Abstract
Significant alterations of intestinal microbiota and anemia are hallmarks of inflammatory bowel disease (IBD). It is widely accepted that iron is a key nutrient for pathogenic bacteria, but little is known about its impact on microbiota associated with IBD. We used a model device to grow human mucosa-associated microbiota in its physiological anaerobic biofilm phenotype. Compared to microbiota from healthy donors, microbiota from IBD patients generate biofilms ex vivo that were larger in size and cell numbers, contained higher intracellular iron concentrations, and exhibited heightened virulence in a model of human intestinal epithelia in vitro and in the nematode Caenorhabditis elegans. We also describe an unexpected iron-scavenging property for an experimental hydrogen sulfide-releasing derivative of mesalamine. The findings demonstrate that this new drug reduces the virulence of IBD microbiota biofilms through a direct reduction of microbial iron intake and without affecting bacteria survival or species composition within the microbiota. Metabolomic analyses indicate that this drug reduces the intake of purine nucleosides (guanosine), increases the secretion of metabolite markers of purine catabolism (urate and hypoxanthine), and reduces the secretion of uracil (a pyrimidine nucleobase) in complex multispecies human biofilms. These findings demonstrate a new pathogenic mechanism for dysbiotic microbiota in IBD and characterize a novel mode of action for a class of mesalamine derivatives. Together, these observations pave the way towards a new therapeutic strategy for treatment of patients with IBD.
Keywords: microbiota biofilms, colitis, iron, hydrogen sulfide, mesalamine, inflammatory bowel disease
INTRODUCTION
Therapeutic limitations, high costs, and adverse effects of currently available drugs for treatment of inflammatory bowel disease (IBD) underscore the need for medications that are more effective and safe. Although anemia is one of the most common extraintestinal complications of IBD, dietary iron supplementation leads to disease exacerbation and a higher risk of infection, possibly through alterations of commensal microbiota and increased abundance of Enterobacteriacae.1–3 Beyond the taxonomic microbiota abnormalities associated with IBD (decreased diversity, increased representation of Proteobacteria), the presence of pathobionts (for example adherent-invasive strains of Escherichia coli) or abnormal host responses toward commensals are suspected.4, 5 A better understanding of functional alterations of gut microbiota during IBD is sorely needed and may pave the way toward more precise therapies.
We hypothesized that microbiota with increased ability to access iron, and subsequently increased virulence, contribute significantly to intestinal inflammation. We thus aimed to evaluate if novel derivatives of a widely used drug for treating inflammation in patients with IBD (ie, hydrogen-sulfide releasing mesalamine) could have a direct and beneficial effect toward dysbiotic microbiota associated with IBD in a biologically relevant model. Microbiota indeed naturally exist as biofilms in the large intestine, both in health6, 7 and during colitis.6, 8, 9 Biofilms are defined as communities of bacteria encased in a complex extracellular matrix composed of polysaccharides, proteins, nucleic acids, and glycolipids.10 We used a well-established model to study multispecies microbiota biofilms obtained from human colonic biopsy tissues ex vivo under anerobic and static conditions.6, 11–13 This model reproduces some of the complexity of microbial-microbial interactions in nature and thus served as an attractive platform to perform preclinical studies of a drug designed to correct dysbiotic behavior associated with IBD.
MATERIALS AND METHODS
Patients
Descending colon biopsies of human donors who had not taken antibiotics in the previous 16 weeks were provided by the Inflammation Tissue Bank of the University of Calgary. Ethics approvals were delivered by the Research Ethics Board and the Calgary Health Region (#REB14-2430 REN1). Clinical characteristics of the donors are presented in Table 1. Biopsies were transported in BBL Port-a-cul (anaerobic transport, BD Bioscience, Mississauga, Canada) from the hospital to the lab and mucosa-associated bacteria were cultured from these biopsies immediately under an anaerobic hood.
TABLE 1:
Characteristics of Patients in the Study
| No. | Age (years ± SEM) |
Sex Ratio (M:F) | Current Medication (no.) | Antibiotic Treatment | |
|---|---|---|---|---|---|
| Healthy Controls | n = 7 | 33 ± 2.2 | 2.5 | None | No |
| Crohn’s Disease | n = 13 | 49 ± 4.1 | 1.4 | Immunotherapy (6) Steroids or mesalamine (3) Others (1) |
No |
| Ulcerative Colitis | n = 11 | 40 ± 4.0 | 1.8 | Immunotherapy (1) Mesalamine (3) Others (1) |
No |
Values are total number of patients (n). F, female; M, male; SEM, standard error of mean.
Drugs
ATB-428 and ATB-429 are compounds consisting of mesalamine covalently linked to an H2S-donor: 4-hydroxythiobenzamide (TBZ) in the case of ATB-428, and (5-(4-hydroxyphenyl)-3H-1,2-dithiole-3-thione, ADT-OH) in the case of ATB-429. These drugs were provided by Antibe Holdings Inc. (Calgary, Canada).
Growth of Biofilms
All steps described herein were performed in anerobic conditions (Bactron II, Sheldon Manufacturing, Oregon, USA) as previously described, 11–13, 49. Colon biopsies were homogenized in a microtube pestle and mucosa-associated microbiota was cultured in tryptic soy broth supplemented with yeast extract (5 g/L, BD Bioscience), L-cysteine (5%, Sigma-Aldrich, Oakville, ON, Canada), hemin (100 mg/L), and menadione (2 mg/L, Sigma-Aldrich). The Calgary Biofilm Device (Innovotech, Edmonton, Canada) features a lid with 96 polystyrene extensions on which biofilms grow from the shear stress generated by agitation.11, 47 Biofilms were generated from identical optical density of bacteria (OD600nm = 0.1) and matured for 3 days under agitation (125 g). Mature biofilms were transferred onto a plate with wells containing fresh media or containing working concentrations of drugs (in broth media as vehicle), and after 24 hours biofilm-dispersed bacteria were collected in the bottom plate. Viability was assessed by colony-forming unit counting in agar plates.
Human Intestinal Epithelial Cells
Human intestinal epithelial cells (Caco-2, ATCC, Manassas, USA) were grown in Dulbecco’s modified Eagle medium (Gibco, Invitrogen) on flat-bottom 6- or 12-well plates for 7 days (Costar, Thermo Fischer) or on polycarbonate 12- or 24-transwells plates for 21 days (Costar). Cells were co-cultured with bacteria (8 × 107 CFU/ml) in Hanks Balanced Salt solution (HBSS, Gibco), in 5% CO2 incubator at 37°C. During co-culture, bacterial growth rate was equivalent in all groups. Alternatively, 1 ml of biofilm spent media (OD600nm = 1) was filter-sterilized through 0.22 µm filters, and cells were stimulated with 10% dilution of spent media in HBSS. Epithelial necrosis was quantified by lactate dehydrogenase activity (LDH) assay (Roche, Mississauga, Canada) and absorbance was read (SpectraMax, Molecular Devices Corporation, Menlo Park, USA). Bacteria adhesion (90 minutes) and invasion (3 hours) protocols were based on standardized antibiotic assay and agar plate counting, using a broad-spectrum cocktail of antibiotics: gentamicin (200 μg/ml), ampicillin (100 µg/ml), and vancomycin (100 µg/ml). For translocation assays, Caco-2 were differentiated on 8 µm transwells and stimulated apically with live bacteria for 5 h. Translocation was assessed by agar plating of the basolateral media and was expressed as a translocation percentage of total bacteria. Total RNA was extracted in Trizol after 6 hours co-culture (Qiagen, Montreal, Canada). Quantitative PCR for CXCL8 mRNA (RT2qPCR Primer, Qiagen) was performed on IQ5 PCR machine (SYBRgreen, BioRad, Mississauga, ON, Canada). Data were analyzed by 2-ΔΔCT method using human B2M (b2-microglobulin, Qiagen) and GAPDH (Qiagen) as control genes and expressed as a fold-change compared to control cells with no bacteria.
Confocal Staining of Biofilms and Epithelial Cells
Biofilms were stained, without fixation, with acridine orange (unspecific biomass stain, BD Bioscience), TO-PRO-3 (DNA-RNA, Invitrogen) and visualized on a confocal microscope (Leica DM IRE2 Microsystems, Richmond Hill, ON, Canada). Total biofilm biomass (acridine orange + TOP-PRO-3 stains) was measured using surface staining measurement in Imaris Bitplane (v7, Concord, MA, USA).
Microbiota 16S Sequencing and Analysis
DNA from biofilm-dispersed bacteria were extracted by a silica-membrane-based purification method according to the manufacturer’s instructions (QIAmp DNA MiniKit, Qiagen), with an additional step of incubation in lysozyme solution (20 mg/ml, 1 hour). Amplicon sequencing libraries were obtained from the V4 region of the 16S SSU rRNA using 515F-806R primers.48 Paired-end amplicons (250 bases) were sequenced on Illumina Mi-Seq (Génome Québec Innovation Centre, Montreal, Canada). Bioinformatics analysis was performed by first removing the primers and quality trimming the reads. UPARSE pipeline was used to generate the OTU (operational taxonomic unit) table and taxonomy was assigned using the RPD database and classifier.49–51 Downstream analysis was done in R using Phyloseq 1.16.2 package.
Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis
Mature biofilms were incubated in fresh media. After 16 hours, spent media was collected and diluted 1:1 in pure methanol. Methanolic extracts were separated by Ultra High-Performance Liquid Chromatography (UHPLC) mass spectrometry (MS), using a hydrophilic interaction liquid chromatography column (Syncronis HILIC, Thermo Fisher, stationary phase).52 Solvent A (20mM ammonium formate pH 3 in H20) and B (MS Grade Acetonitrile with 0.1% formic acid) were used for LC Gradient. High-resolution full-scan MS data were acquired on a Thermo Fisher Scientific Q-Exactive HF mass spectrometer using negative-mode electrospray ionization. Metabolites were identified using retention times of standards using MAVEN freeware.53
Caenorhabditis elegans Model
The Caenorhabditis elegans toxicity model was based on a recently published protocol.54 Briefly, Bristol N2 wild-type C. elegans (Caenorhabditis Genetics Center, University of Minnesota, St. Paul, MN, USA) synchronized in L4 stage were fed in 96-well microtiter plates with biofilm-dispersed bacteria in PBS/S-Basal complete media (OD600nm = 0.1). Plates were incubated with bacteria at 20°C for 72 hours. Worm mortality (no active movement, stiff body structure, and no response to mechanical stimuli) was assessed by an experimented investigator blinded to treatments using a binocular microscope (Leica MZ75 microscope).
Dinitrobenzene Sulfonic Acid (DNBS) Colitis Model
All experiments using animals were approved by the University of Calgary’s Animal Care Committee (certificate #AC13-0067). Colitis was induced by intracolonic instillation of 5 mg of DNBS dissolved in 50% ethanol.55 Control groups were treated similarly with PBS. Mice were given 50 mg/kg of ATB-429 per os in a vehicle of 1% carboxymethylcellulose, twice daily, for 5 days. The severity of colitis was blindly evaluated using previously described endpoints.6, 56 Liver biopsies were collected aseptically, weighed, homogenized, and plated on blood agar for 24 hours. Carnoy’s-fixed mice colon tissues were paraffin-embedded. Visualization of microbiota biofilm was performed by FISH staining (EUB338-Cy3,.6 Images were acquired using a Leica DM IRE2 confocal microscope and analyzed on Fiji freeware (v.1.51).
Iron Quantification
Thirty microliters of detection buffer (5 mM ferrozine, 6.5 mM neocuproine, and 2.5 M ammonium acetate; Sigma-Aldrich) were added to 170 µl of samples, and specific ferrozine-iron absorbance (562 nm) was measured after 30 minutes on a spectrophotometer. Iron concentrations were extrapolated from a standard curve generated with FeCl2. Changes in absorbance of ferrozine-iron complexes after 24 hours were monitored to evaluate the binding ability of chelators.19 Biofilms were incubated 24 hours to ATB-428 (1 mM), ATB-429 (1 mM), or 2,2-bipyridil (1 mM, Sigma Aldrich), after which biofilm-dispersed bacteria were collected, normalized to OD600nm = 1, and lysed by sonication for measurement of intracellular iron. Mice feces were mechanically homogenized in 1 ml of 0.1 mM HCl, and iron concentrations were first measured in the fecal spent media (Fig. S7C). Cellulose debris were discarded after centrifugation. Bacteria left were centrifuged and sonicated for measurement of intracellular iron.
Statistical Analysis
Graphic representation and statistical analysis were performed using GraphPad Prism (v6, La Jolla, USA). One-way ANOVA with Dunnett’s or Fisher’s test and Kruskal–Wallis with Dunn’s test were used accordingly after D’Agostino-Pearson normality test. For multiple variables, we used 2-way ANOVA with Dunnett’s test. An associated P value less than 5% was considered significant. All center values are means for histograms and dot plots, median for box plots. Error bars represent standard error of the mean (histograms, dot plots) or minimum-maximum (box plot).
RESULTS
Microbiota from IBD Patients Generate Larger BioFilms and Exhibit Greater Iron Intake than Microbiota from Healthy Controls
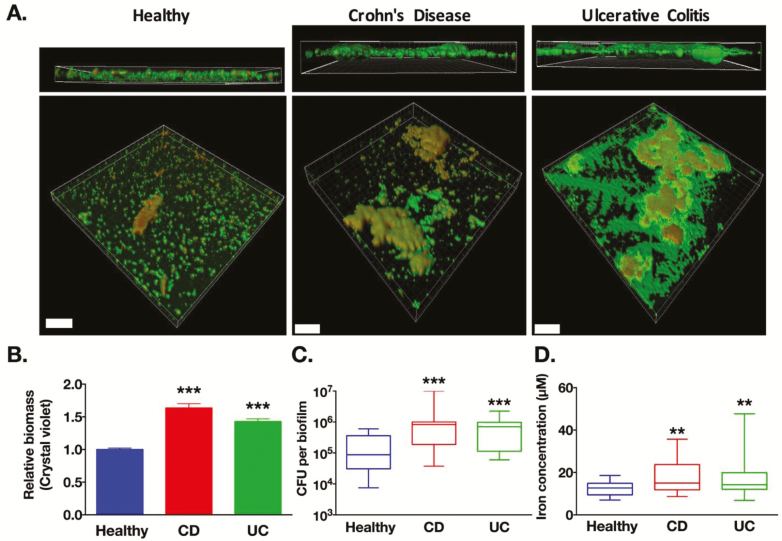
Microbiota biofilms abnormally adhere to the epithelial surface of intestinal tissues in patients with IBD.9, 14, 15 In an attempt to more precisely characterize these biofilms, mucosa-associated microbiota in colonic biopsies from either healthy donors (healthy; n = 7) or IBD patients (Crohn’s disease, CD, n = 13; or ulcerative colitis, UC, n = 11) were first homogenized in modified tryptic soy broth, normalized to cell numbers, and plated onto the Calgary Biofilm Device to allow the growth of bacteria under their natural adherent biofilm phenotype (Fig. S1). All procedures were performed under anaerobic conditions and experimental conditions were kept identical among the different cohorts of patients. Taxonomic identification confirmed that biofilms generated ex vivo from the biopsy samples reproduced a complex community of microbiota containing all major bacterial phyla reported in the human gut (Fig. S2). Confocal laser scanning microscopy allowed 3-dimensional representations of biofilms (Fig. 1A). Total biomass quantifications revealed that IBD biofilms had a significantly greater biomass (bacteria and their extracellular matrix) compared to healthy control biofilms (Fig. 1B). IBD-biofilms contained more viable bacteria compared to healthy control biofilms (Fig. 1C). Pathogenic strains of bacteria (eg, pathogens from Enterobacteriaceae family) are known to have high iron uptake capacity,16 but data regarding human microbiota living as a complex biofilm community are lacking. Bacteria from IBD-biofilms contained significantly higher concentrations of intracellular iron (1.5-fold higher in CD and 1.4-fold in UC) compared to concentrations measured in bacteria from healthy control biofilms (Fig. 1D). Overall, these data demonstrate for the first time that mucosa-associated microbiota from IBD patients have an increased affinity for iron and a greater capacity to grow under a biofilm phenotype compared to microbiota from healthy controls.
FIGURE 1.
Microbiota from IBD patients form larger biofilms than microbiota from healthy controls and contains higher intracellular iron. A, Human microbiota biofilms (healthy; Crohn’s Disease, CD: ulcerative colitis UC) were grown on the Calgary Biofilm Device. Confocal laser scanning microscopy of biofilms was performed, the extracellular matrix of biofilms was stained by acridine orange (green) and bacteria DNA with TO-PRO-3 (red). Scale bars correspond to 70 μm. B, Biofilms from IBD patients had a higher relative biomass compared to healthy biofilms (Crystal violet staining, n = 145 to 313 biofilms per group). C, Biofilms from IBD patients contained higher numbers of colony forming units (CFU per biofilm) compared to biofilms from healthy biofilms (n = 21 to 24 biofilms per group). D, Equivalent numbers of bacteria from IBD biofilms contained higher concentration of intracellular iron compared to healthy biofilms (n = 3 3 biofilms per group). B, 1-way ANOVA followed by Dunnett’s tests. C, D, Kruskal-Wallis followed by Dunn’s tests versus healthy group aP < 0.01, bP < 0.001
Bacteria from IBD-biofilms Have a More Virulent Phenotype
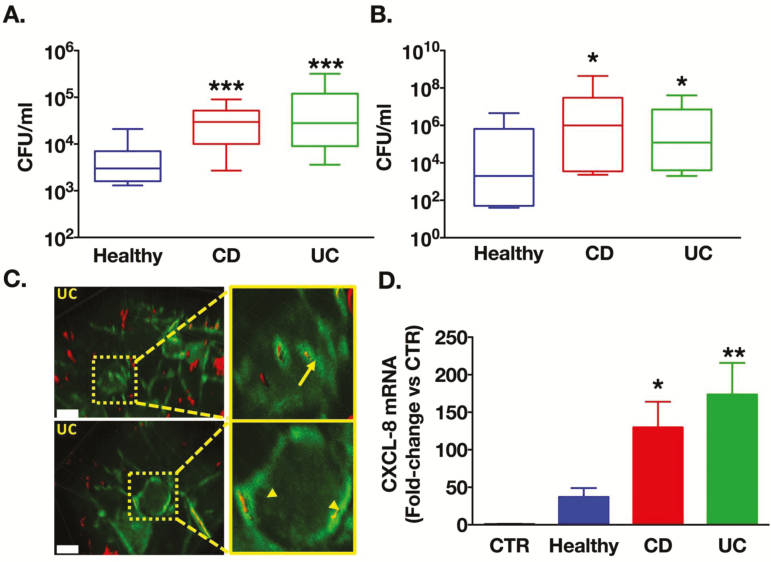
To determine adhesion, invasion and virulence characteristics of IBD microbiota biofilms, we used dispersed bacteria from these biofilms (Fig. S1). Indeed, bacteria released by adherent biofilms have strikingly different characteristics than their sessile counterparts, including elevated antimicrobial resistance and virulence.17 Human intestinal epithelial cells (Caco-2) were co-cultured with identical numbers of these biofilm bacteria (8 × 106 CFU/ml). After 3 hours of co-culture, bacteria from IBD biofilms were 5.6-fold (for CD) and 14-fold (for UC) more invasive into Caco-2 cells compared to healthy controls (1 ± 0.2 × 103 CFU/ml, Figs. 2A, S3A). Finally, bacteria from CD (88-fold) and UC biofilms (13-fold) translocated significantly more across epithelia than bacteria from healthy control biofilms (5 × 105 CFU/ml, Fig. 2B). Confocal imaging revealed that translocation followed both the paracellular (Fig. 2C, upper panel) and transcellular (Fig. 2C, lower panel) pathways. These results suggest that mucosa-associated microbiota from IBD patients, grown under a biofilm phenotype, have increased virulent properties compared to microbiota from healthy controls.
FIGURE 2.
IBD biofilms have increased virulence compared to bacteria from healthy controls biofilms. Caco-2 monolayers were co-cultured with equal number of dispersed bacteria from microbiota biofilms of healthy controls, Crohn’s disease (CD) or ulcerative colitis (UC), n = 4 donors per cohort. A, Bacterial invasion (n = 11 wells per group), and (B) apical-to-basolateral translocation were significantly higher when microbiota originated from IBD biofilms compared to healthy biofilms (n = 21 to 24 transwells per group). C, Transwells of Caco-2 cocultures were fixed and epithelial cytoskeleton was stained with F-actin-FITC antibody (green), and bacteria with universal 16S bacteria probe (fluorescent in situ hybridization). Bacteria staining was contrasted to facilitate visualization. Representative images illustrated bacteria translocating intracellularly (arrow) and paracellularly (arrowheads). Scale bars represent 10 µm. D, Bacteria dispersed from IBD biofilms induced significantly higher expression of CXCL8 mRNA versus biofilm bacteria from healthy controls (n = 11 to 15 wells per group). A, B, Kruskal-Wallis followed by Dunn’s tests (D) 1-way ANOVA with Fisher’s tests.aP < 0.05, bP < 0.01, cP < 0.001
IBD Biofilms Induce Elevated CXCL-8 and Pathogenic Responses Compared to Healthy Biofilms
To evaluate and compare the proinflammatory host response induced by microbiota from IBD patients or healthy controls, we co-cultured Caco-2 cells with bacteria dispersed from their biofilm and measured CXCL8 mRNA transcription. At 6 hours, CXCL8 mRNA was increased versus vehicle (media only) by 130-fold (for CD biofilms) and 174-fold (for UC biofilms), hence significantly more than the 37-fold increase induced by healthy biofilms (Fig. 2D). IBD microbiota also caused significantly more epithelial necrosis after 5 hours than those from healthy controls (Fig. S4A). Filter-sterilized spent media from IBD biofilms significantly increased paracellular permeability to dextran-FITC (Fig. S4B). Bacteria from IBD biofilms induced significantly greater lethality in the nematode Caenorhabditis elegans than bacteria from healthy biofilms (Fig. S3B). Together, these data demonstrate that microbiota biofilms from IBD triggered a more severe inflammatory host response than that caused by microbiota from healthy controls, and induced negative effects in epithelia in vitro and are lethal to C. elegans in vivo.
Hydrogen Sulfide-releasing Mesalamine Derivatives Have Potent Iron-chelating Properties
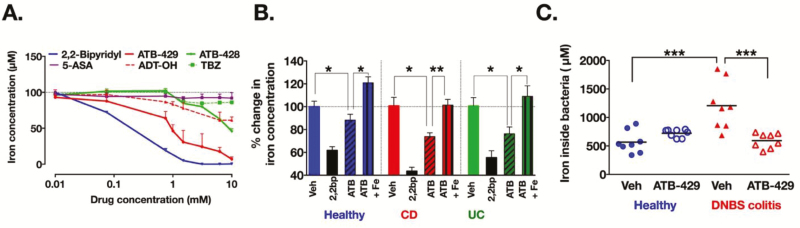
ATB-428 and ATB-429 are novel antiinflammatory drugs consisting of mesalamine (5-aminosalicylate; 5-ASA) covalently linked to 2 different H2S-donors (TBZ or ADT-OH, respectively). Considering the independent interactions of H2S18 and mesalamine19 with iron, we tested the ability of these drugs to chelate iron. Both ATB-428 and ATB-429 (Fig. 3A) effectively chelated iron in a concentration-dependent manner. ATB-429 was more potent than ATB-428 (maximum chelation of 93% for ATB-429 versus 55% for ATB-428), possibly attributable to the more potent iron-scavenging property of ADT-OH (maximum chelation of 39%) versus TBZ (maximum chelation 14%, Fig. 3A). At equimolar concentrations, the 2 drugs had markedly enhanced iron-scavenging properties compared to mesalamine (maximum chelation of 9%). These results demonstrate that ATB-429, in particular, has significant iron-scavenging properties.
FIGURE 3.
Hydrogen sulfide-releasing mesalamine chelates iron and reduces iron intake in microbiota associated with colitis. A, Drugs were dissolved in bacterial media containing 100 µm of FeCl2. After 24 hours, the remaining concentration of free iron was markedly reduced in a positive control iron chelator (2,2-bipyridil) and in ATB-429, ATB-428, and ADT-OH, but not in TBZ and mesalamine (5-ASA). n = 3 to 9 duplicates per concentrations of drugs. B, Biofilms (healthy controls; Crohn’s disease, CD; and ulcerative colitis, UC) were incubated for 24 hours in media containing 2,2-bipyridil (200 µM; iron chelator positive control), ATB-429 (1 mM), or ATB-429 + FeCl2 (1 mM and 100µM, respectively). Concentrations of intracellular iron were reduced in microbiota exposed to positive control (2,2-bipyridil, 2,2bp) or to ATB-429; addition of iron abolished the effect of ATB-429 (n = 12 biofilms per group for 2,2-biprydil, n = 32 biofilms per group for vehicle and ATB-429). C, Intracellular concentrations of iron in fecal microbiota (normalized to 1 g of feces) were significantly higher in microbiota from the colitis group (DNBS) compared to microbiota from control animals (vehicle, veh) and groups treated with ATB-429 (50 mg/kg, twice daily, per os). n = 8 mice per group. B, C, Kruskal-Wallis followed by Dunn’s tests. aP < 0.05, bP < 0.01, cP < 0.001
ATB-429 Limits Iron Intake in Colitis-associated Biofilms and Modifies their Metabolome, But Not Their Composition
Considering the newly revealed iron-chelating properties of ATB-429, and in a lesser way of ATB-428, we exposed human microbiota biofilms to 1 mM of these drugs dissolved in media for 24 hours. ATB-429, but not ATB-428 (Fig. S5A), reduced intracellular iron content in bacteria from biofilms, and this effect was reversed by the addition of iron to the medium (Fig. 3B). At 1 mM, H2S-releasing mesalamine derivatives did not modify the pH of bacterial media (Figs. S5B, S6A), did not affect bacteria survival (Figs. S5C, S6B), and did not significantly alter biofilms overall species composition (Fig. S2C). Biofilms were incubated for 16 hours in fresh medium containing ATB-429 (1 mM), then metabolomic analyses were performed. Purine and pyrimidine metabolites were measured in the spent media because their metabolism relies on intracellular iron intake and is important for host colonization.20–22 Upon exposure to ATB-429, biofilm intake of purine nucleosides (guanosine, Fig. S6C) was reduced, whereas release of urate (Fig. S6D) and hypoxanthine (Fig. S6E), 2 metabolites of purine catabolism, were increased. Biofilm secretion of uracil (Fig. S6F), a pyrimidine nucleobase, was significantly reduced with the addition of ATB-429. The ATB-429 alone (media + ATB-429) had no effect on relative concentrations of metabolites in bacterial media. Together, these data demonstrated that ATB-429 reduces iron intake by colitis-associated microbiota. ATB-429 had a profound effect on microbiota biofilm metabolism, without significantly changing the microbiota species composition.
ATB-429 Reduces the Virulent Properties and Associated Proinflammatory Effects of IBD Biofilms In Vitro and Reduced the Severity of Experimental Colitis in Mice
Using in vitro (Caco-2 cells) and in vivo (experimental colitis in mice) models, additional experiments sought to assess whether the reduction of iron intake in microbiota biofilms by ATB-429 was therapeutically meaningful.
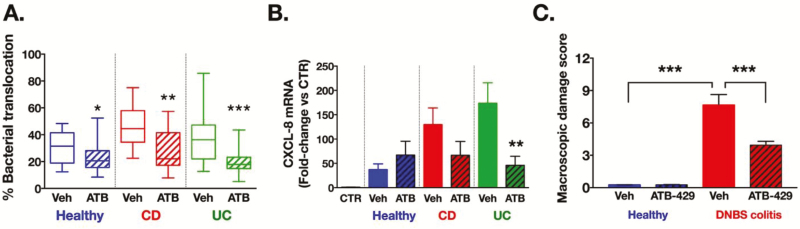
When biofilms were preexposed to ATB-429, bacteria translocated significantly less (60% reduction from healthy and CD, 50% reduction for UC) through epithelial monolayers compared to bacteria from biofilms exposed to vehicle alone (Fig. 4A). Preexposure to ATB-429 also reduced by 51% (for CD) and 27% (for UC) the induction of epithelial CXCL8 mRNA by biofilm bacteria (Fig. 4B). Supplementation of iron (100 µM of FeCl2) partially reversed the effect of ATB-429 (1 mM) by increasing levels of bacterial adherence to Caco-2 (Fig. S4C). These data suggest that ATB-429 is able to directly reduce virulence of gut microbiota grown in their natural state of complex multispecies biofilms through an iron-dependent mechanism.
FIGURE 4.
ATB-429 reduces the virulence of IBD microbiota biofilms and protects from DNBS colitis in mice. Human microbiota biofilms (healthy; Crohn’s disease, CD; and ulcerative colitis, UC) were first incubated with ATB-429 (1 mM) for 24 hours. Caco-2 cells were then co-cultured with equal numbers of biofilm-dispersed bacteria. A, Bacteria from IBD biofilms exposed to ATB-429 had a reduced capacity to translocate across Caco-2 cells monolayers compared to bacteria exposed to vehicle (n = 21 to 28 transwells per group, *versus corresponding vehicle group). B, Induction of CXCL8 mRNA transcription in Caco-2 cells was significantly lower when induced by bacteria dispersed from IBD biofilms exposed to ATB-429 versus bacteria from biofilms exposed to vehicle (n = 8 to 15 wells per group, *versus corresponding vehicle in group). C, Macroscopic damage scores in the colon of mice from animals with colitis were significantly greater than the scores measured in controls (Healthy); treatment with ATB-429 (DNBS/ATB-429) maintained the damage scores at control levels (DNBS/vehicle; n = 8 mice per group. A, B, Kruskal-Wallis followed by Dunn’s tests. C, 1-way ANOVA followed by Fisher’s tests. aP < 0.05, bP < 0.01, cP < 0.001
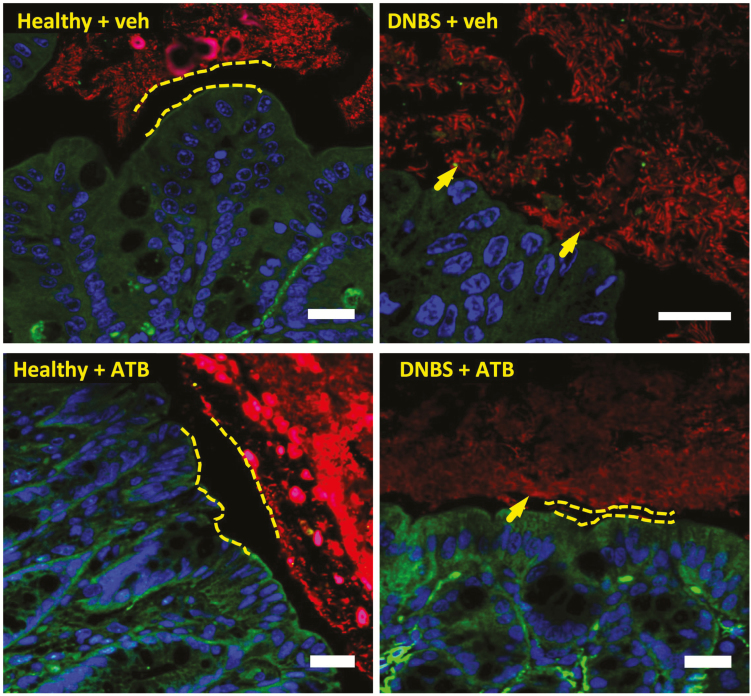
Concentrations of iron in fecal bacteria from mice with experimental colitis (dinitrobenzene sulfonic acid, DNBS) were greater than in fecal bacteria from healthy mice (Fig. 3C), consistent with the effects observed using human IBD microbiota biofilms ex vivo (Fig. 1D). In healthy mice, ATB-429 (50 mg/kg twice daily per os) was well tolerated with no signs of disease (Figs. 4C, S7A). In the DNBS colitis group, coinciding with the decreased concentrations of intracellular iron in fecal bacteria (Fig. 3C), administration of ATB-429 reduced the severity of colitis (Figs. 4C, S7A) and inhibited the translocation of commensal bacteria into the liver (Fig. S7B). Staining of microbiota biofilms in situ revealed that during colitis, bacteria invaded the mucus layer and adhered to the intestinal epithelium (arrow, Fig. 5). Treatment with ATB-429 abolished these abnormalities (Fig. 5). Together, these results indicate that ATB-429 reduced iron intake in colitis-associated microbiota and restored the normal localization of the commensal microbiota biofilm, thus efficiently reducing the severity of experimental colitis.
FIGURE 5.
ATB-429 restores normal gut microbiota biofilm localization. C57Bl/6 mice were given DNBS intracolonically to induce colitis, treated orally with ATB-429 (50 mg/kg, twice daily), and killed after 5 days (n = 8 per group). Intestinal microbiota biofilms were stained by fluorescent in situ hybridization (Eubacteria probe, in red), host nuclei were labeled with To-pro-3 (in blue), and the epithelial host cytoskeleton was stained with an F-actin-FITC antibody (in green). In animals with colitis, bacteria invaded the mucus layer and adhered to epithelium (arrows), whereas in healthy and in ATB-429-treated mice, the microbiota remained separated from the epithelial surface by a normal mucus layer. Scale bars represent 20 µm, dashed lines represent the sterile mucus layer.
DISCUSSION
Mesalamine is a commonly prescribed drug for treating IBD patients. In addition to its well-known effects on the host,23–25 a potential action on microbiota has been scarcely investigated. Indeed, mesalamine concentrations in the lumen of patients can reach 100 mM, thus exceeding by 100-fold concentrations in the host mucosa or in the circulation.26 It can modify bacterial gene expression27 and appears to cause a transient increase in the abundance of Firmicutes in humans.28 It can also suppress polyphosphate accumulation in bacteria,29 which in turn may serve as an intracellular iron chelator. Independently, antiinflammatory properties of H2S have been well established in the gastrointestinal tract and H2S-releasing drugs have become part of extensive research and development programs.30 In an effort to combine beneficial effects of H2S and mesalamine, H2S-releasing mesalamine derivatives have been synthesized and their efficacy in protecting against colitis was demonstrated, but exact mechanisms of action of these novel classes of drugs remain incompletely understood.31 Little is known about the effects of H2S on microbiota. Recent reports demonstrated that dietary-derived H2S donors stimulates the growth of anaerobic microbiota biofilms obtained from healthy human donors.6 These findings motivated our present work to investigate the precise effects of H2S-releasing antiinflammatory drugs on the microbiota living in their natural biofilm phenotype.
Anemia is the most common extraintestinal complication of IBD.32 However, dietary iron supplementation leads to disease exacerbation and a higher risk of infection, possibly through alterations of commensal microbiota.1–3 Whether chronic blood loss associated with IBD could elevate iron concentrations in the intestinal lumen, and thus favor populations of pathogenic bacteria, is unknown. In this study, biofilms grown ex vivo from either CD or UC patients were larger in size than healthy control biofilms (Fig. 1) and exhibited elevated adhesion, invasion and virulence compared to healthy control biofilms. Although iron concentrations in the fecal spent media were identical between healthy and colitis group (Fig. S7C), we observed an increase of intracellular iron in fecal microbiota (mice, Fig. 3C) and in human IBD biofilms (Fig. 1D). It was demonstrated in vitro33 in animal models34 and in humans35 that Proteobacteria can thrive at the expense of other gut bacteria in an iron-rich environment. Proteobacteria abundance was indeed higher in UC biofilms versus healthy biofilms, but also versus CD biofilms, with no differences between healthy and CD (Fig. S2). Because iron is an essential nutrient for several other microorganisms, including enterococci, these findings suggest that a unique taxon could not totally explain increased affinity for iron in IBD-associated microbiota. Elevated biofilm growth and virulence were observed for both CD and UC biofilms, although no major alterations in relative abundance were revealed by 16S taxonomic sequencing. Further whole genome sequencing and transcriptomic analyses thus are warranted to determine whether there is a specific strain-advantage or an iron-favored genotype/phenotype in IBD microbiota biofilms, which will help identify new genetic alterations of these pathogenic IBD biofilms.
The present findings show that ATB-429 does not affect bacterial survival, and, importantly, that it does not modify overall bacterial composition of complex multispecies biofilms from human patients (Fig. S2). Nevertheless, exposure of microbial communities to exogenous H2S might lead to a community shift, as this molecule has been demonstrated to be lethal for several Enterobacteria pathogen,6 and it could modify the ratio of microbes performing anaerobe H2S reduction (sulfate-reducing bacteria among the Firmicutes or in the Archae). However, exposure of pathogenic IBD biofilms to ATB-429 repressed their virulence (adherence, invasion, and translocation), through its iron-scavenging property. Importantly, we demonstrate that exposing bacteria (and not the host cells) to ATB-429 was sufficient to reduce the host inflammatory response toward virulent IBD microbiota. This direct property of ATB-429 could have significant therapeutic advantages in vivo during colitis. Indeed, a significant shift in the amount of Clostridiales such as Ruminococcaceae and Eubacteriaceae, and decreased Enterococcaceae have been reported in animal models given exogenous H2S-donors.36 Apart from its iron-scavenging mechanism of action, we further discovered that bacteria exposed to ATB-429 significantly altered their purine (reduced intake of nucleoside, increased release of xanthine and urate) and pyrimidine metabolism (reduced uracil release). An iron-deprived environment induced by ATB-429 could lead to impaired function of enzymes involved in the catabolism of purine nucleosides (xanthine oxidase and xanthine dehydrogenase) that is regulated by redox mechanisms and the formation of iron-sulfur clusters.37–39 Several species of Enterobacteria (E. coli, Salmonella, Vibrio) as well as Bacillus species (members of Firmicute) need to properly synthesize or incorporate nucleotides from their environment to efficiently colonize and persist in the mouse intestine or to proliferate in the human bloodstream.20–22 Elevated serum levels of urate have been associated with chronic inflammatory and metabolic diseases (hypertension atherosclerosis, diabetes), possibly through its pro and antioxidative properties and its interactions with iron.40 The reduction of uracil release (a pyrimidine nucleobase) observed in ATB-429-treated biofilms does not have obvious links to iron metabolism, suggesting that other metabolic pathways could be altered by this class of drug. Interestingly, increased microbial uracil release was shown to cause chronic inflammation via oxidative damage to the intestinal epithelium.41 The exact meaning of increased urate release in ATB-429-driven iron poor environment remains to be further elucidated. This targeted approach on purine and pyrimidine nucleobases could motivate future studies to investigate the total exometabolome of biofilms treated with ATB-429 drugs in more depth. Other potential consequences of iron deprivation by ATB-429 in gut microbiota biofilms warrant further investigation for translational applications. It is well known that pathogenic bacteria require iron during infection. Local iron deprivation may induce some bacteria to increase their expression of iron-chelating siderophores and/or hijack host iron-specific receptors and transport proteins (eg, heme, transferrin).42 Surrounding low iron concentrations can have either negative (Escherichia coli and Pseudomonas43,44) or positive (Staphylococcus45) influence on the formation of monospecies biofilm, but the influence of iron in the context of polymicrobial gut biofilms needs to be further elucidated. Furthermore, knowing that iron can generate powerful radicals, and that iron chelators are strong antioxidants,46 future research will need to study the potential antioxidant properties of ATB-429, which could have significant implications for its application in other clinical contexts.
The culture of gut mucosal microbiota in their complex, multispecies, anaerobic biofilm phenotype has been a missing link in studies trying to understand the role of microbiota in IBD. Ex vivo models, like the one we used in the present report, help reproduce key components of the microbial-microbial and microbial-host interactions occurring in the gut. Several biases are inevitably associated with these ex vivo models, and some of these include the lack of host components, which are likely to be essential components of the microbial biofilm in vivo (eg, mucus and host DNA). Future research in vivo on molecular characteristics of microbiota biofilms in healthy and disease states, on geolocalization of various bacterial species within the commensal biofilm, and metagenomics will help improve understanding of how gut microbiota may influence health in the gut, and beyond.
In summary, this study offers new evidence that gut microbiota biofilms grown ex vivo from patients with IBD are characterized by increased iron uptake and increased virulent properties compared to biofilms from healthy subjects. The findings also provide proof-of-principle evidence that a new H2S-releasing antiinflammatory drug can suppress iron intake in complex multispecies gut microbiota grown in their natural biofilm phenotype, hence reducing their virulence without modifying overall composition and survival. These are promising observations in our attempts to generate advances towards the development of a novel therapeutic for IBD that will help target both host inflammation and microbiota dysbiosis.
SUPPLEMENTARY DATA
Supplementary data are available at Inflammatory Bowel Diseases online.
ACKNOWLEDGMENTS
The authors are grateful to Dr. R. Leong-Quong (BioCore Facility), Dr. D. Hansen for providing C. elegans, Dr. D.W. Morck for access to the anaerobic hood, Dr. J.J. Harrison for access to the confocal microscope and software, and the Animal Care facility staff. G. Kasendra (translational research technician) helped to provide human biopsies. H2S-mesalamine and H2S-donors were provided by Antibe Holdings Inc., and Innovotech donated the Calgary Biofilm Device.
Author contributions: Study concept and design, acquisition of data and analysis, writing of the paper: Jean-Paul Motta, John L. Wallace, and Andre G. Buret; acquisition of data, critical review of the paper: Thibault Allain, Luke E. Green-Harrison, and Troy Feener; data acquisition and analysis on metabolome, and intellectual input: Ryan A. Groves and Ian A. Lewis; microbiome analysis and statistics: Hena Ramay.; intellectual input, provision of characterized clinical samples: Paul L.Beck. All authors approved the current version of the manuscript.
Conflicts of Interest: John L. Wallace and Andre G. Buret are cofounders of Antibe Holdings Inc., which is developing hydrogen-sulfide releasing anti-inflammatory drugs.
Supported by: Jean-Paul Motta was funded by postdoctoral fellowships from the Alberta-Innovate Health Services (AIHS), University of Calgary Eye’s High Fellowship, Izaak Walton Killam Fellowship, and AgreenSkills Fellowship from the EU. This research was funded by grants to Andre G. Buret and John L. Wallace from the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery and CREATE grants to Andre G. Buret), from Crohn’s and Colitis Canada (Andre G. Buret and John L. Wallace), and the Canadian Institutes of Health Research (John L. Wallace and Andre G. Buret). Metabolomics data were collected by Ryan A. Groves at the Calgary Metabolomics Research Facility, supported by the International Microbiome Centre and the Canada Foundation for Innovation (CFI-JELF 34986); Ian A. Lewis is supported by a Alberta Innovates Traslational Health Chair.
REFERENCES
- 1. Lee T, Clavel T, Smirnov K, et al. . Oral versus intravenous iron replacement therapy distinctly alters the gut microbiota and metabolome in patients with IBD. Gut. 2017;66:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uritski R, Barshack I, Bilkis I, et al. . Dietary iron affects inflammatory status in a rat model of colitis. J Nutr. 2004;134:2251–5. [DOI] [PubMed] [Google Scholar]
- 3. Kortman GA, Raffatellu M, Swinkels DW, et al. . Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol Rev. 2014;38:1202–34. [DOI] [PubMed] [Google Scholar]
- 4. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buret AG. Good bugs, bad bugs in the gut: the role of microbiota dysbiosis in chronic gastrointestinal consequences of infection. Am J Gastroenterol. 2016;3:25–32. [Google Scholar]
- 6. Motta JP, Flannigan KL, Agbor TA, et al. . Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm Bowel Dis. 2015;21:1006–1017. [DOI] [PubMed] [Google Scholar]
- 7. Macfarlane S, Bahrami B, Macfarlane GT. Mucosal biofilm communities in the human intestinal tract. Adv Appl Microbiol. 2011;75:111–143. [DOI] [PubMed] [Google Scholar]
- 8. Macfarlane S, Macfarlane GT. Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl Environ Microbiol. 2006;72:6204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Swidsinski A, Weber J, Loening-Baucke V, et al. . Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. [DOI] [PubMed] [Google Scholar]
- 11. Ceri H, Olson ME, Stremick C, et al. . The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sproule-Willoughby KM, Stanton MM, Rioux KP, et al. . In vitro anaerobic biofilms of human colonic microbiota. J Microbiol Methods. 2010;83:296–301. [DOI] [PubMed] [Google Scholar]
- 13. Beatty JK, Akierman SV, Motta JP, et al. . Giardia duodenalis induces pathogenic dysbiosis of human intestinal microbiota biofilms. Int J Parasitol. 2017;47:311–26. [DOI] [PubMed] [Google Scholar]
- 14. Swidsinski A, Ladhoff A, Pernthaler A, et al. . Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. [DOI] [PubMed] [Google Scholar]
- 15. Barbara G, Scaioli E, Barbaro MR, et al. . Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut. 2017;66:1252–61. [DOI] [PubMed] [Google Scholar]
- 16. Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. Plos Pathog. 2010;6:e1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chua SL, Liu Y, Yam JK, , et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462. [DOI] [PubMed] [Google Scholar]
- 18. Morse JW, Millero FJ, Cornwell JC, et al. . The chemistry of the hydrogen-sulfide and iron sulfide systems in natural-waters. Earth-Sci Rev. 1987;24:1–42. [Google Scholar]
- 19. Dinis TCP, Maderia VM, Almeida LM. Action of phenolic derivatives (acetaminophen, salicylate, and 5-aminosalicylate) as inhibitors of membrane lipid peroxidation and as peroxyl radical scavengers. Arch Biochem Biophys. 1994;315:161–9. [DOI] [PubMed] [Google Scholar]
- 20. Vogel-Scheel J, Alpert C, Engst W, , et al. Requirement of purine and pyrimidine synthesis for colonization of the mouse intestine by Escherichia coli. Appl Environ Microbiol. 2010;76:5181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiang SL, Mekalanos JJ. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. [DOI] [PubMed] [Google Scholar]
- 22. Samant S, Lee H, Ghassemi M, , et al. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 2008;4:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reifen R, Nissenkorn A, Matas Z, et al. . 5-ASA and lycopene decrease the oxidative stress and inflammation induced by iron in rats with colitis. J Gastroenterol. 2004;39:514–9. [DOI] [PubMed] [Google Scholar]
- 24. Graham PM, Li JZ, Dou X, et al. . Protection against peroxynitrite-induced DNA damage by mesalamine: implications for anti-inflammation and anti-cancer activity. Mol Cell Biochem. 2013;378:291–8. [DOI] [PubMed] [Google Scholar]
- 25. Gionchetti P, Guarnieri C, Campieri M, et al. . Scavenger effect of sulfasalazine, 5-aminosalicylic acid, and olsalazine on superoxide radical generation. Dig Dis Sci. 1991;36:174–8. [DOI] [PubMed] [Google Scholar]
- 26. Rousseaux C, Lefebvre B, Dubuquoy L, et al. . Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. J Exp Med. 2005;201:1205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaufman J, Griffiths TA, Surette MG, et al. . Effects of mesalamine (5-aminosalicylic acid) on bacterial gene expression. Inflamm Bowel Dis. 2009;15:985–96. [DOI] [PubMed] [Google Scholar]
- 28. Andrews CN, Griffiths TA, Kaufman J, et al. . Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2011;34:374–83. [DOI] [PubMed] [Google Scholar]
- 29. Dahl JU, Gray MJ, Bazopoulou D, et al. . The anti-inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol. 2017;2:16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallace JL, Wang R. Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat Rev Drug Discov. 2015;14:329–45. [DOI] [PubMed] [Google Scholar]
- 31. Fiorucci S, Orlandi S, Mencarelli A, et al. . Enhanced activity of a hydrogen sulphide-releasing derivative of mesalamine (ATB-429) in a mouse model of colitis. Br J Pharmacol. 2007;150:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stein J, Hartmann F, Dignass AU. Diagnosis and management of iron deficiency anemia in patients with IBD. Nat Rev Gastroenterol Hepatol. 2010;7:599–610. [DOI] [PubMed] [Google Scholar]
- 33. Kortman GA, Dutilh BE, Maathuis AJ, et al. . Microbial metabolism shifts towards an adverse profile with supplementary iron in the tim-2 in vitro model of the human colon. Front Microbiol. 2015;6:1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Werner T, Wagner SJ, Martínez I, et al. . Depletion of luminal iron alters the gut microbiota and prevents Crohn’s disease-like ileitis. Gut. 2011;60:325–33. [DOI] [PubMed] [Google Scholar]
- 35. Jaeggi T, Kortman GA, Moretti D, et al. . Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64:731–42. [DOI] [PubMed] [Google Scholar]
- 36. Blackler RW, Motta JP, Manko A, , et al. Hydrogen sulphide protects against NSAID-enteropathy through modulation of bile and the microbiota. Br J Pharmacol. 2015;172:992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Switzer RL. Discoveries in bacterial nucleotide metabolism. J Biol Chem. 2009;284:6585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elli M, Zink R, Rytz A, et al. . Iron requirement of lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol. 2000;88:695–703. [DOI] [PubMed] [Google Scholar]
- 39. Kelley EE. A new paradigm for XOR-catalyzed reactive species generation in the endothelium. Pharmacol Rep. 2015;67:669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. So A, Thorens B. Uric acid transport and disease. J Clin Invest. 2010;120:1791–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee KA, Kim SH, Kim EK, et al. . Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. 2013;153:797–811. [DOI] [PubMed] [Google Scholar]
- 42. Bullen JJ, Rogers HJ, Spalding PB, , et al. Iron and infection: the heart of the matter. FEMS Immunol Med Microbiol. 2005;43:325–30. [DOI] [PubMed] [Google Scholar]
- 43. Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76:1756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DePas WH, Hufnagel DA, Lee JS, , et al. Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2005;102:11076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jomova K, Valko M. Importance of iron chelation in free radical-induced oxidative stress and human disease. Curr Pharm Des. 2011;17:3460–73. [DOI] [PubMed] [Google Scholar]
- 47. Harrison JJ, Ceri H, Yerly J, et al. . The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the calgary biofilm device. Biol Proced Online. 2006;8:194–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Caporaso JG, Lauber CL, Walters WA, et al. . Ultra-high-throughput microbial community analysis on the illumina HiSeq and MiSeq platforms. Isme J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cole JR, Wang Q, Fish JA, et al. . Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 51. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. Plos One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Buszewski B, Noga S. Hydrophilic interaction liquid chromatography (HILIC)–a powerful separation technique. Anal Bioanal Chem. 2012;402:231–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gerbaba TK, Gupta P, Rioux K, et al. . Giardia duodenalis-induced alterations of commensal bacteria kill Caenorhabditis elegans: a new model to study microbial-microbial interactions in the gut. Am J Physiol Gastrointest Liver Physiol. 2015;308:G550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wallace JL, Keenan CM. An orally active inhibitor of leukotriene synthesis accelerates healing in a rat model of colitis. Am J Physiol. 1990;258:G527–G534. [DOI] [PubMed] [Google Scholar]
- 56. Wallace JL, Le T, Carter L, et al. . Hapten-induced chronic colitis in the rat: alternatives to trinitrobenzene sulfonic acid. J Pharmacol Toxicol Methods. 1995;33:237–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.