Abstract
MicroRNA (miR)-199b-5p has been reported to have a critical role in various types of malignancy. However, the exact function miR-199b-5p in renal cancer remains to be fully elucidated. The present study aimed to detect the expression levels of miR-199b-5p in renal cell carcinoma (RCC) tissues and RCC cell lines, and investigated the effect of miR-199b-5p in vitro with Cell Counting Kit-8, MTT, scratch wound, Transwell and flow cytometric assays. The results demonstrated that the expression levels of miR-199b-5p were significantly downregulated in RCC tissues and cell lines compared with those in paired adjacent normal renal tissues and a reference cell line, respectively. Downregulation of miR-199b-5p by transfection with a synthetic inhibitor promoted cellular proliferation and migration, while reducing the apoptotic rate, indicating that miR-199b-5p may serve as a tumor suppressor in RCC. Further study is required to identify target genes of miR-199b-5p to elucidate the mechanisms underlying the role of miR-199b-5p in the occurrence and development of RCC.
Keywords: microRNA, microRNA-199b-5p, tumor suppressor, renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) accounts for 3% of all malignant tumors; ~270,000 new cases are diagnosed and ~116,000 RCC-associated mortalities occur worldwide per annum (1,2). Furthermore, 25–30% of patients have already developed metastatic disease at the time-point of diagnosis (3). The 5-year survival rate of RCC patients at stage I is ~95%, but that of patients at stage IV is only 20% (4). Thus, early detection and treatment of RCC are important. However, RCC is resistant to conventional chemotherapy and radiotherapy, and there is a lack of effective biomarkers for screening (4,5). Therefore, novel therapeutic methods and biomarkers should be developed to improve RCC diagnosis and treatment.
MicroRNAs (miRNAs/miRs), a class of RNAs of ~20 nucleotides in length, regulate gene expression at the post-transcriptional level and are involved not only in normal biological processes, but also in tumorigenesis (6). Increasing evidence has indicated that the expression of various miRNAs is aberrantly regulated in different cancer types (7). Previous studies have demonstrated that certain miRNAs are involved in RCC. However, the role of miR199b-5p in RCC has remained elusive. In the present study, a series of experiments was performed to investigate the effect of miR-199b-5p on RCC cell lines.
Materials and methods
Specimens and cell lines
RCC tissues and paired adjacent normal tissues (located 2.0 cm outside of the visible RCC lesions) were collected from 42 patients undergoing surgery at Peking University Shenzhen Hospital (Shenzhen, China). None of the patients had received any anti-cancer treatment prior to the surgery. All selected cases have been pathologically diagnosed. All of above tissues were stored at −80°C until the RNA was extracted. Patient characteristics are presented in Table I.
Table I.
Clinicopathological characteristics of patients with RCC.
| Characteristic | Number of cases |
|---|---|
| Mean age, range (year) | 53 (27–72) |
| Sex (male/female) | 33/9 |
| Histological type (clear cell/papillary) | 35/7 |
| Fuhrman grade (I/II/III/IV) | 29/10/2/1 |
| AJCC clinical stage (I/II/III+IV) | 32/9/1 |
RCC, renal cell carcinoma; AJCC, American Joint Committee on Cancer.
The ACHN and 786-O RCC cell lines and the 293T reference cell line were obtained from the American Type Culture Collection (Manassas, VA, USA) and the Type Culture Collection of the Chinese Academy of (Shanghai, China), respectively. The human RCC cell lines (786-O and ACHN) were originally obtained from the American Type Culture Collection (Manassas, VA, USA). The human embryo kidney cell line 293T (293T) was purchased from the Type Culture Collection of the Chinese Academy of Medical Sciences (Shanghai, China). All of them were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), with 10% fetal bovine serum (FBS; GE Healthcare, Little Chalfont, UK), 1% glutamine and 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin; Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in an incubator at 37% in a humidified atmosphere with 5% CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells was isolated with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and purified with the RNeasy Maxi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After determining the RNA concentration with a NanoDrop 2000c (Thermo Fisher Scientific, Inc.), the miScript II RT kit (Qiagen) was applied to reverse-transcribe miRNA to complementary (c)DNA. The expression levels of miR-199b-5p were measured with a miScript SYBR® Green PCR Kit (Qiagen) by real-time qPCR on the Roche Lightcycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland). The 10-µl reaction mixture contained 5 µl 2X QuantiTect SYBR Green PCR Master mix, 3.7 µl RNase-free water, 1 µl cDNA template, 0.4 µl specific miRNA primer and 10X miScript Universal Primer. The forward primer of miR-199b-5p had the sequence 5′-CCCAGUGUUUAGACUAUCUGUUC-3′ and the reverse primer was a universal primer, provided with the miScript SYBR® green PCR Kit. U6 was used as an internal control. The forward primer of U6 was 5′-CTCGCTTCGGCAGCACA-3′ and the reverse primer was 5′-ACGCTTCACGAATTTGCGT-3′. The reaction conditions were as follows: 95°C for 2 min and 40 cycles of 95°C for 10 sec, 55°C for 30 sec and at 72°C for 30 sec. The expression levels of miR-199b-5p in tissues and cell lines were analyzed by the ΔΔCq method (8).
Cell transfection
According to the manufacturer's protocol, the expression levels of miR-199b-5p in ACHN and 786-O cells were transfected with 5 ml miR-199b-5p inhibitor (5′-GAACAGAUAGUCUAAACACUGGG-3′; Shanghai GenePharma, Co., Ltd., Shanghai, China) or inhibitor negative control (5′-CAGUACUUUUGUGUAGUACAA-3′; Shanghai GenePharma, Co., Ltd.) by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM® I Reduced Serum Medium (Gibco; Thermo Fisher Scientific, Inc.). The efficiency of transfection was detected using RT-qPCR after 24 h.
Cell Counting Kit-8 (CCK-8) assay. The proliferation ability of ACHN and 786-O cells was assessed using a CCK-8 assay (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. ACHN or 786-O cells were respectively seeded in each well of a 96-well plate at 5×103 cells/well, cultured for 24 h and then transfected with miR-199b-5p inhibitor or inhibitor negative control (NCin). At 0, 24, 48 and 72, 10 µl CCK-8 stain was added to each well, followed by further culture for 30 min in the dark at 37°C in a humidified atmosphere with 5% CO2 prior to measurement of the optical density (OD) value with an ELISA microplate reader at a wavelength of 450 nm (with 620 nm as the reference wave length).
MTT assay
The number of viable ACHN and 786-O cells was detected using an MTT assay. ACHN or 786-O cells were respectively seeded in each well of a 96-well plate at 5×103 cells/well, cultured for 24 h and then transfected with miR-199b-5p inhibitor or NCin by using Lipofectamine® 2000. After 4 days of incubation, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well of 96-well plate. After culture for 4 h, the medium was discarded and 100 µl dimethylsulfoxide (Sigma-Aldrich; Merck KGaA) was added to each well, followed by incubation under exclusion of light with agitation at room temperature for 10 min. Subsequently, an ELISA microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) was used to measure the OD value of each well at a wavelength of 595 nm (with 620 nm as the reference wavelength).
Transwell migration and invasion assay
The migration and invasion ability of the ACHN and 786-O cells in vitro was measured using a Transwell assay. The Transwell chambers (pore size, 8 µm; cat. no. 3422; BD Biosciences, Franklin Lakes, NJ, USA) with Matrigel® were applied to evaluate the invasion ability, while Transwell chambers without Matrigel® were applied to evaluate the migration ability. After transfection for 24 h, ~2×104 ACHN or 786-O cells were added into each of the upper chambers with serum-free medium, while DMEM with 10% FBS was added to the lower chamber. The chambers were incubated for 48 h at 37°C, and subsequently, the cells that had transgressed through the filter/membrane on the lower side were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet at room temperature for 25 min. The cells in the bottom of the chamber were then counted using a microscope (magnification, ×100).
Scratch wound assay
A scratch wound assay was used to assess the migration ability of the 786O and ACHN cells in vitro. The cells were seeded in a 6-well plate at 1×106 cells/well. After 24 h, they were transfected with miR-199b-5p inhibitor and NCin by using Lipofectamine® 2000 for 24 h, and a vertical line was scratched with a sterile 1-ml pipette tip. Images of the scratches were respectively captured under a microscope (magnification, ×100; Optical digital microscope; Olympus Corporation, Tokyo, Japan) at 0 and 12 h.
Flow cytometric assay
The apoptotic rates of ACHN and 786-O cells in vitro were measured by a flow cytometric assay. Following the manufacturer's protocols, ACHN or 786-O cells were incubated in each well of a 6-well plate at a concentration of 1×106 cells/well and transfected with miR-199b-5p inhibitor or NCin. After 24 h of incubation, the ACHN or 786-O cells were collected, washed twice with cold PBS and then resuspended in a flow cytometry tube in 100 µl 1X binding buffer. Next, 5 µl Annexin V-fluorescein isothiocyanate (Invitrogen; Thermo Fisher Scientific, Inc.) and 5 µl propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.) were added to each sample, followed by incubation under the exclusion of light at room temperature for 15 min. Finally, after addition of 400 µl 1X binding buffer to each tube, the apoptotic rate of the ACHN or 786-O cells was analyzed by flow cytometry (EPICS Xl-4; Beckman-Coulter, Brea, CA, USA). All assays were repeated at least 3 times.
Statistical analysis
Values are expressed as the mean ± standard error. Differences between pairs of groups were analyzed by using Student's t-test, while the paired t-test was used to compare the expression levels of miR-181a-5p in corresponding tumor/normal tissues. Prior to the t-test, a normality test had been performed to confirm the normal distribution of the data. Comparisons between cell lines were performed using one way analysis of variance followed by a Tukey's post-hoc test. The SPSS 23.0 statistical software package (IBM Corp., Armonk, NY, USA) was applied for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-199b-5p is downregulated in RCC tissues and cell lines
The ratios of the expression of miR-199b-5p in RCC tissues vs. adjacent normal tissues are presented in Fig. 1A. As displayed in Fig. 1B the expression levels of miR-199b-5p in RCC tissues were significantly lower than those in adjacent normal tissues (1.000±0.257 vs. 3.572±0.729; P<0.05). The results on renal cell lines suggested that the relative expression levels of miR-199b-5p were higher in 293T (1.000±0.025) than those in 786-O (0.771±0.064; P<0.05) and ACHN cells (0.667±0.063; P<0.01; Fig. 2).
Figure 1.
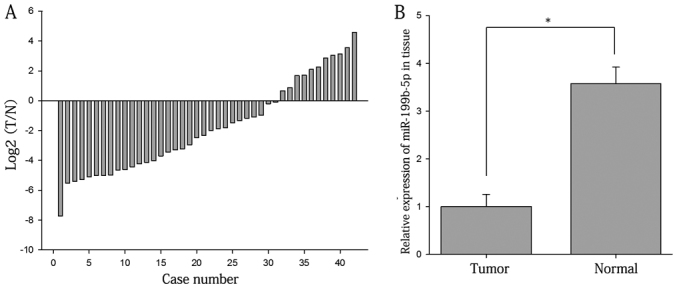
Expression levels of miR-199b-5p in 42 paired tissue samples. (A) Log2 ratios of miR-199b-5p expression in 42 paired T and N tissues. (B) Relative expression of miR-199b-5p in T and N tissues. *P<0.05. miR, microRNA; T, RCC tissues; N, normal renal tissues.
Figure 2.
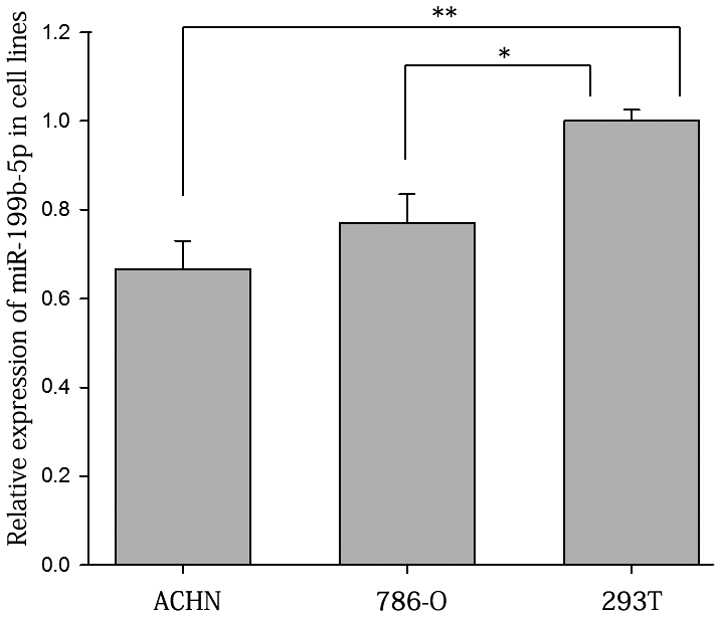
Expression of miR-199b-5p in the ACHN and 786-O renal cell carcinoma cell lines and a reference cell line. *P<0.05, **P<0.01. miR, microRNA.
Cell transfection efficiency
RT-qPCR was used to measure the knockdown efficiency of miR-199b-5p inhibitor compared with NCin. The results indicated that after transfection with miR-199b-5p inhibitor for 24 h, the expression levels of miR-199b-5p in 786-O and ACHN cells were decreased to 60 and 21%, respectively, of those in the NCin-transfected group (P<0.01; Fig. 3).
Figure 3.
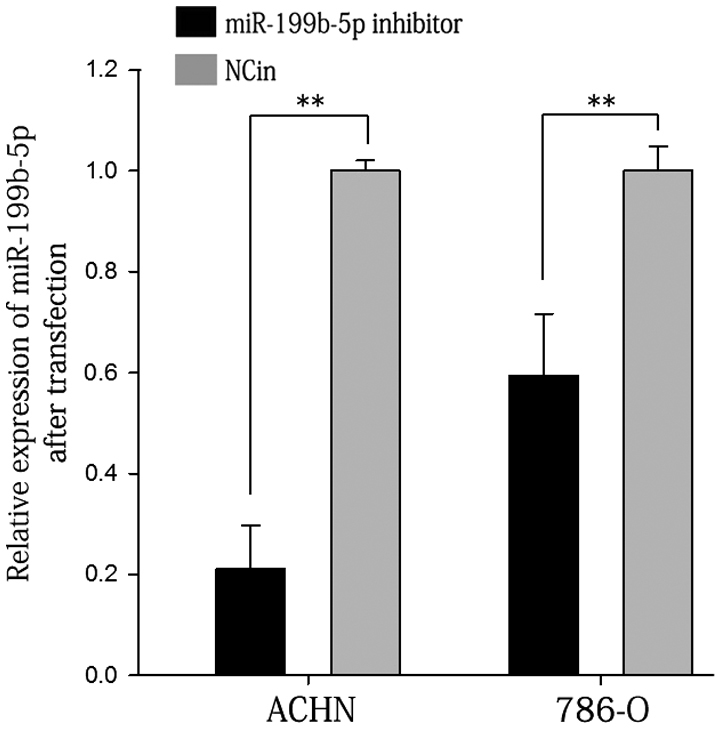
Relative expression of miR-199b-5p after transfection into ACHN and 786-O cells. **P<0.01. miR, microRNA; NCin, negative control of inhibitor.
Inhibition of miR-199b-5p promotes ACHN- and 786-O-cell proliferation
The proliferation ability of the ACHN and 786-O cells was assessed with a CCK-8 assay. The results suggested that the inhibition of miR-199b-5p enhanced the proliferation of the ACHN and 786-O cells. At 24, 48 and 72 h of incubation, respectively, the proliferation rate of the ACHN cells transfected with miR-199b-5p inhibitor was increased by 15.42, 11.80 and 13.47% of that of the control (Fig. 4A), and that of 786-O cells was increased by 17.21, 54.95 and 69.49% (Fig. 4B; P<0.05 or <0.01 for all).
Figure 4.
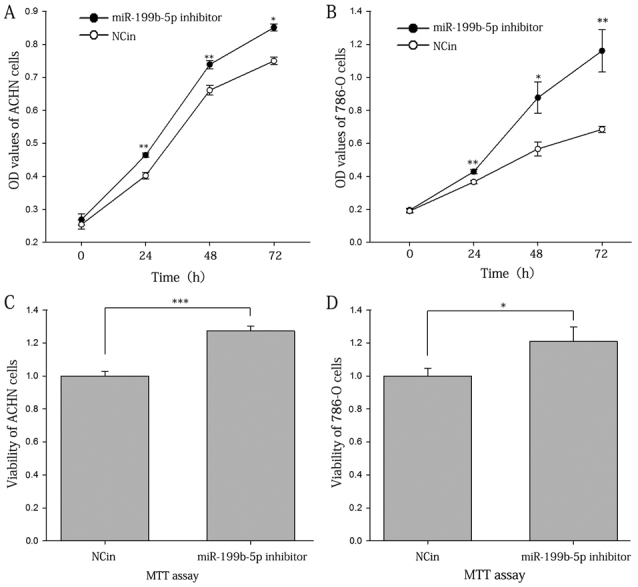
Proliferation of ACHN and 786-O cells transfected with miR-199b-5p inhibitor vs. NCin. (A and B) OD values of ACHN and 786-O cells, respectively, determined by a Cell Counting Kit-8. (C and D) Viability of ACHN and 786-O cells, respectively, as determined by an MTT assay. *P<0.05, **P<0.01, ***P<0.001. OD, optical density; miR, microRNA; NCin, negative control inhibitor.
The number of viable cells was also determined with an MTT assay. The results suggested that after 4 days of incubation, the viability of ACHN and 786-O cells transfected with miR-199b-5p inhibitor were 1.3 and 1.2 times increased compared with that in the NCin-transfected group (P<0.001 and P<0.05; Fig. 4C and D, respectively).
Inhibition of miR-199b-5p promotes ACHN and 786-O cell motility
In order to assess the effect of miR-199b-5p in the motility of ACHN and 786-O cells, a Transwell assay and a scratch wound assay were performed. As presented in Fig. 5A and B, the results of the Transwell migration assay indicated that the migratory ability of ACHN cells transfected with miR-199b-5p inhibitor was increased by 101.39% compared with that in the NCin-transfected group (P<0.001). Furthermore, the migratory ability of 786-O cells transfected with miR-199b-5p inhibitor was increased by 73.60% compared with that in the NCin-transfected group (P<0.01; Fig. 5C and D). The Transwell invasion assay indicated that the invasive ability of 786-O cells was increased by 78.45% following transfection with miR-199b-5p inhibitor (P<0.01; Fig. 5C and D); however, the effect of miR-199b-5p inhibitor on the invasion of ACHN cells was not significant (Fig. 5A and B).
Figure 5.
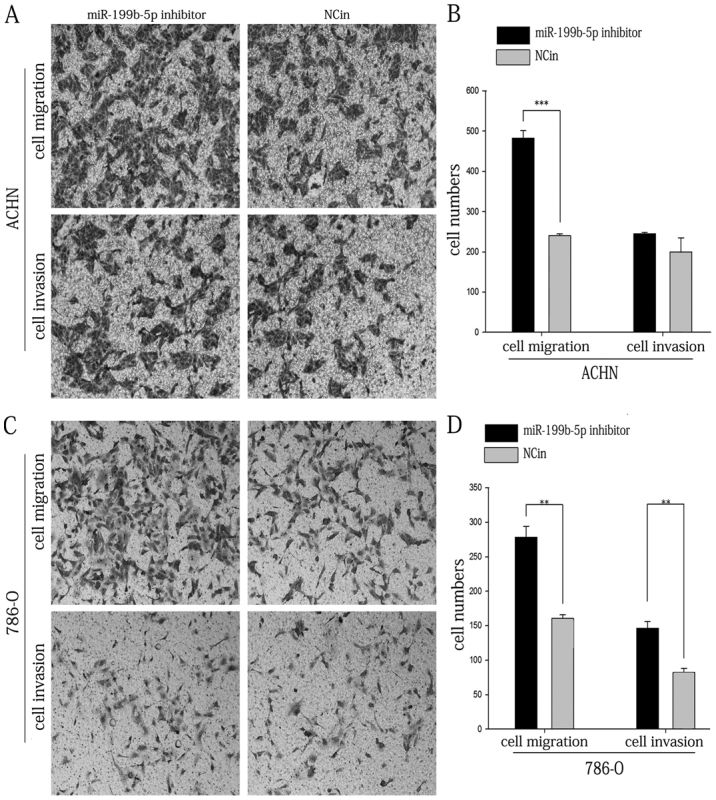
Transwell assay of ACHN and 786-O cells. (A) Representative images of the lower side of the Transwell membranes with invaded and migrated ACHN cells. (B) The numbers of migratory and invasive ACHN cells were quantified. (C) Representative images of the lower side of the Transwell membranes with invaded and migrated 786-O cells (magnification, ×100). (D) The numbers of migratory and invasive 786-O cells were quantified. **P<0.01, ***P<0.001. miR, microRNA; NCin, negative control inhibitor.
The results of the scratch wound assay demonstrated that the migratory ability of ACHN cells transfected with miR-199b-5p inhibitor was increased by 65.00% compared with that in the NCin-transfected group (P<0.001; Fig. 6A and B). Furthermore, 786-O cells exhibited a 213.30% increase in cell migration after transfection with the inhibitor (P<0.001; Fig. 6C and D).
Figure 6.
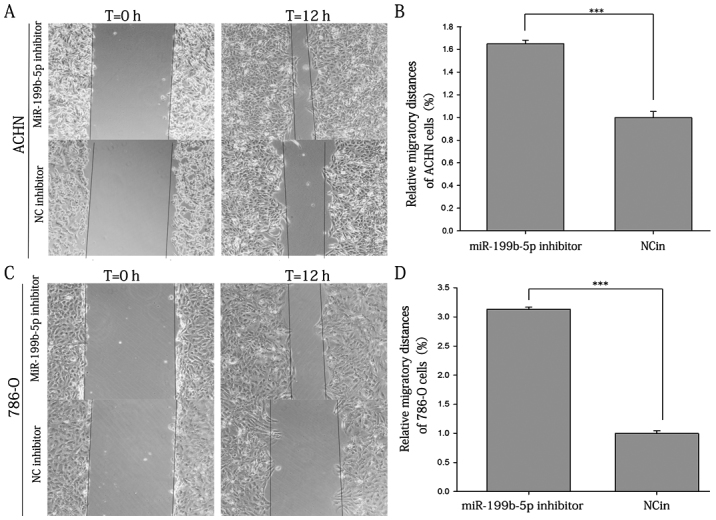
Scratch wound assay of ACHN and 786-O cells. (A) Representative images of ACHN cell migration. (B) Migration distance of ACHN cells transfected with miR-199b-5p inhibitor vs. NCin. (C) Representative images of 786-O cell migration (magnification, ×100). (D) Migration distance of 786-O cells transfected with miR-199b-5p inhibitor vs. NCin. ***P<0.001. miR, microRNA; NCin, negative control inhibitor.
Inhibition of miR-199b-5p reduces the apoptotic rate of ACHN and 786-O cells
A flow cytometric assay was applied to analyze the effect of miR-199b-5p inhibitor on the apoptotic rate of RCC cells. The results indicated that the apoptotic rate of ACHN cells transfected with miR-199b-5p inhibitor or NC in was 13.267±0.433 vs. 19.133±1.161% (P<0.01), while that of 786-O cells was 13.167±0.353 vs. 17.367±0.536% in the miR-199b-5p inhibitor and NCin group, respectively (P<0.01; Fig. 7). Propidium iodide was used to distinguish between viable early cells and necrotic or late apoptotic cells. Therefore, the cells in the upper right quadrant contains necrotic cells and the population in the lower right quadrant of the flow cytometry dot plots represents the early apoptotic cells. The aforementioned data were the average result of three replicates of experimental data. The graph that was selected was typically represented. The above results indicated that inhibition of miR-199b-5p reduced the apoptotic rate of ACHN and 786-O cells in vitro.
Figure 7.
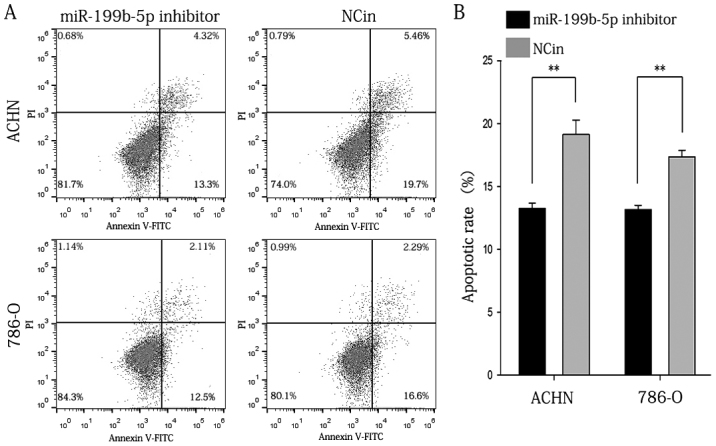
Flow cytometric analysis of apoptosis. (A) Representative flow cytometry dot plots. (B) Apoptotic rate of ACHN and 786-O cells transfected with miR-199b-5p inhibitor compared with that of cells transfected with NCin. The above data were the average result of three replicates of experimental data. The graph chosen was typically represented. **P<0.01. NCin, negative control inhibitor; miR, microRNA; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Discussion
The molecular mechanisms of tumorigenesis and tumor development have remained to be fully elucidated. However, numerous tumor suppressor genes and oncogenes are known to be involved in biological processes of tumorigenesis and tumor development, including the dysregulation of cell proliferation and apoptosis. According to a growing number of studies, certain miRNAs have important functions in numerous types of malignant tumor by regulating the expression of their target genes (9). In malignant tumor cells, the expression levels of certain miRNAs are altered (10). miRNAs are a class of small non-coding and evolutionarily conserved RNAs of 19–25 nucleotides in length, which regulate mRNA expression at the post-transcriptional level (11). According to statistics, ~1,500 miRNA sequences have been identified, and >50% of the genes in the human genome were revealed to be targets of miRNAs (12). This provides a theoretical basis for the potential of miRNAs for use in tumor diagnosis, treatment and prognosis.
A number of studies have suggested that miRNAs have a crucial role in the occurrence and development of malignances (13). For instance, Won et al (14) suggested that miR-199b-5p is involved in Notch signaling pathway in osteosarcoma. Joshi et al (15) reported that low expression levels of miR-199b were associated with imatinib drug resistance in 9q34.1-deleted breakpoint cluster region protein/Abelson murine leukemia viral oncogene homolog 1-positive chronic myeloid leukemia patients. In addition, a study by Fang et al (16) demonstrated that downregulation of miR-199b-5p is correlated with aggressive clinical characteristics of breast cancer. Furthermore, growing evidence has indicated that miR-199b-5p acts as a biomarker in various cancer types, including acute myeloid leukemia (AML) and hepatocellular carcinoma (17,18). Furthermore, a study by Favreau et al (17) indicated that downregulation of miR-199b is correlated with a worse overall survival of AML patients and appears to be a promising prognostic marker for the French-American-British M5 subtype. Apart from the above studies, abnormal expression levels of miR-199b has been reported in other tumor types, including ovarian cancer (19), prostate cancer (20), human osteosarcoma (21) and medulloblastoma (22), while it has largely remained to be elucidated in RCC.
In the present study, the expression levels of miR-199b-5p in RCC tissues and cell lines were quantified by RT-qPCR, revealing that miR-199b-5p was downregulated in RCC. As presented in Fig. 1A, the expression of miR-199b was still upregulated in certain patients; however, this may be due to instrumental measurement errors and the small quantity of specimens used. The present study assessed the function of miR-199b-5p in RCC in vitro by performing a CCK-8, MTT assay, scratch wound, Transwell and flow cytometric assays, which indicated that after transfection with miR-199b-5p inhibitor, RCC cells present with increased cellular proliferation, migration and invasion, but less cellular apoptosis compared with that in the negative control group. The present results demonstrate that miR-199b-5p may serve as a tumor suppressor in RCC by regulating various biological processes, including cell proliferation, migration, invasion and cell apoptosis. It is undeniable that more in-depth experiments, including those aiming to identify cell migration markers as well as specific analysis of cellular key proteins, are lacking in the present study. For example, the relationship between tumor stage and the down-regulation of miRNA-199b-5p may be better assessed by increasing the sample size and analyzing the samples in greater detail. The specific molecular mechanisms of the regulatory effects of miR-199b-5p still require further study. For instance, a subcutaneous tumorigenesis test (23,24) in animals may further verify the tumor biological function of miRNA in vivo in the future. However, Brodaczewska et al (25) demonstrated that in vitro RCC cell lines may be unable to represent the full pathological features of RCC, which is a limitation that requires further study. In addition, 293T cells are only used as a representation of kidney physiology, and whilst they may be used as controls in RCC in vitro studies, caution is required for interpretation (26,27).
In summary, the results of the present study revealed that miR-199b-5p was downregulated in RCC and functioned as a tumor suppressor. Inhibition of miR-199b-5p in RCC promoted cellular proliferation, migration and invasion, while inhibiting cellular apoptosis; however, the specific molecular mechanism remains elusive. In the present study, the expression levels of miR-199b-5p were downregulated in RCC lines, as well as in RCC vs. paired adjacent normal tissues. RCC is not sensitive to traditional radiotherapy and chemotherapy, and the prognosis remains poor; therefore, easily detectable and specific biomarkers for early detection and treatment guidance are required. To further reveal the mechanisms associated with RCC and to facilitate early diagnosis, treatment and prognostication of RCC patients, the search of new biomarkers for renal cell carcinoma is imminent. However, the focus of further research will be on the roles of miR-199b-5p in RCC with regard to its diagnostic and prognostic value for RCC. In addition, further study is essential to identify the molecular mechanisms of miR-199b-5p in the genesis of RCC and its potential in early detection, prognosis prediction and targeted therapy for RCC. Perhaps in the future, circulating miR-199b-5p may be able to reflect the disease status of RCC patients and be of great early diagnostic and prognostic value.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81101922), the Science and Technology Development Fund Project of Shenzhen (grant nos. JCYJ20150403091443329 and JCYJ20170307111334308), the fund of the ‘San-ming’ project of medicine in Shenzhen and the fund of the Guangdong Key medical subject.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LCN and YQL conceived and refined experimental designs; YLL, JQ, JH, PJC collected data; JLX, XG and WJX performed the experiments; YLL and JQ evaluated the data; YLL drafted the manuscript; and YLL and JQ edited the manuscript. All authors read and approved the final manuscript.
Ethical approval and consent to participate
All patients provided written informed consent. The present study was approved by the Ethical Review Committee of Peking University Shenzhen Hospital (Shenzhen, China) and abided by the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu Y, Han X, Yu Y, Ding Y, Ni C, Liu W, Hou X, Li Z, Hou J, Shen D, et al. A genetic polymorphism affects the risk and prognosis of renal cell carcinoma: Association with follistatin-like protein 1 expression. Sci Rep. 2016;6:26689. doi: 10.1038/srep26689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y, Zhang N, Li K, Chen H, Lin X, Yu Y, Gou Y, Hou J, Jiang D, Na R, et al. Genetic scores based on risk-associated single nucleotide polymorphisms (SNPs) can reveal inherited risk of renal cell carcinoma. Oncotarget. 2016;7:18631–18637. doi: 10.18632/oncotarget.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin YW, Lee LM, Lee WJ, Chu CY, Tan P, Yang YC, Chen WY, Yang SF, Hsiao M, Chien MH. Melatonin inhibits MMP-9 transactivation and renal cell carcinoma metastasis by suppressing Akt-MAPKs pathway and NF-κB DNA-binding activity. J Pineal Res. 2016;60:277–290. doi: 10.1111/jpi.12308. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Yun EJ, Chen W, Ding Y, Wu K, Wang B, Ding C, Hernandez E, Santoyo J, Pong RC, et al. Targeting 3-phosphoinositide-dependent protein kinase 1 associated with drug-resistant renal cell carcinoma using new oridonin analogs. Cell Death Dis. 2017;8:e2701. doi: 10.1038/cddis.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 7.Ling H, Girnita L, Buda O, Calin GA. Non-coding RNAs: The cancer genome dark matter that matters! Clin Chem Lab Med. 2017;55:705–714. doi: 10.1515/cclm-2016-0740. [DOI] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Cortés-Sempere M, de Cáceres Ibáñez I. microRNAs as novel epigenetic biomarkers for human cancer. Clin Transl Oncol. 2011;13:357–362. doi: 10.1007/s12094-011-0668-z. [DOI] [PubMed] [Google Scholar]
- 10.Rotomskis A, Margevičiūtė R, Germanavičius A, Kaubrys G, Budrys V, Bagdonas A. Differential diagnosis of depression and Alzheimer's disease with the Addenbrooke's cognitive Examination-revised (ACE-R) BMC Neurol. 2015;15:57. doi: 10.1186/s12883-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roomi MW, Kalinovsky T, Rath M, Niedzwiecki A. In vitro modulation of MMP-2 and MMP-9 in pediatric human sarcoma cell lines by cytokines, inducers and inhibitors. Int J Oncol. 2014;44:27–34. doi: 10.3892/ijo.2013.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawless N, Vegh P, O'Farrelly C, Lynn DJ. The role of microRNAs in bovine infection and immunity. Front Immunol. 2014;5:611. doi: 10.3389/fimmu.2014.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 14.Won KY, Kim YW, Kim HS, Lee SK, Jung WW, Park YK. MicroRNA-199b-5p is involved in the Notch signaling pathway in osteosarcoma. Hum Pathol. 2013;44:1648–1655. doi: 10.1016/j.humpath.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Joshi D, Chandrakala S, Korgaonkar S, Ghosh K, Vundinti BR. Down-regulation of miR-199b associated with imatinib drug resistance in 9q34.1 deleted BCR/ABL positive CML patients. Gene. 2014;542:109–112. doi: 10.1016/j.gene.2014.03.049. [DOI] [PubMed] [Google Scholar]
- 16.Fang C, Wang FB, Li Y, Zeng XT. Down-regulation of miR-199b-5p is correlated with poor prognosis for breast cancer patients. Biomed Pharmacother. 2016;84:1189–1193. doi: 10.1016/j.biopha.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Favreau AJ, McGlauflin RE, Duarte CW, Sathyanarayana P. miR-199b, a novel tumor suppressor miRNA in acute myeloid leukemia with prognostic implications. Exp Hematol Oncol. 2016;5:4. doi: 10.1186/s40164-016-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Song B, Song W, Liu J, Sun A, Wu D, Yu H, Lian J, Chen L, Han J. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1α in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol. 2011;26:1630–1637. doi: 10.1111/j.1440-1746.2011.06758.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. 2014;5:944–958. doi: 10.18632/oncotarget.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang W, Chen X, Nie L, Xu M, Chen N, Zeng H, Zhou Q. MiR199b suppresses expression of hypoxia-inducible factor 1alpha (HIF-1α) in prostate cancer cells. Int J Mol Sci. 2013;14:8422–8436. doi: 10.3390/ijms14048422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng H, Zhang Z, Dai X, Chen Y, Ye J, Jin Z. Increased expression of microRNA-199b-5p associates with poor prognosis through promoting cell proliferation, invasion and migration abilities of human osteosarcoma. Pathol Oncol Res. 2016;22:253–260. doi: 10.1007/s12253-015-9901-3. [DOI] [PubMed] [Google Scholar]
- 22.Andolfo I, Liguori L, De Antonellis P, Cusanelli E, Marinaro F, Pistollato F, Garzia L, De Vita G, Petrosino G, Accordi B, et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, Wang F, Cheng G, Si S, Sun X, Han J, Yu H, Zhang W, Lv Q, Wei JF, Yang H. Wilms' tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J Exp Clin Cancer Res. 2018;37:40. doi: 10.1186/s13046-018-0706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023–1033. doi: 10.1007/s00535-012-0733-6. [DOI] [PubMed] [Google Scholar]
- 25.Brodaczewska K, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cell cancer research. Mol Cancer. 2016;15:83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaravinos A, Pieri M, Mourmouras N, Anastasiadou N, Zouvani I, Delakas D, Deltas C. Altered metabolic pathways in clear cell renal cell carcinoma: A meta-analysis and validation study focused on the deregulated genes and their associated networks. Oncoscience. 2014;1:117–131. doi: 10.18632/oncoscience.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Araújo Júnior RF, Oliveira Leitão AL, de Melo Silveira RF, de Oliveira Rocha HA, de França Cavalcanti P, de Araújo AA. Telmisartan induces apoptosis and regulates Bcl-2 in human renal cancer cells. Exp Biol Med (Maywood) 2015;240:34–44. doi: 10.1177/1535370214546267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


