Abstract
Understanding the genetic architecture of phenotypic traits can provide important information about the mechanisms and genomic regions involved in local adaptation and speciation. Here, we used genotyping-by-sequencing and a combination of previously published and newly generated data to construct sex-specific linkage maps for sockeye salmon (Oncorhynchus nerka). We then used the denser female linkage map to conduct quantitative trait locus (QTL) analysis for 4 phenotypic traits in 3 families. The female linkage map consisted of 6322 loci distributed across 29 linkage groups and was 4082 cM long, and the male map contained 2179 loci found on 28 linkage groups and was 2291 cM long. We found 26 QTL: 6 for thermotolerance, 5 for length, 9 for weight, and 6 for condition factor. QTL were distributed nonrandomly across the genome and were often found in hotspots containing multiple QTL for a variety of phenotypic traits. These hotspots may represent adaptively important regions and are excellent candidates for future research. Comparing our results with studies in other salmonids revealed several regions with overlapping QTL for the same phenotypic trait, indicating these regions may be adaptively important across multiple species. Altogether, our study demonstrates the utility of genomic data for investigating the genetic basis of important phenotypic traits. Additionally, the linkage map created here will enable future research on the genetic basis of phenotypic traits in salmon.
Keywords: condition factor, genotyping by sequencing, linkage map, quantitative trait loci, RAD, sockeye salmon, size, thermotolerance
Understanding the genetic basis of phenotypic traits can provide important insights into how organisms adapt to their environment and if they will be able to adapt to changing environments in the future (Stinchcombe and Hoekstra 2008). A common method used to elucidate the genetic basis of phenotypic traits involves examining genotypes at a large number of markers to identify associations with phenotypes of interest (Lynch and Walsh 1998). This method, termed quantitative trait locus (QTL) analysis, has proven useful in many model organisms, especially those that are agriculturally important (reviewed in Dekkers and Hospital 2002; Wallace et al. 2014). However, QTL analysis has historically been difficult to conduct in nonmodel organisms due the absence of genomic resources and the large number of genetic markers required (Slate 2005).
The proliferation of genomic data provides a potential solution to this limitation (reviewed in Allendorf et al. 2010). Genotyping-by-sequencing (GBS) techniques now make it possible to screen thousands of markers in hundreds of individuals (Tonsor 2012). Additionally, genomic data facilitate the creation of high-density linkage maps that assign markers to specific genomic locations (Davey et al. 2011). These advances have enabled QTL studies in a variety of nonmodel organisms, including cichlid fish (Tropheops sp., Albertson et al. 2014), great tits (Parus major, Santure et al. 2013), and moths (Heliothis sp., Groot et al. 2013).
Salmonids represent ideal candidates for QTL studies due to their cultural and economic importance, but a lack of genomic resources has historically limited QTL studies to 2 species of salmonids commonly used in aquaculture: Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). Previous QTL studies in these species have provided some important insights into the genetic basis of phenotypic traits such as thermotolerance, size, and condition factor (O’Malley et al. 2003; Perry et al. 2005; Reid et al. 2005). However, many of these studies employed a relatively small number of loci (< 300), suggesting that many QTL were not discovered due to inadequate coverage across the genome (Johnston et al. 2014; Santure et al. 2013).
The recent availability of genomic data has facilitated the creation of high density linkage maps for salmonids that provide extensive coverage of the genome and can be used for QTL analysis (see Gutierrez et al. 2014). These modern linkage maps often include thousands of loci mapped in both sexes (Lien et al. 2011; Kodama et al. 2014) and contain both nonduplicated loci and duplicated loci resulting from an ancient whole genome duplication in salmon (Brieuc et al. 2014; Waples et al. 2015). Additionally, since many of these maps are constructed using restriction site associated DNA (RAD) data from the same restriction enzyme (SbfI), maps can be easily aligned to discover orthologous regions and marker overlap between species and studies (Brieuc et al. 2014; Kodama et al. 2014). Loci on these maps can also be aligned to various genomic resources to investigate the functional significance of certain genomic regions (Everett and Seeb 2014; McKinney et al. 2015; Waples et al. 2015). QTL studies using high-density linkage maps have revealed loci associated with growth and life-history type in rainbow trout (Hecht et al. 2012; Miller et al. 2012), thermotolerance and size in Chinook salmon (O. tshawytscha, Everett and Seeb 2014), and ecotype in lake whitefish (Coregonus clupeaformis, Gagnaire et al. 2013a).
Sockeye salmon (O. nerka) is one of the most intensely managed species across the Pacific Rim because of their iconic stature, supporting both native cultures and valuable commercial fisheries (Schindler et al. 2010; Dann et al. 2013); but, the genetic basis of phenotypic traits in this species has rarely been studied. Here, we investigated the genetic basis of 4 phenotypic traits, thermotolerance, length, weight, and condition factor, in anadromous sockeye salmon from southwestern Alaska. Thermotolerance is an important predictor of how sockeye salmon may respond to climate change (Eliason et al. 2011); size-related traits including length and weight are highly correlated with survival and reproductive success in sockeye salmon (Bradford 1995; Quinn 2005); and condition factor is associated with the ability of salmonids to survive stressful environmental conditions (Robinson et al. 2008).
Thermotolerance is an especially important trait given the recent increases in premature mortality that have been experienced by sockeye salmon near the southern extent of their range (reviewed in Hinch et al. 2012; Miller et al. 2014). These increases are likely driven by unusually warm summer temperatures and have resulted in over 95% mortality in some river systems such as the Fraser River in British Columbia. The recent trends in temperature likely foreshadow future environmental conditions emphasizing the importance of understanding thermotolerance in salmon.
Our objectives were to: 1) construct a dense linkage map for sockeye salmon using a combination of newly generated and previously published data (Everett et al. 2012; Limborg et al. 2015), 2) conduct QTL analysis for 4 phenotypic traits in 3 families from a wild population (families from Everett et al. 2012), 3) align our QTL with available genomic resources to find potential genes underlying phenotypic variation, and 4) compare our results to previous studies in closely related species. Our study represents a significant initial step towards understanding the genetic basis of important phenotypic traits in sockeye salmon. Additionally, the genomic resources created here will prove extremely valuable for future research in this and other species.
Materials and Methods
Families Used for Linkage Mapping and QTL Analysis
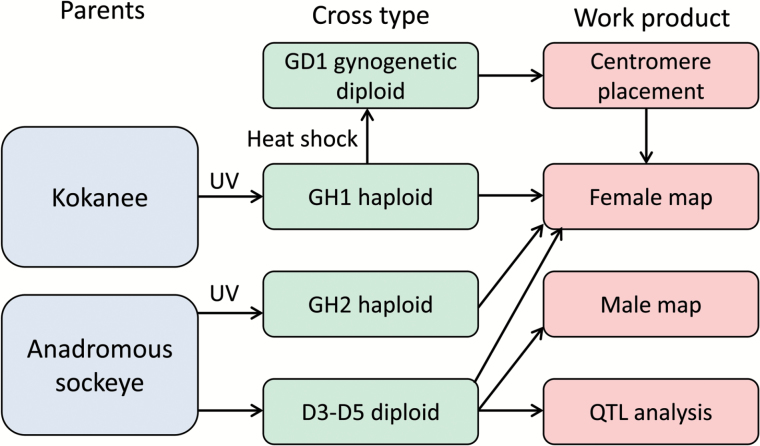
We used a combination of existing and newly generated data from 6 families to generate a dense linkage map and conduct QTL analysis (see Table 1 and Figure 1 for information on each family and an overview of the study design). The families consisted of 2 gynogenetic haploid families (GH1, GH2), 1 gynogenetic diploid family (GD1), and 3 diploid families (D3–D5) (Table 1, Figure 1). These families were created from populations sampled at the northern and southern ends of the species range of sockeye salmon in North America and represent both the anadromous and freshwater resident (kokanee) forms of the species (Table 1). The methods used to create families, preserve samples, and validate ploidy (when applicable) have been described for families GH1, and GD1 by Limborg et al. (2015) and families D3–D5 by Everett et al. (2012). All fish handling and rearing was done following University of Washington Institutional Animal Care and Use Committee protocol 4229-01.
Table 1.
Sampling information for the families used to place markers on the linkage map and conduct QTL analysis. The life history column denotes whether the family was constructed from resident sockeye salmon that remain in freshwater (kokanee) or from anadromous sockeye salmon. The construction of families D3–D5 was described in Everett et al. (2012); RAD sequencing of families D3 and D4 was conducted in the current study and sequencing of family D5 was conducted in Everett et al. (2012). Genotypes from gynogenetic diploids produced from family GH1 (family GD1) were used for centromere placement (see text and Limborg et al. 2015). See Figure 1 for a visualization of the experimental design for this study
| Family | Source | Ploidy | Life history | Sampling location | Number of Individuals | Sequencing methoda | Average no. reads/individual | |
|---|---|---|---|---|---|---|---|---|
| Mapping | QTL analysis | |||||||
| GH1 | (Limborg et al. 2015) | Haploid | Resident | Puget Sound, Washington, USA | 92 | 0 | SE100 | 2 500 000 |
| GD1 | (Limborg et al. 2015) | Diploid | Resident | Puget Sound, Washington, USA | NAb | NAb | NAb | NAb |
| GH2 | This study | Haploid | Anadromous | Bristol Bay, Alaska, USA | 86 | 0 | SE100 | 1 117 053 |
| D3 | (Everett et al. 2012) | Diploid | Anadromous | Bristol Bay, Alaska, USA | 79 | 79 | SE80 | 1 387 758 |
| D4 | (Everett et al. 2012) | Diploid | Anadromous | Bristol Bay, Alaska, USA | 88 | 87 | SE80 | 1 323 787 |
| D5 | (Everett et al. 2012) | Diploid | Anadromous | Bristol Bay, Alaska, USA | 138 | 96 | SE80, SE100, PE80 | 4 671 087 |
aSE100: single-end 100bp Illumina sequencing; SE80: single-end 80bp sequencing; PE80: paired-end 80bp sequencing.
bGenotypes from gynogenetic diploids were available from Limborg et al. (2015) and were used to place centromeres. See Limborg et al. (2015) for information on sample sizes and sequencing.
Figure 1.
Workflow for this study. The study included a haploid and gynogenetic diploid family of kokanee (freshwater resident sockeye salmon) sampled at the southern end of the species range and haploid and diploid families of anadromous sockeye salmon sampled at the northern end of the species range (GH2, D3–D5). Gynogenetic haploids (families GH1, GH2) were created by combining eggs and UV irradiated sperm and gynogenetic diploids (family GD1) were created by heat shocking eggs and UV irradiated sperm after fertilization. Diploids (families D3-D5) were created by mating wild individuals from a single population. The female linkage map included data from all 5 families and centromeres were placed on this map using knowledge of recombination events available from the gynogenetic diploids. The male map was constructed using only the diploid families because gynogenetic haploid families do not include information about recombination events in males. QTL analysis for 4 phenotypic traits was conducted for each of the diploid families (D3–D5). Haploid families were not used for QTL analysis because haploid embryos do not survive past hatch. Additional information on each family including sample size can be found in Table 1. Colors are viewable in the online version of the article.
Family GH2 was created by combining eggs with UV-irradiated sperm following the methods of Thorgaard et al. (1983). Embryos were preserved in 100% ethanol as close to hatch as possible, and DNA from the parents and offspring was isolated using QIAGEN DNAeasy 96 Tissue Kits (Qiagen, Valencia, California). To confirm ploidy, we genotyped the parents and offspring for 96 EST-derived 5′-nuclease assays (Elfstrom et al. 2006; Storer et al. 2012) following the methods of Smith et al. (2011) and Everett and Seeb (2014). Genotypes for these assays were also available for families GH1 and D3–D5 and were used for linkage mapping and QTL analysis.
Restriction Site-Associated DNA (RAD) Sequencing, SNP Discovery, and Genotyping
RAD sequencing was conducted using the enzyme SbfI following the methods of Baird et al. (2008) and Everett et al. (2012). Sequence data was then analyzed with the STACKS software package (version 1.20, Catchen et al. 2011; Catchen et al. 2013) and genotyping methods developed by Waples et al. (2015). Different parameters were used to genotype haploid and diploid individuals (see Supplementary File S1). As a final step before linkage mapping, genotypes were filtered to remove individuals and SNPs with > 20% missing data. Additional information on RAD sequencing and genotyping can be found in Supplementary File S1.
Linkage Mapping
Separate female and male linkage maps were constructed with the program LepMap (Rastas et al. 2013). LepMap is a fast and memory efficient program that utilizes data from multiple families simultaneously to construct consensus linkage maps. Parameters for LepMap analysis were identical to those of McKinney et al. (2015) with 1 exception: the LOD (log10 odds) score limit used to form linkage groups (LGs) in our study was 9.5 for the female map and 4 for the male map. We excluded data from diploid families for markers that were heterozygous for the same alleles in both parents because phase cannot be unambiguously determined in the offspring.
Gynogenetic diploids (half-tetrads) provide information about marker-centromere distances from recombination events during meiosis and facilitate placement of centromeres based on observed heterozygosity (also known as y, Thorgaard et al. 1983). We placed centromeres on the female and male linkage maps using genotype data available from the gynogenetic diploid family GD1 described and genotyped in Limborg et al. (2015). Centromeres on the female map were defined as the region of each LG containing all markers with heterozygosity < 0.1 (Limborg et al. 2015). Centromeres on the male map were defined as the region containing all markers that were found to be centromeric in the female map. A and b Arms for each LG on the female map were arbitrarily assigned based on centromere location and do not correspond to previous studies (Everett et al. 2012; Limborg et al. 2015).
We compared our linkage map to existing maps for sockeye salmon and Chinook salmon to orient our LGs, correlate marker orders between maps, and establish orthologous relationships. First, we compared our map to the map generated by Limborg et al. (2015) and named our LGs based on this map. No alignment step was necessary for this comparison because the locus names were identical across studies. We then compared our map to the map of Everett et al. (2012). Loci shared between studies were identified with BLASTN (parameters: minimum alignment length of 57bp, 95% identity, and no more than 2 mismatching bases). Finally, we aligned our map to a map for Chinook salmon (McKinney et al. 2015) to establish orthologous relationships between the 2 species (BLASTN parameters: minimum alignment length of 80bp, 90% identity, and no more than 4 mismatching bases). Information from this alignment was combined with data presented in Brieuc et al. (2014) and Kodama et al. (2014) to report orthologous relationships among sockeye salmon, Chinook salmon, coho salmon (O. kisutch), rainbow trout, and Atlantic salmon.
Thermal Challenge and Other Phenotypic Data
A thermal challenge was conducted on 96 offspring from families D3–D5 following methods similar to Everett and Seeb (2014). Prior to the thermal challenge, offspring from each family were raised for 30 days post hatch in separate aquaria kept at 11 °C. Water in each aquarium was then gradually replaced with water heated to 29 °C until the temperature reached 25 °C. The first 48 individuals from each family that lost equilibrium were removed and recorded as thermosusceptible. After 48 individuals lost equilibrium (~2h), the remaining 48 individuals were sampled and classified as thermotolerant. Total length and weight were recorded for each individual, and samples were preserved in RNALater (Life Technologies, Carlsbad, California). Condition factor (K), a standardized measure of fish health, was then calculated using the formula K = W/L3 × 105 where W = weight in grams and L = length in mm (Bagenal and Tesch 1978).
Summary statistics for length, weight, and condition factor were calculated separately for thermotolerant and thermosusceptible individuals from each family, as well as the family as a whole. We conducted Student’s t-tests to investigate the hypothesis that phenotypic distributions were significantly different between thermotolerant and thermosusceptible individuals and among families (alpha = 0.05). Finally, we plotted weight versus length for each family and visually examined the distribution of thermotolerant and thermosusceptible individuals in relation to the line of best fit derived for each family.
QTL Analysis
QTL analysis for thermotolerance, length, weight, and condition factor was conducted separately for each diploid family with the R package R/qtl (Broman et al. 2003) and methods similar to Hecht et al. (2012). First, we identified single QTL using the function scanone. We then iteratively ran the scanone function, adding previously identified QTL as cofactors, until no additional QTL were detected. Experiment and LG-wide significance thresholds (alpha = 0.05) were determined with permutation tests (1000 iterations) implemented in scanone. We considered QTL with LOD scores > 3 that were also above the experiment or LG-wide significance threshold as significant (Lander and Kruglyak 1995).
Potential interactions between QTL discovered with scanone were investigated with the addint function. We then refined the positions of significant QTL using the refineqtl function. Finally, we fit a multiple-QTL model including interaction terms for all QTL found for a given phenotypic trait with the fitqtl function. QTL that were not significant in the context of the full model (P > 0.05) were removed and refineqtl and fitqtl were rerun until all QTL included in the full model were significant. The percentage of variation explained (PVE) by each QTL was obtained from the results of fitqtl. Approximate 95% confidence intervals for the position of each QTL were calculated with the LOD drop-off method implemented in the lodint function (1.5 LOD drop, Visscher et al. 1996; Dupuis and Siegmund 1999).
We used orthologous relationships to compare the locations of QTL in sockeye salmon with QTL discovered in other salmonids to investigate whether the same genomic regions influence phenotypic traits across multiple species.
Paired-End Assembly, Alignment to Genomic Resources, and Functional Annotation
We conducted paired-end assemblies for each locus to increase query length for functional annotation and alignment to genomic resources. Paired-end sequences from the 6 parents of families D3–D5 were assembled with the alignment program CAP3 (150bp minimum alignment length, Huang and Madan 1999) following the methods of Etter et al. (2011) and Waples et al. (2015). Consensus sequences for each locus were aligned to Atlantic salmon genome scaffolds (ICSASB_v1; GenBank accession: GCA_000233375.3). Alignments were conducted with the longest sequence available from each locus using BLASTN (parameters: > 90% identity, ≤4 mismatches per 100bp, ≤1 gap per 100bp, and alignment length > 80% of query sequence). Scaffolds were placed on the linkage map if at least 3 loci on the same LG aligned to the scaffold and the order of the loci on the linkage map was concordant with their order on the scaffold. Consensus sequences for each locus were also aligned to all expressed sequence tags (ESTs) for sockeye salmon in the cGrasp database (http://web.uvic.ca/grasp/) using BLASTN (parameters: > 90% identity, ≤4 mismatches per 100bp, ≤1 gap per 100bp, and alignment length > 50% of query sequence). If multiple alignments met these parameters for a single locus, the alignment with the lowest e-value was retained.
Functional annotation was conducted by aligning paired-end consensus sequences for each locus to the Swiss-Prot database using BLASTX. The alignment with the lowest e-value < 10–4 for each locus was accepted as the annotation. Additional annotations were attempted for QTL peak loci that were placed on the Atlantic salmon genome by aligning 100 000bp of 3′ and 5′ flanking sequence for each locus to the Swiss-Prot database using BLASTX and the parameters described above. QTL peak loci that did not directly align to the genome but were found at map locations spanned by a scaffold were aligned to the scaffold with relaxed parameters. If the QTL could be placed in the correct scaffold, annotation was attempted using the methods described above.
In fulfillment of data archiving guidelines for the Journal of Heredity (Baker 2013), all data underlying this study are included as supplementary material or have been deposited in DRYAD or the NCBI short read archive.
Results
Sequencing, SNP Discovery, and Genotyping
RAD sequence data were obtained from 525 individuals across 5 families (Table 1). Sequencing depth varied substantially by family ranging from an average of 1.1 million sequences per individual for family GH2 to 4.7 million sequences per individual for family D5 (excluding low quality individuals). SNP discovery using the RAD data revealed 11 377 polymorphic loci that were genotyped in > 80% of individuals. We added 80 polymorphic 5′-nuclease assays that were genotyped in > 80% of individuals to this dataset. Finally, we removed 34 individuals that were genotyped at < 80% of loci resulting in a final dataset of 491 individuals genotyped at 11 457 loci.
Linkage Mapping
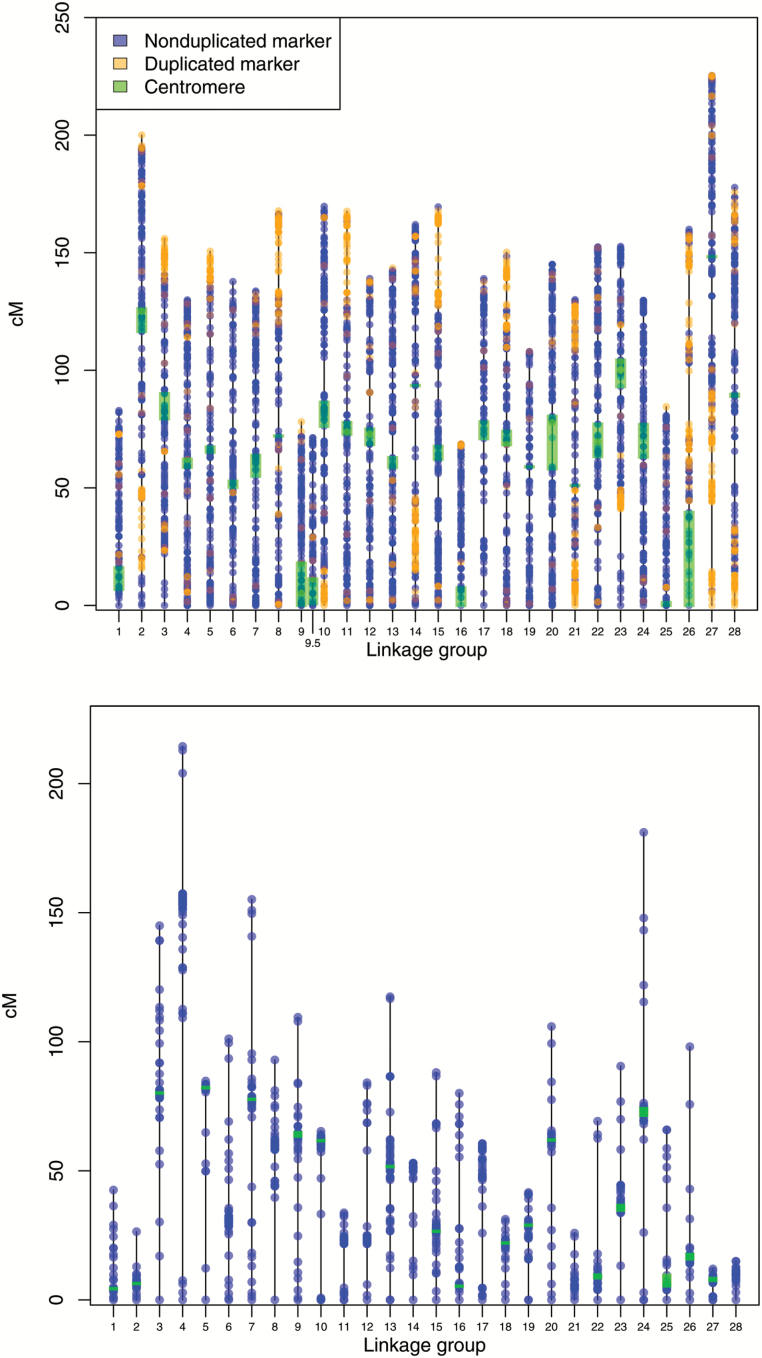
We constructed a female linkage map containing 6322 loci distributed across 29 LGs and a male linkage map containing 2179 loci distributed across 28 LGs (Supplementary Figure S1, Figure 2, Table 2, Supplementary Table S1). The total length of the female map was 4082 cM, and the total length of the male map was 2291 cM. These maps contained 7367 unique loci, with 1143 loci found on both maps. Placement of centromeres using gynogenetic diploids was successful for all LGs in the female map, and we were able to place centromeres on 19 of 28 LGs in the male map using information from markers found to be centromeric on the female map. The female map contained 6 acrocentric and 23 metacentric LGs, and LG type was well conserved between male and female maps (excluding LG So9). LG So9 has been previously identified as the sex chromosomes for sockeye salmon (Limborg et al. 2015) and is composed of 2 pairs of acrocentric chromosomes (X1 and X2) in females. In males, 1 copy each of X1 and X2 are fused into a single metacentric chromosome (Y) resulting in a single copy of both X1, X2, and Y in males (Thorgaard 1978; Faber-Hammond et al. 2012). We designated the 2 acrocentric sex LGs in the female map as So9 and So9.5.
Figure 2.
(a) Female and (b) male linkage maps for sockeye salmon containing 6322 and 2179 loci, respectively. Each dot represents a locus, and darker shading indicate higher marker density. Centromeres were successfully placed on all LGs in the female map and 19 of 28 LGs in the male map. LGs So9 and So9.5 are the sex chromosomes in sockeye salmon and are represented by 2 acrocentric LGs in the female map and a single metacentric LG in the male map (see text for additional information). Colors are viewable in the online version of the article.
Table 2.
Summary of male and female linkage maps for sockeye salmon. Arms for each LG were assigned based on centromere location. LG type denotes acrocentric (A) and metacentric (M) LGs. Orthology support is the number of loci shared between sockeye salmon and Chinook salmon for each orthologous relationship. See Brieuc et al. (2014) and Kodama et al. (2014) for additional information on orthologous relationships between Chinook salmon, coho salmon, rainbow trout, and Atlantic salmon. LG arm designations and cM positions for markers in this map do not correspond to Limborg et al. (2015), but markers are named the same
| Sockeye LG | Length (cM) | # markers | LG type | Sockeye LG arm | Chinook chromosome | Coho LG | Rainbow trout chromosome | Atlantic salmon chromosome | Orthology support | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Female | Male | ||||||||
| So1 | 82.88 | 42.57 | 170 | 44 | A | So1a | Ots26 | Co26 | Omy22 | Ssa21 | 12 |
| So2 | 200.06 | 26.44 | 290 | 71 | M | So2a | Ots07p | Co05a | Omy07p | Ssa17qb | 6 |
| M | So2b | Ots07q | Co05b | Omy07q | Ssa22 | 11 | |||||
| So3 | 155.99 | 144.96 | 288 | 64 | M | So3a | Ots03q | Co02b | Omy03q | Ssa25 | 9 |
| M | So3b | Ots03p | Co02a | Omy03p | Ssa02p | 7 | |||||
| So4 | 129.91 | 214.42 | 272 | 110 | M | So4a | Ots13p | Co17a | Omy18q | Ssa27 | 10 |
| M | So4b | Ots20 | Co23 | Omy05p | Ssa01qb | 8 | |||||
| So5 | 150.6 | 84.79 | 246 | 67 | M | So5a | Ots08q | Co15a | Omy25q(Omy29) | Ssa09qb | 8 |
| M | So5b | Ots14p | Co16b | Omy18p | Ssa16qb | 7 | |||||
| So6 | 137.72 | 101.15 | 192 | 79 | M | So6a | Ots29 | Co11b | Omy15p | Ssa29 | 5 |
| M | So6b | Ots21 | Co19b | Omy14q | Ssa05p | 5 | |||||
| So7 | 133.68 | 155.15 | 235 | 80 | M | So7a | Ots31 | Co14b | Omy14p | Ssa14qb | 5 |
| M | So7b | Ots16q | Co17b | Omy09q | Ssa15qb | 3 | |||||
| So8 | 167.79 | 92.96 | 186 | 60 | M | So8a | Ots15q | Co09b | Omy21q | Ssa07q | 3 |
| M | So8b | Ots15p | Co09a | Omy21p | Ssa07p | 4 | |||||
| So9a | 78.12 | 168 | A | So9a | Ots19 | Co22 | Omy02q | Ssa10qb | 13 | ||
| So9.5a | 71.26 | 116 | A | So9.5a | Ots10q | Co30 | Omy08q | Ssa14qa | 9 | ||
| So10 | 169.69 | 65.33 | 263 | 82 | M | So10a | Ots06q | Co04b | Omy01q | Ssa18qa | 4 |
| M | So10b | Ots30 | Co28 | Omy10p | Ssa04q | 7 | |||||
| So11 | 167.61 | 33.75 | 226 | 67 | M | So11a | Ots13q | Co15b | Omy27 | Ssa20qb | 3 |
| M | So11b | Ots34 | Co12b | Omy10q | Ssa08q | 5 | |||||
| So12 | 139.01 | 84.12 | 232 | 82 | M | So12a | Ots08p | Co14a | Omy25p | Ssa09qa | 12 |
| M | So12b | Ots10p | Co16a | Omy09p | Ssa18qb | 8 | |||||
| So13 | 143.45 | 117.44 | 237 | 123 | M | So13a | Ots18 | Co21 | Omy04q | Ssa06p | 6 |
| M | So13b | Ots12p | Co08a | Omy11p&q | Ssa20qa | 9 | |||||
| So14 | 161.94 | 52.96 | 271 | 91 | M | So14a | Ots23 | Co13b | Omy02p | Ssa05q | 1 |
| M | So14b | Ots14q | Co18a | Omy24 | Ssa09qc | 8 | |||||
| So15 | 169.46 | 88.09 | 293 | 97 | M | So15a | Ots02p | Co01a | Omy17p | Ssa02q | 6 |
| M | So15b | Ots02q | Co01b | Omy17q | Ssa12qb | 3 | |||||
| So16 | 68.61 | 80.06 | 126 | 53 | A | So16a | Ots25 | Co25 | Omy20p+q | SSa08p&Ssa28 | 7 |
| So17b | 138.95 | 60.53 | 173 | 61 | M | So17a | Ots01p | Co10a | Omy04p | Ssa23 | 2 |
| M | So17b | Ots01p | Co10a | Omy04p | Ssa23 | 15 | |||||
| So18c | 150.19 | 31.24 | 239 | 60 | M | So18a | Ots11q | Co07b | Omy19q | Ssa01p | 14 |
| M | So18b | Ots11p | Co07a | Omy19p | Ssa04p | 5 | |||||
| So19 | 107.94 | 41.62 | 156 | 57 | M | So19a | Ots33p | Co29 | OmySex | Ssa11qa | 9 |
| M | So19b | Ots33q | Co29 | OmySex | Ssa11qa | 3 | |||||
| So20 | 144.99 | 105.9 | 227 | 108 | M | So20a | Ots22 | Co24 | Omy16q | Ssa13qa | 7 |
| M | So20b | Ots28 | Co27 | Omy28 | Ssa03p | 9 | |||||
| So21 | 130.01 | 25.88 | 189 | 64 | M | So21a | Ots32 | Co20b | Omy13p | Ssa12qa | 4 |
| M | So21b | Ots27 | Co10b | Omy13q | Ssa06q | 3 | |||||
| So22 | 152.37 | 69.22 | 248 | 82 | M | So22a | Ots16p | Co18b | Omy11p | Ssa19qa | 6 |
| M | So22b | Ots09p | Co06a | Omy12p | Ssa13qb | 9 | |||||
| So23 | 152.67 | 90.55 | 222 | 75 | M | So23a | Ots17 | Co19a | Omy15q | Ssa17qa | 3 |
| M | So23b | Ots24 | Co20a | Omy16p | Ssa19qb | 5 | |||||
| So24 | 129.68 | 181.11 | 221 | 96 | M | So24a | Ots05q | Co13a | Omy05q | Ssa10qa | 9 |
| M | So24b | Ots05p | Co12a | Omy08p | Ssa15qa | 4 | |||||
| So25 | 84.5 | 65.86 | 145 | 63 | A | So25a | Ots06p | Co04a | Omy01p | Ssa16qa | 10 |
| So26 | 159.93 | 98.08 | 170 | 51 | A | So26a | Ots09q | Co06b | Omy12q | Ssa03q | 4 |
| So27 | 225.35 | 12.00 | 281 | 77 | M | So27a | Ots04q | Co03b | Omy06q | Ssa26 | 4 |
| M | So27b | Ots04p | Co03a | Omy06p | Ssa24 | 13 | |||||
| So28 | 177.74 | 14.96 | 240 | 108 | M | So28a | Ots12q | Co08b | Omy26 | Ssa11qb | 3 |
| M | So28b | Ots01q | Co011a | Omy23 | Ssa01qa | 8 | |||||
| Total | 4082 | 2291 | 6322 | 2179 | 353 | ||||||
aLG So9 is the sex chromosome for sockeye salmon and was represented by 2 acrocentric LGs in the female (So9, So9.5) and a single metacentric LG in the male (So9). LG So9 in the male contained 107 markers and was 109.5 cM long. LG So9 is denoted as 9A_(X2) in (Limborg et al. 2015), and So9.5a is denoted as 9B_(X1).
bDesignated as acrocentric in Limborg et al. (2015).
cAssembled as separate LGs in Limborg et al. (2015).
As expected, marker order and LG designations were highly concordant between our map and 2 previous maps for sockeye salmon constructed using families included in this study (Everett et al. 2012; Limborg et al. 2015, data not shown). However, some differences did exist. We identified 2 differences between our map and Everett et al. (2012): 1) all markers that we identified from LG 29 in Everett et al. (2012) were placed on LG So27 in our map and 2) LG 9 in Everett et al. (2012) was composed of 2 separate LGs in our female map. We also identified differences between our map and Limborg et al. (2015): 1) we were able to join LG So18a and So18b from Limborg et al. (2015) into a single metacentric LG, 2) we identified LG So17 as metacentric rather than acrocentric, and 3) we identified 2 additional homeologous relationships in this study that were not identified in Limborg et al. (2015) (So10a-So28b, So27a-So28a, Table 3).
Table 3.
Homeologous LG arms in sockeye salmon, the number of marker pairs supporting each relationship, and corresponding homeologous relationships in other salmonids (Brieuc et al. 2014; Kodama et al. 2014)
| Homeology in sockeye | # marker pairs | Homeology | |||
|---|---|---|---|---|---|
| Chinook salmon | coho salmon | rainbow trout | Atlantic salmon | ||
| So2a-So5b | 9 | Ots07p-Ots14p | Co05a-Co16b | Omy07p-Omy18p | Ssa17qa-Ssa16qb |
| So3b-So14a | 19 | Ots03p-Ots23 | Co02a-Co13b | Omy03p-Omy02p | Ssa02p-Ssa05q |
| So8b-So23a | 9 | Ots15p-Ots17 | Co09a-Co19a | Omy21p-Omy15q | Ssa07p-Ssa17qa |
| So10a-So28b | 6 | Ots06q-Ots01q | Co04b-Co11a | Omy01q-Omy23 | Ssa18qa-Ssa01qa |
| So11b-So18b | 11 | Ots11p-Ots34 | Co07a-Co12b | Omy19p-Omy10q | Ssa04p-Ssa08q |
| So15b-So21a | 17 | Ots02q-Ots32 | Co01b-Co20b | Omy17q-Omy13p | Ssa02q-Ssa12qa |
| So21b-So26 | 10 | Ots09q-Ots27 | Co06b-Co10b | Omy12q-Omy13q | Ssa03q-Ssa06p |
| So27a-So28a | 13 | Ots04q-Ots12q | Co03b-Co08b | Omy06q-Omy26 | Ssa26-Ssa11qa |
We identified orthologous relationships between Chinook and sockeye salmon for all 52 LG arms on our maps and extended these relationships to coho salmon, rainbow trout, and Atlantic salmon (Table 2). Each orthologous relationship between sockeye and Chinook salmon was supported by 1–15 marker pairs (average of 7 pairs per relationship, 353 total markers shared between the 2 species, Table 2). All LG arms in Chinook salmon aligned to a single LG arm in sockeye salmon except for LG So17, where both the a and b arms aligned to a single arm in Chinook salmon (Ots01p). This relationship could be a result of a centromere re-location after the species diverged, but additional information is necessary to validate this finding.
We mapped 1101 potentially duplicated loci on the female linkage map using haploids. High concentrations of duplicated loci were found near the distal ends of 16 LG arms across 14 LGs (Figure 2), and 8 homeologous relationships were identified by mapping 94 duplicated loci that segregated at both paralogs (Table 3). Comparisons with Chinook salmon, coho salmon, rainbow trout, and Atlantic salmon revealed that a conserved orthologous suite of chromosome arms is involved in homeologous pairing across these species (Table 3).
Phenotypic Data
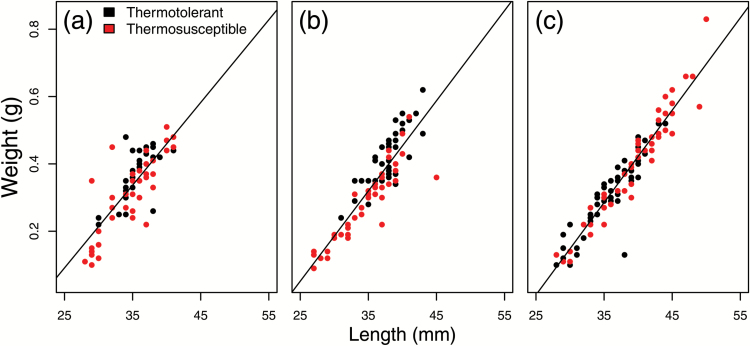
Phenotypic data for thermotolerance, length, weight, and condition factor was obtained from 3 diploid families (Table 4). Distributions of length and condition factor were significantly different among the 3 families (P < 0.05), but distributions of weight were not. Thermotolerant individuals in families D3 and D4 were significantly larger than thermosusceptible individuals (P < 0.05). However, the opposite relationship was present in family D5 (Figure 3, Table 4). Condition factor was higher for thermotolerant individuals in all 3 families but was only significantly higher in families D3 and D4.
Table 4.
Mean and standard deviation (SD) of length, weight, and condition factor in experimental families. Bold values indicate significant differences between thermotolerant and thermosusceptible groups based on a Student’s t-test (P < 0.05). Distributions of length and condition factor were significantly different (P < 0.05) among the 3 families, whereas distributions of weight were not. Combined statistics for both groups are also given
| Phenotype | Family | Thermotolerant | Thermosusceptible | Combined | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | ||
| Length (mm) | D3 | 35.46 | 2.10 | 34.80 | 3.92 | 35.12 | 3.16 |
| Length (mm) | D4 | 37.79 | 2.55 | 34.80 | 4.14 | 36.24 | 3.76 |
| Length (mm) | D5 | 35.94 | 4.16 | 39.56 | 5.52 | 37.75 | 5.19 |
| Weight (g) | D3 | 0.37 | 0.07 | 0.32 | 0.12 | 0.34 | 0.10 |
| Weight (g) | D4 | 0.42 | 0.08 | 0.29 | 0.11 | 0.35 | 0.11 |
| Weight (g) | D5 | 0.31 | 0.12 | 0.41 | 0.17 | 0.36 | 0.15 |
| Condition factor | D3 | 0.82 | 0.11 | 0.72 | 0.19 | 0.77 | 0.17 |
| Condition factor | D4 | 0.77 | 0.10 | 0.66 | 0.10 | 0.71 | 0.11 |
| Condition factor | D5 | 0.64 | 0.11 | 0.62 | 0.07 | 0.63 | 0.10 |
Figure 3.
Visualization of weight versus length relationships for each individual in family (a) D3, (b) D4, and (c) D5. Each dot represents an individual; dots are colored according to thermotolerance. A line of best fit is drawn through each distribution. Colors are viewable in the online version of the article.
QTL Analysis
We conducted QTL mapping for 4 phenotypic traits in 3 diploid families using 3496 unique loci placed on the female linkage map. Family D3 contained 2218 loci suitable for QTL mapping; family D4 contained 2160 loci; and family D5 contained 2212 loci. We identified 26 QTL with peaks at 22 unique genomic positions (Table 5). Of these QTL, 2 were identified as significant at the LG and experiment-wide level, and 24 were identified as significant at the LG level. The percentage of variation explained by each QTL ranged from 6.18 to 34.08%. No significant epistatic interactions were found among QTL (P > 0.1).
Table 5.
Description of 26 significant QTL for 4 phenotypes in 3 diploid families. QTL peak marker is the marker with the highest LOD score for each QTL, cM is the position of the QTL peak marker, 95% CI is the approximate 95% confidence interval for the position of the QTL, PVE is the percentage of variation in the phenotype explained by the QTL, p(F) is the P value of the F statistic in the multiple-QTL model, and Sig denotes whether the QTL was significant at the LG or experiment-wide level (Exp). Gene abbreviations are provided for loci that were annotated directly (bold) or annotated based on flanking sequence from the Atlantic salmon genome (italics). See Supplementary Table S3 for more information on QTL annotations. Marker RAG3 is the 5′-nuclease assay One_RAG-93. Instances where the QTL peak marker location is not included in the 95% CI indicate a lack of confidence in the true QTL location and should be interpreted with caution
| Phenotype | Family | QTL Peak Marker | LG | cM | 95% CI | LOD | PVE | p(F) | Sig | Annotation(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Thermotolerance | D3 | 63 844 | 2 | 171.57 | 0.00–192.07 | 3.35 | 16.75 | 5.10E−04 | LG | KLD7A, MLVCB |
| Thermotolerance | D3 | 36 045 | 11 | 129.89 | 126.7–133.03 | 3.32 | 9.88 | 9.50E−03 | LG | |
| Thermotolerance | D4 | 70 808 | 25 | 48 | 41.28–50.74 | 3.57 | 12.37 | 4.00E−03 | LG | PABP |
| Thermotolerance | D5 | 7896 | 6 | 31.28 | 17.53–90.35 | 3.45 | 7.58 | 8.22E−03 | LG | SMHD3 |
| Thermotolerance | D5 | 87 489 | 13 | 51.86 | 6.13–138.3 | 4.01 | 16.35 | 5.68E−05 | LG | PANTR |
| Thermotolerance | D5 | 85 651 | 19 | 86.91 | 1.71–93.68 | 3.29 | 7.89 | 6.85E−03 | LG | |
| Length | D4 | RAG3 | 9 | 54.19 | 37.36–57.78 | 3.07 | 7.56 | 1.80E−02 | LG | RAG |
| Length | D4 | 76 581 | 26 | 68.59 | 14.44–158.81 | 3.49 | 8.49 | 1.10E−02 | LG | |
| Length | D4 | 767 479 | 28 | 21.56 | 19.73–23.02 | 3.59 | 10.63 | 4.00E−03 | LG | |
| Length | D5 | 44 010 | 7 | 63.04 | 3.61–111.8 | 3.40 | 6.97 | 1.53E−03 | LG | |
| Length | D5 | 84 879 | 11 | 164.78 | 164.48–164.9 | 4.13 | 13.06 | 1.03E−05 | LG | |
| Weight | D3 | 82 443 | 11 | 117.29 | 0.83–167.61 | 3.41 | 11.41 | 1.00E−02 | LG | |
| Weight | D4 | 70 636 | 15 | 116.61 | 112.53–117.01 | 3.67 | 10.85 | 2.20E−03 | LG | |
| Weight | D4 | 2639 | 25 | 78.49 | 54.16–78.92 | 3.09 | 6.18 | 2.70E−02 | LG | |
| Weight | D4 | 76 581 | 26 | 68.59 | 21.35–158.81 | 3.28 | 8.34 | 8.00E−03 | LG | SIA7B |
| Weight | D4 | 767 479 | 28 | 21.56 | 19.73–23.02 | 4.64 | 13.90 | 4.60E−04 | Exp | |
| Weight | D5 | 7896 | 6 | 31.28 | 3.60–126.87 | 3.60 | 6.67 | 4.70E−03 | LG | SMHD3 |
| Weight | D5 | 66 074 | 7 | 94.75 | 3.56–111.8 | 3.11 | 13.99 | 2.53E−05 | LG | BSN |
| Weight | D5 | 84 879 | 11 | 164.78 | 164.48–164.9 | 4.60 | 19.75 | 6.22E−07 | Exp | |
| Weight | D5 | 30 636 | 19 | 11.35 | 7.24–58.94 | 3.40 | 9.96 | 4.13E−04 | LG | |
| Condition factor | D3 | 86 442 | 6 | 53.86 | 50.26–58.87 | 5.56 | 34.08 | 1.33E−07 | LG | |
| Condition factor | D4 | 765 637 | 6 | 4.82 | 1.52–97.22 | 3.45 | 13.72 | 1.60E−03 | LG | |
| Condition factor | D4 | 5975 | 28 | 145.33 | 3.70–68.97 | 3.47 | 6.92 | 3.50E−02 | LG | FAS |
| Condition factor | D5 | 53 011 | 4 | 64.01 | 139.87–145.86 | 3.16 | 15.16 | 5.61E−06 | LG | |
| Condition factor | D5 | 15 419 | 10 | 27.56 | 9.69–82.72 | 4.92 | 33.89 | 5.86E−11 | LG | |
| Condition factor | D5 | 7448 | 20 | 0 | 0.00–144.99 | 3.49 | 9.74 | 2.93E−04 | LG |
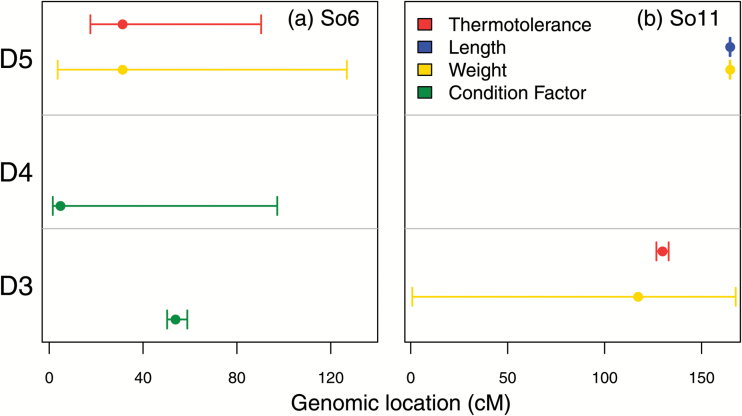
The number of QTL identified varied substantially by family, phenotypic trait, and LG. We identified 4 QTL in family D3, 10 QTL in family D4, and 12 QTL in family D5. The phenotypic trait with the most QTL was weight (9), followed by thermotolerance and condition factor (6), and length (5). Two LGs contained 4 QTL (So6, So11), So28 contained 3 QTL, and the remaining LGs contained 2 or fewer QTL (Figure 4, Supplementary Figure S2).
Figure 4.
Results from QTL analysis for the 2 LGs containing the most QTL, (a) So6 and (b) So11. Bracket lines represent 95% confidence intervals for the location of each QTL, and dots signify the QTL peak. Colors are viewable in the online version of the article.
Genomic regions containing overlapping QTL from different families were generally uncommon, but we did see this pattern on LGs So6 and So11 (Figure 4). So6 contained overlapping QTL for thermotolerance (family D5), weight (family D5), and condition factor (families D3 and D4), and So11 contained overlapping QTL for thermotolerance (family D3), length (family D5), and weight (families D3 and D5). We also found 8 QTL that shared a peak marker with another QTL within the same family. Shared markers were most often associated with QTL for length and weight, but we did find 1 example of a shared peak QTL marker for thermotolerance and weight (locus 7896).
Comparisons of the locations of QTL discovered in this study to QTL found in rainbow trout and Chinook salmon revealed several orthologous regions of interest. For example, the QTL hotspot found on LG So6 in this study corresponded to a region of chromosome Omy14 in rainbow trout that contained multiple QTL related to growth, condition factor, and morphology (Hecht et al. 2012). We also discovered that LG So7, which contained QTL for thermotolerance and weight in this study, was orthologous with regions containing QTL for length and thermotolerance in rainbow trout (Perry et al. 2005) and length in Chinook salmon (Everett and Seeb 2014).
Paired-End Assembly, Alignments to Genomic Resources, and Functional Annotation
Construction of consensus sequences longer than 150bp from PE data was possible for 7146 of 7367 loci (97%, average length 258bp, Supplementary Table S1). Consensus sequences from 480 loci were successfully aligned to the Atlantic salmon genome and used to anchor 97 unique scaffolds spanning approximately 25% of the total female map (Supplementary Tables S1 and S2). Alignment to sockeye salmon ESTs from the cGRASP database was possible for 98 loci (Supplementary Table S1).
Functional annotation from RAD sequence data was successful for 840 of 7,367 loci (11%, Supplementary Table S1). Transposable elements comprised approximately 25% of these annotations; other common functional groups included DNA polymerases and genes involved in regulation of programmed cell death. Annotations were successful for 8 QTL peak markers (9 total annotations, Table 5, Supplementary Table S3). Of these annotations, 5 were derived from RAD sequence data, 3 were obtained using flanking sequence from the Atlantic salmon genome, and 1 was obtained from previous annotation of a 5′-nuclease assay. Notable annotations included a QTL for condition factor that aligned to a gene involved in fatty acid synthesis (FAS, locus 5975) and a QTL for length that aligned to a gene involved in metabolism and biosynthesis (RAG, locus One_RAG3-93).
Discussion
Linkage Mapping and Alignment to Genomic Resources
The first objective of this study was to create a dense linkage map for sockeye salmon that could be used for QTL analysis. Our linkage map was constructed from a combination of newly acquired data along with raw reads from 2 previous mapping studies (Everett et al. 2012; Limborg et al. 2015) and contains more than double the number of markers of those maps. Additionally, our map includes data from both the northern and southern extremes of the species’ range in North America and represents both anadromous and non-anadromous life-history types.
The female map was approximately twice as long as the male map, and markers on the male map tended to group towards the centromeres. Similar results have been well-documented in salmonids and are thought to occur because of sex-specific differences in the distribution of recombination sites across chromosomes (Lien et al. 2011; Everett et al. 2012; Kodama et al. 2014). We did not merge sex-specific maps because of these differences and, instead, utilized the denser female map for QTL analysis and alignments to genomic resources. The elevated recombination found in the telomeric regions in males makes the male map an important resource for future studies attempting to order telomeric markers or genome scaffolds (Lien et al. 2011).
Mapping of duplicated loci on the female map using haploids revealed patterns of homeology similar to previous studies (reviewed in Allendorf et al. 2015). High concentrations of duplicated loci were found in the telomeric regions of 8 pairs of homeologous chromosomes, and these chromosomes were orthologous with chromosomes involved in homeologous pairing in other species (Brieuc et al. 2014; Kodama et al. 2014; McKinney et al. 2015). It is important to note that we were able to locate the 6 pairs of homeologous chromosomes described in Limborg et al. (2015) as well as 2 additional pairs that have not been previously described. This finding provides further evidence for the existence of a conserved set of 8 homeologous chromosome arms containing high concentrations of duplicated loci across all salmonids (reviewed in Allendorf et al. 2015).
Alignment of our mapped loci to existing genomic resources provided important functional annotations and allowed us to anchor our map in the context of 4 other salmonids. Functional annotations were similar to past RAD studies in salmonids and included a high proportion of transposable elements (Everett et al. 2012; Everett and Seeb 2014; Larson et al. 2014). Notably, transposable elements comprised approximately 25% of annotations, but only 10% of annotations for duplicated loci (c.f., McKinney et al. 2015; Waples et al. 2015). Transposable elements are hypothesized to facilitate differentiation between homeologs during rediploidization; this may explain the reduced frequencies of transposable elements in duplicated regions that have not been fully rediplodized (McKinney et al. 2015; Waples et al. 2015). Alignment with RAD-derived linkage maps from other species revealed orthologous relationships for all 52 LG arms in sockeye salmon. The success of these alignments highlights the utility of comparing findings from RAD studies across species and demonstrates an advantage for the continued use of SbfI or other enzymes with restriction sites that overlap with SbfI to ensure the compatibility of future studies.
We were able to successfully anchor 97 scaffolds from the Atlantic salmon draft genome to our map, covering about 25% of the total map length. The continuous sequence provided by these scaffolds represents an excellent tool for functional annotation and exploration of genomic regions that are proximate to loci of interest (Allendorf et al. 2010). However, we were unable to anchor scaffolds to a large portions of our linkage map, likely due to the draft nature of the genome assembly, the marker density of our linkage map, and sequence divergence between sockeye and Atlantic salmon.
Phenotypic Variation in Experimental Families
Significant variation in size and condition factor existed among all 3 families in our study. This variation is likely a result of genetic rather than environmental effects because the families were raised in similar environments and these traits have been shown to have high heritability in other salmonids (reviewed in Garcia et al. 2008).
Thermotolerant individuals had higher condition factors in all 3 families and were larger in 2 of the 3 families. A positive correlation between condition factor and temperature tolerance was also demonstrated in cutthroat trout (O. clarki, Robinson et al. 2008). These results suggest that aspects of body composition that are correlated with condition factor, such as lipid and protein content, may be important components of temperature tolerance in salmonids (Robinson et al. 2008). Significant correlations between size and upper temperature tolerance have also been observed in rainbow trout (Perry et al. 2005), but these correlations were not consistent among experimental families and appeared to be related to parental effects. A more comprehensive study of size and thermotolerance across 5 species of Pacific salmon also found no consistent correlation between these traits (Brett 1952). Taken together, these results suggest that body size is unlikely to be an accurate predictor of thermotolerance, which may explain the inconsistent trend between these 2 traits in our families.
QTL Analysis
We identified 26 QTL related to 4 phenotypic traits in 3 experimental families of anadromous sockeye salmon. Each trait displayed between 0 and 4 QTL within each family. Across families, each trait contained at least 1 QTL that explained > 10% of the phenotypic variation. Prevailing theory suggests that most continuous phenotypic traits such as those that we examined are likely controlled by many genes of small effect (Roff 2007). The fact that we found relatively few QTL for each trait with generally large effect sizes appears to contrast this theory. However, it is important to note that the experimental design and relatively small sample sizes used in this study likely prevented us from discovering the majority of small-effect QTL related to each trait. Classical QTL studies generally employ a multigenerational design and sample sizes of more than 300 individuals per family to maximize their power to detect QTL and accurately estimate QTL effect sizes (Beavis et al. 1994; Xu 2003; Erickson et al. 2004). Our study design employing ~100 individuals from F1 families derived from a wild population undoubtedly limited our power to detect QTL, but this design also provides a cost effective and practical template to discover large-effect QTL in wild populations of non-model organisms.
The distribution of QTL across the genome in our study was non-random and was characterized by a few regions containing high concentrations of QTL related to multiple phenotypic traits interspersed within large regions containing very few QTL. Past studies have suggested that regions containing large numbers of QTL (QTL hotspots) are likely involved in the early stages of speciation as well as the evolution of different life history types (Via and West 2008; Hecht et al. 2012; Gagnaire et al. 2013a). Extensive life history diversity exists in populations of sockeye salmon from our study system, including the presence of 3 distinct ecotypes associated with environment used for spawning (Hilborn et al. 2003). These ecotypes are characterized by differences in a number of traits, such as size (Quinn et al. 2001) and condition factor (WAL, personal observation), and experience substantially different temperature regimes, likely leading to adaptive differences in thermotolerance. Although our families were derived from a single ecotype (stream type), the QTL hotspots that we discovered appear to control at least some of the variation in traits that distinguish ecotypes, providing evidence that these hotspots may be involved in local adaptation and the formation of distinct ecotypes. Future research should focus on investigating the co-location of QTL hotspots with loci displaying signatures of divergent selection among ecotypes to further investigate this hypothesis (c.f., Via and West 2008; Gagnaire et al. 2013b).
We found 4 pairs of QTL that shared a peak marker with another QTL within the same family. All but 1 of these pairs contained QTL associated with length and weight, an anticipated result given the high degree of correlation between these 2 size-related traits. The remaining pair was associated with thermotolerance and weight. A QTL affecting thermotolerance and size was also discovered in rainbow trout and was hypothesized to be the result of either pleiotropy or linkage disequilibrium (Perry et al. 2005). Pleiotropy occurs when a single gene influences multiple seemingly unrelated phenotypic traits and may explain the results we observed. It is also possible that linkage disequilibrium between 2 proximate genes related to thermotolerance and size may be responsible for these results.
No QTL peak markers were replicated across families. This may be the result of different segregation patterns in these families and/or low power to detect small effect QTL due to sample size limitations. However, we did observe overlapping confidence intervals for QTL related to the same traits across families, providing strong evidence for the existence of the QTL.
Comparing the locations of single QTL and QTL hotspots across related species can provide important information about the genetic architecture of phenotypic traits (Reid et al. 2005). We found 1 genomic region that contained multiple QTL in sockeye salmon and rainbow trout and another region that contained multiple QTL in sockeye salmon, Chinook salmon, and rainbow trout. These regions represent ideal candidates for future research on the genetic basis of phenotypic traits in salmonids. Additionally, these results further illustrate the importance of using a conserved RAD-seq protocol among studies to accumulate evidence of orthology.
Functional annotation of QTL can potentially be used to identify the genes underlying phenotypic variation in traits of interest (Pavlidis et al. 2012). We were able to annotate about a third of our QTL, and several of these QTL annotated to genes with plausible connections to the phenotypic traits examined (Table 5, Supplementary Table S3). For example, the QTL for condition factor on LG So28 annotated to a gene involved in fatty acid synthesis and the QTL for length on LG So9 annotated to a gene involved in metabolism and biosynthesis. However, it is important to note that the traits we examined are likely controlled by many genes with obscure roles and that “storytelling” using functional annotations should be approached with caution (Pavlidis et al. 2012). Nevertheless, information about genes found in genomic regions of interest is vital for increasing our understanding of the genetic architecture of phenotypic traits and guiding future research (Allendorf et al. 2010). It is also important to note that alignments to the Atlantic salmon genome proved helpful for annotating additional QTL. As this resource improves, it should be possible to annotate a much larger proportion of QTL discovered with RAD data.
Conclusions
We successfully constructed the densest linkage map to date for sockeye salmon and used this map to detect QTL for 4 phenotypic traits. QTL were distributed non-randomly across the genome and often colocalized within QTL hotspots. These hotspots may be important for adaptation and represent ideal candidates for future studies seeking to understand processes of local adaptation in salmonids. For example, these QTL hotspots could be examined in thermotolerant populations to improve our understanding of the genetic basis of thermotolerance and help to identify thermotolerant populations for conservation. Comparison of our results with QTL studies in rainbow trout and Chinook salmon revealed several regions with overlapping QTL for similar phenotypic traits. These results provide evidence that the genetic basis of some phenotypic traits is similar across species and emphasize the importance of additional interspecies comparisons.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
Funding was provided by grants from the Gordon and Betty Moore Foundation and the Bristol Bay Regional Seafood Development Association. W.A.L. was supported by the H. Mason Keeler Endowment for Excellence and a US National Science Foundation Graduate Research Fellowship (grant # DGE-0718124). M.T.L. was supported by the Danish Council for Independent Research’s career program Sapere Aude (Grant # 12-126687).
Conflict of interest
The authors declare no conflict of interest.
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.5n0v4. Raw sequences reads deposited in the NCBI sequence read archive: SRP064002.
Supplementary Material
Acknowledgments
We thank the researchers from the Alaska Salmon Program at the University of Washington, especially Daniel Schindler, Jackie Carter, and Chris Boatright, for assisting with sample collection and rearing experimental families. We also thank Jon Wittock and Sewall Young for helping to rear families, Carita Pascal for her excellent laboratory assistance, and Kristen Gruenthal for her editorial comments.
References
- Albertson RC, Powder KE, Hu Y, Coyle KP, Roberts RB, Parsons KJ. 2014. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 23:5135–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf FW, Bassham S, Cresko WA, Limborg MT, Seeb LW, Seeb JE. 2015. Effects of crossovers between homeologs on inheritance and population genomics in polyploid-derived salmonid fishes. J Hered. 106:217–227. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Hohenlohe PA, Luikart G. 2010. Genomics and the future of conservation genetics. Nat Rev Genet. 11:697–709. [DOI] [PubMed] [Google Scholar]
- Bagenal TB, Tesch FW. 1978. Age and growth. In: Bagenal T.B., editor. Methods for assessment of fish production in freshwater, 3rd edition Oxford, UK: Blackwell Scientific Publication; p. 101–136. [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3:e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Beavis WD, et al. 1994. Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny. Crop Sci. 34:882–896. [Google Scholar]
- Bradford MJ. 1995. Comparative review of Pacific salmon survival rates. Can J Fish Aquat Sci. 52:1327–1338. [Google Scholar]
- Brett JR. 1952. Temperature tolerance in young Pacific salmon, genus Oncorhynchus. J Fish Res Board Canada. 9:265–323. [Google Scholar]
- Brieuc MS, Waters CD, Seeb JE, Naish KA. 2014. A dense linkage map for Chinook salmon (Oncorhynchus tshawytscha) reveals variable chromosomal divergence after an ancestral whole genome duplication event. G3 (Bethesda). 4:447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19:889–890. [DOI] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22:3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, et al. 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3: Genes Genomes Genetics. 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann TH, et al. 2013. Exploiting genetic diversity to balance conservation and harvest of migratory salmon. Can J Fish Aquat Sci. 70:785–793. [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 12:499–510. [DOI] [PubMed] [Google Scholar]
- Dekkers JC, Hospital F. 2002. The use of molecular genetics in the improvement of agricultural populations. Nat Rev Genet. 3:22–32. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Siegmund D. 1999. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 151:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfstrom CM, Smith CT, Seeb JE. 2006. Thirty-two single nucleotide polymorphism markers for high-throughput genotyping of sockeye salmon. Mol Ecol Notes. 6:1255–1259. [Google Scholar]
- Eliason EJ, Clark TD, Hague MJ, Hanson LM, Gallagher ZS, Jeffries KM, Gale MK, Patterson DA, Hinch SG, Farrell AP. 2011. Differences in thermal tolerance among sockeye salmon populations. Science. 332:109–112. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Fenster CB, Stenøien HK, Price D. 2004. Quantitative trait locus analyses and the study of evolutionary process. Mol Ecol. 13:2505–2522. [DOI] [PubMed] [Google Scholar]
- Etter PD, Preston JL, Bassham S, Cresko WA, Johnson EA. 2011. Local de novo assembly of RAD paired-end contigs using short sequencing reads. PLoS One. 6:e18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett MV, Miller MR, Seeb JE. 2012. Meiotic maps of sockeye salmon derived from massively parallel DNA sequencing. BMC Genomics. 13:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett MV, Seeb JE. 2014. Detection and mapping of QTL for temperature tolerance and body size in Chinook salmon (Oncorhynchus tshawytscha) using genotyping by sequencing. Evol Appl. 7:480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber-Hammond J, Phillips RB, Park LK. 2012. The sockeye salmon neo-Y chromosome is a fusion between linkage groups orthologous to the coho Y chromosome and the long arm of rainbow trout chromosome 2. Cytogenet Genome Res. 136:69–74. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Pavey SA, Bernatchez L. 2013a. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis). Mol Ecol. 22:3036–3048. [DOI] [PubMed] [Google Scholar]
- Gagnaire PA, Pavey SA, Normandeau E, Bernatchez L. 2013b. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by RAD sequencing. Evolution. 67:2483–2497. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Afeche SC, Scialfa JH, do Amaral FG, dos Santos SH, Lima FB, Young ME, Cipolla-Neto J. 2008. Insulin modulates norepinephrine-mediated melatonin synthesis in cultured rat pineal gland. Life Sci. 82:108–114. [DOI] [PubMed] [Google Scholar]
- Groot AT, Staudacher H, Barthel A, Inglis O, Schöfl G, Santangelo RG, Gebauer-Jung S, Vogel H, Emerson J, Schal C, et al. 2013. One quantitative trait locus for intra- and interspecific variation in a sex pheromone. Mol Ecol. 22:1065–1080. [DOI] [PubMed] [Google Scholar]
- Gutierrez AP, Lubieniecki KP, Fukui S, Withler RE, Swift B, Davidson WS. 2014. Detection of quantitative trait loci (QTL) related to grilsing and late sexual maturation in Atlantic salmon (Salmo salar). Mar Biotechnol (NY). 16:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht BC, Thrower FP, Hale MC, Miller MR, Nichols KM. 2012. Genetic architecture of migration-related traits in rainbow and steelhead trout, Oncorhynchus mykiss. G3 (Bethesda). 2:1113–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilborn R, Quinn TP, Schindler DE, Rogers DE. 2003. Biocomplexity and fisheries sustainability. Proc Natl Acad Sci USA. 100:6564–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinch SG, Cooke SJ, Farrell AP, Miller KM, Lapointe M, Patterson DA. 2012. Dead fish swimming: a review of research on the early migration and high premature mortality in adult Fraser River sockeye salmon Oncorhynchus nerka. J Fish Biol. 81:576–599. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Orell P, Pritchard VL, Kent MP, Lien S, Niemelä E, Erkinaro J, Primmer CR. 2014. Genome-wide SNP analysis reveals a genetic basis for sea-age variation in a wild population of Atlantic salmon (Salmo salar). Mol Ecol. 23:3452–3468. [DOI] [PubMed] [Google Scholar]
- Kodama M, Brieuc MS, Devlin RH, Hard JJ, Naish KA. 2014. Comparative mapping between Coho Salmon (Oncorhynchus kisutch) and three other salmonids suggests a role for chromosomal rearrangements in the retention of duplicated regions following a whole genome duplication event. G3 (Bethesda). 4:1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 11:241–247. [DOI] [PubMed] [Google Scholar]
- Larson WA, Seeb LW, Everett MV, Waples RK, Templin WD, Seeb JE. 2014. Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol Appl. 7:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Gidskehaug L, Moen T, Hayes BJ, Berg PR, Davidson WS, Omholt SW, Kent MP. 2011. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics. 12:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg MT, Waples RK, Allendorf FW, Seeb JE. 2015. Linkage Mapping Reveals Strong Chiasma Interference in Sockeye Salmon: Implications for Interpreting Genomic Data. G3 (Bethesda). 5:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates. [Google Scholar]
- McKinney GJ, et al. 2015. An integrated linkage map reveals candidate genes underlying adaptive variation in Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol Resour, doi:10.1111/1755-0998.12479 [DOI] [PubMed] [Google Scholar]
- Miller KM, Teffer A, Tucker S, Li S, Schulze AD, Trudel M, Juanes F, Tabata A, Kaukinen KH, Ginther NG, et al. 2014. Infectious disease, shifting climates, and opportunistic predators: cumulative factors potentially impacting wild salmon declines. Evol Appl. 7:812–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Brunelli JP, Wheeler PA, Liu S, Rexroad CE, 3rd, Palti Y, Doe CQ, Thorgaard GH. 2012. A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol Ecol. 21:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley KG, Sakamoto T, Danzmann RG, Ferguson MM. 2003. Quantitative trait loci for spawning date and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J Hered. 94:273–284. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 29:3237–3248. [DOI] [PubMed] [Google Scholar]
- Perry GM, Ferguson MM, Sakamoto T, Danzmann RG. 2005. Sex-linked quantitative trait loci for thermotolerance and length in the rainbow trout. J Hered. 96:97–107. [DOI] [PubMed] [Google Scholar]
- Quinn TP. 2005. The behavior and ecology of Pacific salmon and trout. Seattle: University of Washington Press. [Google Scholar]
- Quinn TP, et al. 2001. Influence of breeding habitat on bear predation and age at maturity and sexual dimorphism of sockeye salmon populations. Can J Zool. 79:1782–1793. [Google Scholar]
- Rastas P, Paulin L, Hanski I, Lehtonen R, Auvinen P. 2013. Lep-MAP: fast and accurate linkage map construction for large SNP datasets. Bioinformatics. 29:3128–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DP, Szanto A, Glebe B, Danzmann RG, Ferguson MM. 2005. QTL for body weight and condition factor in Atlantic salmon (Salmo salar): comparative analysis with rainbow trout (Oncorhynchus mykiss) and Arctic charr (Salvelinus alpinus). Heredity (Edinb). 94:166–172. [DOI] [PubMed] [Google Scholar]
- Robinson ML, et al. 2008. Fulton’s body condition factor K correlates with survival time in a thermal challenge experiment in juvenile Lahontan cutthroat trout (Oncorhynchus clarki henshawi). J Thermal Biol. 33:363–368. [Google Scholar]
- Roff DA. 2007. A centennial celebration for quantitative genetics. Evolution. 61:1017–1032. [DOI] [PubMed] [Google Scholar]
- Santure AW, De Cauwer I, Robinson MR, Poissant J, Sheldon BC, Slate J. 2013. Genomic dissection of variation in clutch size and egg mass in a wild great tit (Parus major) population. Mol Ecol. 22:3949–3962. [DOI] [PubMed] [Google Scholar]
- Schindler DE, Hilborn R, Chasco B, Boatright CP, Quinn TP, Rogers LA, Webster MS. 2010. Population diversity and the portfolio effect in an exploited species. Nature. 465:609–612. [DOI] [PubMed] [Google Scholar]
- Slate J. 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol Ecol. 14:363–379. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Pascal CE, Grauvogel Z, Habicht C, Seeb JE, Seeb LW. 2011. Multiplex preamplification PCR and microsatellite validation enables accurate single nucleotide polymorphism genotyping of historical fish scales. Mol Ecol Resour. 11(Suppl 1):268–277. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity (Edinb). 100:158–170. [DOI] [PubMed] [Google Scholar]
- Storer CG, et al. 2012. Rank and order: evaluating the performance of SNPs for individual assignment in a non-model organism. PLoS One. 7:e49018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgaard GH. 1978. Sex-chromosomes in sockeye salmon - y - autosome fusion. Can J Genetics Cytol. 20:349–354. [DOI] [PubMed] [Google Scholar]
- Thorgaard GH, Allendorf FW, Knudsen KL. 1983. Gene-centromere mapping in rainbow trout: high interference over long map distances. Genetics. 103:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsor SJ. 2012. Population genomics and the causes of local differentiation. Mol Ecol. 21:5393–5395. [DOI] [PubMed] [Google Scholar]
- Via S, West J. 2008. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol Ecol. 17:4334–4345. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Thompson R, Haley CS. 1996. Confidence intervals in QTL mapping by bootstrapping. Genetics. 143:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JG, Larsson SJ, Buckler ES. 2014. Entering the second century of maize quantitative genetics. Heredity (Edinb). 112:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RK, Seeb LW, Seeb JE. 2015. Linkage mapping with paralogs exposes regions of residual tetrasomic inheritance in chum salmon (Oncorhynchus keta). Mol Ecol Resour, doi:10.1111/1755-0998.12394 [DOI] [PubMed] [Google Scholar]
- Xu S. 2003. Theoretical basis of the Beavis effect. Genetics. 165:2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.5n0v4. Raw sequences reads deposited in the NCBI sequence read archive: SRP064002.