Abstract
Background
Animal mitochondrial genomes typically encode 37 genes: 13 proteins, 22 tRNAs and two rRNAs. However, many species represent exceptions to that rule. Bivalvia along with Nematoda and Platyhelminthes are often suspected to fully or partially lack the ATP synthase subunit 8 (atp8) gene. This raises the question as to whether they are really lacking this gene or is this maybe an annotation problem? Among bivalves, Mytilus edulis has been inferred to lack an ATP8 gene since the characterization of its mitochondrial genome in 1992. Even though recent bioinformatic analyses suggested that atp8 is present in Mytilus spp., due to high divergence in predicted amino acid sequences, the existence of a functional atp8 gene in this group remains controversial.
Results
Here we demonstrate that M. edulis mitochondrial open reading frames suggested to be atp8 (in male and female mtDNAs) are actively translated proteins. We also provide evidence that both proteins are an integral part of the ATP synthase complex based on in-gel detection of ATP synthase activity and two-dimensional Blue-Native and SDS polyacrylamide electrophoresis.
Conclusion
Many organisms (e.g., Bivalvia along with Nematoda and Platyhelminthes) are considered to be lacking certain mitochondrial genes often only based on poor similarity between protein coding gene sequences in genetically closed species. In some situations, this may lead to the inference that the ATP8 gene is absent, when it is in fact present, but highly divergent. This shows how important complementary role protein-based approaches, such as those in the present study, can provide to bioinformatic, genomic studies (i.e., ability to confirm the presence of a gene).
Keywords: ATP8, Western blot, Bivalvia, Blue Native, Mitochondrial DNA, Doubly uniparental inheritance
Introduction
The blue mussel Mytilus edulis is one of the bivalve species possessing the unusual system of doubly uniparental inheritance (DUI) of mitochondria (Skibinski, Gallagher & Beynon, 1994; Boore, Medina & Rosenberg, 2004). Contrary to strictly maternal inheritance (SMI) of mtDNA found in other animal species, male Mytilus spp. mussels have two different mitogenomes. One is inherited from the father (M-type mtDNA; located mainly in male germ line cells), and the second from the mother (F-type mtDNA; located in female germ line cells and in somatic cells of both sexes) (Zouros et al., 1994). Estimated K2P genetic distance (Kimura two-parameter corrected for multiple substitutions) between M and F mtDNAs reaches 0.245 (Zbawicka, Burzyński & Wenne, 2007), and while the presence of two mitogenomes (heteroplasmy) has been demonstrated several times using DNA-based methods (Garrido-Ramos et al., 1998; Dalziel & Stewart, 2002; Zbawicka, Skibinski & Wenne, 2003; Obata et al., 2006; Kyriakou, Zouros & Rodakis, 2010), proteomic approaches have not been much explored yet (Chakrabarti et al., 2006). Table 1 shows mean pairwise distance between male and female M. edulis mitochondrial proteins, which reaches 0.386 for ATP8.
Table 1. Mean pairwise distance between male and female M. edulis mitochondrial proteins (GenBank HM489874 and MF407676) (Zbawicka, Burzyński & Wenne, 2007; Kumar, Stecher & Tamura, 2016).
| Protein | Mean pairwise distance |
|---|---|
| COX1 | 0.047 |
| COX3 | 0.093 |
| COX2 | 0.112 |
| ND3 | 0.112 |
| CYTB | 0.140 |
| ND4 | 0.143 |
| ATP6 | 0.155 |
| NAD4L | 0.172 |
| ND1 | 0.186 |
| ND5 | 0.196 |
| ND2 | 0.210 |
| ND6 | 0.221 |
| *ATP8 | 0.386 |
Notes.
The atp8 gene lacks annotation.
M. edulis was the first bivalve species with a nearly completely sequenced mitogenome (Hoffmann, Boore & Brown, 1992). The F-type mtDNA was annotated with 37 genes: two ribosomal RNAs, 23 tRNAs (with an additional tRNAmet gene) and only 12 protein-coding genes, i.e., the mitogenome was lacking the atp8 gene (GenBank Acc. No. AY484747 (Hoffmann, Boore & Brown, 1992)). This publication created a belief that bivalves might lack atp8, although a few bivalvian mitogenomes published later had this gene annotated. Putative atp8 gene in Mytilus F and M mitochondrial genomes was identified in 2010 (Breton, Stewart & Hoeh, 2010; Smietanka, Burzyński & Wenne, 2010). Difficulties with the annotation of this gene most probably originated from the interspecies differences in coded protein amino acid sequence.
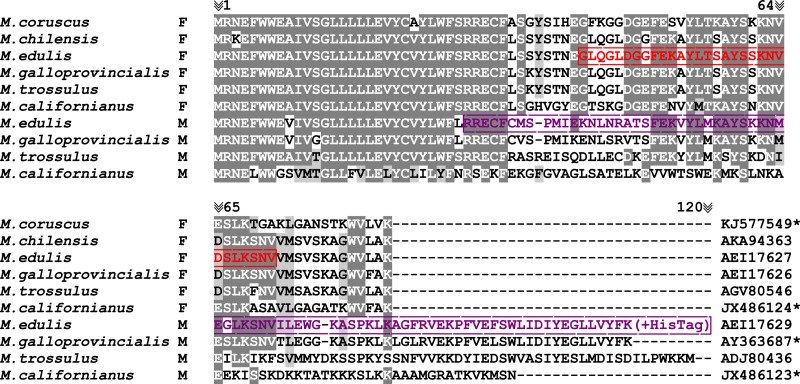
ATP8 is a part of the non-catalytic hydrophobic membrane component (Fo) of the ATP synthase (F1Fo) complex (Rak, Gokova & Tzagoloff, 2011; Lee et al., 2015). It is a short protein composed of 37–70 amino acids (6,784 out of 7,301 ATP8 protein sequences deposited in Genbank RefSeq database), and 94% of known sequences (i.e., 6,873 out of 7,301) start with a conserved MPQ tripeptide and possess one predicted transmembrane domain (Papakonstantinou et al., 1996; Gissi, Iannelli & Pesole, 2008). The predicted ATP8 in Mytilus mitochondrial genomes also possesses one predicted transmembrane domain but lacks the MPQ sequence at the N-terminus of the peptide and the length of the protein varies from 84 aa in the F-type mtDNA to 106–128 aa in the M-type mtDNA (Fig. 1).
Figure 1. Mytilus. spp. ATP8 protein alignment.
JX486123; JX486124; KJ577549; AY363687, *Protein sequences extracted from mtDNAs without annotated atp8 gene. Alignment colored with BoxShade ExPASy online tool with 0.6 shading parameter (grey color). Red color represents FATP8-antigen peptide sequence used for immunisation; violet color represents MATP8-antigen protein sequence used for immunisation (Mizi et al., 2005; Ort & Pogson, 2007; Smietanka, Burzyński & Wenne, 2010; Lee & Lee, 2014; Sańko & Burzyński, 2014; Gaitán-Espitia et al., 2016; Zbawicka, Wenne & Burzyński, 2014).
Bioinformatic analyses of Mytilus spp. mitogenomes (Breton, Stewart & Hoeh, 2010; Smietanka, Burzyński & Wenne, 2010) have shown that this “putative atp8 gene” in both F and M mtDNAs possesses a pattern of evolution expected for a protein-coding gene evolving under purifying selection (i.e., the 3rd>1st>2nd codon pattern of evolution). Furthermore, both F and M sequences are actively transcribed in Mytilus species (based on EST sequences), and comparison of protein hydropathy predictions with ATP8 proteins from other bivalves revealed similar profiles (Breton, Stewart & Hoeh, 2010; Smietanka, Burzyński & Wenne, 2010). However, due to its high divergence in predicted amino acid sequences compared to other ATP8, the existence of a functional atp8 gene in Mytilus spp. remains controversial. In fact, some authors have suggested that sequences annotated as atp8 in Mytilus spp. could represent a pseudogene (Uliano-Silva et al., 2016).
Here we demonstrate that the open reading frames (ORFs) suggested to be atp8 in both M. edulis F and M mtDNAs are actively expressed and effectively represent parts of the ATP synthase complex.
Materials and Methods
Western blot
Living specimens of M. edulis mussels were bought at a local market in January and September 2016 (Collecting area: the Oosterschelde and the Waddenzee bay; Netherlands). The sex of each individual was determined by examination of gonadal tissues for the presence of sperm or eggs under light microscope. Ten pairs of male and female individuals where checked during the course of the experiment. Mantle with gonads, gill, foot and hepatic gland tissues of male and female specimens were carefully sectioned with sterile scalpels to minimize possibility of cross contamination of tissue samples and subsequently washed in sterile water and stored frozen at −20 °C for further analyses. Approximately 100 mg of every tissue type were suspended in 1 ml of Radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, pH 7,6) supplemented with 5 µl of protease inhibitors cocktail (Sigma-Aldrich, St. Louis, MO, USA), homogenized at 20,000 RPM for 20 s (Heidolph SilentCrusher M; tool 8F; Sigma-Aldrich, St. Louis, MO, USA) and sonicated (Vibra-Cell; Sonics, Newton, CT, USA) for 20 s; amplitude 50%. Samples were kept on ice during the whole protein isolation process. Isolates were then centrifuged at 15,000× g for 4 min, to separate insoluble residues, transferred into new vials and stored frozen or used immediately in further steps. 40–60 µg of crude protein isolates per sample were separated by SDS-PAGE electrophoresis (5% stacking gel; 10% separating gel in 6 M urea to protect low molecular weight bands from diffusing and smearing during electrophoresis; 30 min at 80 V and 1 h at 150 V) with cooling (BlueStar; DNAGdansk; Gdańsk, Poland) and transferred to membrane (OWL electroblotting semidry system). Due to the low molecular weight of targeted proteins (FATP8 9,5∼kDa; MATP8∼13 kDa), 0.2 µm PVDF or nitrocellulose membranes (GE Healthcare, Little Chalfont, UK) were used and electroblotting time did not exceed 30 min at 200 mV (longer transfer times were causing over transfer of small proteins through membranes onto the Whatman filter papers).
The remaining procedures were performed in the standard manner: membrane blocking in 4% low fat dry milk in Phosphate-buffered saline buffer for 1 h, overnight incubation with primary polyclonal antibody dilution 1:5,000 at 4 °C, rinsing with 0.05% Tween-20 in PBS 3 × 5 min, incubation with anti-rabbit secondary HRP-conjugated monoclonal antibody dilution 1:10,000 for 1 h (Sigma-Aldrich, St. Louis, MO, USA), colorimetric immunodetection with DAB (SigmaAldrich) or DAB supplemented with Co or Ni ions. The whole procedure was repeated at least 6 times with different pairs of male and female specimens and all tested specimens gave coherent results.
Polyclonal antibodies were custom-made by GenScript. It is worth mentioning that due to the high similarity of the M and FATP8 protein N-terminus sequences, their low antigenicity and predicted difficulties with solubility, the N-terminal hydrophobic domain of both M and FATP8 proteins has been removed from the sequences at the antibody designing step (Fig. 1). The MATP8 antigen was acquired through expression in bacterial host with HisTag on C-term of the peptide and the FATP8 peptide was chemically synthetized and purified before immunisation. Antibodies were acquired from rabbits after a triple immunization procedure.
BN-PAGE/SDS-PAGE
Small specimens of M. edulis were sexed and sectioned for mitochondrial isolation (the whole body, around 300 mg, without the hepatic gland was used for the females and the ripe mantle/gonad tissues were used for the male individuals). Tissues were homogenised 10 s at 4,000 RPM (Heidolph SilentCrusher M; tool 8F), with 1.5 ml of 440 mM sucrose, 1 mM ethylenediaminetetraacetic acid, 20 mM 3-(N-morpholino) propanesulfonic acid, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate, pH 7.2 and subsequently centrifugated at 5,000× g for 10 min. The supernatant was discarded and the remaining disrupted tissue pellet was suspended in 1.5 ml isolation buffer (1 M aminocaproic acid, 50 Mm Bis-Tris, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate pH 7.0) and homogenised 10 s at 20,000 RPM (Dabbeni-sala et al., 1997). Whole mitochondria were collected by differential centrifugation with the first centrifugation at 5,000× g for 20 min at 4 °C to remove unbroken cells and bigger cell fragments, followed by a second centrifugation of the remaining supernatant (25,000× g, 20 min, 4 °C) to pellet mitochondria and other small cell organelles. The resultant mitochondrial pellets were then solubilized in 100 µl 19:1 solution of isolation buffer and Triton X-100. After 5 min incubation, samples were centrifuged again at 25,000× g for 10 min at 4 °C. Typical Blue Native 4–15% gradient gel (Eubel, Braun & Millar, 2005; Wittig, Braun & Scha, 2006; Fiala, Schamel & Blumenthal, 2011) was substituted with discontinuous gradient gel (layers of different percentage acrylamide gels 4/5/6/8/10/15%). Before loading, every 15 µl of sample was supplied with 5 µl of 10% Coomassie G-250.
In-gel visualisation of Complex V (ATP synthase) was performed through overnight incubation of Blue Native gel in 35 mM Tris Base, 276 mM Glycine, 14 mM MgCl2, 0.2% Pb(NO3)2 and 8 mM ATP pH∼7.8 as in (Dabbeni-sala et al., 1997). Gel fragments containing stained ATPase complex were then cut out, incubated 45min in SDS-PAGE running buffer to dissociate and denature ATP synthase complexes to individual subunits and loaded horizontally on standard 10% (5% stacking) SDS-PAGE gel. Electrophoresis and western blot procedure were performed as described above. Enzymatic detection was completed according to manufacturer’s protocol with Ultra-Sensitive ABC Peroxidase Rabbit IgG Staining Kit (Thermo Fisher, Waltham, MA, USA). Remaining Blue Native gel fragments were slightly decolorized by longer (1–2 h) incubation in SDS containing Tris-glycine buffer and electroblotted together with gels after two dimensional electrophoresis (presence of Coomassie G-250 in Blue Native gels hinders later detection due to the high binding affinity to the hydrophobic blotting membranes). Two-dimensional BN/SDS PAGE procedure was repeated with at least 6 male and 6 female specimens.
Results
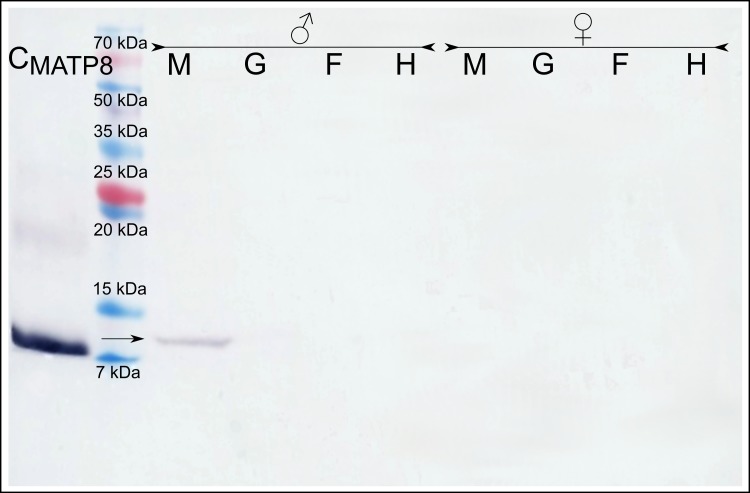
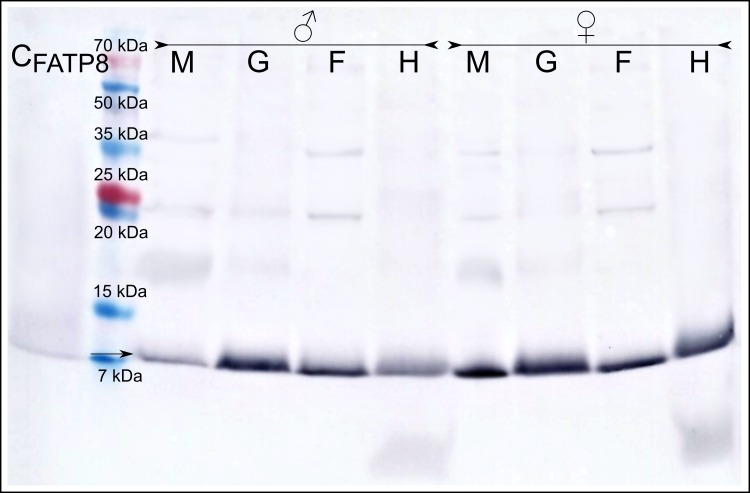
Immunodetection with the anti-MATP8 antibody gave positive results only in male mantle/gonad tissue (Fig. 2). The size of the signal generated was highly similar to the predicted molecular weight of MATP8 (∼13.5 kDa) and also highly specific with no additional protein bands. No signal was visible in other male and female tissue samples. Contrary to anti-MATP8, anti-FATP8 antibody gave signal for every tissue (mantle, gill, foot, hepatic gland) both for male and female specimens (Fig. 3). This signal also corresponded to the predicted protein molecular weight of FATP8 (∼10 kDa). The signal for FATP8 in male mantle/gonad tissue was visibly weaker than the signals for other tissues. The anti-FATP8 antibody was also less specific then the anti-MATP8. Non-specific bands were detectable on levels corresponding for proteins larger than 15 kDa.
Figure 2. Male ATP8: tissue segregation.
CMATP8- positive control; M-mantle tissue where gonads are localized in Mytilus spp.; G-gills; F-foot; H-hepatic gland, arrow indicates detected male version of ATP8 protein. The signal detected in male mantle/gonad tissue highly corelates with predicted 13.5 kDa molecular weight of MATP8 protein. As expected (Skibinski, Gallagher & Beynon, 1994), MATP8 is absent in all other tissues.
Figure 3. Female ATP8: tissue segregation.
CFATP8-positive control; M-mantle with gonads; G-gills; F-foot; H-hepatic gland, arrow indicates detected female version of ATP8 protein. Signal for FATP8 (predicted molecular weight 10 kDa) is present in all tissues both in male and female individuals. FATP8 and MATP8 (Fig. 2) are both present in the male mantle tissue containing inseparable gonads and somatic cells.
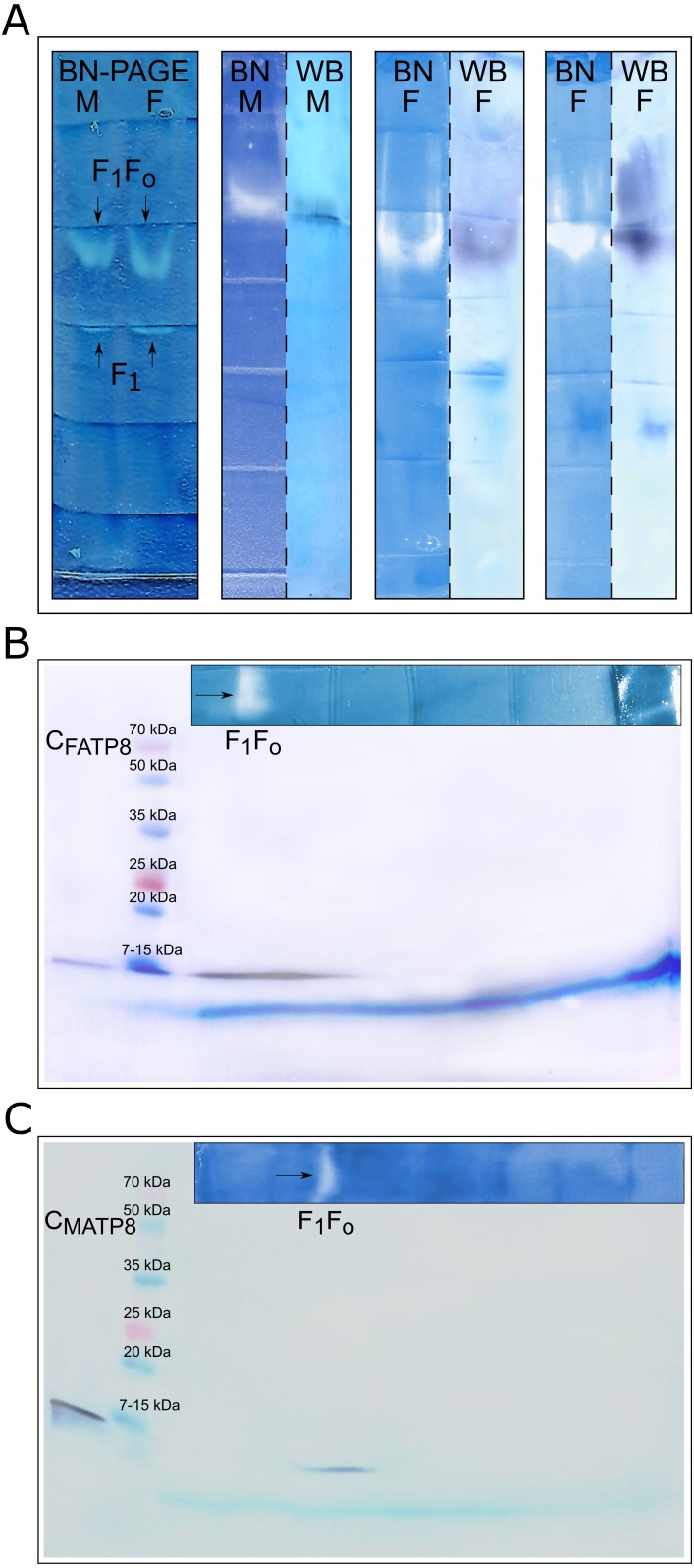
In-gel detection (Fig. 4) of F and M ATP synthase activity (after Blue Native electrophoresis) indicated the presence of one or two white lead phosphate precipitates (depending on the sample). The higher band corresponded to the whole Complex V (F1Fo), whereas the lower band corresponded to the dissociated catalytic part (F1). Blue Native gel parts blotted after slight decolorization from Coomassie G-250 and dissociation in SDS containing Tris-glycine buffer resulted with signals matching in-gel activity spots. Both male and female mitochondrial isolates separated by two-dimensional BN-PAGE/SDS-PAGE electrophoresis also gave positive results. Signals on the membrane were present for separated parts of the whole F and M ATP synthase complexes (F1Fo) at expected molecular weight positions. No nonspecific bands were observed.
Figure 4. ATP8 as an integral part of the ATPase complex.
(A) Enzymatic staining of Blue native gels and immunodetection of F1Fo and F1 (catalytic) parts of ATPase complex: white and white-blue bands represent in-gel localization of ATP synthase complex after enzymatic staining; dark purplish blue bands show immunodetection of MATP8 (in M), FATP8 (in F) as well as colocalization of those proteins with ATP synthase complex (white bands in BN); BN-Blue native polyacrylamide gel enzymatic staining, WB-Western blot immunodetection, F-female specimen, M-male specimen. (B) Immunodetection of ATP8 female version after two-dimensional SDS-PAGE electrophoresis: CFATP8-control, F1Fo-whole ATP synthase complex. (C) Immunodetection of ATP8 male version after two-dimensional SDS-PAGE electrophoresis: CMATP8-control, F1Fo-whole ATP synthase complex.
Discussion
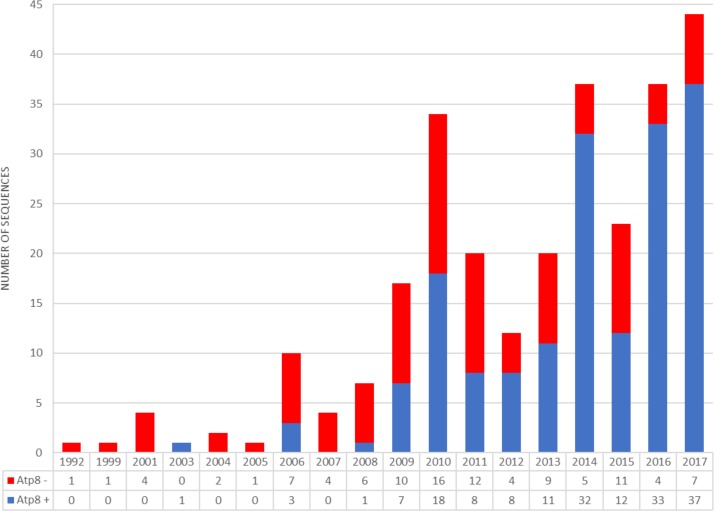
Before 2009, only five out of 31 bivalve mitochondrial genomes deposited in GenBank have featured atp8 (Serb & Lydeard, 2003; Dreyer & Steiner, 2006). Since 2009, a constant increase in the number of bivalve mitogenomes annotated with atp8 has been observed (Fig. 5; Data S1). However, at the end of 2017 only 62% of all bivalvian mtDNAs available in GenBank had an annotated atp8 gene and only a small amount (if any at all) of old submissions appeared to have been corrected. Although the percentage of annotated atp8 in bivalve mitogenomes published between 2010 and 2017 equals 77% ( Data S1), there still remain some scientists publishing bivalve mtDNAs without this gene, even within the genus Mytilus (e.g., M. coruscus Lee & Lee, 2014), where amino acid sequences for this protein are very similar among species (Fig. 1). As mentioned above, some authors have suggested that sequences annotated as atp8 in Mytilus spp. could represent a pseudogene (Uliano-Silva et al., 2016).
Figure 5. Number of bivalvian mitochondrial genomes with and without annotated atp8 gene deposited since 1992 in GenBank database.
In the present study, protein products coded by both M and F mitochondrial ORFs suggested to be atp8 were detected in male and female M. edulis mussels. The female FATP8 was present in every studied tissue (male and female mantle, gill, foot, hepatic gland). In contrast, MATP8 was shown to be present only in male mantle/gonad, a result that was expected because male mitochondrial genome in Mytilus spp. has been observed predominantly in gonad tissues and is the only mtDNA present in sperm cells (Skibinski, Gallagher & Beynon, 1994). There are reports suggesting leakage of small amounts of M mtDNA to somatic tissues (Garrido-Ramos et al., 1998; Dalziel & Stewart, 2002; Zbawicka, Skibinski & Wenne, 2003; Obata et al., 2006; Kyriakou, Zouros & Rodakis, 2010). However, the western blot technique is less sensitive than the polymerase chain reaction (PCR), and this could explain why we did not detect the MATP8 in somatic tissues. Also, our results should not be extrapolated to all bivalvian species e.g., Venerupis philipinarium, where gonad is located within main body of the clams and signal from male mtDNA is predominant in most of male somatic tissues (Ghiselli, Milani & Passamonti, 2011).
Question: Is this protein active?
In-gel detection of ATP synthase activity and two-dimensional Blue Native and SDS polyacrylamide electrophoreses suggested that proteins detected by anti-MATP8 and anti-FATP8 antibodies are integral parts of the ATPase complex in M. edulis. Straightforward blotting of Blue Native gels also supported these results.
Conclusions
Based on protein sequence similarities (Fig. 1) and the results above, we consider it likely that active M and FATP8 proteins are present not only in M. edulis but through the genus Mytilus. Even though this gives us no right to claim that the whole Bivalvia class possesses an atp8 gene, we strongly encourage scientists to focus more attention on the subject of presence and absence of this gene in bivalve mitochondrial genomes. Especially because similar atp8 annotation problems have been observed in other organisms (flatworms (Nickisch-Rosenegk, Brown & Boore, 2000; Egger, Bachmann & Fromm, 2017) and nematodes (Hyman, 1988; Lavrov & Brown, 2001)) and because only proteomic based experimental approaches are capable of unambiguously resolving issues concerning “uncertain” protein genes.
Supplemental Information
List of accession numbers.
Funding Statement
This work was supported by the Polish National Science Center (NCN) grant no. UMO-201 2/06/M/NZ3/00051 to Artur Burzyński. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Marek Lubośny performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Aleksandra Przyłucka performed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Beata Śmietanka and Artur Burzyński conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Sophie Breton conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Bivalvian Mitochondrial DNA sequences are available at GenBank with accession numbers: AB055624-NC_036488. See Supplemental Information 1 for a full list of accession numbers.
The raw data are included in the Figures in the manuscript.
References
- Boore, Medina & Rosenberg (2004).Boore JL, Medina M, Rosenberg LA. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the Bivalve Mytilus edulis. Molecular Biology and Evolution. 2004;21:1492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- Breton, Stewart & Hoeh (2010).Breton S, Stewart DT, Hoeh WR. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: Mytilidae): identification of the “missing” ATPase 8 gene. Marine Genomics. 2010;3:11–18. doi: 10.1016/j.margen.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Chakrabarti et al. (2006).Chakrabarti R, Walker JM, Stewart DT, Trdan RJ, Vijayaraghavan S, Curole JP, Hoeh WR. Presence of a unique male-specific extension of C-terminus to the cytochrome c oxidase subunit II protein coded by the male-transmitted mitochondrial genome of Venustaconcha ellipsiformis (Bivalvia: Unionoidea) FEBS Letters. 2006;580:862–866. doi: 10.1016/j.febslet.2005.12.104. [DOI] [PubMed] [Google Scholar]
- Dabbeni-sala et al. (1997).Dabbeni-sala F, Farmacologia D, Scienze D, Padova U. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis. 1997;18:2059–2064. doi: 10.1002/elps.1150181131. [DOI] [PubMed] [Google Scholar]
- Dalziel & Stewart (2002).Dalziel AC, Stewart DT. Tissue-specific expression of male-transmitted mitochondrial DNA and its implications for rates of molecular evolution in Mytilus mussels (Bivalvia: Mytilidae) Genome. 2002;45:348–355. doi: 10.1139/G01-159. [DOI] [PubMed] [Google Scholar]
- Dreyer & Steiner (2006).Dreyer H, Steiner G. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica—and the first record for a putative Atpase subunit 8 gene in marine bivalves. Frontiers in Zoology. 2006;3:13. doi: 10.1186/1742-9994-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger, Bachmann & Fromm (2017).Egger B, Bachmann L, Fromm B. Atp8 is in the ground pattern of flatworm mitochondrial genomes. BMC Genomics. 2017;18:414. doi: 10.1186/s12864-017-3807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel, Braun & Millar (2005).Eubel H, Braun H, Millar AH. Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods. 2005;1:11. doi: 10.1186/1746-4811-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala, Schamel & Blumenthal (2011).Fiala GJ, Schamel WWA, Blumenthal B. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. Journal of Visualized Experiments. 2011;48:e2164. doi: 10.3791/2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitán-Espitia et al. (2016).Gaitán-Espitia JD, Quintero-galvis JF, Mesas A, Elía GD. Mitogenomics of southern hemisphere blue mussels (Bivalvia: Pteriomorphia): insights into the evolutionary characteristics of the Mytilus edulis complex. Scientific Reports. 2016;6:26853. doi: 10.1038/srep26853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Ramos et al. (1998).Garrido-Ramos MA, Stewart DT, Sutherland BW, Zouros E. The distribution of male-transmitted and female-transmitted mitochondrial DNA types in somatic tissues of blue mussels: implications for the operation of doubly uniparental inheritance of mitochondrial DNA. Genome. 1998;41:818–824. doi: 10.1139/g98-081. [DOI] [Google Scholar]
- Ghiselli, Milani & Passamonti (2011).Ghiselli F, Milani L, Passamonti M. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae) Molecular Biology and Evolution. 2011;28:949–961. doi: 10.1093/molbev/msq271. [DOI] [PubMed] [Google Scholar]
- Gissi, Iannelli & Pesole (2008).Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Hoffmann, Boore & Brown (1992).Hoffmann RJ, Boore JL, Brown WM. A novel mitochondrial genome organization for the blue mussel, Mytilus-Edulis. Genetics. 1992;131:397–412. doi: 10.1093/genetics/131.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman (1988).Hyman BC. Nematode mitochondrial DNA: anomalies and applications. Journal of Nematology. 1988;20:523–531. [PMC free article] [PubMed] [Google Scholar]
- Kumar, Stecher & Tamura (2016).Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakou, Zouros & Rodakis (2010).Kyriakou E, Zouros E, Rodakis GC. The atypical presence of the paternal mitochondrial DNA in somatic tissues of male and female individuals of the blue mussel species Mytilus galloprovincialis. BMC Research Notes. 2010;3:222. doi: 10.1186/1756-0500-3-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov & Brown (2001).Lavrov DV, Brown WM. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics. 2001;157:621–637. doi: 10.1093/genetics/157.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2015).Lee J, Ding S, Walpole TB, Holding AN, Montgomery MG, Fearnley IM, Walker JE. Organization of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. Jurnal of Biological Chemistry. 2015;290:13308–13320. doi: 10.1074/jbc.M115.645283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee & Lee (2014).Lee Y, Lee Y. The F type mitochondrial genome of hard-shelled mussel: Mytilus coruscus (Mytiloida, Mytilidae) Mitochondrial DNA. 2014;27:624–625. doi: 10.3109/19401736.2014.908375. [DOI] [PubMed] [Google Scholar]
- Mizi et al. (2005).Mizi A, Zouros E, Moschonas N, Rodakis GC. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Molecular Biology and Evolution. 2005;22:952–967. doi: 10.1093/molbev/msi079. [DOI] [PubMed] [Google Scholar]
- Nickisch-Rosenegk, Brown & Boore (2000).Nickisch-Rosenegk M Von, Brown WM, Boore JL. Complete sequence of the mitochondrial genome of the tapeworm Hymenolepis diminuta: gene arrangements indicate that platyhelminths are eutrochozoans. Molecular Biology and Evolution. 2000;18:721–730. doi: 10.1093/oxfordjournals.molbev.a003854. [DOI] [PubMed] [Google Scholar]
- Obata et al. (2006).Obata M, Kamiya C, Kawamura K, Komaru A. Sperm mitochondrial DNA transmission to both male and female offspring in the blue mussel Mytilus galloprovincialis. Development, Growth & Differentiation. 2006;48:253–261. doi: 10.1111/j.1440-169x.2006.00863.x. [DOI] [PubMed] [Google Scholar]
- Ort & Pogson (2007).Ort BS, Pogson GH. Molecules of the California Sea Mussel, Mytilus californianus. Genetics. 2007;177:1087–1099. doi: 10.1534/genetics.107.072934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papakonstantinou et al. (1996).Papakonstantinou T, Law RHP, Nesbitt WS, Nagley P, Devenish RJ. Molecular genetic analysis of the central hydrophobic domain of subunit 8 of yeast mitochondrial ATP synthase. Current Genetics. 1996;30:12–18. doi: 10.1007/s002940050094. [DOI] [PubMed] [Google Scholar]
- Rak, Gokova & Tzagoloff (2011).Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. The EMBO Journal. 2011;30:920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sańko & Burzyński (2014).Sańko TJ, Burzyński A. Co-expressed mitochondrial genomes: recently masculinized, recombinant mitochondrial genome is co-expressed with the female—transmitted mtDNA genome in a male Mytilus trossulus mussel from the Baltic Sea. BMC Genetics. 2014;15:28. doi: 10.1186/1471-2156-15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serb & Lydeard (2003).Serb JM, Lydeard C. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca) Molecular Biology and Evolution. 2003;20:1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- Skibinski, Gallagher & Beynon (1994).Skibinski DOF, Gallagher C, Beynon CM. Sex-limited mitochondrial DNA transmission in the marine mussel. Genetics. 1994;138:801–809. doi: 10.1093/genetics/138.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smietanka, Burzyński & Wenne (2010).Smietanka B, Burzyński A, Wenne R. Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: rapidly evolving atp8. Journal of Molecular Evolution. 2010;71:385–400. doi: 10.1007/s00239-010-9393-4. [DOI] [PubMed] [Google Scholar]
- Uliano-Silva et al. (2016).Uliano-Silva M, Americo JA, Costa I, Schomaker-Bastos A, De Freitas Rebelo M, Prosdocimi F. The complete mitochondrial genome of the golden mussel Limnoperna fortunei and comparative mitogenomics of Mytilidae. Gene. 2016;577:202–208. doi: 10.1016/j.gene.2015.11.043. [DOI] [PubMed] [Google Scholar]
- Wittig, Braun & Scha (2006).Wittig I, Braun H, Scha H. Blue native PAGE. Nature Protocols. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Zbawicka, Burzyński & Wenne (2007).Zbawicka M, Burzyński A, Wenne R. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene. 2007;406:191–198. doi: 10.1016/j.gene.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zbawicka, Skibinski & Wenne (2003).Zbawicka M, Skibinski DOF, Wenne R. Doubly uniparental transmission of mitochondrial DNA length variants in the mussel Mytilus trossulus. Marine Biology. 2003;142:455–460. doi: 10.1007/s00227-002-0969-4. [DOI] [Google Scholar]
- Zbawicka, Wenne & Burzyński (2014).Zbawicka M, Wenne R, Burzyński A. Mitogenomics of recombinant mitochondrial genomes of Baltic Sea Mytilus mussels. Molecular Genetics and Genomics. 2014;289:1275–1287. doi: 10.1007/s00438-014-0888-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros et al. (1994).Zouros E, Ball AMYO, Saavedra C, Freeman KR. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of accession numbers.
Data Availability Statement
The following information was supplied regarding data availability:
Bivalvian Mitochondrial DNA sequences are available at GenBank with accession numbers: AB055624-NC_036488. See Supplemental Information 1 for a full list of accession numbers.
The raw data are included in the Figures in the manuscript.