Abstract
In plants, the most widely used cytological techniques to assess parental genome contributions are based on in situ hybridization (FISH and GISH), but they are time-consuming and need specific expertise and equipment. Recent advances in genomics and molecular biology have made PCR-based markers a straightforward, affordable technique for chromosome typing. Here, we describe the development of a molecular assay that uses single-copy conserved ortholog set II (COSII)-based single nucleotide polymorphisms (SNPs) and the high-resolution melting (HRM) technique to assess the chromosome dosage of interspecific hybrids between a Solanum phureja–S. tuberosum diploid (2n = 2x = 24) hybrid and its wild relative S. commersonii. Screening and analysis of 45 COSII marker sequences allowed S. commersonii-specific SNPs to be identified for all 12 chromosomes. Combining the HRM technique with the establishment of synthetic DNA hybrids, SNP markers were successfully used to predict the expected parental chromosome ratio of 5 interspecific triploid hybrids. These results demonstrate the ability of this strategy to distinguish diverged genomes from each other, and to estimate chromosome dosage. The method could potentially be applied to any species as a tool to assess paternal to maternal ratios in the framework of a breeding program or following transformation techniques.
Keywords: COSII, haplotype, SNPs, Solanum commersonii, Solanum tuberosum
Interspecific hybridization is an important component of many plant breeding programs. It can be achieved by either sexual crossing or by protoplast fusion. From a breeding standpoint, hybridization provides the basis for developing new genetic diversity and further selecting individuals by combining the best characteristics from their parents. Following hybridization, offspring with unbalanced parental chromosome contributions can be obtained, such as triploids and aneuploids. Despite their unusual chromosome number, they may display traits with significant impact on breeding. For instance, triploid hybrids may allow the development of new seedless commercial varieties as reported in banana (Raboin et al. 2005), watermelon (Barham et al. 2003), citrus (Ye et al. 2009), grape (Sun et al. 2011a), and papaya (Sun et al. 2011b). In addition, triploids can be produced to overcome incompatibility barriers between incongruent species (Carputo and Barone 2005; Schatlowski and Kӧhler 2012). Knowing the chromosome number is an important prerequisite for breeding decisions. Currently, the most commonly used techniques to assess variations in chromosome number are based upon root tip counts, flow cytometry, or fluorescence in situ hybridization (FISH) (Henry et al. 2010). The last-named technique makes use of chromosome-specific fluorophore-labeled probes to detect DNA sequences (Pita et al. 2014). A special type of FISH, genomic in situ hybridization (GISH), is based on the use of total genome as a labeled probe and helps to discriminate parental genomes in natural and artificial hybrids (Chester et al. 2010). Although in situ hybridization techniques are reliable, they are time-consuming, chromosome-size/shape-dependent and need specific instruments.
Recent developments in genomics and molecular marker techniques suggest that the use of PCR-based markers can be a straightforward, affordable strategy for chromosome typing. Fulton et al. (2002) developed universal primers in solanaceous species with single-copy conserved ortholog set II (COSII) and Wu et al. (2006) successfully applied them to study synteny among tomato, aubergine, pepper, and tobacco (Wu et al. 2009a, 2009b, 2010). Even in crops that have not yet been sequenced, it may be feasible to develop universal markers based on orthologous genes coming from the most closely related sequenced plant species (Liu et al. 2013). Single nucleotide polymorphisms (SNPs) are the most abundant DNA markers in plant genomes and therefore represent ideal instruments to detect a high level of polymorphisms. A recent paper by Tonosaki et al. (2014) showed their ability to identify alien chromosomes in 2 monosomic addition lines of Brassica.
Although SNPs can be screened through a variety of technologies, high-resolution melting (HRM) analysis has the advantages of increased simplicity and rapid turnaround time (Wu et al. 2009c). Briefly, after PCR amplification, the HRM instrument heats the samples to denature DNA and monitors the pattern of fluorescence reduction due to the release of an intercalating dye (Reed and Wittwer 2004). Data outputs are the dissociation or melting curves that plot the reduction in fluorescence against the increase in temperature. SNPs in amplicon sequences can result in different melting curves. HRM analysis to detect SNPs is very efficient in terms of sensitivity, cost, time, and labor (Ujino-Ihara et al. 2010). In potato, De Koeyer et al. (2010) used HRM for genotyping diploid and tetraploid materials. Zhou et al. (2014) proposed HRM analysis in humans as an alternative to karyotyping for rapid diagnosis of aneuploidy syndromes. Plants provide an excellent opportunity for a genome-wide investigation of aneuploid syndromes: sample size is not limited, phenotypes can be described and assessed in detail, and plant aneuploid populations provide a complex mixture of viable karyotypes (Henry et al. 2010).
Among cultivated plants, the tetraploid (2n = 4x = 48) potato Solanum tuberosum is receiving much attention in terms of applying new technologies for genetic and breeding studies. Its genome sequence has been released (Potato Genome Sequence Consortium, 2011), facilitating potato functional and comparative genomics (Spooner and Salas 2006). The potato has more cultivated and wild relatives than any other crop, some of which have been used as source of genes and allelic diversity both in conventional breeding and genetic engineering programs (Carputo et al. 2005). Among wild potatoes, diploid S. commersonii has received much attention due to its noteworthy resistance traits. It has also been the first potato relative whose genome was sequenced (Aversano et al. 2015). A breeding scheme based on the production of triploid bridges was developed to introgress its genes into the cultivated gene pool (Carputo et al. 1997). The triploid state of bridge genotypes produced and their genomic constitution make this material suitable for setting up a reliable and efficient method to monitor chromosome dosages. This seems very interesting in a crop like the potato, a species where small chromosomes make it difficult to determine chromosome dosages.
In this study, we propose a PCR-based method for chromosome dosage estimation in potato hybrids through the 1) identification of SNPs among single-copy COSII markers, and 2) the application of HRM to differentiate each chromosome rapidly and quantitatively. We give an example of the effectiveness of our method by assessing the genomic ratio of the entire chromosome set of triploid (2n = 3x = 36) hybrids using 3 different species, diploid S. commersonii and S. phureja (2n = 2x = 24) and S. tuberosum (2n = 4x = 48). The distinctive features of our method are critically discussed.
Materials and Methods
Plant Materials
The material used consisted of 5 potato triploid (2n = 3x = 36) hybrids with S. commersonii and S. tuberosum in their pedigree. These triploids (MCA1, MCB1, MCB3, MCB10, and MCC1) were obtained from 4x × 2x crosses between a tetraploid (2n = 4x = 48) clone of S. commersonii and S. phureja-S. tuberosum diploid (2n = 2x = 24, clone UP88P5) hybrids (Carputo et al. 1995). This study also included S. commersonii (PI 243503, clone cmm1T), S. tuberosum cv. Blondy, a S. phureja-S. tuberosum hybrid (clone UP88P5), S. phureja (clone IVP35), useful for the identification of S. commersonii-specific SNPs and the set-up of 3 reference samples. The reference samples were obtained by mixing DNA from S. commersonii (clone cmm1T) and S. tuberosum cv. Blondy in 2:1, 1:2 and 1:1 ratios.
COSII Marker Selection
Eighty-seven single-copy COSII markers with a fragment length ranging between 500 and 2100bp were selected from the SOL genomics network database (Fernandez-Pozo et al. 2015). Total DNA was extracted from 100mg of leaf material using the Qiagen Plant DNeasy Kit according to the manufacturer’s instructions (Qiagen) and quantified using spectrophotometric (Nanodrop 1000, Thermo Fisher Scientific) and fluorometric (Qubit 2.0, Life Technologies) techniques. PCR reactions were conducted in a volume of 25 µL containing 30ng of genomic DNA, 0.2 µM of each primer, dNTPs 0.8mM, MgCl2 3mM, recombinant Taq polymerase 0.06U (Invitrogen Life Technologies) and 1X PCR buffer. Thermal cycling conditions were as follows: 4min at 94 °C; 35 cycles of 45s at 94 °C, 1min at annealing temperature (Supplementary Table 1) and 2min at 72 °C, with a final extension step of 10min at 72 °C. Monomorphic (1 amplicon) markers were selected using agarose gel electrophoresis and then, for each chromosome, 1 monomorphic COSII marker was selected and amplified in S. commersonii (clone cmm1T), S. tuberosum cv. Blondy, a S. phureja-S. tuberosum hybrid (clone UP88P5) and S. phureja (clone IVP35). The amplicons were purified using the High Pure PCR Product purification Kit (Roche Applied Science) according to the manufacturer’s instructions and sequenced (Eurofins MWG Operon).
Sequence Alignment and SNP Discovery
Each COSII marker sequence obtained was analyzed and aligned using SeqScape Software v2.5 (Applied Biosystems). The alignment was used to discover S. commersonii-specific SNPs. Primers flanking S. commersonii-specific SNPs were designed using Primer3 web version 4.0.0 (Untergrasser et al. 2012) for HRM analysis. The parameters for primer design included an amplicon containing a number of SNPs no higher than 4 and ranging from 80 to 150bp.
HRM Analysis
Real time PCR reaction and HRM technology were conducted with Rotor-Gene™ 6000® (Corbett, Life Science), in a volume of 25 µL containing 40ng of genomic DNA, 0.4 µM of each primer, dNTPs 0.8 µM, Taq polymerase 0.026U (TakaRa Shuzo), 1X PCR buffer and 1X EvaGreen Dye (Biotium). Thermal cycling conditions were as follows: 30s at 95 °C; 40 cycles of 10s at 95 °C, 30s at primer melting temperature, and 30s at 72 °C. Samples were melted from 65 to 90 °C with a melting rate of 0.1 °C s−1. All the HRM data collected were analyzed by the dedicated HRM software (Rotor-Gene 6000 Series Software 1.7). Normalization of HRM curves was followed by calculation of genotype confidence percentages (GCPs) (value attributed to each genotype with respect to another one, with a value of 100 indicating an exact match) were calculated. The reference genotype was specified and the fluorescence readings for the melt curve were recovered (ref_fluorescence). For each sample, the fluorescence readings for the melt curve were also retrieved (sample_fluorescence). For each reading/point X in the respective melt curves, an error value was calculated as the square of the difference between the readings: Error(X) = [(sample_fluorescence(X) − ref_fluoresence(X)]2. The error values were then summed over the entire set of readings. The percentage of confidence (confidence percentage) was subsequently calculated as: confidence percentage = C_spread_factor (−C_sharpness_factor × sum_error) where C_spread_factor = 1.05, C_sharpness_factor = 0.02. The percentage of confidence above 90% is recommended by the technical support of the manufacturer. The GCPs calculation was based on a single reading of the reference sample. HRM analyses were carried out in triplicates.
Data Archiving
In fulfilment of data archiving guidelines (Baker 2013), we have deposited the primary data underlying these analyses as follows:
COSII markers alignments: Dryad
Results and Discussion
Discover of S. commersonii-specific SNPs
Previous studies in mammals, birds, and insects have shown that anchoring primers in conserved orthologs can provide useful markers for comparative mapping (Lyons et al. 1997; Smith et al. 2000; Chambers et al. 2003). In plants, the availability of whole genome sequences and large expressed sequence tag (EST) databases for a growing number of species provides the potential to align DNA sequences from multiple species and to identify a COS of single-copy genes (Wu et al. 2009a, 2009b, 2010; Kuhn et al. 2012; Nowak et al. 2012). In Solanaceae, COSII markers have been shown to be randomly distributed along the whole genome (Fulton et al. 2002) and mostly present as single copies (Lindqvist-Kreuze et al. 2013). Such features make them useful tools for comparative genomics studies.
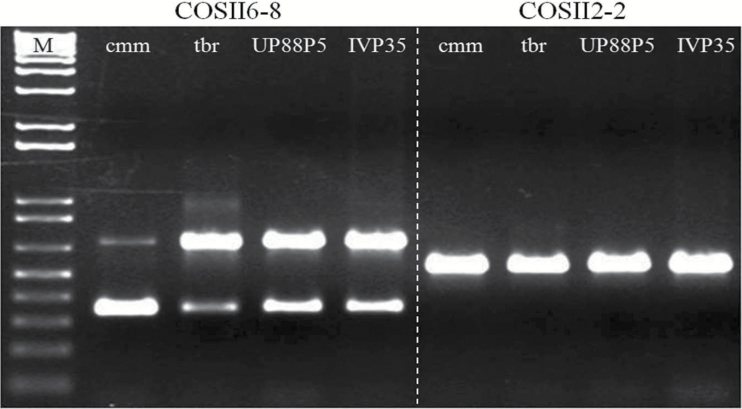
We identified 87 COSII markers distributed across the S. commersonii genome, with an average of 7 markers per chromosome (Supplementary Table 1). Among them, 45 with a size smaller than 750bp were amplified in S. commersonii clone cmm1T (cmm), S. tuberosum cv. Blondy (tbr), S. phureja-S. tuberosum hybrid (phu-tbr, UP88P5) and S. phureja (phu, clone IVP35) (Supplementary Table 1). Given the number of bands amplified, each COSII was classified as “1 amplicon,” “multiple amplicons,” or “null” (no amplicon). Out of 45 markers tested, 24 were “1 amplicon,” 14 “multiple amplicons,” and 7 “null.” An example of these markers is reported in Figure 1. The “1 amplicon” COSIIs were sequenced and aligned to identify polymorphisms (Supplementary Figure 1). Sequences obtained in cmm1T were also used to confirm their uniqueness in the genome of S. commersonii. Blastn alignments against cmm1T genome sequence (Aversano et al. 2015) revealed a single hit for each of these markers and allowed us to select 1 marker per chromosome. Out of 264 polymorphisms, 6 insertions, 6 deletions, and 252 SNPs were found, with an average of 21 SNPs per COSII marker (Supplementary Table 2). A fraction of them (66%) was identified as the S. commersonii-specific SNP set and was used as target to design primers for HRM analysis. All of them were tested but only primers reported in Table 1 were considered as the optimized set of SNPs specific to S. commersonii chromosomes.
Figure 1.
One and multiple amplicon COSII electrophoresis patterns. Electrophoresis profiles of COSII markers of chromosomes VI (multiple amplicon) and II (1 amplicon) on 4 genotypes. M = 1kb plus DNA ladder; cmm = Solanum commersonii (clone cmm1T); tbr = S. tuberosum cv. Blondy; UP88P5 = S. phureja-S. tuberosum hybrid (clone UP88P5); IVP35 = S. phureja (clone IVP35).
Table 1.
Solanum commersonii-specific markers
| Chromosome | ID | Primer sequences | Primer length, bp | Annealing temperature, °C | Sequence length, bp | SNPs, No. |
|---|---|---|---|---|---|---|
| 1 | COSII1-4c | 5′-TTGGGATTGAGGTGTGGTTC/ 5′-TCCCACCCATAATAAAGAACA |
20/21 | 61 | 144 | 4 |
| 2 | COSII2-2c | 5′-TCCCTGGTAAACAAAATGAGC/ 5′-AAGATCCTGAAATTCGATCCAT |
22/21 | 59 | 140 | 2 |
| 3 | COSII3-5b | 5′-CCAAAATGTTAACCAGATTCCT/ 5′-GCAGTTTGTTTTTCCTTTTAGTG |
22/23 | 57 | 139 | 1 |
| 4 | COSII4-6c | 5′-ATTCCTGAGACCAAATCAATTTC/ 5′-TTGGTCAAGACATCCTATTATTC |
23/23 | 58 | 115 | 1 |
| 5 | COSII5-3c | 5′-TGCCTATTGGATTCAAGCAA/ 5′-GCATGGTCTCCATCTTAGTGAA |
22/20 | 59 | 100 | 3 |
| 6 | COSII6-5d | 5′-ATCATCTTCAACAAGTGAAGTTG/ 5′-AAGCAATCAAGAAATTAGCAGTC |
23/23 | 57 | 90 | 4 |
| 7 | COSII-7-2a | 5′-CATCCCTTGTCGCTTTTCC/ 5′-AGCAACATGAAACACGTAAAC |
19/22 | 58 | 107 | 1 |
| 8 | COSII 8-6e | 5′-GACGTGTTTTCTCATCTTGACA/ 5′-GGCATAGGCAGAGTGACATAC |
22/21 | 58 | 130 | 1 |
| 9 | COSII 9-4d | 5′-ACATCTCCTCCTTTTCTCCG/ 5′-TTCATGCCCAGATGCAGCAG |
20/20 | 59 | 132 | 3 |
| 10 | COSII10-2a | 5′-AAATCAGGAATTTGCATGAC/ 5′-GTGTTCACCTCGTTGACAAA |
20/20 | 57 | 139 | 1 |
| 11 | COSII11-5a | 5′-CTCTAAACGTTTGGCCATT/ 5′-CCTAAATTCAGTTGTTAAAG |
19/20 | 54 | 158 | 4 |
| 12 | COSII12-8a | 5′-GAACATGTATATATGATTG/ 5′-GTAACTGCATAACGAATT |
19/18 | 53 | 121 | 3 |
Chromosome, COSII marker ID, primer sequence and length, annealing temperature, sequence length and number of SNPs are reported.
Assessment of Chromosome Dosage Using HRM
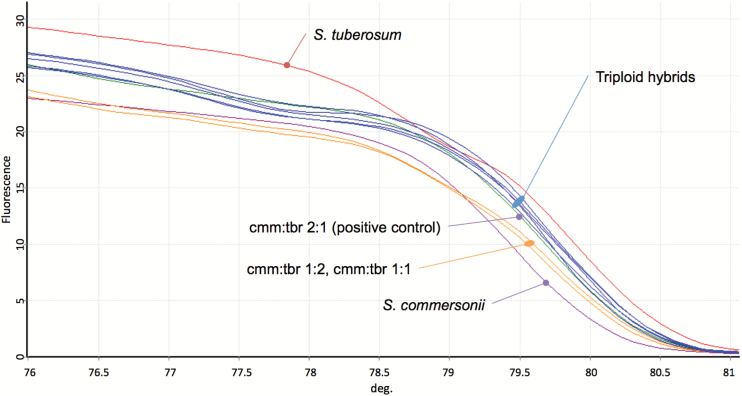
Solanum commersonii specific SNPs for all 12 chromosomes were analyzed using the HRM technique. HRM assays were carried out on 10 genotypes, including the 2 parents, 5 triploid hybrids, and 3 reference samples. The reference samples are an equimolar DNA mixture simulating triploid and diploid chromosome set ratios, the establishment of these samples proving a pivotal step for the experiment. The reference synthetic mixtures were produced using 2 parental genome DNAs, namely S. commersonii (clone cmm1T) and S. tuberosum cv. Blondy. Three S. commersonii:S. tuberosum (cmm:tbr) chromosome set ratios were simulated: 2:1, 1:2 and 1:1. Based on their pedigree (Carputo et al. 1995), for all 5 triploid hybrids, a 2:1 cmm:tbr ratio was expected. The similarity between the HRM profile of the reference and the HRM profile of the triploid samples was based on a percentage of similarity score, called the GCP. GCP values reflect the similarity of a reference sample to the other samples included in the analysis. When the GCP value was higher than 90, a sample was considered to have the same chromosome dosage as the reference. An example of these results is reported in Figure 2 and Supplementary Figure 2, where normalized HRM curves and GCPs for COSII9-4d are shown. The results suggest that all triploids analyzed with the COSII9-4d marker have a chromosome dosage similar to the synthetic triploid cmm:tbr 2:1, considered in this study as the positive control. A GCP value >90 was detected for most the comparisons between the triploid HRM profile and the cmm:tbr 2:1 reference samples, indicating that for all 12-chromosome triplets the triploid hybrids harbor 2 S. commersonii homologues and 1 S. tuberosum homeologous (Table 2). In the comparisons with the other reference samples (cmm:tbr 1:1 and 1:2) GCP values were always < 90 (not shown). The results were 100% consistent with the expected parental chromosome dosage of the triploid hybrids, strengthening our confidence in our strategy to distinguish diverged genomes from each other, as well as to differentiate each of the nonhomologous chromosomes within a genome.
Figure 2.
HRM curve of COSII9-4d HRM analysis. Normalized melting plots obtained with HRM analysis set up on chromosome IX. Samples are Solanum commersonii (cmm1T), S. tuberosum cv. Blondy (tbr), 5 triploid hybrids, the reference DNA mix (cmm:tbr 2:1, 1:1 and 1:2) and the negative control (water). All triploids show the same shape of the synthetic DNA mixture with a 2:1 cmm:tbr ratio.
Table 2.
GCPs values of 5 triploids in all chromosomes
| Hybrid | Chromosome | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | |
| MCA1 | 91.3 | 94.0 | 99.2 | 91.4 | <90 | 94.6 | <90 | 92.5 | <90 | 91.4 | 96.6 | 96.3 |
| MCB1 | 96.6 | 95.5 | 93.2 | <90 | 97.5 | 97.6 | 92.1 | 90.4 | 92.0 | 93.0 | 91.0 | 93.5 |
| MCB3 | 95.9 | 91.8 | 95.1 | 94.0 | 99.5 | 90.6 | 95.9 | 92.6 | 94.5 | 93.2 | 96.9 | 97.7 |
| MCB10 | 90.2 | 91.3 | 97.7 | 95.5 | 95.7 | 90.1 | 95.6 | 96.8 | 95.3 | 96.2 | 98.3 | 97.0 |
| MCC1 | 98.0 | 96.4 | <90 | 90.5 | 96.3 | 90.2 | 96.3 | 95.6 | 92.2 | 94.0 | 93.9 | 95.2 |
GCPs values obtained from HRM analysis of 5 triploids in all chromosomes using the synthetic triploid cmm:tbr 2:1 as reference.
The present method can be applied to study various genetic materials and has several attractive features. In sexual hybrids it may allow estimation of parental chromosome-specific dosage with a view to studying chromosome inheritance and its possible phenotypic effects, especially in crosses involving closely related species. Similarly, in interspecific somatic hybrids this method may facilitate the assessment of parental chromosome contribution. Our approach cannot be applied to estimate chromosome dosage after the F1 generation since this could lead to false estimates due to potential meiotic recombination between homologous or homeologous chromosomes. However, it does enable allele dosage in advanced generations to be followed. Indeed, compared to qPCR, it confers the benefit of discriminating different alleles using the polymorphisms (SNPs) identified between the parents. For example, Milner et al. (2014) proposed a method able to establish a correlation between the dosage of a homologous transgene in T1 lines and its phenotypic effects.
The method presented here is easy to apply, since amplification and HRM analysis are performed as a single-step, closed tube process. Alternative analyses, such as pyrosequencing and SSR, have some disadvantages, which limit their application. In particular, pyrosequencing requires specialized equipment and reagents, whereas SSR-HRM, as far as we know, are not yet available in potato and therefore its set-up may result more laborious than the approach here described. In the COSII-HRM analysis, the most important step is the preliminary determination of genomic DNA concentration. Nowadays, several techniques are available to obtain very accurate estimations of double-stranded DNA (dsDNA) concentration, as UV and fluorescence spectrometry, able to detect also “low molecular weight” dsDNA (∼150bp PCR products). The only manipulations required are DNA extraction and PCR reaction set-up. The method is highly sensitive, robust and has the potential to be widely applied to nonmodel species for marker development. Indeed, having included the necessary references DNA mixes simulating the chromosome dosages of interest, individuals showing matching melting patterns can be considered to have the same dosage. This method is promising especially for species with large numbers of small chromosomes in which alternative methods of chromosome dosage determination are more cumbersome.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/
Supplementary Material
Acknowledgments
We are grateful to R. Garramone for helping in sampling, the public-private genomic platform GenoPOM for the sequencing analyses and Dr. Marina Iovene for critically reviewing the manuscript. We thank Mark Walters for editing the manuscript.
References
- Aversano R, Contaldi F, Ercolano MR, Grosso V, Iorizzo I, Tatino F, Xumerle L, Dal Molin A, Avanzato C, Ferrarini A, et al. 2015. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell. 27:954–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Barham R, Barham W, McCuiston FT, Juarez B, Tolla B. 2003. Method of producing seedless watermelon. US Patent US2003/0163852 A1. [Google Scholar]
- Carputo D, Aversano R, Frusciante L. 2005. Breeding potato for quality traits. Acta Hort. 684:55–64. [Google Scholar]
- Carputo D, Barone A. 2005. Ploidy level manipulations in potato through sexual hybridisation. Ann Appl Biol. 146:71–79. [Google Scholar]
- Carputo D, Barone A, Cardi T, Sebastiano A, Frusciante L, Peloquin SJ. 1997. Endosperm balance number manipulation for direct in vivo germplasm introgression to potato from a sexually isolated relative (Solanum commersonii Dun.). Proc Natl Acad Sci U S A. 94:12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carputo D, Cardi T, Frusciante L, Peloquin SJ. 1995. Male fertility and cytology of triploid hybrids between tetraploid Solanum commersonii (2n=4x=48, 2EBN) and Phureja-Tuberosum haploid hybrids (2n=2x=24, 2EBN). Euphytica. 83:123–129. [Google Scholar]
- Chambers EW, Lovin DD, Severson DW. 2003. Utility of comparative anchor-tagged sequences as physical anchors for comparative genome analysis among the Culicidae. Am J Trop Med Hyg. 69:98–104. [PubMed] [Google Scholar]
- Chapman MA, Chang J, Weisman D, Kesseli RV, Burke JM. 2007. Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theor Appl Genet. 115:747–755. [DOI] [PubMed] [Google Scholar]
- Chester M, Leitch AR, Soltis PS, Soltis DE. 2010. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridisation). Genes (Basel). 1:166–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koeyer D, Douglass K, Murphy A, Whitney S, Nolan L, Song Y, De Jong W. 2010. Application of high-resolution DNA melting for genotyping and variant scanning of diploid and autotetraploid potato. Mol Breed. 25:67–90. [Google Scholar]
- Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD. 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell. 14:1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L. 2010. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics. 186:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DN, Don Livingstone, Main D, Zheng P, Saski C, Feltus FA, Mockaitis K, Farmer AD, May GD, Schnell RJ, et al. 2012. Identification and mapping of conserved ortholog set (COS) II sequences of cacao and their conversion to SNP markers for marker-assisted selection in Theobroma cacao and comparative genomics studies. Tree Genet Genome. 8:97–111. [Google Scholar]
- Lindqvist-Kreuze H, Cho K, Portal L, Rodríguez F, Simon R, Mueller LA, Spooner DM, Bonierbale M. 2013. Linking the potato genome to the conserved ortholog set (COS) markers. BMC Genet. 14:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Guo X, Wu J, Chen GB, Ying Y. 2013. Development of universal genetic markers based on single-copy orthologous (COSII) genes in Poaceae. Plant Cell Rep. 32:379–388. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O’Brien SJ. 1997. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet. 15:47–56. [DOI] [PubMed] [Google Scholar]
- Milner SG, Ferradini N, Nicolia A, Veronesi F, Salvi S, Rosellini D. 2014. Copy number estimation of a plant-derived selectable marker gene by high resolution melting analysis: a tool to simplify transgenic plant breeding. Crop Sci. 54:1133–1138. [Google Scholar]
- Nowak MD, Davis AP, Yoder AD. 2012. Sequence data from new plastid and nuclear COSII regions resolves early diverging lineages in Coffea (Rubiaceae). Syst Bot. 37:995–1005. [Google Scholar]
- Pita M, Orellana J, Martínez-Rodríguez P, Martínez-Rodríguez Á, Fernández-Calvín B, Bella J. 2014. FISH methods in cytogenetic studies. In: Stockert JC, Espada J, Blázquez-Castro A, editors. Functional analysis of DNA and chromatin. Vol. 1094 New York: Springer Protocols; p. 109–135. [Google Scholar]
- Potato Genome Sequence Consortium 2011. Genome sequence and analysis of the tuber crop potato. Nature. 475:189–195. [DOI] [PubMed] [Google Scholar]
- Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40:e115. [DOI] [PMC free article] [PubMed]
- Raboin L-M, Carreell F, Noyer J-L, Baurens J-P, Horry J-P, Bakry F, Du Montcel1 HT, Ganry J, Lanaud C, Pierre JL. 2005. Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol Breed. 16:333–341. [Google Scholar]
- Reed GH, Wittwer CT. 2004. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem. 50:1748–1754. [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Köhler C. 2012. Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J Exp Bot. 63:6059–6067. [DOI] [PubMed] [Google Scholar]
- Smith E, Shi L, Drummond P, Rodriguez L, Hamilton R, Powell E, Nahashon S, Ramlal S, Smith G, Foster J. 2000. Development and characterization of expressed sequence tags for the turkey (Meleagris gallopavo) genome and comparative sequence analysis with other birds. Anim Genet. 31:62–67. [DOI] [PubMed] [Google Scholar]
- Spooner DM, Salas A. 2006. Structure, biosystematic, and genetic resource. In: Gopal J, Khurana SMP, editors. Handbook of potato production, improvement, and post-harvest management. New York: Haworth Press Inc; p. 1–39. [Google Scholar]
- Sun D, Lu X, Liang G, Guo Q, Mo Y, Xie J. 2011b. Production of triploid plants of papaya by endosperm culture. Plant Cell Tiss. 104:23–29. [Google Scholar]
- Sun L, Zhang GJ, Yan AL, Xu HY. 2011a. The study of triploid progenies crossed between different ploidy grapes. Afr J Biotechnol. 10:5967–5971. [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, et al. 2015. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 43:D1036-D1041. [DOI] [PMC free article] [PubMed]
- Tonosaki K, Akaba M, Bang SW, Kitashiba H, Kaneko Y, Nishio T. 2014. The use of species-specific DNA markers for assessing alien chromosome transfer in Brassica rapa and Brassica oleracea-monosomic additions of Raphanus sativus. Mol Breed. 34:1301–1311. [Google Scholar]
- Ujino-Ihara T, Taguchi Y, Moriguchi Y, Tsumura Y. 2010. An efficient method for developing SNP markers based on EST data combined with high resolution melting (HRM) analysis. BMC Res Notes. 3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, Tanksley SD. 2009a. A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet. 118:1279–1293. [DOI] [PubMed] [Google Scholar]
- Wu F, Eannetta NT, Xu Y, Plieske J, Ganal M, Pozzi C, Bakaher N, Tanksley SD. 2010. COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor Appl Genet. 120:809–827. [DOI] [PubMed] [Google Scholar]
- Wu F, Eannetta NT, Xu Y, Tanksley SD. 2009b. A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor Appl Genet. 118:927–935. [DOI] [PubMed] [Google Scholar]
- Wu F, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD. 2006. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics. 174:1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SB, Tavassolian I, Rabiei G, Hunt P, Wirthensohn M, Gibson JP, Ford CM, Sedgley M. 2009c. Mapping SNP-anchored genes using high-resolution melting analysis in almond. Mol Genet Genomics. 282:273–281. [DOI] [PubMed] [Google Scholar]
- Ye W, Qin Y, Ye Z, da Silva JAT, Zhang L, Wu X, Lin S, Hu G. 2009. Seedless mechanism of a new mandarin cultivar ‘Wuzishatangju’(Citrus reticulata Blanco). Plant Sci. 177:19–27. [Google Scholar]
- Zhou Y, Xiao L, Wu Q, Zhang K, Guo Q. 2014. Rapid prenatal diagnosis of common numerical chromosomal abnormalities by high-resolution melting analysis of segmental duplications. Genet Test Mol Biomarkers. 18:141–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.