Abstract
Background
Adalimumab is an anti-TNF biologic drug that is efficacious in the treatment of psoriasis. However, the effect of adalimumab on genome-wide gene expression changes in skin and peripheral blood is not well characterized.
Methods
Thirty adult subjects with > 10% body surface area of chronic plaque psoriasis were recruited for the study. Lesional skin and peripheral blood mononuclear cell samples prior to and one month following treatment with adalimumab were collected. The skin samples were analyzed using genome-wide RNAseq, and the blood samples were analyzed using genome-wide Affymetrix microarrays. Data preprocessing and analysis were conducted using the EdgeR and Affy packages in R/Bioconductor.
Results
In the skin, paired analysis before and after treatment revealed changes in pathways important to epidermal development and keratinocyte differentiation. Such important genes as keratin 6A and 6B, tubulin B6, desmocollin and desmoglein 3 were among the top differentially expressed genes. In peripheral blood, pathways involved in hematopoetic cell lineage and immune response were found to be differentially expressed, including genes such as the Fc receptor-like A and 5, as well as immunoglobulin heavy chains. Using a principal components approach, we show that expression of genes in post-treatment skin more closely resembles that of healthy controls.
Conclusion
Treatment of psoriasis with adalimumab appears to be associated with modulation of keratinocyte and epidermal proliferation in the skin and with immunologic changes in the blood. We discuss the ramifications of these findings for the treatment for psoriasis.
Introduction
Psoriasis is a chronic systemic inflammatory disease with cutaneous manifestations known to have an immune-mediated etiology. It is characterized by hyperproliferative epidermis and a mixed cutaneous lymphocytic infiltrate.1 This disease can have severely negative impact on patient quality of life.2–5
Though traditional systemic therapies have been shown to be effective for psoriasis, they are also associated with abundant side effects.2 Because of the limitations of traditional systemic therapies, newer biologic agents have shown promise in targeting specific cytokines in the inflammatory cascade thought to precipitate psoriasis. Of these biologic agents, therapies targeting tumor necrosis factor (TNF) have shown particular efficacy.6 TNF is a cytokine that plays multiple roles in the development and maintenance of psoriasis, include recruiting T cells to the skin and increasing the proliferation of keratinocytes.7
Adalimumab is an IgG1 isotype of a human monoclonal antibody that binds specifically with high affinity to human TNF.8 Adalimumab blocks the interaction of TNF with the p55 and p75 cell surface TNF receptors, thereby neutralizing TNF-mediated pathways.9 It has been shown to be efficacious in treating moderate to severe psoriasis in randomized controlled trials.6,10–12 Adalimumab may even be more effective than other systemic therapies, such as etanercept.13–15
Recently, Bose et al. (2013) examined the effect of anti-TNF therapy in skin and blood by cytokine assay and RTPCR. Surprisingly, they found that cytokines such as IL17, IL10 and IFN-gamma increased their expression in the peripheral blood following therapy. Additionally, the proliferative response of CD4+ T cells to TCR stimulation was enhanced. In the skin, in contrast, the expression of Th17/Th1 cytokine and early inflammatory genes were decreased in expression following treatment. The authors asserted that inhibition of the CCR7/CCL19 axis in lesional skin is an important event for the efficacy of treating psoriasis. These results have been replicated by other authors.16
In another study using RTPCR to investigate the effect of adalimumab on the key drivers in the pathogenesis of psoriasis, markers for innate immunity and epidermal differentiation and proliferation in psoriatic skin were rapidly restored to normal levels. In contrast, the genes of the adaptive immune system normalized in a delayed fashion.17
Genetic expression responses in patients with psoriasis to other immunologic agents have also been investigated, with varying effects found on dendritic cell activation, regulation of cytokine and chemokine expression, and innate and adaptive immunity.18,19 However, no studies to date have examined the effect of adalimumab treatment in the skin in relation to that in the blood using genome-wide expression analysis. Thus, we aimed to identify the genetic pathways affected by adalimumab treatment in the skin, then to compare these expression changes to those in blood mononuclear cells.
Methods
Participants and samples
Thirty adult subjects with chronic plaque psoriasis were recruited from the University of California San Francisco (UCSF) Dermatology Department. A board certified dermatologist confirmed the diagnosis of psoriasis. The participants were required to have affected body surface area > 10% and to not already be on systemic medications for their psoriasis. The demographics and clinical response of the participants with psoriasis are outlined in Table 1. All participants provided written informed consent for participating in the study. The study was approved by the UCSF institutional review board.
Table 1.
Demographics. Age, sex, BMI, and PASI score prior to (PASI 0) and following (PASI 1) treatment with adalimumab of each participant in the study.
| Study ID | AGE | SEX | BMI | PASI 0 | PASI 1 |
|---|---|---|---|---|---|
| HUM-001 | 48 | Male | 27.9 | 15.8 | 8.4 |
| HUM-002 | 46 | Male | 22.3 | 15.8 | 5.0 |
| HUM-003 | 39 | Male | 31.6 | 17.2 | 9.4 |
| HUM-004 | 53 | Male | NA | 8.0 | 2.8 |
| HUM-005 | 38 | Male | 28.1 | 12.1 | 9.5 |
| HUM-006 | 40 | Male | 27.3 | 17.9 | 8.6 |
| HUM-007 | 30 | Male | 41.6 | 17.2 | 16.4 |
| HUM-008 | 25 | Male | 20.5 | 22.5 | 11.3 |
| HUM-009 | 34 | Male | NA | 13.3 | 2.0 |
| HUM-010 | 34 | Male | 25.1 | 18.3 | 17.1 |
| HUM-011 | 34 | Male | 38.9 | 16.3 | 11.9 |
| HUM-012 | 25 | Female | 31.4 | 13.8 | 7.1 |
| HUM-013 | 30 | Female | NA | 22.6 | 13.2 |
| HUM-014 | 34 | Female | 25.4 | 8.5 | 2.9 |
| HUM-015 | 57 | Female | 27.5 | 13.7 | 8.4 |
| HUM-016 | 22 | Male | 29.8 | 14.6 | 8.3 |
| HUM-017 | 31 | Female | 34.6 | 19.3 | 13.3 |
| HUM-018 | 25 | Male | 29.8 | 13.2 | 1.3 |
| HUM-019 | 47 | Male | 31.7 | 15.6 | 8.9 |
| HUM-020 | 30 | Female | 32.5 | 16.4 | 9.2 |
| HUM-021 | 28 | Female | 24.0 | 13.6 | 8.3 |
| HUM-022 | 56 | Male | NA | 12.9 | 7.0 |
| HUM-023 | 32 | Male | 41.0 | 12.5 | 9.7 |
| HUM-024 | 35 | Male | 25.1 | 9.7 | 5.0 |
| HUM-025 | 43 | Male | 43.0 | 20.1 | 10.9 |
| HUM-026 | 39 | Male | 28.7 | 13.3 | 5.2 |
| HUM-027 | 41 | Female | 25.1 | 12.0 | 3.6 |
| HUM-029 | 46 | Male | 39.5 | 13.2 | 8.9 |
| HUM-030 | 24 | Male | 28.4 | 15.2 | 6.9 |
| HUM-031 | 24 | Female | 33.8 | 30.4 | 20.1 |
Five millimeter punch biopsies were taken from the edge of a psoriatic plaque of each patient. Two peripheral blood samples and 2 skin biopsies were taken from each participant, the first prior to the initiation of adalimumab and the second one month after starting treatment. In total, there were 30 psoriasis patients who provided blood samples, and of these patients 18 also provided skin samples. Sixteen normal skin samples were obtained from healthy control surgical discard specimens.
Skin sample RNA-seq analysis
The collected skin samples were stored in RNALater at −80ºC. The Bio-Gen Pro 200 homogenizer was used to homogenize the samples. The Qiagen RNeasy mini kit was used to extract whole RNA. The Agilent 2100 Bioanalyzer was used to assess RNA quality.
The Ribo-Zero Gold rRNA removal kit was used to remove ribosomal RNA. cDNA synthesis and library preparation was completed using ScriptSeq Complete kits (Human/Mouse/Rat) from Epicentre. The Agilent 2100 Bioanalyzer was again used to assess the quality and quantity of the library.
An Illumina HiSeq 2500 machine was used for RNA sequencing, using four samples per flow cell. An average of 52.3 million 101 bp paired end reads per sample were obtained, and FastQC was used to validate data quality. Trimmomatic (version 0.3) was applied for quality and adapter trimming. Tophat2 (version 2.0.9) was used to align reads to the human genome (hg19). Each library had an average of 34.8 million aligned paired ends (minimum of 25.3 million, maximum of 61.4 million).
Count data was generated by HTSeq-Count (v0.6.0) and analyzed in EdgeR for differential expression. Genes were filtered by count number >100 in at least 2 samples. A paired analysis design was applied in the linear model, and the genes showing a main effect of treatment were identified. Genes with an FDR < 0.05 were considered differentially expressed.
Blood sample microarray analysis
Peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples. Total RNA was extracted using the Qiagen RNeasy mini kit. The Agilent 2100 Bioanalyzer was used to assess the quality of the RNA. cDNA synthesis and array hybridization were performed as previously described.20
The resulting Affymetrix CEL files were preprocessed using the Affy package in Bioconductor. Cluster dendrograms, boxplots and density plots were used for quality control. One sample, 31, was a clear outlier and was removed from the analysis. Background removal was accomplished using the RMA method. Quantile normalization and PM correction with the Pmonly function was performed. Summarization was completed using the Median Polish technique.20 The microarray data also showed batch effects despite normalization, and so batch adjustment was conducted using the ComBat bioconductor package.21
Differential expression analysis was performed using the Limma package. A paired analysis design was applied in the linear model, and the main effect of treatment was queried. Genes with an adjusted p-value < 0.05 were considered differentially expressed.
Pathway analysis
The differential expression gene lists were submitted to DAVID bioinformatics Resources22 for analysis. Pathways, functional annotations, gene ontology results were considered to be significant if FDR < 0.05 and p < 0.05.
Principal components analysis
To gain a better overall picture of gene cluster expression changes, principal components analysis was conducted using the genes differentially expressed in the skin between controls and cases prior to treatment. After performing quality control to remove two outlier samples and any genes with no variance, EdgeR was applied to identify the main effect of disease, comparing patients prior to treatment and controls. Principal components analysis was performed. All analyses were done using R 3.2.0 (http://www.r-project.org).
Results
Skin RNAseq analysis reveals modulation of genes important in epidermal growth and differentiation following treatment with adalimumab
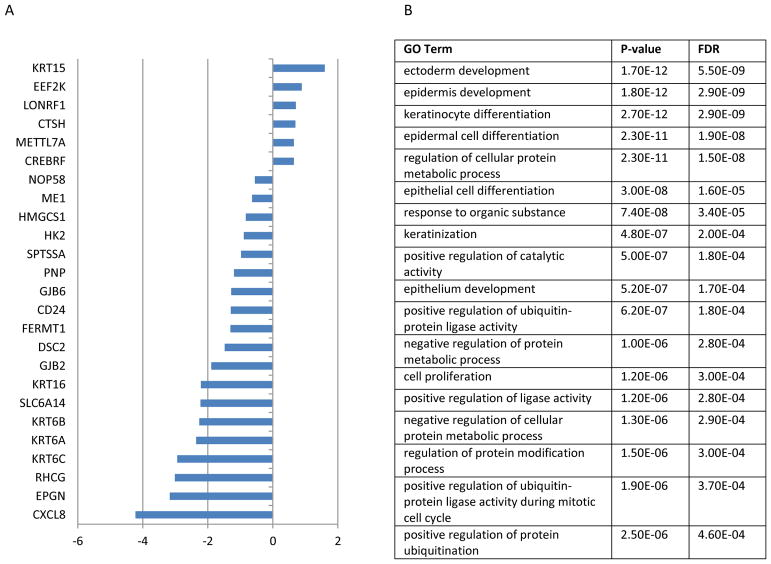
1138 genes were differentially expressed between the pre-treatment and post-treatment groups in a paired analysis (Figure 1). Among the most highly differentially expressed genes were KRT6C (logFC=−2.9, FDR=1.6E-11), KRT6B (logFC=−2.3, FDR=3.0E-11), KRT15 (logFC=1.6, FDR=1.0E-9), SLC6A14 (logFC=−2.2, FDR=7.5E-10), and LONRF1 (logFC=−0.71, FDR=7.5E-10). Prior analyses have also identified KRT (keratin) family and LONRF1 expression differences in psoriatic skin.23,24
Figure 1.
Differentially expressed skin genes and GO pathways after adalimumab treatment identified by RNAseq. A) Log 2 fold change of top 25 differentially expressed genes in the skin is listed. B) GO pathway term, p-value, false discovery rate (FDR) identified by DAVID enrichment analysis based on gene list.
Many gene ontology terms associated with epidermal growth and differentiation were enriched. They included ectoderm development (FDR=5.5E-9), epidermis development (FDR=2.9E-9), keratinocyte differentiation (FDR=2.9E-9), epidermal cell differentiation (FDR=1.9E-8), regulation of cellular protein metabolic process (FDR=1.5E-8), and epithelial cell differentiation (FDR=1.6E-5). These observations compliment the observation that active psoriatic lesions have increased numbers of EGF / TGF alpha receptors, which are thought to regulate the formation of lesions.25 In addition, the proteasomal pathway was enriched within this gene set (FDR=1.4E-5). Increased proteasomal activity, regulated post-transcriptionally, has previously been associated with psoriasis severity.26
The functional categories of genes were enriched in phosphoproteins (FDR=8.5E-55), acetylation (FDR= 5.0E-50), cytoplasm (FDR=9E-32), nucleotide-binding (FDR=3.9E-15) and ATP-binding (FDR=3.9E-15). The protein domains identified included the Tailless complex polypeptide 1 chaperone (FDR=5.0E-5), the Cpn60/TCP-1 Chaperonin (FDR=3.9E-5), TCP-1 Chaperonin (FDR=2.0E-4). Chaperonin is a highly conserved protein that regulates protein folding. Recombinant chaperonin has been found to improve severe plaque psoriasis,27 possibly through our identified pathways.
Blood microarray analysis reveals modulation of immunoglobulin-related immune response
79 genes were differentially expressed in the paired analysis seeking to identify the main effect of treatment on blood gene expression (Figure 2). Such immunologic genes as Fc receptor-like A and Fc receptor-like 5 were differentially expressed. Fc receptors are expressed in keratinocytes,28 and have been thought to mediate the epidermal immune response in psoriasis.29 A number of immunoglobulin heavy chains were also differentially expressed (IGHimmV3-30 (LogFC=−0.7, Adj p-value=6.6E-4), IGHV3-43 (LogFC=−1.0, Adj p-value=2.2E-4), IGHV3-11 (LogFC=−1.0, Adj p-value=2.9E-3), IGHV3-53 (LogFC=−0.8, Adj p-value=2.9E-3), IGHV4-61 (LogFC=−0.8, Adj p-value=5.7E-4), IGHV3-74 (LogFC=−0.8, Adj p-value=1.9E-3)). These expression differences may contribute to the psoriasis phenotype, as immunoglobulin heavy chain polymorphisms have previously been associated with psoriasis.30
Figure 2.
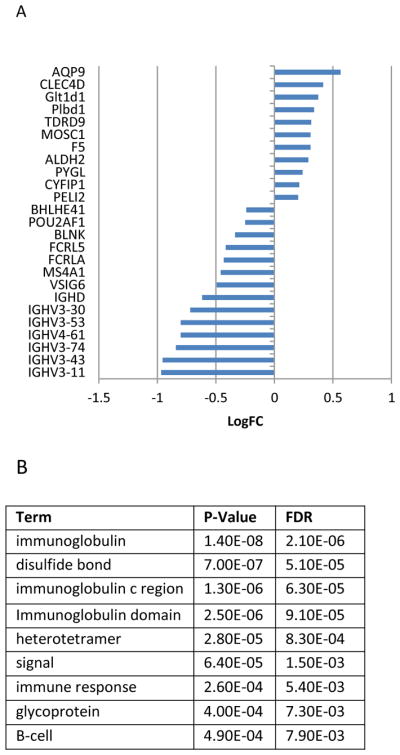
Differentially expressed peripheral blood mononuclear cell (PBMC) genes and Protein information resource (PIR) enrichment after adalimumab treatment identified by expression microarray. A) Log 2 fold change of top 25 differentially expressed genes in PBMCs is listed. B) PIR terms, p-values and false discovery rate (FDR) identified by DAVID enrichment analysis based on gene list.
Important pathways associated with the differentially expressed genes included hematopoetic cell lineage (FDR=6.7E-3). Many functional categories in the immune response domain, including immunoglobulin (FDR=2.1E-6), immunoglobulin C region (FDR=6.3E-5), immunoglobulin domain (FDR=2.5E-6), immune response (FDR=5.4E-3), and B-cell (FDR=7.9E-3) could be used to classify the differentially expressed genes. These pathways and functional categories have been well-documented to play a role in the uncontrolled proliferation of keratinocytes and recruitment of T cells to the skin.31
In the protein domain category, the differentially expressed genes showed strong enrichment in the domains of immunoglobulin-like (FDR=1.6E-6), immunoglobulin V-set (FDR=2.1E-6), immunoglobulin-like fold (FDR=1.6E-6), immunoglobulin V-set subgroup (FDR=9.3E-5), immunoglobulin major histocompatibility complex (FDR=1.3E-4), and immunoglobulin C1-set (FDR=4.8E-2). Intriguingly, prior studies have shown that immunoglobulin-like proteins are expressed in unique and specific patterns in normal and psoriatic skin, with altered protein expression patterns in psoriatic skin.32
Comparison of pre-treatment psoriatic skin, post-treatment psoriatic skin, and healthy control skin by principal components analysis
There were 2207 genes found to be differentially expressed when comparing healthy control skin and pre-treatment psoriatic skin. Among the top differentially expressed genes were Sox9 (logFC=2.4, FDR=1.1E-64), TMEM56 (logFC=5.7, FDR=8.6E-56), FADS2 (logFC=7.3, FDR=4.1E-58), FOXC1 (logFC=3.1, FDR=2.6E-54), and PLBD1 (logFC=−2.6, FDR=3.3E-54). Sox 9 is expressed in the basal layer of normal human epidermis, regulates epidermal keratinocyte proliferation and pro-survival and is known to be overexpressed in psoriasis.33 TMEM56 and FADS2 have both been previously associated with psoriasis.34,35
Overall, GO analysis of this gene list revealed genes important in epidermis development (FDR=2.1E-19), ectoderm development (FDR=3.3E-19), keratinocyte differentiation (2.8E-9), epidermal cell differentiation (FD=3.1E-8) and keratinization (FDR=7.2E-7). Consistent with these findings, psoriasis has been shown to be mediated by hyperproliferation with incomplete differentiation of keratinocytes.36
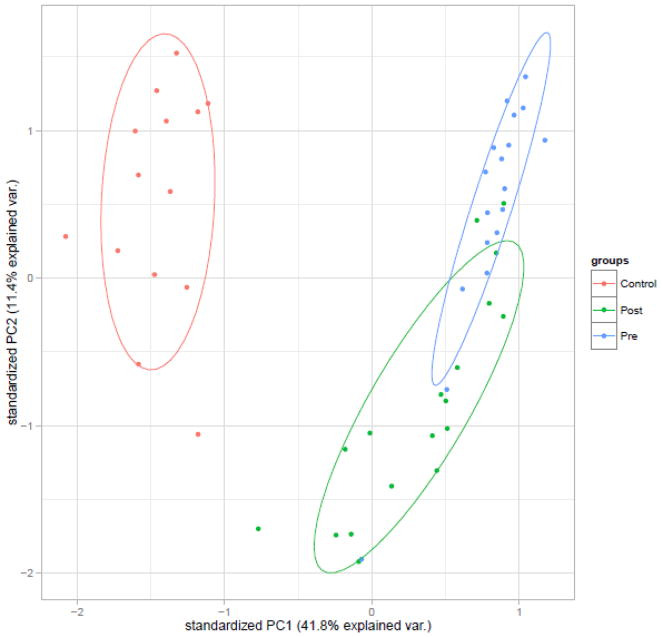
Results of the principal components analysis are shown in Figure 3. Principal component 1 explained 41.8% of the variance in the dataset, and principal component 2 explained 11.4% of the variance in the dataset. Following treatment, the expression of the differentially expressed genes in the psoriasis group deviated less from that of controls than prior to treatment. Similar to other studies, clinical improvement in our sample is correlated with normalization of aberrantly expressed genes.37
Figure 3.
PCA plot of differentially expressed genes comparing psoriasis patients prior to treatment and control RNA-seq expression.
Discussion
In this study, we have shown that the effect of adalimumab treatment differs in its effect on the gene expression profiles of the skin and PBMCs. In skin, pathways modulating the differentiation and proliferation of epidermal keratinocytes were normalized following treatment. In contrast, pathways modulating immunoglobulin-mediated immune pathways were more prominently affected in the blood. Together, these results demonstrate that adalimumab treatment modulates different pathways in skin compared to blood.
Our global RNAseq results in the skin offer the molecular basis of clinical improvement with adalimumab therapy. We corroborate the results of Henriks et al. (2014), suggesting that in the skin, markers of epidermal differentiation and proliferation as well as innate immunity were normalized following treatment with adalimumab. Mechanistically, inhibition of the CCR7/CCL19 axis by TNF blockade is an important early event that leads to clinical remission.38 Other inflammatory markers, including other IL17-responsive genes, have also been shown to decrease with effective therapy.39–41 In the skin, IL17 induces expression modulation through C/CAAT enhancer-binding (C/EBP)-beta, a transcription factor that is preferentially expressed in differentiated keratinocytes.42 Thus, it is possible that modulation of these immunologic pathways ultimately normalizes the expression of epidermal proliferation and differentiation factors, including those in the proteasomal pathway.
Our PBMC array results further expand upon the results of prior studies. Using qPCR, Balato et al. (2013) identified that adalimumab therapy normalized aberrantly expressed markers of the Th17 pathway in both the blood and skin. Furthermore, Bose et al., (2011) found that anti-TNF therapy actually increased the expression of such cytokines as IL10, IL17, IFN-gamma and TCR-mediated CD4 T-cell activation markers in the blood using RTPCR and cytokine assays. Generally, our results suggest that treatment downregulates and normalizes immunoglobulin-mediated responses in the blood. Perhaps the final downstream effect of these changes is modulation of the Th17 pathway that effects immunologic change in the skin, or this B cell-mediated pathway is in fact a parallel process to the Th17 pathway, leading to the epidermal changes outlined above.
To this point, it remains unknown whether humoral immunity is directly involved in the pathogenesis of psoriasis, but multiple observations have linked aberrant humoral immunity with psoriasis.43,44 It is known that patients with psoriasis have increased serum IgG and IgA levels as well as decreased B-cell response to PWM and PHA.45
Conclusion
In sum, we have shown that the potent TNF inhibitor adalimumab has distinct biological effects in psoriatic skin compared to blood. It will be important for future mechanistic studies to examine whether modulation of the immune system in the blood contributes to the decreased cell proliferation observed in the epidermis.
Contributor Information
Maggie Chow, University of California, San Francisco School of Medicine, Kaiser Permanente LA Medical Center.
Kevin Lai, University of California, San Francisco, Department of Dermatology.
Richard Ahn, University of California, San Francisco, Department of Dermatology.
Rashmi Gupta, University of California, San Francisco, Department of Dermatology.
Sarah Arron, University of California, San Francisco, Department of Dermatology.
References
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Stern RS, Nijsten T, Feldman SR, Margolis DJ, Rolstad T. Psoriasis is common, carries a substantial burden even when not extensive, and is associated with widespread treatment dissatisfaction. J Investig Dermatol Symp Proc. 2004;9(2):136–139. doi: 10.1046/j.1087-0024.2003.09102.x. [DOI] [PubMed] [Google Scholar]
- 3.Rapp SR, Feldman SR, Exum ML, Fleischer AB, Reboussin DM. Psoriasis causes as much disability as other major medical diseases. J Am Acad Dermatol. 1999;41(3):401–407. doi: 10.1016/S0190-9622(99)70112-X. [DOI] [PubMed] [Google Scholar]
- 4.Finlay AY, Coles EC. The effect of severe psoriasis on the quality of life of 369 patients. [Accessed February 12, 2015];Br J Dermatol. 1995 132(2):236–244. doi: 10.1111/j.1365-2133.1995.tb05019.x. http://www.ncbi.nlm.nih.gov/pubmed/7888360. [DOI] [PubMed] [Google Scholar]
- 5.Krueger G, Koo J, Lebwohl M, Menter A, Stern RS, Rolstad T. The impact of psoriasis on quality of life: Results of a 1998 National Psoriasis Foundation Patient-Membership Survey. Arch Dermatol. 2001;137(3):280–284. http://www.scopus.com/inward/record.url?eid=2-s2.0-0035069606&partnerID=tZOtx3y1. [PubMed] [Google Scholar]
- 6.Menter A, Tyring SK, Gordon K, et al. Adalimumab therapy for moderate to severe psoriasis: A randomized, controlled phase III trial. J Am Acad Dermatol. 2008;58(1):106–115. doi: 10.1016/j.jaad.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Kristensen M, Chu CQ, Eedy DJ, Feldmann M, Brennan FM, Breathnach SM. Localization of tumour necrosis factor-alpha (TNF-α) and its receptors in normal and psoriatic skin: Epidermal cells express the 55-kD but not the 75-kD TNF receptor. Clin Exp Immunol. 1993;94(2):354–362. doi: 10.1111/j.1365-2249.1993.tb03457.x. http://www.scopus.com/inward/record.url?eid=2-s2.0-0027490541&partnerID=tZOtx3y1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabrese LH. Molecular differences in anticytokine therapies. [Accessed March 11, 2015];Clin Exp Rheumatol. 2003 21(2):241–248. http://www.ncbi.nlm.nih.gov/pubmed/12747285. [PubMed] [Google Scholar]
- 9.Gordon KB, Langley RG, Leonardi C, et al. Clinical response to adalimumab treatment in patients with moderate to severe psoriasis: double-blind, randomized controlled trial and open-label extension study. J Am Acad Dermatol. 2006;55(4):598–606. doi: 10.1016/j.jaad.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Leonardi C, Langley RG, Papp K, et al. Adalimumab for Treatment of Moderate to Severe Chronic Plaque Psoriasis of the Hands and Feet. Arch Dermatol. 2011;147(4):429. doi: 10.1001/archdermatol.2010.384. [DOI] [PubMed] [Google Scholar]
- 11.Saurat J-H, Stingl G, Dubertret L, et al. Efficacy and safety results from the randomized controlled comparative study of adalimumab vs. methotrexate vs. placebo in patients with psoriasis (CHAMPION) Br J Dermatol. 2008;158(3):558–566. doi: 10.1111/j.1365-2133.2007.08315.x. [DOI] [PubMed] [Google Scholar]
- 12.Gordon KB, Langley RG, Leonardi C, et al. Clinical response to adalimumab treatment in patients with moderate to severe psoriasis: double-blind, randomized controlled trial and open-label extension study. J Am Acad Dermatol. 2006;55(4):598–606. doi: 10.1016/j.jaad.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Strober BE, Poulin Y, Kerdel FA, et al. Switching to adalimumab for psoriasis patients with a suboptimal response to etanercept, methotrexate, or phototherapy: efficacy and safety results from an open-label study. J Am Acad Dermatol. 2011;64(4):671–681. doi: 10.1016/j.jaad.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Van Lümig PPM, Lecluse LLA, Driessen RJB, et al. Switching from etanercept to adalimumab is effective and safe: results in 30 patients with psoriasis with primary failure, secondary failure or intolerance to etanercept. Br J Dermatol. 2010;163(4):838–846. doi: 10.1111/j.1365-2133.2010.09950.x. [DOI] [PubMed] [Google Scholar]
- 15.Bissonnette R, Bolduc C, Poulin Y, Guenther L, Lynde CW, Maari C. Efficacy and safety of adalimumab in patients with plaque psoriasis who have shown an unsatisfactory response to etanercept. J Am Acad Dermatol. 2010;63(2):228–234. doi: 10.1016/j.jaad.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 16.Balato A, Schiattarella M, Di Caprio R, et al. Effects of adalimumab therapy in adult subjects with moderate-to-severe psoriasis on Th17 pathway. J Eur Acad Dermatology Venereol. 2014;28(8):1016–1024. doi: 10.1111/jdv.12240. [DOI] [PubMed] [Google Scholar]
- 17.Hendriks AGM, van der Velden HMJ, Wolberink EAW, et al. The effect of adalimumab on key drivers in the pathogenesis of psoriasis. Br J Dermatol. 2014;170(3):571–580. doi: 10.1111/bjd.12705. [DOI] [PubMed] [Google Scholar]
- 18.Quaglino P, Bergallo M, Ponti R, et al. Th1, Th2, Th17 and regulatory T cell pattern in psoriatic patients: modulation of cytokines and gene targets induced by etanercept treatment and correlation with clinical response. Dermatology. 2011;223(1):57–67. doi: 10.1159/000330330. [DOI] [PubMed] [Google Scholar]
- 19.Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, Gudjonsson JE. Early tissue responses in psoriasis to the antitumour necrosis factor-α biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br J Dermatol. 2014;171(1):97–107. doi: 10.1111/bjd.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. [Accessed January 26, 2015];Nucleic Acids Res. 2003 31(4):e15. doi: 10.1093/nar/gng015. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=150247&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Grennan K, Badner J, et al. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. In: Kliebenstein D, editor. PLoS One. 2. Vol. 6. 2011. p. e17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Quaranta M, Knapp B, Garzorz N, et al. Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci Transl Med. 2014;6(244):244ra90. doi: 10.1126/scitranslmed.3008946. [DOI] [PubMed] [Google Scholar]
- 24.Piruzian ES, Sobolev VV, Abdeev RM, et al. Study of Molecular Mechanisms Involved in the Pathogenesis of Immune-Mediated Inflammatory Diseases, using Psoriasis As a Model. [Accessed March 19, 2015];Acta Naturae. 2009 1(3):125–135. http://www.researchgate.net/publication/225079719_Study_of_Molecular_Mechanisms_Involved_in_the_Pathogenesis_of_Immune-Mediated_Inflammatory_Diseases_using_Psoriasis_As_a_Model. [PMC free article] [PubMed] [Google Scholar]
- 25.Nanney LB, Yates RA, King LE. Modulation of epidermal growth factor receptors in psoriatic lesions during treatment with topical EGF. [Accessed March 19, 2015];J Invest Dermatol. 1992 98(3):296–301. doi: 10.1111/1523-1747.ep12497963. http://www.ncbi.nlm.nih.gov/pubmed/1545139. [DOI] [PubMed] [Google Scholar]
- 26.Henry L, Le Gallic L, Garcin G, et al. Proteolytic activity and expression of the 20S proteasome are increased in psoriasis lesional skin. Br J Dermatol. 2011;165(2):311–320. doi: 10.1111/j.1365-2133.2011.10447.x. [DOI] [PubMed] [Google Scholar]
- 27.Williams B, Vanags D, Hall S, et al. Efficacy and safety of chaperonin 10 in patients with moderate to severe plaque psoriasis: evidence of utility beyond a single indication. Arch Dermatol. 2008;144(5):683–685. doi: 10.1001/archderm.144.5.683. [DOI] [PubMed] [Google Scholar]
- 28.Cauza K, Grassauer A, Hinterhuber G, et al. FcgammaRIII expression on cultured human keratinocytes and upregulation by interferon-gamma. J Invest Dermatol. 2002;119(5):1074–1079. doi: 10.1046/j.1523-1747.2002.19527.x. [DOI] [PubMed] [Google Scholar]
- 29.Livden JK. Fc gamma receptors on keratinocytes in psoriasis. [Accessed March 18, 2015];Arch Dermatol Res. 1988 280(1):12–17. doi: 10.1007/BF00412682. http://www.ncbi.nlm.nih.gov/pubmed/2451478. [DOI] [PubMed] [Google Scholar]
- 30.Sakkas LI, Marchesoni A, Kerr LA, et al. Immunoglobulin heavy chain gene polymorphisms in Italian patients with psoriasis and psoriatic arthritis. [Accessed March 18, 2015];Br J Rheumatol. 1991 30(6):449–450. doi: 10.1093/rheumatology/30.6.449. http://www.ncbi.nlm.nih.gov/pubmed/1684125. [DOI] [PubMed] [Google Scholar]
- 31.Bhalerao J. The genetics of psoriasis: a complex disorder of the skin and immune system. Hum Mol Genet. 1998;7(10):1537–1545. doi: 10.1093/hmg/7.10.1537. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson T, Mark EB, Henriksson R, Hedman H. Redistribution of LRIG proteins in psoriasis. J Invest Dermatol. 2008;128(5):1192–1195. doi: 10.1038/sj.jid.5701175. [DOI] [PubMed] [Google Scholar]
- 33.Shi G, Sohn K-C, Li Z, et al. Expression and functional role of Sox9 in human epidermal keratinocytes. PLoS One. 2013;8(1):e54355. doi: 10.1371/journal.pone.0054355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suarez-Farinas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger J. Expanding the Psoriasis Disease Profile: Interrogation of the Skin and Serum of Patients with Moderate-to-Severe Psoriasis. J Invest Dermatol. 2015 Jun; doi: 10.1038/jid.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudjonsson JE, Ding J, Li X, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol. 2009;129(12):2795–2804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaštelan M, Prpić-Massari L, Brajac I. Apoptosis in psoriasis. Acta Dermatovenerologica Croat. 2009;17(3):182–186. [PubMed] [Google Scholar]
- 37.Zaba LC, Cardinale I, Gilleaudeau P, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204(13):3183–3194. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosè F, Petti L, Diani M, et al. Inhibition of CCR7/CCL19 axis in lesional skin is a critical event for clinical remission induced by TNF blockade in patients with psoriasis. Am J Pathol. 2013;183(2):413–421. doi: 10.1016/j.ajpath.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 39.Russell CB, Rand H, Bigler J, et al. Gene expression profiles normalized in psoriatic skin by treatment with brodalumab, a human anti-IL-17 receptor monoclonal antibody. J Immunol. 2014;192(8):3828–3836. doi: 10.4049/jimmunol.1301737. [DOI] [PubMed] [Google Scholar]
- 40.Lemster BH, Carroll PB, Rilo HR, Johnson N, Nikaein A, Thomson AW. IL-8/IL-8 receptor expression in psoriasis and the response to systemic tacrolimus (FK506) therapy. Clin Exp Immunol. 2008;99(2):148–154. doi: 10.1111/j.1365-2249.1995.tb05525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubert J, Reiniche P, Fogel P, et al. Gene expression profiling in psoriatic scalp hair follicles: clobetasol propionate shampoo 0.05% normalizes psoriasis disease markers. J Eur Acad Dermatol Venereol. 2010;24(11):1304–1311. doi: 10.1111/j.1468-3083.2010.03637.x. [DOI] [PubMed] [Google Scholar]
- 42.Chiricozzi A, Nograles KE, Johnson-Huang LM, et al. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PLoS One. 2014;9(2):e90284. doi: 10.1371/journal.pone.0090284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurmin V, Mediwake R, Fernando M, Whittaker S, Rustin MHA, Beynon HLC. Psoriasis: response to high-dose intravenous immunoglobulin in three patients. [Accessed August 17, 2015];Br J Dermatol. 2002 147(3):554–557. doi: 10.1046/j.1365-2133.2002.04753.x. http://www.ncbi.nlm.nih.gov/pubmed/12207600. [DOI] [PubMed] [Google Scholar]
- 44.Dass S, Vital EM, Emery P. Development of psoriasis after B cell depletion with rituximab. Arthritis Rheum. 2007;56(8):2715–2718. doi: 10.1002/art.22811. [DOI] [PubMed] [Google Scholar]
- 45.Zarrabeitia MT, Fariñas MC, Rodríguez-Valverde V, Riancho JA, Llaca HF. T and B cell function in psoriasis and psoriatic arthropathy. [Accessed August 17, 2015];Allergol Immunopathol (Madr) 17(3):155–159. http://www.ncbi.nlm.nih.gov/pubmed/2816658. [PubMed] [Google Scholar]