Abstract
Sporadic early-onset Alzheimer’s disease (sEOAD) exhibits the symptoms of late-onset Alzheimer’s disease but lacks the familial aspect of the early-onset familial form. The genetics of Alzheimer’s disease (AD) identifies APOE ε4 to be the greatest risk factor; however, it is a complex disease involving both environmental risk factors and multiple genetic loci. Polygenic risk scores (PRSs) accumulate the total risk of a phenotype in an individual based on variants present in their genome. We determined whether sEOAD cases had a higher PRS compared to controls. A cohort of sEOAD cases was genotyped on the NeuroX array, and PRSs were generated using PRSice. The target data set consisted of 408 sEOAD cases and 436 controls. The base data set was collated by the International Genomics of Alzheimer’s Project consortium, with association data from 17,008 late-onset Alzheimer’s disease cases and 37,154 controls, which can be used for identifying sEOAD cases due to having shared phenotype. PRSs were generated using all common single nucleotide polymorphisms between the base and target data set, PRS were also generated using only single nucleotide polymorphisms within a 500 kb region surrounding the APOE gene. Sex and number of APOE ε2 or ε4 alleles were used as variables for logistic regression and combined with PRS. The results show that PRS is higher on average in sEOAD cases than controls, although there is still overlap among the whole cohort. Predictive ability of identifying cases and controls using PRSice was calculated with 72.9% accuracy, greater than the APOE locus alone (65.2%). Predictive ability was further improved with logistic regression, identifying cases and controls with 75.5% accuracy.
Keywords: Polygenic risk score (PRS), Sporadic early-onset Alzheimer’s disease (sEOAD), Genotyping, NeuroX, NeuroChip
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia, characterized by the deterioration of memory, language, visuo-spatial skills, and behavior (Budson and Kowall, 2011). Dementia currently affects an estimated 46.8 million people globally (Prince et al., 2015). Hallmarks of AD were originally identified postmortem from histopathological signs of neuritic plaques, composed of amyloid-β, and neurofibrillary tangle formation; postmortem examination of brain tissue for these hallmarks remains the most definitive diagnosis of AD. Clinical diagnosis is accurately verified in more than 85% of cases (Naj and Schellenberg, 2016).
AD can be categorized based on the age of onset, where presentation of symptoms in individuals before the age of 65 years are classified as early-onset Alzheimer’s disease (EOAD), whereas late-onset Alzheimer’s disease (LOAD) classifies individuals with onset over 65 years (Barber et al., 2017; Wingo et al., 2012). LOAD has a heritability estimated to be around 70%, lower than estimates of heritability for EOAD, which vary between 80% and 100% (Barber et al., 2017; Wingo et al., 2012). An estimated 10% of EOAD cases have a familial aspect and are subsequently classified as early-onset familial AD (EOFAD). Autosomal dominant variants in the genes amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) have been discovered to increase amyloid-β production, increasing the risk of EOFAD (Liu et al., 2013; Wingo et al., 2012). The remaining early-onset cases, classed as sporadic (sEOAD), are thought to be predominantly polygenic. The accumulation of variants which independently increase the risk of LOAD may lead to sEOAD at an earlier stage of life (Barber et al., 2017; Wingo et al., 2012).
The association of the APOE gene has been the most consistent observation in AD genetics with the presence of an APOE ε4 allele significantly more common among individuals diagnosed with AD, whereas the ε2 allele is considered protective (Liu et al., 2013; Naj and Schellenberg, 2016). Through genome-wide association studies (GWASs), around 20 genetic loci had been discovered, which affect risk of LOAD (Lambert et al., 2013). Follow-up studies based on the GWAS have identified other potential candidate AD risk genes not previously identified, including the TRIP4, SPPL2A (Ruiz et al., 2014), and ABI3 genes (Sims et al., 2017). Next-generation sequencing has also enabled the identification of rare variants, one of the most consistent being the R47H variant in the TREM2 gene locus (Guerreiro et al., 2013; Jonsson et al., 2013) which affect the risk of AD previously not identified in GWAS (Giri et al., 2016; Lambert et al., 2013; Naj and Schellenberg, 2016). Although most studies utilize Caucasian populations, further risk variants have been identified through next-generation sequencing in African-American individuals within the gene AKAP9 (Giri et al., 2016; Logue et al., 2014). Conversely, protective variants have also been identified including a small coding deletion (rs10553596) within the CASP7 gene associated with reduced incidence of AD among individuals with the APOE ε4ε4 genotype in 4 independent imputed data sets (Ayers et al., 2016). Further protective rare variants have also been identified by imputation of previous data sets such as the PLCG2 gene (Sims et al., 2017).
Following on from these studies, Marden and colleagues (Marden et al., 2014, 2016) sought to determine if a summative analysis of GWAS variants would be able to predict a dementia probability score. An AD genetic risk score was calculated by multiplying each individual GWAS allele effect size using the beta coefficients obtained from a previous data set. This type of analysis demonstrated that AD genetic risk score could predict LOAD phenotype (Chouraki et al., 2016; Desikan et al., 2017; Sleegers et al., 2015; Verhaaren et al., 2013; Xiao et al., 2015; Yokoyama et al., 2015), mild cognitive impairment conversion to LOAD (Adams et al., 2015; Rodriguez-Rodriguez et al., 2013), hippocampal cortical thickness (Harrison et al., 2016; Sabuncu et al., 2012), hippocampal volume (Lupton et al., 2016), cerebrospinal fluid biomarkers (Martiskainen et al., 2015), and plasma inflammatory biomarkers (Morgan et al., 2017). This approach has been expanded to include further polymorphisms of smaller but important effect sizes to develop a polygenic risk score (PRS) (Euesden et al., 2015). This is an improvement on previous tests as they do not perform well when nonassociated single nucleotide polymorphisms (SNPs) are included (Basu et al., 2011; Chapman and Whittaker, 2008) and is considered to find SNPs of disease relevance that have too small an effect size to be identified conventionally (Pan et al., 2015).
In a recent study, polygenic scores were calculated for a cohort of LOAD cases and controls: the study used genotype information of the cohort to identify common variants that affect the risk of developing AD and used polygenic scores to form a risk prediction model (Escott-Price et al., 2015). By producing a model which identifies individuals with a high PRS, the potential for early screening, diagnosis, and determination of disease severity becomes possible (Euesden et al., 2015).
In this study, we have used genotype information generated on the NeuroX chip to generate a PRS in sEOAD. The NeuroX is a customized genotyping array built on the foundation of the Infinium HumanExome BeadChip v1.1, with additional custom content (Illumina, 2012). The array is designed to collect genotype information at markers across the entire genome. The HumanExome BeadChip foundation is made up of 242,901 markers, identifying variants in a series of metabolic, cancerous, diabetic, and psychiatric disorders (Barber et al., 2017; Nalls et al., 2015). The custom content includes 24,706 markers from candidate loci associated with neurological diseases such as AD, frontotemporal dementia, Parkinson’s disease, multiple system atrophy, amyotrophic lateral sclerosis, myasthenia gravis, Charcot-Marie-Tooth, and progressive supranuclear palsy (Nalls et al., 2015).
To calculate a PRS, we have used the software package, PRSice, which utilizes genotype information from individuals in a target data set based on the effect scores of SNPs from a second data set, termed the base data set. The program uses R to define parameters and PLINK for the computational analysis (Purcell et al., 2007; R Core Team, 2013). PRSice is a command line program that allows specific parameters to be considered when generating PRS. The output files of the analysis include a list of individuals’ scores at the best-fit threshold for predicting disease risk and a list of each tested threshold with its corresponding Nagelkerke’s R2 value, quantifying the level of predictability using that threshold (Eusden et al., 2015).
Linkage disequilibrium (LD) is a common problem when SNPs are scored based on their weighted effect and frequency when comparing cases and controls of a disease. The alleles of 2 SNPs present on the same chromosome can be commonly inherited together, and the recurrence of particular alleles at loci is an indicator of the degree of LD between SNPs (Bush and Moore, 2012). Given 2 SNPs in tight LD, both could be perceived as contributing to the disease risk in a functional haplotype; however, it may be that only 1 polymorphism is responsible for the phenotypic effect.
The aim of this study was to genotype sEOAD cases and controls to generate a PRS based on the genotype information of SNPs identified, and then using the estimated cumulative effect size the SNPs have on disease risk, to determine the predictability of the PRS at predicting cases versus controls.
2. Methods
2.1. Samples
The cohort genotyped consisted of 451 sEOAD cases (48.6% female) and 528 controls (51.3% female). sEOAD cases were screened for known disease causing variants within exons 16 and 17 of APP as well as variants in genes PSEN1 and PSEN2 to minimize inclusion of EOFAD cases. The diseased individuals had a documented or predicted age of onset of ≤65 years. Diagnosis of definite or probable sEOAD had met guidelines set by the National Institute of Neurological and Communicative Disorders and Stroke, the Alzheimer’s disease and Related Disorders Association, and the Consortium to Establish a Registry for Alzheimer’s disease. APOE ε status was determined for all individuals. At least 1 APOE ε4 allele was present in 57.6% of cases, with 22.3% of which being homozygotes (n = 58); 22.7% of controls harbored at least 1 ε4 allele, 9 control samples were ε4 homozygotes. These samples are described in greater detail in the article by Barber et al. (2017). Full details of the samples used in this study are outlined in Table 1. Experimental procedures were completed with informed consent, with approval from local ethics committee (Nottingham Research Ethics Committee 2 (REC reference 04/Q2404/130) and completed in accordance with approved guidelines. A standard phenol chloroform DNA extraction method was used on 2 mL of blood or 100 mg of brain tissue. DNA quality was assessed using gel electrophoresis, and quantity was determined by NanoDrop 3300 spectrometry (Barber et al., 2017).
Table 1.
Demographics of the sEOAD and controls cohort
| A | |||||
|---|---|---|---|---|---|
|
| |||||
| Center | N | Mean age at onset |
Females (%) | APOE ε4+ (%) | APOE ε4ε4 (%) |
| Bristol | 21 | 53.3 | 11 (52.4) | 10 (47.6) | 3 (14.3) |
| Manchester | 328 | 57.1 | 152 (46.3) | 194 (59.1) | 47 (14.3) |
| Nottingham | 26 | 58.2 | 12 (46.2) | 11 (42.3) | 1 (3.8) |
| Oxford | 33 | 55.6 | 19 (57.6) | 19 (57.6) | 3 (9.1) |
| All sEOAD cases | 408 | 56.8 | 194 (47.5) | 234 (57.4) | 54 (13.2) |
| B | |||||
|---|---|---|---|---|---|
|
| |||||
| Center | N | Mean age at death |
Females (%) | APOE ε4+ (%) | APOE ε4ε4 (%) |
| UCL | 436 | 77.2 | 256 (58.7) | 104 (23.9) | 9 (2.2) |
(A) The sEOAD cases were recruited from 4 centers within the UK, and the number of individuals from each center is outlined in the table. All cases had a documented or calculated age at onset below 65 years. The number of females from each center is recorded with the percentage per center together with the percentage of individuals from each center harboring at least 1 APOE ε4 allele (ε4+) along with the number and percentage of individuals from each center with the ε4ε4 genotype. (B) All controls were recruited from a single center in the UK (UCL, London); the number of individuals is given in the table. The mean age at death for controls is given with the number and proportion of females. The number of individuals harboring at least 1 APOE ε4 allele and the number and proportion of controls with the ε4ε4 genotype are also included.
Key: sEOAD, sporadic early-onset Alzheimer’s disease; UCL, University College London.
2.2. NeuroX array
Clustering and the first stage of quality control (QC) were completed in Illumina GenomeStudio 2011.1. GenomeStudio took raw fluorescent signal results and formed clusters of the individual genotypes for each SNP. A cluster file, provided by the Cohorts for Heart and Aging Research in Genomic Epidemiology, was used to assist in forming the cluster boundaries for most SNPs present on the chip, and the remaining SNPs are allocated automatically by the program (Barber et al., 2017).
The SNPs were assessed on how well clusters formed in GenomeStudio: clusters are expected to localize at single points with high intensity, to form in certain locations based on the allele present and whether the genotype is heterozygous, not form too close to one another, and to not be too wide or elliptical. SNPs were grouped into each nonautosomal chromosome while all autosomal SNPs were assessed together. Clustering of SNPs in genes of interest such as APOE were also assessed (Barber et al., 2017).
Once QC was completed for clustering, the resulting data set underwent final QC, SNPs and individuals were assessed using PLINK (Purcell et al., 2007). Individuals with a sample call rate below 95% were removed, likely as a result of poor DNA quality, as well as SNPs with a call rate below 90% as that could be due to probe design issues. PLINK was used to calculate ancestry information, Hardy-Weinberg equilibrium (HWE), relatedness, and heterozygosity (Barber et al., 2017). Adherence to HWE was identified, and SNPs which did not meet HWE (p < 1.2E-06) were removed. Individuals sharing more than 18.75% identity by state (equivalent to second or third cousins) were removed to distinguish between relatives with atypical heterozygosity and outliers in populations. Individuals with a heterozygosity greater than or less than 3 times the standard deviation were removed as indicators of cross-contamination or inbreeding, respectively. Univariate logistic regression was performed where the outcome variable was disease status (case vs. control), and all SNPs with a p-value below this corrected threshold were removed. The final target data set contained genotype information for 265,049 SNPs of 408 cases (48.0% female) and 436 controls (58.6% female) (Barber et al., 2017).
2.3. Polygenic risk scoring
PRS calculated using PRSice required SNP information (SNP coordinate, affected allele, reference allele, p-value, and effect size as either odds ratio or θ) from an independent cohort, to act as a base data set (Eusden et al., 2015). The base data set was collated by the International Genomics of Alzheimer’s Project (IGAP) consortium, with association data for 7,055,881 SNPs from 17,008 LOAD cases and 37,154 controls. The data were accumulated as a meta-analysis of GWASs performed by Genetic and Environmental Risk for Alzheimer’s Disease, European Alzheimer’s Disease Initiative, Cohorts for Heart and Aging Research in Genomic Epidemiology, and Alzheimer’s Disease Genetics Consortium (Lambert et al., 2013). There is no equivalent data available for sEOAD due to the lower frequency of the disease and its diagnosis; however, the shared phenotype between the 2 forms of AD may be caused by variants which affect the risk of developing AD common to both LOAD and sEOAD.
PRSice initially identified all SNPs common between the base and target data set; PRSs were calculated by ordering all SNPs in the base data set by association tested p-value; SNPs present within the p-value threshold defined by the user were used to provide an accumulative risk score for individuals in the target data set, based on the alleles present at each SNP. The PRSs calculated were compared between sEOAD cases and controls, and the ability to successfully identify cases and controls was determined by Nagelkerke’s R2 value: the threshold which contains SNPs that produce the greatest Nagelkerke’s R2 value is the best-fit threshold for analysis.
PRSice was set to calculate PRSs for all individuals in the cohort at each p-value threshold in increments of 1000th between 10−3 and 1. Uninformative SNPs determined to be in strong LD (r2 > 0.8) within a linkage block when compared to the index SNP were removed. We tested a range of r2 from 0.2 to 0.9 and selected 0.8 as this gave the best predictive model—Nagelkerke’s value of 0.169 for r2 < 0.2 versus 0.209 for r2 < 0.8.
2.4. Statistical analyses
Using the best-fit model, as identified by PRSice by the greatest Nagelkerke’s R2 value, the scores for each individual were analyzed in SPSS to calculate the sensitivity and specificity of the model. The predictability of the model at correctly identifying cases and controls was calculated from the area under the receiver operating characteristic curve (AUC).
The results produced by PRSice were further analyzed by decile scoring as carried out by Escott-Price et al. (2015). Decile scoring is an alternative to quartile and percentile scoring and provides further detail of trends in the data. Decile ranges were determined by segmenting the range of PRS into tenths and counting the number of cases and controls within each decile. Average scores of cases and controls within each decile were also calculated.
2.5. Polygenic risk score of the APOE locus
The APOE region is known to contain SNPs which affect the risk of LOAD; the presence of the APOE ε4 allele correlates with a high risk of AD. To ensure coverage of the entire APOE locus with nearby genes, a 500 kb region surrounding the APOE gene was isolated in the analysis by extracting the SNPs within this region from the NeuroX data set to produce an alternative target data set (Karolchik et al., 2004). The locus was identified as chr19:45,160,844- 45,660,844 (GRCh37) (Kent et al., 2002). This altered version of the target data set, carrying genotype information for 198 SNPs (including rs7412 but not including rs429358 as this failed QC) within the APOE region, was also tested using PRSice to calculate risk scores.
Additional cohort information is traditionally found to also be associated with AD risk; age, sex, and number of APOE ε2 and/or ε4 alleles were also integrated into the analysis. Logistic regression was performed on the APOE locus, PRS including the APOE locus, variables relevant to the analysis, and the combination of relevant variables with individual scores. Age was excluded as a variable as all cases were below age 65 years, whereas healthy controls were over age 65 years at the time of sampling. The AUC was calculated to determine whether accuracy of the model at predicting disease status improved with the inclusion of these demographic variables in the model. Hosmer-Lemeshow p-value is a result used to identify the goodness-of-fit of regression models; a nonsignificant value is considered a good model. Nagelkerke’s R2 value was calculated to compare models for the best fit; the greater the R2 value, the better fit the model had for prediction.
3. Results
In this study, sEOAD cases and controls were genotyped on the NeuroX array; the array results were clustered and subjected to QC to produce a target data set. This, along with the base data set provided by the IGAP consortium, was used to generate PRS for all cases and controls using PRSice.
PRSice provides the Nagelkerke’s R2 scores produced at every p-value threshold tested and the number of SNPs used to calculate the scores. A total of 28,538 SNPs were common between the target data set and the base data set. The p-value threshold with the highest Nagelkerke’s R2 defines the best-fit for the data set for identifying cases and controls. The best-fit threshold used association data from 9434 SNPs with a p-value ≤ 0.302 and produced the highest Nagelkerke’s R2 value of 0.209. A range of scores at different p-value thresholds with their corresponding R2 values and number of SNPs included is presented in Table 2.
Table 2.
Nagelkerke’s R2 values at varying p-value thresholds
| p-value threshold | Nagelkerke’s R2 | Number of SNPs |
|---|---|---|
| ≤0.001 | 0.149 | 141 |
| ≤0.002 | 0.154 | 203 |
| ≤0.005 | 0.163 | 355 |
| ≤0.010 | 0.172 | 546 |
| ≤0.020 | 0.176 | 930 |
| ≤0.050 | 0.181 | 2022 |
| ≤0.100 | 0.193 | 3595 |
| ≤0.200 | 0.204 | 6545 |
| ≤0.302 | 0.209 | 9434 |
| ≤0.500 | 0.207 | 14,995 |
| ≤1.00 | 0.203 | 28,538 |
The table lists some of the p-value thresholds tested by PRSice and their corresponding Nagelkerke’s R2 value, along with the number of SNPs used in calculating PRS. A total of 28,438 SNPs were common between the NeuroX array and the SNPs collected by the IGAP consortium. The greatest R2 value was at the threshold of p ≤ 0.302 and used variant information from 9434 SNPs.
Key: SNP, single nucleotide polymorphism.
The sensitivity and specificity of the best PRSice model was calculated in SPSS. Of the 408 cases in the NeuroX data set, 59.1% were correctly identified as cases, and 72.9% of the 436 controls were correctly identified as controls. The greatest predictive ability, AUC, of the PRS calculated by PRSice for this cohort was 72.9%. The value of Nagelkerke’s R2 calculated in SPSS, 0.209, was identical to the value obtained in PRSice, an indicator of reproductive power.
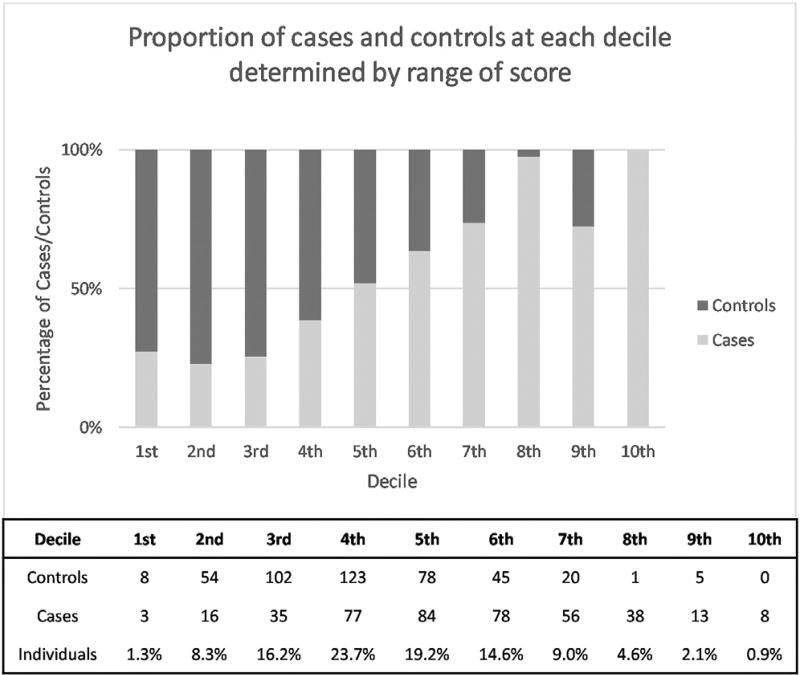
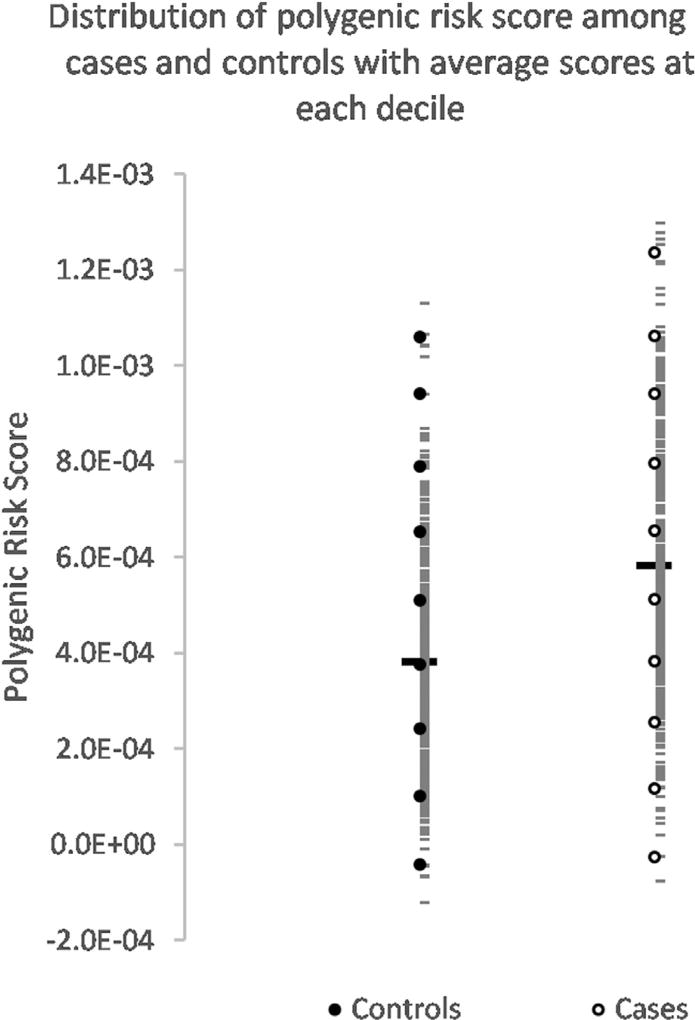
The average PRS for controls was 3.8E-04 ± 6.75E-04 and 5.8E- 04 ± 6.9E-04 for cases—using an unpaired t-test on these PRS values gives a t-value of 12.33 with p < 0.0001. Decile scoring was also used to visualize the pattern of PRS distribution between cases and controls. Each decile covered 1/10th of the score; however, the proportion of individuals within each decile varied. The first 4 deciles have a majority of controls, with fewer controls having high PRS compared to cases. A PRS >0.00045 would determine an individual to more likely be a case than control. The details of decile scoring are displayed in Fig. 1 along with the distribution of scores for cases and controls, with the identification of the average score at each decile presented in Fig. 2.
Fig. 1.
Proportion of diagnosed sEOAD cases and controls at each decile determined by a range of score. The figure breaks down the range of PRS into deciles. The range of scores which make up each decile are depicted, as well as the number of cases and controls, and the percentage of individuals which fall into each decile. Controls are right skewed, whereas cases demonstrate left skewness. These figures were produced from PRS of 408 sEOAD cases and 436 controls. The embedded table lists the decile ranges with the number of cases and controls in each decile along with the proportion of the cohort which make up each decile. Abbreviations: PRS, polygenic risk score; sEOAD, sporadic early-onset Alzheimer’s disease.
Fig. 2.
Distribution of polygenic risk score among sEOAD cases and controls with average scores at each decile. The range of PRS obtained for cases and controls is distributed into deciles. The range of coverage of each decile is shown in the bar plot together with the proportion of cases and controls which make up each decile. The average scores for all cases and controls are indicated by the thick bar, while the short horizontal bars show PRS for each individual. Average scores at each decile are indicated as hollow circles for cases and filled circles for controls. Abbreviations: PRS, polygenic risk score; sEOAD, sporadic early-onset Alzheimer’s disease.
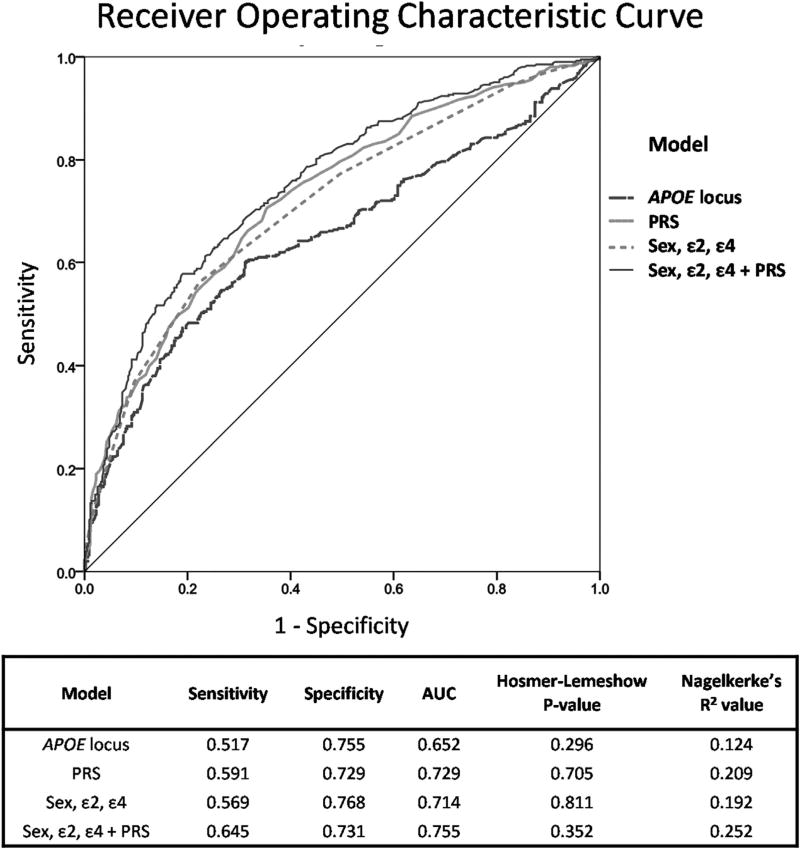
A 500 kb region surrounding the APOE gene was identified, and the SNPs within the locus were isolated in the target data set; PRS was calculated for individuals using association data from the IGAP consortium. Of the 198 SNPs present on the NeuroX array within this region, 31 were common with the base data set. The Nagelkerke’s R2 value corresponding with the best-fit threshold of p ≤ 0.001 was 0.124 using association data from 28 SNPs; using only the APOE locus, cases and controls of AD can be predicted with an AUC of 65.2%. Linear regression was completed o n the APOE locus data set and compared to the PRS model as shown in Fig. 3. The APOE model identified controls with a specificity similar to the PRS model, calculated as 75.5% and 72.9%, respectively; however, the ability to identify cases was not as accurate as the PRS model, with a sensitivity of 51.7% compared to 59.1%.
Fig. 3.
Results of logistic regression with an area under the receiver operating characteristic curve (AUC) for alternative risk-scoring models in sEOAD. For this analysis, the APOE locus was defined as a 500 kb region surrounding the APOE gene, and the scores produced by PRSice for this model are based on the SNPs within that region; PRS represents the score produced for all SNPs present on both the NeuroX array and in the base data set. The relevant variables included sex together with the number of APOE ε2 allele and/or APOE ε4 allele. As shown in the table, a nonsignificant Hosmer-Lemeshow p-value suggests that the model is suitable for using as a predictive tool. Nagelkerke’s R2 can also be used to identify the best model for risk prediction; the higher the value of R2 the greater the predictive accuracy of each model. This approach identified sex, ε2, ε4 + PRS as the best model for calculating risk in our sEOAD cohort as the largest AUC value is produced from the combination of variables. Abbreviations: PRS, polygenic risk score; sEOAD, sporadic early-onset Alzheimer’s disease; SNPs, single nucleotide polymorphisms.
Variable information can also impact an individuals’ risk of developing AD: identifying an individuals’ APOE ε status is common in diagnosis, while gender needs to be controlled for whenever possible. A combination of these variables using logistic regression produced the best model for identifying controls, with a specificity of 76.8% and sensitivity of 56.9%, most likely due to the protective effect of the ε2 allele. However, combining these 3 variables (ε2, ε4, and sex) with individuals’ PRS produced a model with the best overall predictive ability of 75.5%, together with sensitivity of 64.5% and specificity of 73.1%. Results of logistic regression analysis for each risk-scoring model are depicted in Fig. 3.
4. Discussion
A cohort of sEOAD cases and controls were genotyped on the NeuroX array, and this information was used to generate PRS in PRSice using SNP association data from the IGAP consortium as the training set. The resulting risk model could successfully recognize an individual to either have sEOAD or be a healthy control with 72.9% accuracy. The addition of variables (number of APOE ε2 alleles, APOE ε4 alleles, and sex) in logistic regression improved the predictability to 75.5%.
There was a significantly higher average PRS in cases than controls (p < 0.0001) with most individuals having a PRS above 0. Decile scoring showed most controls were within the lower deciles, with the absence of controls at the highest decile. For this analysis, the base data set we used was formed from LOAD cases rather than sEOAD; however, we do not perceive this as an issue since the pathogenic mechanisms for both sEOAD and LOAD are likely shared (Barber et al., 2017).
The APOE locus (500 kb region centered on APOE) had a predictive accuracy of 65.2%, which confirms the high-risk contribution from known variants within this locus. Our analysis demonstrates that additional genetic variation across the rest of the genome also influences the risk of sEOAD. The NeuroX array that we have used genotypes SNPs from regions across the entire genome together with custom content which includes genes associated with several other neurological diseases including AD. These types of arrays provide a practical means to obtain greater accuracy of predictive ability in complex diseases.
The presence of controls with high PRS suggests that individuals can have SNPs that are associated with a greater risk of sEOAD but they may not develop AD. This would support the idea that in addition to risk variants there are uncharacterized protective variants in the genome that modify an individual’s risk of getting the disease. The 5 controls present within the ninth decile had a high PRS; these individuals might have gone on to develop AD—the average age at death of these individuals was below 80 years. In addition, a high PRS could indicate risk for AD in later life or the risk of other neurological diseases that correlate with AD. Low PRS could be indicative of neuroprotection; however, low scores were also found in some of our sEOAD cases. Healthy controls with high PRS and cases with low PRS are possible indicators of missing heritability or as yet unknown environmental factors affecting the risk of developing AD.
The predictability of disease as estimated by the AUC derived from ε2, ε4, and sex was 71.4%. The AUC for the best model included these variables, but the addition of PRS increased the predictive ability by more than 3%. More extensive genotyping and additional information collected about individuals’ lifestyles could further improve the predictability of AD risk. Further improvements of genetic-based prediction models could increase the predictability to the point where at-risk individuals are readily identified and potentially stratified using genetic testing.
The NeuroX array we have used in this study was the first version of an array specifically designed for neurological diseases. The custom content contributed 4401 variants within the PRS threshold we have utilized, which accounted for 17.3% of the markers used to generate the scores. An increase in the number of markers to include a greater range of genetic variants associated with AD will undoubtedly lead to the generation of improved scores. Several of the loci identified more recently were not present on the first iteration of the NeuroX array. Increased coverage, such as that available from the latest version of the NeuroX chip version 2 (Blauwendraat et al., 2017), could provide additional information for generating more accurate risk scores thereby providing a better predictive model.
In the study performed by Escott-Price et al. (2015), an AUC of 78.2% was achieved using association data from 87,605 SNPs combined with covariate information for sex, age, and APOE. The study produced a set of scores with more variability, due to more SNPs in common between both data sets, although the indicator for a good model is ultimately determined by the Nagelkerke’s R2 value. A more diverse set of scores for AD could lead to the ability of identifying specific groups within the disease cohort and introduce treatment plans according to the variants identified (Eusden et al., 2015). In our study using a much reduced number of pathologically confirmed sEOAD cases (n = 408), we have obtained comparable PRS (AUC of 75.5%) to the original study of Escott-Price et al. (2015) generated for 3049 LOAD cases and 1554 controls. This demonstrates the increased power that can be realized using pathologically confirmed tissue in comparison to clinically defined samples. In a more recent study, Escott-Price et al. (2017a) used a modified approach to calculate the maximum possible predictive power (AUCmax) thereby improving the AUC produced previously from a value of 78.2% to 82%. More recently, Escott-Price et al. (2017b) have performed PRS analysis on pathologically confirmed samples and found improved scores compared with the previous study on clinically diagnosed cases (Escott-Price et al. (2015)).
Other studies in AD using SNP scoring to generate risk scores have used SNPs with greater effect size as a means to reduce the number of SNPs required to calculate risk as discussed in the Introduction. For example, a genome-wide risk score has been calculated previously from the effect scores of just 31 SNPs and genotypes of the APOE ε2 and ε4 alleles (Desikan et al., 2017). Using a model with tens of SNPs compared to thousands would provide a more cost-effective approach to screen for AD in individuals. Alternatively, identifying the variants which increase phenotypic risk in an individual using a risk score model could be used to form a more effective symptom-specific treatment plan. The ultimate driver will be the SNP set which provides the greatest prediction irrespective of SNP number.
Acknowledgments
The ARUK Consortium members are Peter Passmore, David Craig, Janet Johnston, Bernadette McGuinness, Stephen Todd, Reinhard Heun, Heike Kölsch, Patrick G. Kehoe, Emma R.L.C. Vardy, Nigel M. Hooper, Stuart Pickering-Brown, Julie Snowden, Anna Richardson, Matthew Jones, David Neary, Jennifer Harris, James Lowe, A. David Smith, Gordon Wilcock, Donald Warden, and Clive Holmes.
The work of Jose Bras and Rita Guerreiro is funded by fellowships from Alzheimer’s Society. This work was partially funded by Alzheimer’s Research UK ARUK-PG2014-2, Alzheimer’s Society, and an anonymous donor.
The authors thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i-Select chips were funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. GERAD was supported by the Medical Research Council (grant no 503480), Alzheimer’s Research UK (grant no 503176), the Wellcome Trust (grant no 082604/2/07/Z), and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant no 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, part of the Department of Health and Human Services; project ZO1 AG000950.
Footnotes
Disclosure statement
The authors have no actual or potential conflicts of interest.
References
- Adams HH, de Bruijn RF, Hofman A, Uitterlinden AG, van Duijn CM, Vernooij MW, Koudstaal PJ, Ikram MA. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimers Dement. 2015;11:1277–1285. doi: 10.1016/j.jalz.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Ayers KL, Mirshahi UL, Wardeh AH, Murray MF, Hao K, Glicksberg BS, Li S, Carey DJ, Chen R. A loss of function variant in CASP7 protects against Alzheimer’s disease in homozygous APOE ε4 allele carriers. BMC Genomics. 2016;17:445. doi: 10.1186/s12864-016-2725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber I, Braae A, Clement N, Patel T, Guetta-Baranes T, Brookes K, Medway C, Chappell S, Guerreiro R, Bras J, Hernandez D, Singleton A, Hardy J, Mann D, ARUK Consortium. Morgan K. Mutational analysis of sporadic early-onset Alzheimer’s disease using the NeuroX array. Neurobiol. Aging. 2017;49:215.e1–215.e8. doi: 10.1016/j.neurobiolaging.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Pan W, Shen X, Oetting WS. Multilocus association testing with penalized regression. Genet. Epidemiol. 2011;35:755–765. doi: 10.1002/gepi.20625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwendraat C, Faghri F, Pihlstrom L, Geiger JT, Elbaz A, Lesage S, Corvol J, May P, Ryten M, Ferrari R, Bras J, Guerreiro R, Williams J, Sims R, Lubbe S, Hernandez DG, Mok KY, Robak L, Campbell RH, Rogaeva E, Traynor BJ, Chia R, Chung SJ, International Parkinson’s Disease Genomics Consortium (IPDGC) COURAGE-PD Consortium. Hardy JA, Brice A, Wood NW, Houlden H, Shulman JM, Morris HR, Gasser T, Krüger R, Heutink P, Sharma M, Simón Sánchez J, Nalls MA, Singleton AB, Scholz SW. NeuroChip, an updated version of the NeuroX genotyping platform to rapidly screen for variants associated with neurological diseases. Neurobiol. Aging. 2017;57:247.e9–247.e13. doi: 10.1016/j.neurobiolaging.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budson A, Kowall N. The Handbook of Alzheimer’s Disease and Other Dementias. Wiley-Blackwell; Chichester, UK: 2011. Preface XV-XV. [Google Scholar]
- Bush W, Moore J. Chapter 11: genome-wide association studies. PLoS Comput. Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Whittaker J. Analysis of multiple SNPs in a candidate gene or region. Genet. Epidemiol. 2008;32:560–566. doi: 10.1002/gepi.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L, Jakobsdottir J, Mukherjee S, Adams HH, Choi SH, Larson EB, Fitzpatrick A, Uitterlinden AG, de Jager PL, Hofman A, Gudnason V, Vardarajan B, Ibrahim- Verbaas C, van der Lee SJ, Lopez O, Dartigues JF, Berr C, Amouyel P, Bennett DA, van Duijn C, DeStefano AL, Launer LJ, Ikram MA, Crane PK, Lambert JC, Mayeux R, Seshadri S. Evaluation of a genetic risk score to improve risk prediction for Alzheimer’s disease. J. Alzheimers Dis. 2016;53:921–932. doi: 10.3233/JAD-150749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, Thompson WK, Besser L, Kukull WA, Holland D, Chen C, Brewer JB, Karow DS, Kauppi K, Witoelar A, Karch CM, Bonham LW, Yokoyama JS, Rosen HJ, Miller BL, Dillion WP, Wilson DM, Hess CP, Pericak-Vance M, Haines JL, Farrer LA, Mayeux R, Hardy J, Goate AM, Hyman BT, Schellenberg GD, McEvoy LK, Andreassen OA, Dale AM. Genetic assessment of age-associated Alzheimer’s disease risk: development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258. doi: 10.1371/journal.pmed.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Myers AJ, Huentelman M, Hardy J. Polygenic risk score analysis of pathologically confirmed Alzheimer disease. Ann. Neurol. 2017b;82:311–314. doi: 10.1002/ana.24999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol. Aging. 2017a;49:214.e7–214.e11. doi: 10.1016/j.neurobiolaging.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, GERAD/PARADES IGAP consortia. Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert J, van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P, Williams J. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis C, O’Reilly P. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri M, Zhang M, Lü Y. Genes associated with Alzheimer’s disease: an overview and current status. Clin. Interv. Aging. 2016;11:665–681. doi: 10.2147/CIA.S105769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, George-Hislop P, Singleton A, Hardy J, The Alzheimer Genetic Analysis Group TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison TM, Mahmood Z, Lau EP, Karacozoff AM, Burggren AC, Small GW, Bookheimer SY. An Alzheimer’s disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro. 2016;3 doi: 10.1523/ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina. HumanExome BeadChips. [Accessed February 2017];Data Sheet: DNA Analysis. 2012 Available at: www.smd.qmul.ac.uk/gc/Services/InfiniumArrays/datasheet_humanexome_beadchips.pdf.
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The USCS Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Morón FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fiévet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossù P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease Alzheimer’s Disease Genetic Consortium Cohorts for Heart and Aging Research in Genomic Epidemiology. Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nöthen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer’s disease: risk, mechanisms, and therapy. Nat. Rev. Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Schu M, Vardarajan BN, Farrell J, Bennett DA, Buxbaum JD, Byrd GS, Ertekin-Taner N, Evans D, Foroud T, Goate A, Graff-Radford NR, Kamboh MI, Kukull WA, Manly JJ, Alzheimer’s Disease Genetics Consortium. Haines JL, Mayeux R, Pericak-Vance MA, Schellenberg GD, Lunetta KL, Baldwin CT, Fallin MD, Farrer LA. Two rare AKAP9 variants are associated with Alzheimer’s disease in African Americans. Alzheimers Dement. 2014;10:609–618. doi: 10.1016/j.jalz.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton MK, Strike L, Hansell NK, Wen W, Mather KA, Armstrong NJ, Thalamuthu A, McMahon KL, de Zubicaray GI, Assareh AA, Simmons A, Proitsi P, Powell JF, Montgomery GW, Hibar DP, Westman E, Tsolaki M, Kloszewska I, Soininen H, Mecocci P, Velas B, Lovestone S, Brodaty H, Ames D, Trollor JN, Martin NG, Thompson PM, Sachdev PS, Wright MJ. The effect of increased genetic risk for Alzheimer’s disease on hippocampal and amygdala volume. Neurobiol. Aging. 2016;40:68–77. doi: 10.1016/j.neurobiolaging.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden JR, Mayeda ER, Walter S, Vivot A, Tchetgen Tchetgen EJ, Kawachi I, Glymour MM. Using an alzheimer disease polygenic risk score to predict memory decline in black and white Americans over 14 years of follow-up. Alzheimer Dis. Assoc. Disord. 2016;30:195–202. doi: 10.1097/WAD.0000000000000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden JR, Walter S, Tchetgen Tchetgen EJ, Kawachi I, Glymour MM. Validation of a polygenic risk score for dementia in black and white individuals. Brain Behav. 2014;4:687–697. doi: 10.1002/brb3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiskainen H, Helisalmi S, Viswanathan J, Kurki M, Hall A, Herukka SK, Sarajarvi T, Natunen T, Kurkinen KM, Huovinen J, Makinen P, Laitinen M, Koivisto AM, Mattila KM, Lehtimaki T, Remes AM, Leinonen V, Haapasalo A, Soininen H, Hiltunen M. Effects of Alzheimer’s diseaseassociated risk loci on cerebrospinal fluid biomarkers and disease progression: a polygenic risk score approach. J. Alzheimers Dis. 2015;43:565–573. doi: 10.3233/JAD-140777. [DOI] [PubMed] [Google Scholar]
- Morgan AR, Touchard S, O’Hagan C, Sims R, Majounie E, Escott-Price V, Jones L, Williams J, Morgan BP. The correlation between inflammatory biomarkers and polygenic risk score in Alzheimer’s disease. J. Alzheimers Dis. 2017;56:25–36. doi: 10.3233/JAD-160889. [DOI] [PubMed] [Google Scholar]
- Naj AC, Schellenberg GD. Genomic variants, genes, and pathways of Alzheimer’s disease: an overview. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016;174:5–26. doi: 10.1002/ajmg.b.32499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Bras J, Hernandez DG, Keller MF, Majounie E, Renton AE, Saad M, Jansen I, Guerreiro R, Lubbe S, Plagnol V, Gibbs JR, Schulte C, Pankratz N, Sutherland M, Bertram L, Lill CM, DeStefano AL, Faroud T, Eriksson N, Tung JY, Edsall C, Nichols N, Brooks J, Arepalli S, Pilner H, Letson C, Heutink P, Martinez M, Gasser T, Traynor BJ, Wood N, Hardy J, Singleton AB. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol. Aging. 2015;36:1605-e7–1605.e12. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Chen YM, Wei P. Testing for polygenic effects in genome-wide association studies. Genet. Epidemiol. 2015;39:306–316. doi: 10.1002/gepi.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M, Wimo A, Guerchet M, Ali G, Wu Y, Prina M. The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost, and Trends. Alzheimer’s Disease International; London: 2015. World Alzheimer’s Report 2015. [Google Scholar]
- Purcell SM, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker P, Daly M, Sham P. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Accessed February 2017]. Available at: http://www.R-project.org/ [Google Scholar]
- Rodriguez-Rodriguez E, Sanchez-Juan P, Vazquez-Higuera JL, Mateo I, Pozueta A, Berciano J, Cervantes S, Alcolea D, Martinez-Lage P, Clarimon J, Lleo A, Pastor P, Combarros O. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer’s disease. J. Neural Transm. (Vienna) 2013;120:807–812. doi: 10.1007/s00702-012-0920-x. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Heilmann S, Becker T, Hernandez I, Wagner H, Thelen M, Mauleon A, Rosende-Roca M, Bellenguez C, Bis JC, Harold D, Gerrish A, Sims R, Sotolongo-Grau O, Espinosa A, Alegret M, Arrieta JL, Lacour A, Leber M, Becker J, Lafuente A, Ruiz S, Vargas L, Rodriguez O, Ortega G, Dominguez M, IGAP. Mayeux R, Haines JL, Pericak-Vance MA, Farrer LA, Schellenberg GD, Chouraki V, Launer LJ, van Duijn C, Seshradi S, Atunez C, Breteler MM, Serrano-Rios M, Jessen F, Tarraga L, Nothen MM, Maier W, Boada M, Ramirez A. Follow-up of loci from the International Genomics of Alzheimer’s Disease Project identifies TRIP4 as a novel susceptibility gene. Transl. Psychiatry. 2014;4:e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb. Cortex. 2012;22:2653–2661. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nöthen MM, White CC, Hamilton-Nelson KL, Epelbaum J, Maier W, Choi SH, Beecham GW, Dulary C, Herms S, Smith AV, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sánchez-Juan P, Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank-García A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henández I, Barber RC, Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel-Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Longstreth WT, Jr, Fairchild TJ, Bossù P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris J, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairén MD, Hodges A, Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleó A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J, ARUK Consortium. Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin LW, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Morris JC, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult-Stählbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu CK, GERAD/PERADES, CHARGE, ADGC, EADI. Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Mann D, Tsolaki M, Clarimón J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Goate AM, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues JF, DeStefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Launer LJ, Escott-Price V, Mayeux R, Deleuze JF, Amin N, Holmans PA, Pericak-Vance MA, Amouyel P, van Duijn CM, Ramirez A, Wang LS, Lambert JC, Seshadri S, Williams J, Schellenberg GD. Rare coding variants in PLCG2, ABI3 and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, Struyfs H, Van Dongen J, Vermeulen S, Engelborghs S, Vandenbulcke M, Vandenberghe R, De Deyn PP, Van Broeckhoven C. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Abeta42. Alzheimers Dement. 2015;11:1452–1460. doi: 10.1016/j.jalz.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Verhaaren BF, Vernooij MW, Koudstaal PJ, Uitterlinden AG, van Duijn CM, Hofman A, Breteler MM, Ikram MA. Alzheimer’s disease genes and cognition in the nondemented general population. Biol. Psychiatry. 2013;73:429–434. doi: 10.1016/j.biopsych.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Wingo TS, Lah JJ, Levey AI, Cutler DJ. Autosomal recessive causes likely in early-onset Alzheimer’s disease. Arch. Neurol. 2012;69:59. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Liu ZJ, Tao S, Sun YM, Jiang D, Li HL, Chen H, Liu X, Lapin B, Wang CH, Zheng SL, Xu J, Wu ZY. Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the Han Chinese population. Oncotarget. 2015;6:36955–36964. doi: 10.18632/oncotarget.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama JS, Bonham LW, Sears RL, Klein E, Karydas A, Kramer JH, Miller BL, Coppola G. Decision tree analysis of genetic risk for clinically heterogeneous Alzheimer’s disease. BMC Neurol. 2015;15:47. doi: 10.1186/s12883-015-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]