 1995; 39:59-66. [DOI] [PubMed] [Google Scholar]
- 4.Mandl KD, Kohane IS. Escaping the EHR Trap — The Future of Health IT. N Engl J Med 2012; 366:2240-2242. [DOI] [PubMed] [Google Scholar]
- 5.Segal MM, Williams MS, Gropman AL et al. Evidence-Based Decision Support for Neurological Diagnosis Reduces Errors and Unnecessary Workup. J Child Neurol. 2014; 29:487-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal MM, Abdellateef M, El-Hattab AW et al. Clinical pertinence metric enables hypothesis-independent genome-phenome analysis for neurological diagnosis. J Child Neurol. 2015; 30:881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulse NC, Wood GM, Lam S, Segal MM. Integrating a Diagnostic Decision Support Tool into an Electronic Health Record and Relevant Clinical Workflows through Standards-Based Exchange. Proceedings of the American Medical Informatics Association 2014 Annual Fall Symposium; 1431. [Google Scholar]
- 8.Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. J Am Med Inform Assoc. 2016; February 17 pii: ocv189. doi: 10.1093/jamia/ocv189. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandelowski M. Combining qualitative and quantitative sampling, data collection, and analysis techniques in mixed-method studies. Res Nurs Health. 2000; 23:246-55. [DOI] [PubMed] [Google Scholar]
- 10.Segal MM. Genome-Phenome analyzer and methods of using same. US Patent 9,524,373, issued 2016. [Google Scholar]
- 11.Stuckey H, Williams JL, Fan AL et al. Enhancing genomic laboratory reports from the patients’ view: A qualitative analysis. Am J Med Genet A. 2015. 167A:2238-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams JL, Rahm AK, Stuckey H et al. Enhancing genomic laboratory reports: A qualitative analysis of provider review. Am J Med Genet A. 2016; 170:1134-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen JL. Educational strategies to promote clinical diagnostic reasoning. N Engl J Med 2006; 355:2217-25. [DOI] [PubMed] [Google Scholar]
- 14.Segal MM. Systems and methods for diagnosing medical conditions. US Patent 6,754,655 issued June 22, 2004. [Google Scholar]
- 15.Segal MM Genome interpretation: Clinical correlation is recommended. Appl Translat Genomics. 2015; 6: 26-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balogh EP, Miller BT, Ball JR. Improving Diagnosis in Health Care National Academies Press; 2015. [PubMed] [Google Scholar]
- 17.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009; 42:760-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace E, Lowry J, Smith SM, Fahey T. The epidemiology of malpractice claims in primary care: a systematic review. BMJ Open. 2013; 3: e002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal MM, Schiffmann R. Decision support for diagnosis: Coevolution of tools and resources. Neurology 2012; 78:1. [DOI] [PubMed] [Google Scholar]
- 20.Balka E. Ghost charts and shadow records: implication for system design. Stud Health Technol Inform. 2010;160:686-90. [PubMed] [Google Scholar]
- 21.Segal MM, Athreya B, Son MB, Tirosh I, Hausmann JS, Ang EY, Zurakowski D, Feldman LK, Sundel RP. Evidence-based decision support for pediatric rheumatology reduces diagnostic errors. Pediatric rheumatology online journal. 2016; 14(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]


