Abstract
BamA is an essential component of the β-barrel assembly machine (BAM) responsible for insertion and folding of β-barrel Outer Membrane Proteins (OMPs) in Gram negative bacteria. BamA is an OMP itself, and its β-barrel transmembrane domain is thought to catalyze OMP insertion and folding, although the molecular mechanism remains poorly understood. Crystal structures of BamA and complementary molecular dynamics simulations have shown that its β-barrel seam (the interface between the first and last barrel strands) is destabilized. This has led to mechanistic models where the BamA barrel seam functions as a lateral gate that opens and successively accepts β-hairpins from a nascent OMP such that a nascent barrel can bud from BamA. Consistent with this model, disulfide locking of the BamA barrel seam is lethal in E. coli. Here we show that disulfide locking of the BamA barrel has no effect on its ability to catalyze folding of a model OMP into liposomes. However, disulfide trapping experiments indicate that the BamA barrel is highly dynamic in the liposome membranes, with the β-strands at the barrel seam undergoing “register sliding” by more than 14Å both up and down the membrane. Remarkably, these extreme dynamics were also observed in the BamA barrel in the context of the native E. coli outer membrane. These results are consistent with a model where the BamA barrel dynamics induce defects in the outer membrane that facilitate insertion of nascent OMPs.
Graphical Abstract
The outer membrane of Gram-negative bacteria is an essential “organelle” that acts as a molecular sieve to protect against lipophilic agents and antibiotics, while allowing essential functions such as nutrient/waste exchange.1 The outer membrane structure is unique, with an asymmetric lipid bilayer containing Lipopolysaccharide in the outer leaflet.2 Furthermore, the trans-membrane proteins of the outer membrane are almost exclusively β-barrel in structure, compared to the strictly α-helical nature of proteins found in the inner membrane. Also, unlike most α-helical inner membrane proteins that fold co-translationally, outer membrane proteins (OMPs) fold post-translationally3–6.
After being synthesized in the cytoplasm, OMPs are translocated into the periplasm through the SEC translocon5, 7. In the periplasm, OMPs are bound by chaperones SurA, Skp and DegP before insertion and folding into the outer membrane catalyzed by the β-Barrel Assembly Machine (BAM), an essential protein complex that is required for proper assembly of all OMPs8. The BAM complex is composed of BamA, an essential β-barrel protein with an N-terminal periplasmic domain onto which the associated lipoproteins (BamB-E) assemble9–12. In E. coli, BamD is the only essential lipoprotein of the complex, but deletion of any of the other lipoproteins lead to outer membrane permeability phenotypes9, 12.
While much effort has been put into understanding the BAM complex genetically and structurally, its mechanism of action is still elusive. BamA is the central component of the complex and the current mechanistic hypotheses are BamA centric. Crystal structures where the BamA barrel is present indicate that the barrel seam, where the first and last strands hydrogen bond to close the barrel, is destabilized13–16. One model suggests that the destabilized BamA seam could work as a lateral gate that would open to accept β-hairpins of a nascent OMP and, through successive rounds, help a new barrel “bud” into the membrane.13, 17 It has also been suggested that BamA could create local distortions in the membrane to facilitate insertion and folding of nascent OMPs17–20. Lateral gating is becoming a recurring theme in hydrophobic transport as it is thought to mediate α-helical membrane protein insertion by the SEC translocon, OMP biogenesis by TamA21, 22 and transport of hydrophobic small molecules by FadL 23.
The report that disulfide locking of the BamA barrel seam proved to be lethal in E. coli was interpreted as evidence supporting the lateral gating model. 24 However, given the lethality of the disulfide mutations, the direct impact of BamA barrel locking on OMP folding could not be assessed. Recent studies have reported that BamA alone is capable of catalyzing the folding of OMPs into liposomes in vitro.18, 20, 25 Using disulfide locked BamA, we show that lateral gate opening is not required to accelerate folding of the model substrate OmpX in this assay. Nevertheless, we find that the BamA barrel seam undergoes large conformational changes while inserted in the membrane, as cysteines offset by four residues in the barrel seam are capable of forming disulfide bonds. Furthermore, this extreme barrel dynamics is also observed in outer membrane embedded BamA in vivo.
MATERIALS AND METHODS
Cloning, Mutagenesis, Expression and purification of outer membrane proteins for in vitro assays
The gene for OmpX was PCR amplified from E. coli genomic DNA using primers upper and lower 2122 (Table S1), which also introduced NdeI and XmaI sites to the 5′ and 3′ ends respectively. The PCR product was then digested with NdeI and XmaI and ligated into the pMS174 vector (a pET41b derivative modified to incorporate additional restriction sites) to generate plasmid pMS1228 encoding mature OmpX with no signal sequence for expression into inclusion bodies. Similarly, the gene coding for wild-type mature BamA (no signal sequence) was PCR amplified and cloned into pMS174 to generate pMS1224. Cys-less BamA was made by mutating two native cysteines to serines (BamA(C690S-C700S)) using the QuikChange (Agilent Technologies) mutagenesis protocol with primers upper and lower 846 and 847 (Table S1). All other BamA mutations were made on the cys-less background using the same protocol and primers listed in Table S1. The resulting plasmids are described in Table S2.
For expression into inclusion bodies, BL21(DE3) cells (Thermo) were transformed with the appropriate plasmid and plated on kanamycin plates. Single colonies were used to inoculate 100mL of LB media supplemented with kanamycin and grown overnight at 37° as starter cultures. 500mL of LB/kanamycin cultures were inoculated with 5mL of starter cultures and grown at 37° until the cultures reached OD=0.5–0.6. Protein expression was induced with 1mM isopropyl-beta-D-thiogalactoside (IPTG, GoldBio) and the cultures incubated for 3–4 additional hours before harvesting by centrifugation. Cell pellets were resuspended in 50mM Tris pH 8, 0.5mg/mL lysozyme and protease inhibitors cocktail (Roche) (25mL per 500mL culture), snap frozen in liquid nitrogen and stored at −70° until use.
To purify inclusion bodies, frozen cell suspensions were thawed in a 37° water bath and then supplemented with Benzonase (Sigma) before lysis by sonication. Insoluble material was pelleted by centrifugation at 5500xg for 30 minutes at 4°, decanted and resuspended in 50mM Tris pH 8, 0.5mg/mL lysozyme and 0.5% Triton X-100. Resuspended pellets were briefly sonicated and re-pelleted. Pellets were washed twice with 10mM Tris pH 8, 1mM EDTA resulting in purified inclusion bodies that were stored at −20° until further use.
For protein purification, inclusion bodies were solubilized in urea buffer (8M urea, 20mM Tris pH 8), incubated at room temperature for 15 minutes before centrifuging at 10,000×g for 5 minutes to remove debris and insoluble material. Supernatants were filtered through a 0.45um filter and loaded onto a 6mL Q Sepharose fast flow column (GE) at room temperature. For all cysteine mutants, all urea buffers contained 5mM β-mercapto-ethanol (BME). OmpX was eluted with urea buffer + 100mM NaCl. BamA constructs were eluted with urea buffer + 150mM NaCl. Fractions were analyzed by SDS-PAGE and fractions with protein were pooled, concentrated and buffer exchanged with urea buffer until the salt concentration is less than 1mM. For all cysteine mutants, BME was exchanged for 2mM TCEP. All proteins were concentrated to 50μM and frozen at −70° until use.
Omp folding into LUVs and folding kinetics
Refolding experiments were carried out essentially as described by Gessman et al.20. Briefly, 0.21 mg of phosphatidylethanolamine (850700, Avanti Polar Lipids) and 0.906 mg of phosphatidlycholine (850325, Avanti Polar Lipids) were dissolved in chloroform, added to a glass tube and dried under argon to create a thin lipid film. Lipids were desiccated overnight to remove residual chloroform and stored at −20° under argon until use. To make liposomes, lipid films were hydrated with 200uL of degassed 20mM borate pH 10 for a final lipid concentration of 10mM. The suspended lipids were then extruded 25 times through a 0.1um filter using a manual extruder (Avanti Polar Lipids).
Liposomes were first incubated for 1 hour with 2mM TCEP (reducing conditions) or 2mM CuSO4 (oxidizing conditions). To measure the intrinsic rate of OmpX folding, urea denatured OmpX was diluted into a refolding buffer such that final concentrations were 4 μM OmpX, 3.2 mM lipids (liposomes), 20mM borate pH 10, 2mM EDTA, 1M urea and 2mM TCEP (reducing conditions) or 2mM CuSO4 (oxidizing conditions). The refolding reactions were incubated at room temperature and samples taken at time points between 5 and 600 seconds. Refolding was quenched by addition of 4x SDS-PAGE loading buffer and samples were either kept on ice (unboiled) or incubated at 99° for 5 minutes (boiled). All samples were run on TGX Mini-Protean BioRad 4–20% precast gels with cold 1X SDS-Tris-Glycine running buffer.
For BamA accelerated OmpX folding, urea denatured BamA was diluted into a refolding buffer such that final concentrations were to 4μM BamA, 3.2 mM lipids (liposomes), 20mM borate pH 10, 2mM EDTA, 1M urea and 2 mM TCEP and incubated for 2 hours at room temperature with constant stirring. This procedure incorporates BamA into liposomes with the POTRA domain facing out (Figure S1). The reactions were then split in half and incubated for an additional hour in 2mM TCEP (reducing conditions) or 4 mM CuSO4 (oxidizing conditions). Buffer and urea denatured OmpX were then added to these suspensions such that the final concentrations were 2μM BamA, 2μM OmpX, 20mM borate pH 10, 1.6mM lipids (liposomes), 2mM EDTA, 1M urea and 2mM TCEP (reducing conditions) or 2mM CuSO4 (oxidizing conditions). The reactions were incubated at room temperature and samples taken and analyzed as described above.
All SDS-PAGE gels were stained with Coomassie blue, scanned and band intensities quantified using ImageJ. The intensity of the OmpX folded band over the intensity of the boiled OmpX band was used to determine fraction folded. Plots of fraction folded vs. time were fitted using to the equation y=y0-A−kt using Kaleidagraph.
Probing seam dynamics in vitro with offset double-cysteine BamA mutants
BamA G433C-T809C, or G431C-Q803C or G429C-F802C were expressed and refolded into liposomes for 2 hours as described above. CuSO4 was then added to a final concentration of 4mM and incubated for 1 hour. Samples were taken before and after the addition of copper sulfate, immediately mixed with 4x SDS-PAGE loading buffer and either kept on ice (unboiled), boiled in the presence or absence of 360mM BME, or boiled in the presence of 40mM n-ethylmaleimide (NEM) (quenched). All samples were analyzed on a 4–20% TGX Mini-Protean gel with cold 1x SDS-Tris-Glycine running buffer.
Probing seam dynamics in vitro with offset double-cysteine BamA mutants
Wild type bamA with a 6xHis tag inserted between the signal sequence and the mature N-terminus cloned into the pZS21 vector26 (a generous gift from Dr. Silhavy) was used to subclone, from the plasmids described above, a fragment of bamA harboring the cys-less mutations C690S-C700S (pMS950), the cys-less mutations and G433C-T809C (pMS1341), or the cys-less mutations and G431C-Q803C (pMS980) (see Table S2 for details). These plasmids drive constitutive expression of BamA mutants with its native signal sequence and a His tag at the N-terminus of the mature protein.
E. coli JCM-1669 (containing a deletion of the endogenous bamA gene and an insertion of wild-type bamA under the arabinose promoter) cells were transformed with plasmids pMS1341 or pMS980 and plated on kanamycin/0.1% arabinose plates. Single colonies were used to inoculate into 5mL of LB containing 0.01% arabinose and kanamycin and incubated overnight at 37°. These cultures were used to inoculate 100mL of LB supplemented with kanamycin and 0.01% arabinose, such that genomic, wild type BamA is expressed in addition to the plasmid driven expression of the mutants. Cultures were grown to OD = 0.5–06, pelleted by centrifugation and resuspended in 100mL of 20mM Tris pH 7.5. The cell suspensions were split in half. One half was treated with 0.4mM NEM for 30 minutes followed by harvesting by centrifugation. The other half was treated with 0.1mM CuSO4 for 30 minutes, pelleted by centrifugation, resuspended in fresh 20mM Tris pH 7.5 and treated with 0.4mM NEM for 30 minutes before being pelleted again. For purification of His-tagged BamA mutants, cell pellets were resuspended in 2mL of BugBuster (Novagen) and incubated for 20 minutes before being spun at 16,000xg for 20 minutes to remove debris and insoluble material. The pH of the supernatant was adjusted to ~ 8 with NaOH and then added to 200uL of nickel NTA-agarose beads (Quiagen) equilibrated in equilibration buffer (50mM Tris pH 8, 150mM NaCl, 0.5% Triton X-100). After a 15 minute incubation at room temperature, the beads were washed three times with 1mL of equilibration buffer and bound proteins eluted with equilibration buffer supplemented with 500mM imidazole. Elution fractions were mixed with SDS-PAGE loading buffer with or without 25mM dithiothreitol (DTT) and subjected to SDS-PAGE on an 8% gel. The gels were blotted to a PVDF membrane (Milipore) and probed with a polyclonal rabbit antibody raised against BamA (Cocalico Biologicals) (1:20,000 serum dilution). The blots were analyzed using a goat-anti-rabbit secondary antibody conjugated to Horse Radish Peroxidase (Pierce) (1:50,000 antibody dilution) and Western Lightning reagents (Perkin-Elmer).
RESULTS
Barrel locked BamA accelerates OmpX folding into liposomes
An in vitro OMP folding assay was implemented to directly test the impact of BamA barrel-locking on the ability of BamA to fold and insert a model OMP. When urea denatured OMPs are diluted into a solution containing liposomes, they spontaneously fold into the liposome membrane.27 The yield and rate of folding, however, is strongly dependent on the liposome lipid composition. 18, 20 We implemented a folding system, developed by Fleming and coworkers, with liposomes composed of 80% PC and 20% PE.20 This creates a lipid mix with native and non-native E. coli head-groups where the model protein OmpX folds spontaneously at an intrinsic rate, which is accelerated if BamA (but not other OMPs) is pre-folded into the liposomes.20 The reaction can be easily followed by the OMPs “heat modifiability”, where unboiled samples display differential mobility for the folded and unfolded fractions when run using semi-native SDS-PAGE.28
Figure 1A shows the folding rates of OmpX, under oxidizing and reducing conditions, into PE/PC “empty” liposomes (intrinsic rates, gray lines) and liposomes containing a cysteine-free (Cys-less) mutant of BamA (BamA accelerated rates, blue lines). The OmpX folding rate acceleration is similar for cys-less BamA and wild-type BamA (Figure S2), and consistent with rate acceleration reported in the literature.18, 20, 25 As expected, the redox conditions make little difference in the intrinsic folding rate as well as the cys-less BamA accelerated rate.
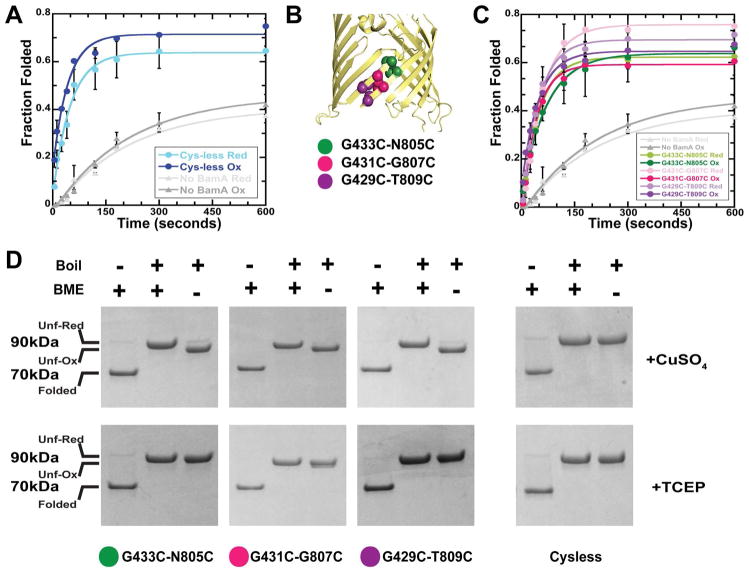
Figure 1.
BamA barrel locking and acceleration of OmpX folding. (A) Time course of folding of urea-denatured OmpX into “empty” liposomes (gray lines) or liposomes with Cys-less BamA embedded in their membranes (blue lines) under reducing (Red) or oxidizing (Ox) conditions. (B) Cartoon representation of the BamA barrel structure highlighting the positions of the cysteine pairs that, when oxidized, lock the barrel at the top (green), middle (pink) or bottom (purple). (C) Time course of folding of urea denatured OmpX into liposomes with BamA double-cysteine mutants embedded in their membranes under reducing (light green, pink and purple) or oxidizing (dark green, pink and purple) conditions. (D) Folding and oxidation state of BamA double-cysteine mutants. Cys-less BamA and the indicated double-cysteine mutants were folded into liposomes and the folding state determined in a semi-native SDS-PAGE (− Boil lanes). Folded BamA has an apparent molecular weight of 70kDa (Folded) whereas unfolded BamA (+ Boil, + BME lanes, Unf-Red) has a molecular weight of 90kDa. Double-cysteine BamA mutants treated with CuSO4 oxidize, forming a lariat structure with a faster electrophoretic mobility than the fully denatured and reduced BamA (compare + Boil, +CuSO4, − BME lanes, Unf-Ox with + Boil, +CuSO4 +BME lanes, Unf-Red). BamA mutants kept in the presence of TCEP do not oxidize and thus do not show the faster migrating species (compare +Boil, +TCEP, −BME lanes with +Boil, +TCEP, +BME lanes). Cys-less BamA cannot oxidize and does not show a faster migrating species under oxidizing conditions (+Boil, +CuSO4, −BME).
The effect of disulfide-locking the BamA barrel on its ability to accelerate OmpX folding was then tested using the in vitro assay. Pairs of cysteine residues were introduced in cys-less BamA such that the BamA barrel could be disulfide locked at three different positions: 1) at the top (G433C-N805C), 2) in the middle (G431C-G807C) and 3) at the bottom (G429C-T809C) (Figure 1B). These BamA mutants were purified under denaturing conditions and allowed to fold for 2 hours into liposomes either in the presence or absence of (2mM tris(2-carboxyethyl)phosphine (TCEP)) to, respectively, prevent or allow formation of disulfides. The samples without TCEP where then incubated for an additional hour in the presence 2mM CuSO4 to further promote formation of disulfides, whereas the samples in TCEP remained under reducing conditions. As shown in Figure 1C, the OmpX folding activities of the BamA mutants under reducing conditions matched well with the cys-less control, as expected (Figure 1C, light green, light pink and light purple lines, compare to Figure 1A blue lines). Surprisingly, however, the activity of the mutants under oxidizing conditions was similar to those under reducing conditions (Figure 1B, dark green, dark pink and dark purple lines). This indicates that BamA can accelerate OmpX folding into liposomes even when its barrel is locked.
Control experiments show that the β-barrel domains of all BamA mutants are properly folded under both reducing and non-reducing conditions as determined by their heat modifiability in semi-native SDS-PAGE. Figure 1D shows that unboiled samples (− Boil) display a band running with an apparent 70kDa molecular weight that corresponds to folded BamA, whereas boiling of the sample (+ Boil) results in shifting to its correct molecular weight of approximately 90kDa. The BamA β-barrel domains are indeed disulfide-locked under oxidizing conditions. Whereas fully unfolded and reduced BamA runs on SDS-PAGE at approximately 90kDa (Unf-Red, Figure 1D, +Boil +BME lanes) disulfide formation between the first and last strand of the BamA barrel leads to a lariat structure upon denaturation by boiling with a faster electrophoretic mobility than the fully denatured and reduced proteins (Unf-Ox, Figure 1D, −BME lanes, top panel). BamA mutants maintained in the presence of 2mM TCEP remain reduced, as seen by the lack of a faster migrating species in the −BME lanes (Figure 1C, −BME lanes, lower panel). Similarly, there is no faster migrating species in the cys-less mutant, confirming that this band corresponds the oxidized form of the barrel. Finally, the faster migrating species are also observed when the BamA mutants are folded and oxidized with CuSO4 followed by treatment with N-ethyl-maleimide (NEM) to block any free cysteines (Figure S3). This demonstrates that the disulfides are formed during the incubation with CuSO4 and the BamA barrels are locked during OmpX folding. These control experiments further support the conclusion that barrel-locked BamA can accelerate OmpX folding into liposomes.
BamA barrel seam is highly dynamic in liposome membranes
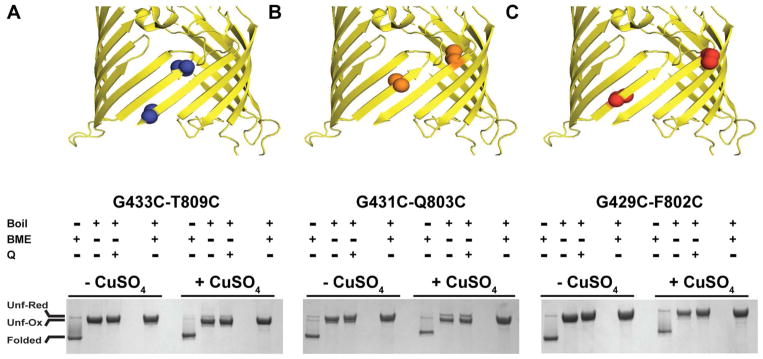
Two sets of “offset” double cysteine BamA mutants were designed to directly probe, by disulfide trapping, the dynamics of the BamA barrel seam while embedded in a membrane. When modeled into the structure of E. coli BamA (PDBID 4N75) the Cβ of the cysteine pairs G433C-T809C and G431C-Q803C face the inside of the BamA barrel, but are 14.6 and 13.8 Å apart and thus too far apart to form disulfides (Figure 2A, B, cartoon structures). However, disulfide bonds efficiently formed when both BamA mutants were first folded into PE/PC liposomes followed by 1 hour incubation under oxidizing conditions (CuSO4). As shown in Figure 2A and B, oxidized samples run on non-reducing SDS-PAGE (+Boil, +CuSO4, −BME lanes) display bands that migrate faster than the fully denatured and reduced species (+Boil, +CuSO4, +BME lanes), consistent with disulfide-linked lariat structures. This indicates that the β-strands at the BamA barrel seam are highly dynamic, undergoing “register sliding” in both directions such that the offset cysteines can align to form disulfides. The disulfides are not forming due to misfolding of the BamA barrel, as unboiled samples of the mutants display the heat-modifiable bands characteristic of folded BamA with an apparent 70 kDa molecular weight (Figure 2A and B, − Boil lanes). Furthermore, most of the disulfides are not forming during BamA folding. This is demonstrated by the absence of the characteristic faster migrating lariat band under non-oxidizing conditions for G433C-T809C (Figure 2A, −CuSO4, +Boil, −BME) and only a small fraction corresponding to less than 50% of the protein displays the lariat band for G431C-Q803C (Figure 2B, −CuSO4, +Boil, −BME). Conversely, the disulfide linked lariat bands are efficiently formed in both mutants during incubation with CuSO4 (Figure 2A and B, + CuSO4, +Boil, −BME). To rule out the possibility that the disulfides are forming during boiling for SDS-PAGE, samples of the BamA mutants were treated with a SDS sample buffer containing a large excess of NEM to quench any free cysteines. These samples were indistinguishable from those without NEM (Figure 2A and B, compare +Boil, −BME, +/− Q). As a negative control, the BamA mutant G429C-F802C was also tested for disulfide formation. In this mutant, the cysteine residues are facing the inside (G429C) and outside (F802C) of the barrel (Figure 2C, cartoon) and would thus not be expected to form disulfides even during register sliding of the barrel β-strands. As shown in Figure 2C, whereas the G429C-F802C barrel folds properly (−Boil lanes), no fast-migrating lariat bands were observed under any condition. Taken together, these experiments demonstrate that the β-barrel of BamA is highly dynamic in liposome membranes, undergoing register shifts of the seam formed by the first and last strands in both directions.
Figure 2.
BamA barrel dynamics in liposome membranes. Two sets of offset double cysteine mutants, G433C-T809C (A) and G431C-Q803C (B), and a control pair facing in opposing directions, G429C-F802C (C) were folded into liposomes and the folding state determined in a semi-native SDS-PAGE (− Boil lanes, Folded). Folded BamA has an apparent molecular weight of 70kDa whereas unfolded BamA (+ Boil, + BME lanes, Unf-Red) has a molecular weight of 90kDa. The offset cysteine pairs are able to oxidize after incubation with CuSO4 forming a lariat structure with a faster electrophoretic mobility than the fully denatured and reduced BamA (compare A, B, + CuSO4, + Boil, −BME lanes, Unf-Ox with A, B, + CuSO4, + Boil, +BME lanes, Unf-Red). Samples boiled in the presence of excess NEM to quench free cysteines show identical oxidation to those without, indicating oxidation is not occurring during boiling (A, B + Q lanes). The control mutant does not show any oxidation under any condition, indicating the barrel cannot oxidize (C, + Boil, −BME lanes).
BamA barrel seam is highly dynamic in E. coli cells
The experiments described above reveal extreme dynamics in the BamA barrel seam in the context of PE/PC liposome membranes. However, both the acyl chains and the phosphocholine head groups of these lipids are not native to E. coli membranes. Therefore, the mutants G433C-T809C and G431C-Q803C were expressed in JCM-166 E. coli cells in the presence of arabinose to probe the BamA barrel dynamics by disulfide trapping in the context of the native outer membrane bilayer.
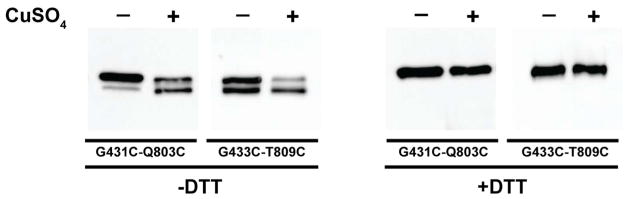
Cells expressing His-tagged mutant BamA from a low copy number plasmid were grown to mid-log phase and then harvested and treated with NEM to block free cysteines, preserve the oxidation that occurred in the cells and prevent oxidation during sample processing. After solubilization and purification on a nickel NTA column to remove endogenous, wild-type BamA, the oxidation state of the BamA mutants was tested by non-reducing SDS-PAGE followed by Western blotting probed with a BamA antibody. Both double-cysteine BamA mutants were able to form disulfides during cell growth albeit oxidation was more efficient for G433C-T809C than it was for G431C-Q803C (Figure 3). This is evidenced by the presence of a doublet containing fully denatured and reduced BamA as well as a faster migrating species due to the disulfide linked lariat structure (Figure 3, −DTT panel, −CuSO4 lanes). The doublet resolved to a single, reduced band, when samples were treated with DTT (Figure 3, +DTT panel, −CuSO4 lanes). Treatment of intact cells with CuSO4 enhances disulfide formation although oxidation remains more efficient for G433C-T809C than G431C-Q803C (Figure 3, −DTT panel, +CuSO4 lanes). These experiments demonstrate that the BamA barrel seam formed by the first and last strand is also highly dynamic in the context of the native E. coli outer membrane. Furthermore, consistent with disulfide formation, the G433C-T809C and G431C-Q803C mutants are not able to complement depletion of wild type BamA (Figure S4).
Figure 3.
BamA barrel dynamics in E. coli cells. His-tagged variants of BamA G431C-Q803C or G433C-T809C were expressed in JCM-166 cells and then treated with NEM (−CuSO4 lanes) or with CuSO4 followed by NEM (+CuSO4 lanes). His-tagged BamA was purified by metal affinity chromatography and analyzed western blot probed with an anti-BamA antibody. Formation of disulfides in E. coli cells is revealed by a lariat structure with a faster electrophoretic mobility than the fully denatured and reduced BamA (compare samples + and − DTT). Formation of disulfides is enhanced by treating the cells with CuSO4 (compare −DTT, +CuSO4 lanes with −DTT, −CuSO4 lanes).
DISCUSSION
OMPs can fold spontaneously, with the correct orientation, into model lipid bilayers without help from extrinsic factors.27, 29, 30 They display large ΔG of folding indicating that membrane insertion and folding are very favorable.19 However, the folding rate is highly dependent on the properties of the lipid bilayer with the polar head groups posing a high kinetic barrier for OMP membrane insertion and folding.19, 20 The BAM complex is the catalyst that lowers this kinetic barrier allowing OMP folding, and its exclusive presence in the outer membrane ensures the strict segregation of OMPs from the inner membrane.20 The BamA subunit, with its soluble POTRA, and membrane embedded β-barrel domains, has a central role in the BAM mechanism. Crystal structures revealed unusually sparse contacts between the first and last strand of the BamA barrel suggesting that this seam may open during BamA mediated OMP folding.13 This idea is supported by molecular dynamics simulations of a BamA barrel in non-native lipids as well as recent crystal and EM structures of detergent solubilized BAM complexes.13–16, 31 These results have led to mechanistic models of BamA-mediated OMP folding in which the barrel seam acts as a lateral gate.13, 17 In these models, β-hairpins from folding OMPs are sequentially accommodated in the lateral gate leading to nascent barrels budding from BamA. Consistent with this model, locking that BamA barrel seam by disulfide crosslinking is lethal to E. coli.24 However, an in vitro system is required to directly test the effect of barrel seam locking on the ability of BamA to accelerate OMP folding.
An in vitro system requires incorporation of BamA into bilayers such as liposomes. However, BamA cannot be refolded into liposomes composed of native E. coli lipids, which predominantly contain 16 carbon acyl chains and phophoethanolamine (PE) as well as mono and di-phosphoglycerol (PG) head groups.32 Conversely, BamA can be readily refolded into liposomes composed of lipids with 10 carbon (C10) acyl chains and phosphocholine (PC) head groups, although these head groups are not present in E. coli membranes.20 Fleming and coworkers thus developed a liposome system with a mix of “host” (PC) and “native-guest” (PE) C10 lipids where BamA and model OMPs can be refolded. The system enables measuring the folding of model OMPs such as OmpX into “empty” liposomes (the intrinsic folding rate) and into BamA containing liposomes (the BamA accelerated rate). BamA in which two cysteines located in an extracellular barrel loop were mutated to serine to generate a cysteine-free variant (cys-less BamA) was utilized in the experiments presented here. Cys-less BamA is able to complement BamA depletion in vivo, indicating that the cysteines are not essential for activity.33 Furthermore, wild-type and cys-less BamA produce similar rate accelerations of OmpX folding in vitro (Figure S2). This cys-less BamA mutant was then used as a platform to introduce pairs of cysteines to disulfide-lock the BamA barrel.
As expected, double cysteine BamA mutants maintained under reducing conditions do not form disulfides and remain capable of accelerating OmpX folding into liposomes. Figure 1C shows that the mutants also remained active after incubation under oxidizing conditions that lead to barrel locking disulfides. Control experiments confirm that mutant barrels are correctly folded, and that the disulfides are indeed formed as designed (Figure 1D). These results are similar for three independent mutants that lock the BamA barrel in three different positions. Therefore, these results effectively rule out a lateral-gate-insertion/nascent barrel budding model as the mechanism for BamA acceleration of OmpX folding into the tested liposomes.
The rate of unassisted OMP folding into bilayers is increased by perturbations that affect the lipid packing. These include reduced hydrophobic thickness and high membrane curvature.34–36 Consistent with this idea, bilayers with PC head-groups, which have lower packing density than PE or PG lipids37, 38 support fast folding of several models OMPs.29, 39–41 Furthermore, unassisted OMP folding is greatly accelerated in membranes at the phase transition temperature, suggesting that “membrane defects” at the boundaries between gel and liquid phases facilitate OMP insertion and folding.34 Therefore, it has been proposed that the role of BamA may be to induce these membrane defects or perturbations to catalyze OMP folding.19 Indeed, molecular dynamics simulations of BamA in dimyristoyl-PE show a large decrease in membrane thickness and disturbed lipid packing in the region next to the BamA barrel seam.13 In this context, a highly dynamic BamA barrel seam may help induce membrane defects to accelerate OMP folding.
To directly probe the BamA barrel seam dynamics in a membrane, cysteine residues were introduced in the first and last strand of the BamA barrel but “out of register”—i.e., at positions approximately 14Å apart in a regular BamA barrel conformation that would not allow disulfide formation (Figure 2). When these mutants were incorporated into C10 PC/PE liposomes, both double cysteine mutants formed disulfides under oxidizing conditions. Control experiments show that the barrels are correctly folded before the disulfides form and that oxidation occurs during incubation with CuSO4 and not during sample processing (Figure 2). These experiments show that the BamA barrel undergoes large conformational changes while embedded in the membrane with the β-strands at the barrel seam shifting the register in both directions by at least 14 Å. These large shifts are not due to the barrels being inserted in the relatively thin and loosely packed membrane of C10 PC/PE liposomes. When the mutants were expressed in E. coli cells, disulfide formation was observed during growth under non-reducing conditions and was strongly enhanced by post-harvest incubation with CuSO4. This indicates that the disulfides formed while BamA is membrane embedded. Recent structures of detergent solubilized BAM complexes also illustrate the extreme dynamics of the BamA barrel, showing the seam in an open conformation and striking distortion of the tilting angle of the first strands of the barrel. However, the results presented here are not simply validating the conformations observed in the structures. For example, the α–carbons of residue pairs G431-Q803 and G433-T809 are 17.3Å and 25.7Å apart respectively in the open barrel conformations of BamA (using a composite of PDBID 5D0Q15 and PDBID 5EKG31 as G433 is not modeled in 5EKG, and T809 is not modeled in 5D0Q), while the pairs are 13.9Å and 6.8Å apart respectively in the closed barrel conformations observed in 5D0O.15. These distances are too big to allow efficient disulfide formation. Thus, our results report distinct conformational dynamics with displacements of the first and last strands both up and down the membrane plane.
The data presented here is consistent with a model where BamA accelerates OMP folding into liposomes by creating membrane defects rather than a lateral-gating/barrel-budding mechanism. In the relatively thin and loosely packed membrane of C10 PC/PE liposomes, barrel locked BamA is still able to create these defects and accelerate OMP folding. Furthermore, the BamA barrel is extremely dynamic while embedded both into liposomes and in its native outer membrane. The β-strands at the barrel seam undergo dramatic register shifts both up and down the membrane plane with displacements of at least 14Å. We propose that this is necessary to create local defects in the outer membrane to facilitate OMP insertion and folding. This model is consistent with the observation that barrel locked BamA is lethal for E. coli as a locked barrel may not be able to create a defect in the notoriously tightly packed outer membrane. Furthermore, BAM complex lipoproteins are required for efficient OMP folding in vivo, with BamD being essential for viability. Recently, a 23-amino acid deletion mutant of LptD (a large OMP that requires the BAM complex for folding) was characterized as a folding intermediate stalled in the BAM complex.42 The LptD mutant appears to stall as an open barrel at the membrane interface that interacts with both BamA and BamD. The study suggests that OMP folding may begin in the periplasm, and once the first and last strands of the nascent barrel are able to pair, membrane insertion occurs.15 Our results are consistent with displacements of the BamA barrel strands out of the membrane, which may allow interaction with nascent OMPs engaged with BAM lipoproteins in the periplasm at the membrane interface.
Supplementary Material
Figure S1. Reconstituted BamA is in a POTRA domain out orientation.
Figure S2. BamA barrel locking and acceleration of OmpX folding.
Figure S3. Barrel locking BamA disulfides are not formed during sample processing.
Figure S4. BamA disulfide mutants do not complement BamA depletion.
Table S1. Cloning and mutagenesis primers.
Table S2. Plasmids.
Table S3. kobs of OmpX folding.
Acknowledgments
Funding Sources
This work was supported in part by NIH grant AI060841 (Sousa). P.A.D. was supported in part by NIH Training Grant GM08759.
The authors would like to thank Dr. Jansen for the initial cloning of BamA cysteine mutants and Ashley Plummer for invaluable assistance in implementing the in vitro reconstituted system of OMP folding.
ABBREVIATIONS
- BAM
β-barrel assembly machine
- OMP
outer membrane protein
- BME
β-mercaptoethanol
- TCEP
tris(2-carboxyethyl)phosphine
- NEM
n-ethylmaleimide
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- DTT
dithiothreitol
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- PG
phosphatidylglycerol
Footnotes
Author Contributions
P.A.D. and M.C.S. designed research; P.A.D. performed research; P.A.D. and M.C.S. analyzed data and wrote the manuscript.
References
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May KL, Silhavy TJ. Making a membrane on the other side of the wall. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 4.Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 6.Selkrig J, Leyton DL, Webb CT, Lithgow T. Assembly of beta-barrel proteins into bacterial outer membranes. Biochim Biophys Acta. 2014;1843:1542–1550. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 8.Plummer AM, Fleming KG. From Chaperones to the Membrane with a BAM! Trends Biochem Sci. 2016;41:872–882. doi: 10.1016/j.tibs.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 11.Voulhoux R, Tommassen J. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol. 2004;155:129–135. doi: 10.1016/j.resmic.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. Structural insight into the biogenesis of beta-barrel membrane proteins. Nature. 2013;501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Commun. 2016;7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 16.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, Huang Y. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 17.Noinaj N, Rollauer SE, Buchanan SK. The beta-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr Opin Struct Biol. 2015;31:35–42. doi: 10.1016/j.sbi.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel GJ, Kleinschmidt JH. The lipid bilayer-inserted membrane protein BamA of Escherichia coli facilitates insertion and folding of outer membrane protein A from its complex with Skp. Biochemistry. 2013;52:3974–3986. doi: 10.1021/bi400103t. [DOI] [PubMed] [Google Scholar]
- 19.Fleming KG. A combined kinetic push and thermodynamic pull as driving forces for outer membrane protein sorting and folding in bacteria. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci USA. 2014;111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruss F, Zahringer F, Jakob RP, Burmann BM, Hiller S, Maier T. The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol. 2013;20:1318–1320. doi: 10.1038/nsmb.2689. [DOI] [PubMed] [Google Scholar]
- 22.Heinz E, Stubenrauch CJ, Grinter R, Croft NP, Purcell AW, Strugnell RA, Dougan G, Lithgow T. Conserved Features in the Structure, Mechanism, and Biogenesis of the Inverse Autotransporter Protein Family. Genome Biol Evol. 2016;8:1690–1705. doi: 10.1093/gbe/evw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral opening and exit pore formation are required for BamA function. Structure. 2014;22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plummer AM, Fleming KG. BamA Alone Accelerates Outer Membrane Protein Folding In Vitro through a Catalytic Mechanism. Biochemistry. 2015;54:6009–6011. doi: 10.1021/acs.biochem.5b00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 27.Surrey T, Jahnig F. Kinetics of folding and membrane insertion of a beta-barrel membrane protein. J Biol Chem. 1995;270:28199–28203. doi: 10.1074/jbc.270.47.28199. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Mizushima S. Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J Biochem. 1976;80:1411–1422. doi: 10.1093/oxfordjournals.jbchem.a131414. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt JH, Tamm LK. Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. [DOI] [PubMed] [Google Scholar]
- 30.Kleinschmidt JH, den Blaauwen T, Driessen AJ, Tamm LK. Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 31.Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cronan JE, Jr, Weisberg LJ, Allen RG. Regulation of membrane lipid synthesis in Escherichia coli. Accumulation of free fatty acids of abnormal length during inhibition of phospholipid synthesis. J Biol Chem. 1975;250:5835–5840. [PubMed] [Google Scholar]
- 33.Warner LR, Gatzeva-Topalova PZ, Doerner PA, Pardi A, Sousa MC. Flexibility in the Periplasmic Domain of BamA Is Important for Function. Structure. 2017;25:94–106. doi: 10.1016/j.str.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danoff EJ, Fleming KG. Membrane defects accelerate outer membrane beta-barrel protein folding. Biochemistry. 2015;54:97–99. doi: 10.1021/bi501443p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plummer AM, Gessmann D, Fleming KG. The Role of a Destabilized Membrane for OMP Insertion. Methods Mol Biol. 2015;1329:57–65. doi: 10.1007/978-1-4939-2871-2_5. [DOI] [PubMed] [Google Scholar]
- 36.Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 37.Boggs JM. Lipid Intermolecular Hydrogen-Bonding - Influence on Structural Organization and Membrane-Function. Biochimica Et Biophysica Acta. 1987;906:353–404. doi: 10.1016/0304-4157(87)90017-7. [DOI] [PubMed] [Google Scholar]
- 38.Murzyn K, Rog T, Pasenkiewicz-Gierula M. Phosphatidylethanolamine-phosphatidylglycerol bilayer as a model of the inner bacterial membrane. Biophys J. 2005;88:1091–1103. doi: 10.1529/biophysj.104.048835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pocanschi CL, Apell HJ, Puntervoll P, Hogh B, Jensen HB, Welte W, Kleinschmidt JH. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J Mol Biol. 2006;355:548–561. doi: 10.1016/j.jmb.2005.10.060. [DOI] [PubMed] [Google Scholar]
- 40.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huysmans GH, Baldwin SA, Brockwell DJ, Radford SE. The transition state for folding of an outer membrane protein. Proc Natl Acad Sci USA. 2010;107:4099–4104. doi: 10.1073/pnas.0911904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. Characterization of a stalled complex on the beta-barrel assembly machine. Proc Natl Acad Sci USA. 2016;113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Reconstituted BamA is in a POTRA domain out orientation.
Figure S2. BamA barrel locking and acceleration of OmpX folding.
Figure S3. Barrel locking BamA disulfides are not formed during sample processing.
Figure S4. BamA disulfide mutants do not complement BamA depletion.
Table S1. Cloning and mutagenesis primers.
Table S2. Plasmids.
Table S3. kobs of OmpX folding.