Abstract
Lutjanidae is a family of primarily marine and carnivorous fishes distributed in the Atlantic, Indian, and Pacific oceans, with enormous economic and ecological importance. In order to better clarify the conservative chromosomal evolution of Lutjanidae, we analyzed the evolutionary dynamics of 5 repetitive DNA classes in 5 Lutjanus and in 1 Ocyurus species from the Western Atlantic. The ribosomal 18S sites were generally located in a single chromosome pair, except for L. jocu and L. alexandrei where they are found in 2 pairs. In turn, the 5S rDNA sites are unique, terminal and nonsyntenic with the 18S rDNA sites. In 3 species analyzed, H3 hisDNA genes were found in 1 chromosomal pair. However, while L. jocu presented 2 H3 sites, O. chrysurus showed a noteworthy dispersion of this gene in almost all chromosomes of the karyotype. Retrotransposons Rex1 and Rex3 do not exhibit any association with the explosive distribution of H3 sequences in O. chrysurus. The low compartmentalization of Rex elements, in addition to the general nondynamic distribution of ribosomal and H3 genes, corroborate the karyotype conservatism in Lutjanidae species, also at the microstructural level. However, some “disturbing evolutionary waves” can break down this conservative scenario, as evidenced by the massive random dispersion of H3 hisDNA in the genome of O. chrysurus. The implication of the genomic expansion of H3 histone genes and their functionality remain unknown, although suggesting that they have higher evolutionary dynamics than previously thought.
Keywords: hisDNA, karyotype evolution, Lutjanus, multigenic family, Ocyurus, retrotransposons
The Lutjanidae family is composed of medium-sized to large predator fishes that play an important role in the marine ecosystem (España 2003). Its species are distributed throughout the Atlantic, Indian, and Pacific oceans (Nelson 2006), being primarily associated with reef environments (Resende et al. 2003). A number of species have been significantly overfished, prompting preservation efforts (Lorenzen et al. 2010;Saillant et al. 2013).
This family is subdivided into 4 subfamilies and composes 17 genera and around 120 species. Lutjaninae represents the most diverse subfamily, with 6 genera, with Lutjanus being the most speciose genus with more than 70 species (Gold et al. 2011; Nelson 2006). A number of Lutjaninae genera are monotypic and, among them, the Ocyurus genus has been questioned based on molecular data that strongly suggest its inclusion in the Lutjanus genus (Gold et al. 2011).
Chromosomal data of Lutjanidae have been reported for a few Indo-Pacific (Raghunath and Prasad 1980), Pacific (Ueno and Ojima 1992), Caribbean (Nirchio et al. 2009), and South Atlantic species (Rocha and Molina 2008). In this family, a remarkable karyotype conservatism is found between various genera (Arai 2011), including karyotypes with 2n = 48 acrocentric chromosomes where the position and frequency of Ag-NORs are the main cytotaxonomic characteristics until now evidenced (Rocha and Molina 2008). Thus, the available data are basically focused on the karyotype structure, restricting further considerations regarding microstructural differentiations of chromosomes and phylogenetic relationships among species.
In such groups with structurally homogeneous karyotypes, the identification of chromosomal differences poses a challenge. In this sense, the analysis of the heterochromatin composition, through the chromosomal mapping of repetitive elements, represents an important tool for clarifying the dynamic processes involved with their karytoype differentiation (Costa et al. 2013; Costa et al. 2014).
In fact, a number of repetitive sequences have been extensively analyzed in Atlantic marine fish chromosomes. Among these are 18S and 5S ribosomal genes and histones H1, H2B-H2A, and H3 (Motta Neto et al. 2011a;Motta Neto et al. 2011b;Lima-Filho et al. 2012;Silva et al. 2013; Calado et al. 2014). These multigenic families are vital for eukaryotes (Jordan et al. 2002; Roehrdanz et al. 2010), for which integrated genomic arrangements has been gathered (Andrews et al. 1987; Roehrdanz et al. 2010). Dynamic processes of karyotype evolution may include the intense participation of transposable elements (TEs) (Cioffi et al. 2010), whose evolutionary role may be interconnected with different repetitive sequences and needs to be better investigated in fishes.
In this study, we analyzed the chromosomal distribution of 5S and 18S ribosomal DNA sequences, H3 histone and the retrotransposable elements Rex1 and Rex3 in 5 Lutjaninae species (L. analis, L. synagris, L. jocu, L. alexandrei, and Ocyurus chrysurus). We aimed to highlight their karyotype differentiation at the microstructural level.
Material and Methods
Specimens, Chromosomal Preparations, and Banding Procedures
Cytogenetic analyses used juvenile specimens from the species L. analis (Cuvier in Cuvier and Valenciennes, 1828) (4 males and 6 immature), L. synagris (Linnaeus, 1758) (5 females, 1 male, and 3 immatures), L. jocu (Bloch and Schneider, 1801) (2 females, 2 males, and 2 immature), collected in Natal (5°45′37.08″S; 35°12′19.61″W), and L. alexandrei (Moura and Lindeman 2007) (7 females, 2 males) and Ocyurus chrysurus (Bloch, 1791) (5 females, 3 males), collected in Touros (5°12′49.53″S; 35°26′2.58″W), both in Rio Grande do Norte state on the northeast coast of Brazil.
Specimens were submitted to in vivo mitotic stimulation by intramuscular application of Munolan, an attenuated association of bacterial and fungal antigens, for 24h (Molina 2001;Molina et al. 2010). After this period, the animals were anesthetized with clove oil and sacrificed to remove kidney tissue. Metaphase chromosomes were obtained from cell suspensions, using in vivo interruption of the mitotic cycle, according to methodology proposed by Gold et al. (1990). A volume of 80 µl of cell suspension was dripped onto a slide covered with a film of distilled water heated to 60 °C. Chromosomes were stained with a solution of 5% Giemsa, and diluted in phosphate buffer (pH 6.8) for 8min to determine the diploid chromosome number (2n) and the composition of the karyotype.
Hybridization with Chromosome Probes
5S and 18S rDNA probes were obtained by PCR from nuclear DNA of Ocyurus chrysurus, using primers A 5′-TAC GCC CGA TCT CGT CCG ATC-3′ and B 5′-CAG GCT GGT ATG GCC GTA AGC-3′ (Pendás et al. 1994), and NS1 5′-GTA GTC ATA TGC TTG TCT C-3′ and NS8 5′-TCC GCA GGT TCA CCT ACG GA-3′ (White et al. 1990), respectively. One probe contained a copy of 5S DNAr of approximately 200 pb and the second 18S DNAr of around 1400 pb. The 5S DNAr probe was marked with biotin-14-dATP and the 18S DNAr with digoxigenin-1-dUTP, both by nick translation, following manufacturer’s recommendations (Roche, Mannheim, Germany).
Probe amplification of the H3 histone gene was obtained from the genomic DNA of O. chrysurus, using primers H3F 5′-ATG GCT CGT ACC AAG CAG ACV GC-3′ and H3R 5′-ATA TCC TTR GGC ATR ATR GTG AC-3′, designated from the histone genes of the mollusc Mytilus edulis (Albig et al. 2003), amplified according to Giribert and Distel (2003). The H3 probes were marked with digoxigenin-11-dUTP by nick translation, following manufacturer’s recommendations (Roche, Mannheim, Germany). Sequences of retrotranspons Rex1 and Rex3 were amplified from the DNA mold of O. chrysurus, using primers Rex1 F 5′-TTC TTC AGT GCC TTC AAC ACC -3′ and Rex1 R (Nirchio et al. 2009), designated to amplify Rex1 segments corresponding to encoding domains 3–7 of the reverse transcriptase (RT) gene. Primers Rex3 F 5′- CGG TGA YAA AGG GCA GCC CTG - 3′ and Rex3 R 5′- TGG CAG ACN GGG GTG GTG GT-3′, for Rex3, were used to amplify encoding domains 1, 2, 2A, A, and B of the RT gene, both designated from Xiphophorus genes (Volff et al. 1999;Volff et al. 2000) and amplified according to Valente et al. (2011).
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) was performed following the procedure described by Pinkel et al. (1986). Metaphase chromosomes were treated with RNAse (20 µg/ml in 2× SSC) at 37 °C for 1h and with pepsin (0.005% in 10mM HCl) at 37 °C for 10min, fixed with 1% formaldehyde for 10min and then dehydrated in an alcohol series (70%/85%/100%) for 5min each. The slides with metaphase chromosomes were incubated in 70% formamide/2× SSC at 72 °C, for 5min. The hybridization solution (50% formamide, 2×SSC and 10% dextran sulfate) and the denatured probe (5ng/µl), with a final volume of 30 µl, were deposited on the slide and hybridization performed for 16h at 37 °C.
Post-hybridization rinses were performed with 15% formamide/0.2×SSC at 42 °C, for 20min, followed by washings in 0.1× SSC at 60 °C for 15min and in 0.5%/4× SSC Tween 20 for 5min at room temperature. The hybridization signs of the probes were detected using rhodamine-conjugated antidigoxigenin (Vector, Burlingame) for 18S rDNA, H3 hisDNA, and Rex3 probes and FITC-conjugated streptavidin (Vector, Burlingame) for 5S rDNA and Rex1 probes. The chromosomes were counterstained with Vectashield/DAPI (1.5 µg/ml) (Vector, Burlingame).
Karyotypic Analyses
Chromosome types were identified according to the position of the centromere (Levan et al. 1964) and they were organized in order of decreasing size. An ideogram of the chromosome pairs bearing ribosomal sequences (18S and 5S rDNA) and histones H3 was performed using Photoshop CS5 software.
Cytogenetic data were interpreted based on the phylogenetic proposal put forth by Gold et al. (2011) for Atlantic snappers, whose clades A, B, and C were summarized, highlighting the species analyzed in this study. Inclusion of L. alexandrei in clade C in the present study is due to the reinterpretation of the taxonomic status of the specimen cited as L. cf. apodus by Gold et al. (2011). Lutjanus apodus and L. griseus have no confirmed distribution on the Brazilian coast, where most of their descriptions were defined as belonging to the endemic species L. alexandrei (Moura and Lindeman 2007). Information on the position of ribosomal cistrons in L. sebae (Guo et al. 2011), L. russelli (Ueno and Ojima 1992), L. griseus (Nirchio et al. 2008), and R. aurorubens (Nirchio et al. 2009) were included, in order to conduct a more thorough analysis of the phylogenetic diversity of the main ribosomal genes in the subfamily Lutjaninae.
Data Archiving
In fulfillment of data archiving guidelines (Baker 2013), we have deposited the micrographs of Figures 1, 2, and 3 underlying these analyses in Dryad.
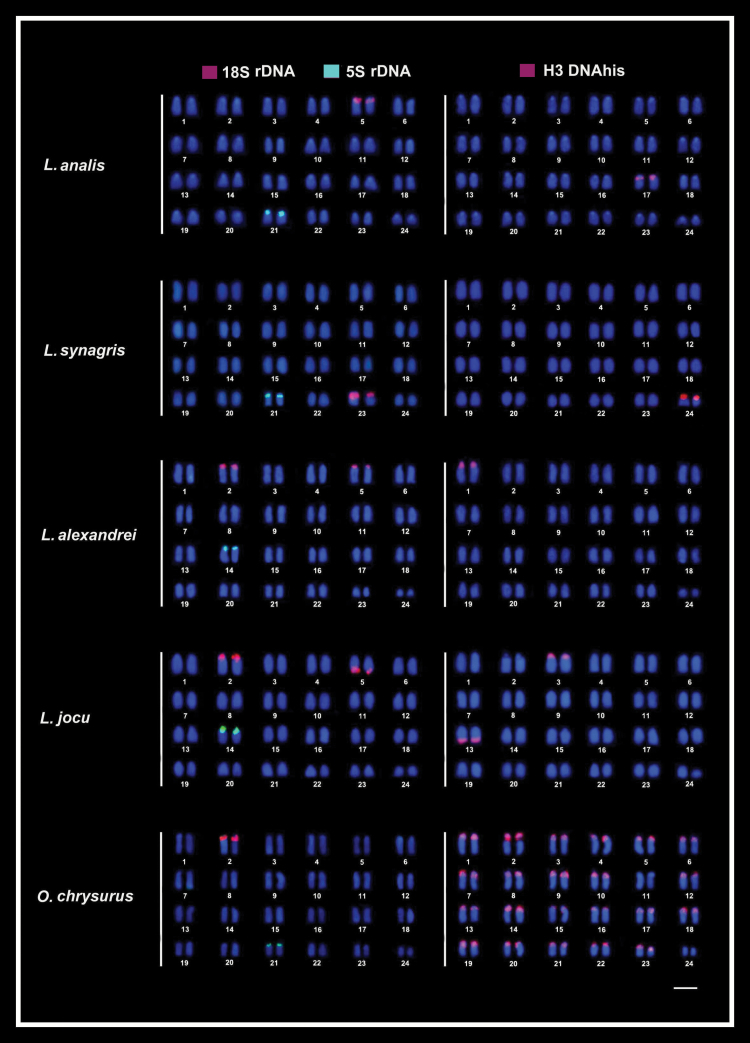
Figure 1.
Fluorescence in situ hybridization (FISH) in the metaphase chromosomes of snappers species highlighting the chromosomal location of 18S rDNA (red) and 5S rDNA (green) sites (left column) and of hisDNA H3 (red) sites (right column). Chromosomes were counterstained with DAPI (blue). Bar = 5 µm.
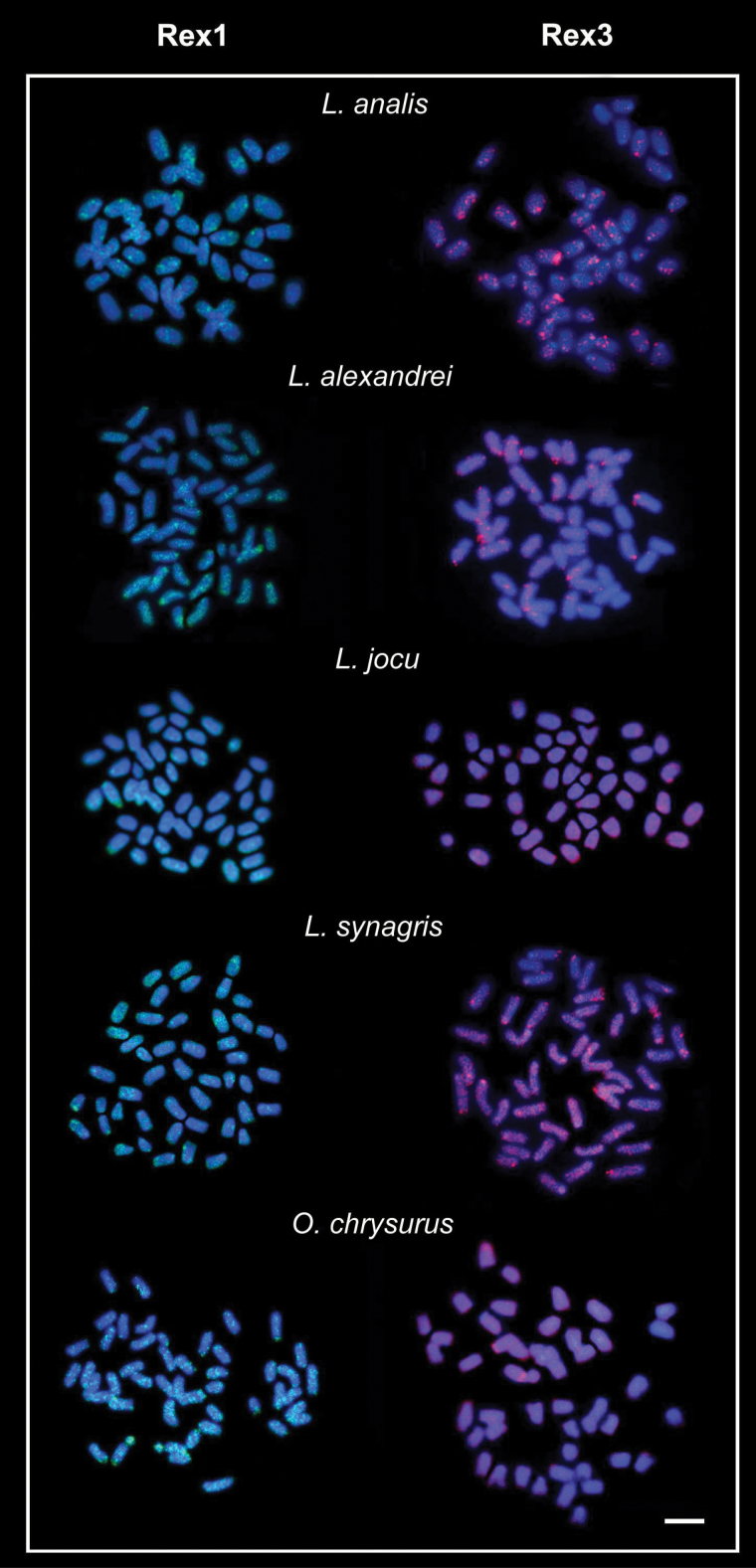
Figure 2.
Fluorescence in situ hybridization (FISH) in the metaphase chromosomes of snappers species, hybridized with Rex1 and Rex3 probes. Chromosomes were counterstained with DAPI. Bar = 5µm.
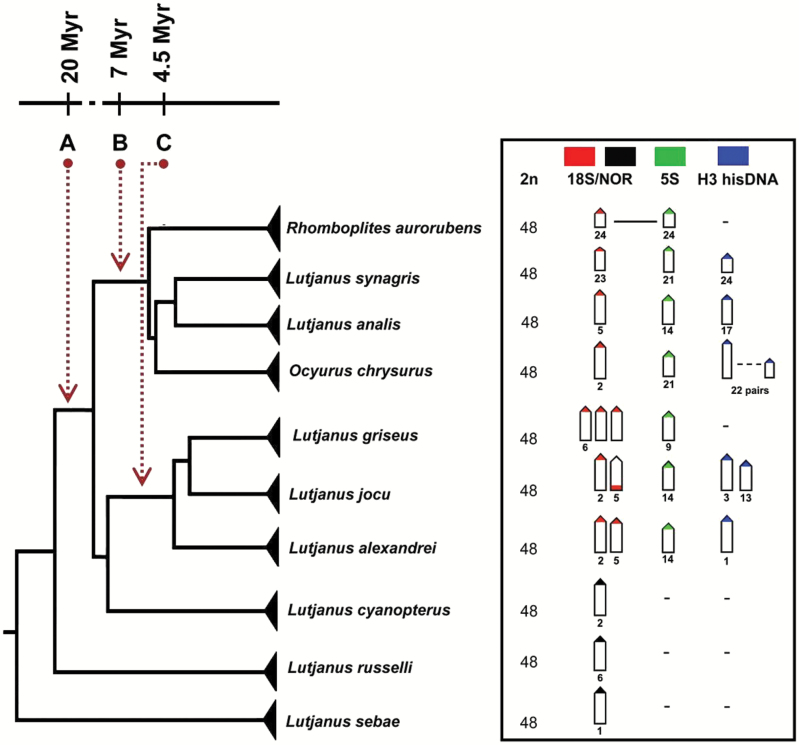
Figure 3.
Phylogenetic distribution of ribosomal and hisDNA H3 sites in Lutjanidae species. The phylogeny and molecular clock of the divergent clades A, B, and C were modified from Gold et al. (2011).
Results
All the 5 species exhibited the same karyotype, with 2n = 48 and composed of exclusively acrocentric chromosomes, corroborating earlier data obtained for species from the Brazilian coast (Rocha and Molina 2008).
18S rDNA sequences were located in the terminal region of the short arms of chromosomal pairs 5, 23, and 2 in L. analis, L. synagris, and O. chrysurus, respectively. In L. alexandrei and L. jocu these sites were identified in the terminal regions of pairs 2 and 5 (Figure 1). Concerning the sites of pair 2, they are both present in the short arms. However, in respect to pair 5, this gene is located in the short arms in L. alexandrei and in the long arms in L. jocu. 5S ribosomal DNA sequences were observed in only one locus in all species analyzed, being mapped in the terminal position of the short arm of pair 14 in L. alexandrei and L. jocu, and in the same position in the pair 21 in L. analis, L. synagris, and O. chrysurus (Figure 1).
Histone H3 sequences were mapped in a single chromosome pair in L. analis (short arm of pair 17), L. synagris (short arm of pair 24), and L. alexandrei (short arm of pair 1), and in 2 pairs in L. jocu (short arm of pair 3 and long arm of pair 13). However, in O. chrysurus, the hisDNA H3 sites notably occur in the centromeric and pericentromeric regions of 22 chromosome pairs, except in pairs 11 and 24 (Figure 1), and colocated with the 18S rDNA in pair 2 and with the 5S rDNA sites in pair 21.
Retrotransposons Rex1 and Rex3 were preferentially dispersed in the chromosomes of all species but with notably accumulation in some specific chromosomal regions (Figure 2). These sites are coincident with some previously characterized chromosomal heterochromatic regions (Rocha and Molina 2008).
Discussion
Phylogenetic and Temporal Distribution Patterns of rDNA Genes in Lutjanidae
Cytogenetic traits are significantly more informative when analyzed under a phylogenetic context. In this respect, we analyzed the chromosomal patterns of the studied snappers species in relation to the phylogenetic hypothesis for Atlantic Lutjaninae proposed by Gold et al. (2011), where the 5 species analyzed are included in the clade A and their subclades B and C. Clade B, with an age of approximately 7 myr, includes the species L. synagris, L. analis, O. chrysurus and in a more basal condition, Rhomboplites aurorubens. Clade C, with an age of approximately 4.5 myr includes the species L. alexandrei, L. jocu, and L. griseus (Figure 3). The inclusion of Ocyurus among the species of Lutjanus reinforces the need for synonymization of these genera (Gold et al. 2011). This condition is supported by molecular (Sarver et al. 1996;Gold et al. 2011) and interspecific hybridization data, as well as comparisons between larval phases (Domeier and Clarke 1992; Loftus 1992;Clarke et al. 1997).
Phylogenetic comparisons of ribosomal sites in more Lutjaninae basal clades suggest that a single rDNA locus is an ancestral condition. Indeed, a single large chromosome pair bearing these sequences is an ancient trait found in some more basal species, such as L. sebae (Guo et al. 2011) and L. russelli (Ueno and Ojima 1992), belonging to clade A with divergence estimated at more than 20.7 myr (Gold et al. 2011). Accordingly, single 18S and 5S rDNA sites are conserved in species of clade B, where they are not syntenic and localized among large or small chromosome pairs, with exception of R. aurorubens that exhibits a variant condition, with the colocalization of 18S and 5S rDNA sites in a single chromosome pair (Figure 3). Thus, the evolutionary dynamics of 18S and 5S rDNA sites among the species of the clade B is characterized by a number conservatism, but with some changes in their distribution in the chromosomes.
On the contrary, species from Clade C demonstrate a derived condition, with multiple 18S rDNA sites in their karyotype, thus diverging from the basal pattern of Lutjanidae. Given the estimate of this clade’s origin, this synapomorphy may be at least 4.5 myr. However, the presence of 18S rDNA sites in 2 large chromosome pairs in L. cyanopterus (Costa, unpublished data), whose divergence precedes clade C, suggests that this condition may be even more ancient. A compared analysis between the number of active nucleolar organizing regions (Ag-NORs) and 18S rDNA sites evidence that not all of the rDNA mapped by FISH have a functional activity. So, whereas in L. jocu the 18S rDNA bearing pairs are coincident with the Ag-NORs ones (Rocha and Molina 2008), the other 2 species, L. alexandrei and L. griseus (Nirchio et al. 2008; Rocha and Molina 2008; present study) show a higher number of 18S rDNA sites than Ag-NORs, suggesting some level of gene regulation or evolutionary transitions to extra sites. The increase of the 18S ribosomal genes in L. jocu, L. alexandrei, and L. griseus represents important features considering the small chromosomal diversification of this species. Indeed, the different numbers and positions of rDNA sites in the karyotypes make them effective cytotaxonomic markers for Lutjanidae species (Rocha and Molina 2008).
Concerning 5S rDNA sequences, they are unique and phylogenetically conserved among species of clades A, B, and C (Figure 3). In L. analis, L. synagris, and O. chrysurus, they are located in small chromosome pairs (pairs 14, 21, and 21, respectively), and so being homeologous between the last 2 species. On the other hand, in L. alexandrei and L. jocu, these sites are located in a medium-sized chromosome pair, with also a homeologous condition putatively reported to pair 14.
It has been accumulated evidence on the role of repetitive DNAs in the structural and functional organization of the genome (Liu et al. 2001; Li et al. 2002; Parise-Maltempi et al. 2007), as well as chromosomal rearrangements and karyotypic variations in many organisms (Kidwell 2002), including fish (Jacobina et al. 2012; Molina and Galetti 2002). In Lutjanidae, 5S and 18S rDNAs display non-syntenic arrangements, which is also the most common condition in vertebrates, indicating independent evolution of these loci (Lucchini et al. 1993; Suzuki et al. 1996;Martins and Wasko 2004). Collinear or equilocal arrangements of 5S and 45S rDNA loci are infrequent in fishes (Pendás et al. 1994;Fujiwara et al. 1998; Calado et al. 2014), although occurring in Lutjanidae, Rhomboplites aurorubens (Nirchio et al. 2009). The syntenic association of 18S and 5S sites in species of Perciformes that exhibit conserved karyotypes, such as Lutjanidae and Gerreidae (Calado et al. 2014), suggests that these regions may represent the main source for karyotype diversification in some species.
Inter and Intraspecific Cytogenetic Divergences Among Populations of Atlantic Snappers
Given the symmetrical karyotypes of Lutjanidae species, interpopulation comparisons must emphasize only conspicuous chromosomal divergences and minimize minor variations due to technical artifacts. Comparisons of 18S and 5S rDNA sites distribution between Brazilian and Caribbean L. analis populations do not indicate karyotypic variation, both in the number or location of these genes (Nirchio et al. 2008; Rocha and Molina 2008). On the other hand, chromosomal polymorphism due to centric fusion (2n = 48/46) in a number of L. synagris individuals from Venezuela (Nirchio et al. 2008) was not identified in individuals from Brazilian coast (Rocha and Molina 2008; this study), suggesting some degree of genetic structuring between these populations.
The impossibility in identifying the chromosome pairs in all-acrocentric karyotypes hinders precise population comparisons. However, the marked difference in size between the 5S rDNA-bearing pair in O. chrysurus and L. synagris individuals from Brazilian (this study) and Venezuelan (Nirchio et al. 2008, 2009) populations indicate real divergences between them. Indeed, phylogeographic analyses have demonstrated that both species show significant population structuring in Caribbean region (Karlsson et al. 2009; Saillant et al. 2012), and that O. chrysurus exhibits high population substructuring between this region and Brazilian coast (Vasconcellos et al. 2008). In this case, the cytogenetic divergences can be attributed to Amazon barrier (≈10 m.a), that restricts gene flow among individuals from these regions. In fact, this biogeographic barrier has been related to the cytogenetic divergences between Haemulidae populations (Motta Neto et al. 2012) and sister species of Grammatidae (Molina et al. 2012), also belonging to these 2 Atlantic regions.
HisDNA H3 Distribution in Lutjanidae
Functionally, hisDNA (H1, H2A, H2B, H3, and H4) encodes a family of highly conserved small basic proteins that exhibit heterogeneity in the pattern of genome organization (Sellos et al. 1990;Cabrero et al. 2009; Eirín-López et al. 2009). These proteins play an important role in the structural organization of chromatin, in DNA packaging in the cell nucleus, as well as in the regulation of gene expression (Sellos et al. 1990;Csink and Henikoff 1998; Chioda et al. 2002). Although the distribution of this family in fish chromosomes is still little known, their presence in 1 chromosomal pair seems to be an ancestral condition for these genes, in spite of growing evidence of their evolutionary flexibility. Although primary hisDNA has a single locus in salmonids, some other genes exhibit greater diversity (Connor et al. 1984). Likewise, hisDNA H1 may occupy a single or 2 loci in some fish species (Hashimoto et al. 2011; Lima-Filho et al. 2012).
H3 genes have been more extensively analyzed than other histone genes, and are conservatively distributed in a single locus in chromosomes of L. analis, L. synagris, and L. alexandrei. However, despite being numerically conserved, these genes are apparently located in non-homeologous chromosome pairs, indicating their genomic reorganization. The diversified distribution of H3 hisDNA in 2 loci in L. jocu and in 22 loci in O. chrysurus indicates a derived condition of these genes. The presence of multiple H3 loci in species from clades B and C suggests that the dispersion of these genes were stochastic and independent. The occurrence of multiple H3 loci is compatible with duplication mechanisms in the genome, allowing the diversification, inactivation or even the elimination of gene copies (González-Romero et al. 2008; Eirín-López et al. 2009; Costa et al. 2014). A similar evolutionary process also seems to be involved in the diversification of H2B–H2A histones in the fish Rachycentron canadum (Costa et al. 2014).
Ocyurus chrysurus exhibits an exclusive and surprisingly divergent evolutionary dynamics related to H3 hisDNA. In contrast to the other species, it displays these sequences spread in almost all chromosomes of the karyotype (Figure 1), being colocalized with large centromeric heterochromatic blocks (Rocha and Molina 2008). In these heterochromatin-rich regions, the H3 sequences seem to have undergone an exclusive evolutionary mechanism. Detection of this level of sequence dispersion is rare in fishes, and could be promoted by the action of TEs (Costa et al. 2013; 2014).
In fact, TEs account for a large portion of the eukaryote genome and are considered a dynamic reservoir of sequences responsible for the structural and functional evolution of many genes, acting in epigenetic regulation, recruitment of chromatin remodeling factors, and chromosome structure (Aparicio et al. 2002; Volff et al. 2003;Böhne et al. 2008). Indeed, a number of sequences with noteworthy genomic expansion may be associated with TEs (Costa et al. 2015). However, the reduced colocalization of Rex elements in multiple domains of H3 hisDNA does not support the role of these retrotransposons in the spreading of the hisDNA sites in O. chrysurus. Nevertheless, the action of other TEs in this process cannot be ruled out.
Colocalization of rDNA and hisDNA Sequences
Ribosomal sites associated with heterochromatic regions are well known in fishes, and have been identified in many Perciformes species with conserved karyotypes, such as Haemulidae (Motta Neto et al. 2011a;Motta Neto et al. 2011b) and Lutjanidae (Rocha and Molina 2008;Nirchio et al. 2009). Even though the evolutionary pattern and phylogenetic distribution of ribosomal genes are relatively well known in fishes (Gornung 2013), their colocalization and interaction with other genes is still under study (Costa et al. 2014). In the species analyzed, the mapping of hisDNA H3 sequences and 18S and 5S ribosomal genes indicates a preferably nonsintenic condition among them. However, in O. chrysurus, the expansion of H3 sequences promoted the overlapping of the 18S and 5S rDNA with H3 sequences. In this species, the presence of single, and therefore functional ribosomal sites, clearly demonstrates that this association does not interfere in their functionality. In several animal groups, including fishes, regular interspersions of histone and rDNA genes have been identified (Costa et al. 2015). It has been suggested that the association between the quintet of histones and rRNA genes may represent the largest group of associated repeated genes (Roehrdanz et al. 2010), representing potential regions of chromosomal diversification. Thus, disruptive expansions of repetitive sequences in chromosomes of species that share conserved karyotypes may have significant evolutionary importance. Extending these analyses to other phylogenetic constructions can clarify the selective or neutral role of hisDNA/rDNA associations.
Final Considerations
Extensively conserved karyotypes in some fish groups contrasts with the phyletic diversification in several Perciformes families (Molina 2007). As now evidenced for Lutjanidae, this karyotype conservation (2n = 48 acrocentric chromosomes) can also be significantly maintained at the microstructural level, concerning the ribosomal and histone genes and heterochromatin distribution, even after long periods of diversification (>20 myr). However, some “disturbing evolutionary waves” can break down this “calm sea of karyotypic conservation”. Indeed, evolutionary escapes can randomly occur, such as the dispersion of H3 sequences in O. chrysurus (clade B), or be phylogenetically shared, such as multiple 18S sites in Lutjanus species from clade C. In such a case, it is being evidenced good chromosomal markers reinforcing the close relationship between these species. The implication of the huge genomic expansion of H3 histone sequences and their functionality in O. chrysurus remain unknown, although showing their potential dynamism in stochastic evolutionary processes. The cytogenetic mapping of new repetitive sequences and functional genes in Atlantic snappers will be important to understand the functional interactions of H3/18S rDNA and H3/5S rDNA sequences, the disruptive role of complex heterochromatins and their implication in karyotype evolution.
Funding
National Counsel of Technological and Scientific Development (CNPq), Coordination for the Improvement of Higher Education Personnel (CAPES), Federal University of Rio Grande do Norte and Federal University of São Carlos.
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.hn34m
Acknowledgments
We thank Chico Mendes Institute for Biodiversity Conservation (ICMBio) for permits to collect specimens, and 3 anonymous reviewers for their valuable critiques.
References
- Albig W, Warthorst U, Drabent B, Prats E, Cornudella L, Doenecke D. 2003. Mytilus edulis core histone genes are organized in two clusters devoid of linker histone genes. J Mol Evol. 56:597–606. [DOI] [PubMed] [Google Scholar]
- Andrews MT, Vaughn JC, Perry BA, Bagshaw JC. 1987. Interspersion of histone and 5S RNA genes in Artemia. Gene. 51:61–67. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 297:1301–1310. [DOI] [PubMed] [Google Scholar]
- Arai R. 2011. Fish Karyotypes: a check list. Tokyo: Springer. [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 16:203–215. [DOI] [PubMed] [Google Scholar]
- Cabrero J, López-León MD, Teruel M, Camacho JP. 2009. Chromosome mapping of H3 and H4 histone gene clusters in 35 species of acridid grasshoppers. Chromosome Res. 17:397–404. [DOI] [PubMed] [Google Scholar]
- Calado LL, Bertollo LA, Cioffi MB, Costa GW, Jacobina UP, Molina WF. 2014. Evolutionary dynamics of rDNA genes on chromosomes of the Eucinostomus fishes: cytotaxonomic and karyoevolutive implications. Genet Mol Res. 13:9951–9959. [DOI] [PubMed] [Google Scholar]
- Chioda M, Eskeland R, Thompson EM. 2002. Histone gene complement, variant expression, and mRNA processing in a urochordate Oikopleura dioica that undergoes extensive polyploidization. Mol Biol Evol. 19:2247–2260. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Martins C, Bertollo LA. 2010. Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol Biol. 10:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ME, Domeier ML, Laroche WA. 1997Development of larvae and juveniles of the mutton snapper (Lutjanus analis), lane snapper (Lutjanus synagris) and yellowtail snapper (Lutjanus chrysurus). B Mar Sci. 61:511–537. [Google Scholar]
- Connor W, Mezquita J, Winkfein RJ, States JC, Dixon GH. 1984. Organization of the Histone Genes in the Rainbow-Trout (Salmo gairdnerii). J Mol Evol. 20:227–235. [DOI] [PubMed] [Google Scholar]
- Costa GW, Cioffi MB, Bertollo LAC, Molina WF. 2013. Transposable elements in fish chromosomes: a study in the marine cobia species. Cytogenet Genome Res. 141:126–132. [DOI] [PubMed] [Google Scholar]
- Costa GWWF, Cioffi MB, Bertollo LAC, Molina WF. 2014. Unusual dispersion of histone repeats on the whole chromosomal complement and their colocalization with ribosomal genes in Rachycentron canadum (Rachycentridae, Perciformes). Cytogenet Genome Res. 144:1–6. [DOI] [PubMed] [Google Scholar]
- Costa GWWF, Cioffi MB, Bertollo LAC, Molina WF. 2015. Structurally complex organization of repetitive DNAs in the genome of Cobia (Rachycentron canadum). Zebrafish. 12:215–220. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. 1998. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 14:200–204. [DOI] [PubMed] [Google Scholar]
- Lucchini SD, Nardi I, Barsacchi G, Batistoni R, Andronico F. 1993. Molecular cytogenetics of the ribosomal (18S + 28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome. 36:762–773. [DOI] [PubMed] [Google Scholar]
- Domeier ML, Clarke ME. (1992) A Laboratory Produced Hybrid between Lutjanus synagris and Ocyurus chrysurus and a Probable Hybrid between L. griseus and O. chrysurus (Perciformes, Lutjanidae). B Mar Sci. 50:501–507. [Google Scholar]
- Eirín-López JM, Romero RG, Dryhurst D, Méndez J, Ausió J. (2009) Long-Term evolution of histone families: Old notions and new insights into their mechanisms of diversification across eukaryotes. In: Pontarotti P, editor. Evolutionary biology: concept, modeling and application. Berlin: Springer; p. 139–162. [Google Scholar]
- España HP. (2003) Ecological importance of snappers in the stability of modeled coastal ecosystems. Ecol Model. 168:13–24. [Google Scholar]
- Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida MC. 1998. Chromosomal localization and heterochromatin association of ribosomal RNA gene loci and silver-stained nucleolar organizer regions in salmonid fishes. Chromosome Res. 6:463–471. [DOI] [PubMed] [Google Scholar]
- Giribert G, Distel D. 2003. Bivalve phylogeny and molecular data. In: Lydeard C, Lindberg DR, editors. Molecular systematics and phylogeography of molluscks. Washington: (DC: ): Smithsonian Books; p. 45–90. [Google Scholar]
- Gold JR, Li YC, Shipley NS, Powers PK. 1990. Improved methods for working with fish chromosomes with a review of metaphase chromosome banding. J Fish Biol. 37:563–575. [Google Scholar]
- Gold JR, Voelker G, Renshaw MA. 2011. Phylogenetic relationships of tropical western Atlantic snappers in subfamily Lutjaninae (Lutjanidae: Perciformes) inferred from mitochondrial DNA sequences. Biol J Linn Soc. 102:915–929. [Google Scholar]
- González-Romero R, Ausió J, Méndez J, Eirín-López JM. 2008. Early evolution of histone genes: prevalence of an ‘orphon’ H1 lineage in protostomes and birth-and-death process in the H2A family. J Mol Evol. 66:505–518. [DOI] [PubMed] [Google Scholar]
- Gornung E. 2013. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet Genome Res. 141:90–102. [DOI] [PubMed] [Google Scholar]
- Guo ML, You XX, Su YQ, Ding SX, Wang J. 2011. Studies on chromosome karyotype, Ag-NORs and C-banding patterns of Lutjanus sebae. J Trop Oceanogr. 30:91–95. [Google Scholar]
- Hashimoto DT, Ferguson-Smith MA, Rens W, Foresti F, Porto-Foresti F. 2011. Chromosome mapping of H1 histone and 5S rRNA gene clusters in three species of Astyanax (Teleostei, Characiformes). Cytogenet Genome Res. 134:64–71. [DOI] [PubMed] [Google Scholar]
- Jacobina UP, Vicari MR, Bertollo LA, Molina WF. 2012. Discriminatory profile of rDNA sites and trend for acrocentric chromosome formation in the genus Trachinotus Lacépède, 1801 (Perciformes, Carangidae). Comp Cytogenet. 6:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Wolf YI, Koonin EV. 2002. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson S, Saillant E, Gold JR. 2009. Population structure and genetic variation of lane snapper (Lutjanus synagris) in the northern Gulf of Mexico. Mar Biol. 156:1841–1855. [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 115:49–63. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220. [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 11:2453–2465. [DOI] [PubMed] [Google Scholar]
- Lima-Filho PA, Cioffi MD, Bertollo LAC, Molina WF. 2012. Chromosomal and morphological divergences in Atlantic populations of the frillfin goby Bathygobius soporator (Gobiidae, Perciformes). J Exp Mar Biol Ecol. 434:63–70. [Google Scholar]
- Liu D, Mack A, Wang RC, Galli M, Belk J, Ketpura NI, Crawford NM. 2001. Functional dissection of the cis-acting sequences of the arabidopsis transposable element Tag1 reveals dissimilar subterminal sequence and minimal spacing requirements for transposition. Genetics. 157:817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus WF. 1992. Lutjanus ambiguus (Poey), a Natural Intergeneric Hybrid of Ocyurus chrysurus (Bloch) and Lutjanus synagris (Linnaeus). B Mar Sci. 50:489–500. [Google Scholar]
- Lorenzen K, Leber KM, Blankenship HL. 2010. Responsible approach to marine stock enhancement: An update. Rev Fish Sci. 18:189–210. [Google Scholar]
- Martins C, Wasko AP. 2004. Organization and evolution of 5S ribosomal DNA in the fish genome. In: Williams CR, editor. Focus on genome research. Hauppauge: (NY: ): Nova Science Publishers; p. 335–363. [Google Scholar]
- Molina WF. 2001. An alternative method for mitotic stimulation in fish cytogenetics. Chromosome Sci. 5:149–152. [Google Scholar]
- Molina WF. 2007. Chromosome changes and stasis in marine fish groups. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG, editors. Fish Cytogenet. Boca Raton: (FL: ): CRC Press; p. 69–110. [Google Scholar]
- Molina WF, Alves DE, Araújo WC, Martinez PA, Silva MF, Costa GW. 2010. Performance of human immunostimulating agents in the improvement of fish cytogenetic preparations. Genet Mol Res. 9:1807–1814. [DOI] [PubMed] [Google Scholar]
- Molina WF, da Costa GWWF, Cioffi MB, Bertollo LAC. 2012. Chromosomal differentiation and speciation in sister-species of Grammatidae (Perciformes) from the Western Atlantic. Helgol Mar Res. 66:363–370. [Google Scholar]
- Molina WF, Galetti PM. 2002. Robertsonian rearrangements in the reef fish Chromis (Perciformes, Pomacentridae) involving chromosomes bearing 5s rRNA genes. Genet Mol Biol. 25:373–377. [Google Scholar]
- Motta Neto CC, Cioffi MB, Bertollo LAC, Molina WF. 2011a. Extensive chromosomal homologies and evidence of karyotypic stasis in Atlantic grunts of the genus Haemulon (Perciformes). J Exp Mar Biol Ecol. 401:75–79. [Google Scholar]
- Motta Neto CC, Cioffi MB, Bertollo LAC, Molina WF. 2011b. Molecular cytogenetic analysis of Haemulidae fish evolutionary conservation. J Exp Mar Biol Ecol. 407:97–100. [Google Scholar]
- Motta Neto CC, Lima-Filho PA, Araújo WC, Bertollo LAC, Molina WF. 2012. Differentiated evolutionary pathways in Haemulidae (Perciformes): karyotype stasis versus morphological differentiation. Rev Fish Biol Fisher. 22:457–465. [Google Scholar]
- Moura RL, Lindeman KC. 2007. A new species of snapper (Perciformes: Lutjanidae) from Brazil, with comments on the distribution of Lutjanus griseus and Lutjanus apodus. Zootaxa. 1422:31–43. [Google Scholar]
- Nelson JS. 2006. Fishes of the world. 4th edn Hoboken: (NJ: ): John Wiley & Sons, Inc. [Google Scholar]
- Nirchio M, Oliveira C, Ferreira DC, Rondon R, Pérez JE, Hett AK, Rossi AR, Sola L. 2009. Cytogenetic characterization of Rhomboplites aurorubens and Ocyurus chrysurus, two monotypic genera of Lutjaninae from Cubagua Island, Venezuela, with a review of the cytogenetics of Lutjanidae (Teleostei: Perciformes). Neotrop Ichthyol. 7:587–594. [Google Scholar]
- Nirchio M, Rondon R, Oliveira C, Ferreira IA, Martins C, Perez J, Sola L, Rossi AR. 2008. Cytogenetic studies in three species of Lutjanus (Perciformes: Lutjanidae: Lutjaninae) from the Isla Margarita, Venezuela. Neotrop Ichthyol. 6:101–108. [Google Scholar]
- Parise-Maltempi PP, Martins C, Oliveira C, Foresti F. 2007. Identification of a new repetitive element in the sex chromosomes of Leporinus elongatus (Teleostei: Characiformes: Anostomidae): new insights into the sex chromosomes of Leporinus. Cytogenet Genome Res. 116:218–223. [DOI] [PubMed] [Google Scholar]
- Pendás AM, Moran P, Freije JP, Garcia-Vazquez E. 1994. Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet Cell Genet. 67:31–36. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 83:2934–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunath P, Prasad R. 1980. Chromosomes of six marine percoids from the Indian Sea. Indian Biol. 11:9–12. [Google Scholar]
- Resende SM, Ferreira BP, Thierry F. 2003. A pesca de lutjanídeos no nordeste do Brasil: Histórico das pescarias, características das espécies e relevância para o manejo. Bol Téc Cepene. 11:257–270. [Google Scholar]
- Rocha EC, Molina WF. 2008. Cytogenetic analysis in western Atlantic snappers (Perciformes, Lutjanidae). Genet Mol Biol 31:461–467. [Google Scholar]
- Roehrdanz R, Heilmann L, Senechal P, Sears S, Evenson P. 2010. Histone and ribosomal RNA repetitive gene clusters of the boll weevil are linked in a tandem array. Insect Mol Biol. 19:463–471. [DOI] [PubMed] [Google Scholar]
- Saillant EA, Leclercq E, Bardon-Albaret A, Sarkisian B, et al. 2013. Development of aquaculture of the red snapper Lutjanus campechanus: research on larval nutrition. Proc 65th Gulf Caribb Fish Inst. 65–72: 352–355. [Google Scholar]
- Saillant EA, Renshaw MA, Cummings NJ, Gold JR. 2012. Conservation genetics and management of yellowtail snapper, Ocyurus chrysurus, in the US Caribbean and South Florida. Fish Manag Ecol. 19:301–312. [Google Scholar]
- Sarver SK, Freshwater DW, Walsh P. 1996. Phylogenetic relationships of western Atlantic snappers (family Lutjanidae) based on mitochondrial DNA sequences. Copeia. 1996:715–721. [Google Scholar]
- Sellos D, Krawetz SA, Dixon GH. 1990. Organization and complete nucleotide sequence of the core-histone-gene cluster of the annelid Platynereis dumerilii. Eur J Biochem. 190:21–29. [DOI] [PubMed] [Google Scholar]
- Silva DM, Pansonato-Alves JC, Utsunomia R, Daniel SN, Hashimoto DT, Oliveira C, Porto-Foresti F, Foresti F. 2013. Chromosomal organization of repetitive DNA sequences in Astyanax bockmanni (Teleostei, Characiformes): dispersive location, association and co-localization in the genome. Genetica. 141:329–336. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Sakurai S, Matsuda Y. 1996. Rat 5S rDNA spacer sequences and chromosomal assignment of the genes to the extreme terminal region of chromosome 19. Cytogenet Cell Genet. 72:1–4. [DOI] [PubMed] [Google Scholar]
- Ueno K, Ojima Y. 1992. Notes on the chromosomes of Girella melanichthys and Lutjanus russelli (Pisces, Perciformes). Chromosome Inform Serv. 52:3–5. [Google Scholar]
- Valente GT, Mazzuchelli J, Ferreira IA, Poletto AB, Fantinatti BEA, Martins C. 2011. Cytogenetic mapping of the retroelements Rex1, Rex3 and Rex6 among cichlid fish: new insights on the chromosomal distribution of transposable elements. Cytogenet Genome Res. 133:34–42. [DOI] [PubMed] [Google Scholar]
- Vasconcellos AV, Vianna P, Paiva PC, Schama R, Sole-Cava A. 2008. Genetic and morphometric differences between yellowtail snapper (Ocyurus chrysurus, Lutjanidae) populations of the tropical West Atlantic. Genet Mol Biol. 31:308–316. [Google Scholar]
- Volff JN, Bouneau L, Ozouf-Costaz C, Fischer C. 2003. Diversity of retrotransposable elements in compact pufferfish genomes. Trends Genet. 19:674–678. [DOI] [PubMed] [Google Scholar]
- Volff JN, Körting C, Schartl M. 2000. Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol Biol Evol. 17:1673–1684. [DOI] [PubMed] [Google Scholar]
- Volff JN, Körting C, Sweeney K, Schartl M. 1999. The non-LTR retrotransposon Rex3 from the fish Xiphophorus is widespread among teleosts. Mol Biol Evol. 16:1427–1438. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press Inc; p. 315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.hn34m