Abstract
We have screened sporadic early-onset Alzheimer’s disease (sEOAD, n = 408) samples using the NeuroX array for known causative and predicted pathogenic variants in 16 genes linked to familial forms of neurodegeneration. We found 2 sEOAD individuals harboring a known causative variant in PARK2 known to cause early-onset Parkinson’s disease; p.T240M (n = 1) and p.Q34fs delAG (n = 1). In addition, we identified 3 sEOAD individuals harboring a predicted pathogenic variant in MAPT (p.A469T), which has previously been associated with AD. It is currently unknown if these variants affect susceptibility to sEOAD, further studies would be needed to establish this. This work highlights the need to screen sEOAD individuals for variants that are more classically attributed to other forms of neurodegeneration.
Keywords: Alzheimer’s disease, Parkinson’s disease, Sporadic, Early-onset, NeuroX, Screening
1. Introduction
Alzheimer’s disease (AD) is the commonest form of dementia in the world. AD and other dementias were the fourth leading cause of death in high-income countries in 2012 (WHO, 2012). Sporadic early-onset Alzheimer’s disease (sEOAD) has a disease onset ≤65 years of age, and these individuals do not harbor a known causative variant, the remaining sporadic cases are classified as late-onset Alzheimer’s disease (LOAD). Both forms of AD have a complex etiology with heritability estimated to be 92%–100% for sEOAD (Wingo et al., 2012) and 70% for LOAD (Gatz et al., 2006). Given the difference in heritability and age of onset between sEOAD and LOAD, it is likely that sEOAD patients have a more penetrant genetic etiology and thus provide a good cohort to explore the genetics of AD.
Many types of dementia have a neuropathological or clinical crossover, for example, both Parkinson’s disease with dementia (PDD) and Parkinson’s disease without dementia (PD) have alpha-synuclein deposits in the brain and a similar clinical presentation to dementia with Lewy bodies (DLB) (Jellinger, 2014). Mixed dementia has features linked to more than 1 type of dementia, for example AD with cerebrovascular lesions or AD with Lewy bodies (Jellinger, 2014). It is not surprising then that some genetic loci identified thus far are associated with multiple types of dementias; the commonest example is that of APOE ε4, which is associated with AD (Corder et al., 1993), posterior cortical atrophy (Carrasquillo et al., 2014) and DLB (Bras et al., 2014). It is believed that sporadic AD could be in part due to the aggregate of multiple causative variants; therefore, it is easy to imagine that different types of dementia have overlapping genetics; whereby a portion of variants that contribute to 1 dementia are also seen to contribute to a different type of dementia. Alternatively, different types of dementia could be a result of pleiotropy, for example, it has recently been reported that the alpha-synuclein gene (SNCA) is associated with DLB, but this is a different haplotype to that associated with PDD (Bras et al., 2014).
The NeuroX is a customized Illumina HumanExome DNA microarray; the first version of the chip contains 267,607 markers, most of which genotype rare missense variants, notably most of the genes in the human genome have at least 1 variant genotyped. The NeuroX includes standard content (242,901) designed by Illumina together with custom content (24,706) designed to be “neuro-specific”. The custom content was selected to genotype specific variants and genes linked to neurologic diseases, the inclusion criterion was determined using literature searches and genotyping data available before 2014. A more descriptive explanation of the NeuroX can be found in the consortia’s published paper (Nalls et al., 2015). The NeuroX provides a convenient approach to screen for causative variants, and test for genetic crossover and pleiotropy among neurologic diseases.
We report the screening of 408 sEOAD individuals with the aim to identify causative or predicted pathogenic variants in 16 selected genes using the NeuroX. These 16 genes are linked to familial forms of neurodegeneration including AD (APP, PSEN1, and PSEN2), frontotemporal dementia and amyotrophic lateral sclerosis (C9orf72, CHMP2B, FUS, GRN, MAPT, TARDBP, and VCP), Parkinson’s disease (LRRK2, PARK2, PARK7, PINK1, and SNCA), and prion disease (PRNP); all have pathogenic variants highlighted in freely accessible online databases.
2. Materials and methods
Methods were conducted according to the manufacturer’s instructions unless otherwise stated. All sEOAD samples were first screened for known causative variants in APP exons 16 and 17 by Sanger sequencing, followed by PSEN1 and PSEN2 using the NeuroX data. Individuals harboring a causative variant in either of these genes were removed prior to this analysis (Barber et al., 2016).
2.1. Samples
sEOAD individuals (n = 408) had an age of disease onset ≤65 years of age (Table 1). For 28 individuals where age at onset (AAO) was not documented, it was derived assuming 8 years disease duration from age at death (Ryman et al., 2014).
Table 1.
Demographics of the sporadic early-onset Alzheimer’s disease (sEOAD) cohort
| Centre | N | Mean AAO (± SD) | Females (%) | APOE ε4+ (%) | APOE ε4 MAF | APOE ε4ε4 (%) | Definite | Probable |
|---|---|---|---|---|---|---|---|---|
| Bristol | 21 | 53.3 (5.3) | 9 (42.9) | 10 (47.6) | 0.31 | 3 (14.3) | 21 | 0 |
| Manchester | 328 | 57.1 (5.5) | 156 (47.6) | 196 (59.8) | 0.58 | 46 (14.0) | 53 | 275 |
| Nottingham | 26 | 58.2 (6.3) | 12 (46.2) | 11 (42.3) | 0.23 | 1 (3.8) | 5 | 21 |
| Oxford | 33 | 55.6 (4.2) | 19 (57.6) | 19 (57.6) | 0.33 | 3 (9.1) | 24 | 9 |
| All | 408 | 56.8 (5.5) | 196 (48.0) | 236 (57.8) | 0.53 | 53 (13.0) | 103 | 305 |
The cohort contains individuals from multiple centres; each centre is represented 1 per row. The number of individuals (N) from each centre is given along with the mean age of onset with standard deviation (Mean AAO [± SD]), this is followed by the number and percentage of female individuals per centre (Females [%]), the number and percentage of individuals harboring at least one APOE ε4 allele (APOE ε4+ [%]), APOE ε4 minor allele frequency (APOE ε4 MAF), and number and percentage of individuals with APOE ε4ε4 genotype (APOE ε4ε4). The final columns state the number of individuals classed as either postmortem verified (Definite) or probable (Probable) AD according to Neurological and Communicative Disorders and Stroke and the Alzheimer’s disease and Related Disorders Association (NINCDS-ADRDA) and Consortium to Establish a Registry for Alzheimer’s disease (CERAD) guidelines.
Key: AAO, age at onset; N, number of individuals; SD, standard deviation.
sEOAD individuals were diagnosed as either definite or probable AD according to NINCDS-ADRDA (National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s disease and Related Disorders Association), and CERAD (Consortium to Establish a Registry for Alzheimer’s disease) guidelines. All samples used in this study were received with informed consent, and experimental procedures were approved by the local ethics committee, Nottingham Research Ethics Committee 2 (REC reference 04/Q2404/130). All experimental procedures were conducted in accordance with approved guidelines.
DNA was extracted from blood or brain tissue using a standard phenol chloroform extraction method. DNA quality and quantity was assessed by gel electrophoresis and NanoDrop 3300 spectrometry, respectively.
2.2. Quality control of NeuroX data
Quality control (QC) of the NeuroX intensity data was conducted in Genome Studio Version 2011.1 using the genotyping module Version 1.9.4. Markers on the Y chromosome were excluded from SNP statistics of female samples using the inbuilt option, this was to ensure variants on the Y chromosome were not incorrectly labeled as having a low call rate. Standard content was clustered using the CHARGE cluster file Version 1.0 (Grove et al., 2013). The following QC procedures in Genome Studio were conducted using only the best quality samples (≥99% call rate). Standard content with call rate <100% and all custom content were clustered using Genome Studio’s clustering algorithm. This was followed by manual assessment and clustering of all nonautosomal markers, and manual assessment and clustering of autosomal markers matching any of the following criteria: ≤99% call rate, excess heterozygote calls, excess heterozygote calls relative to expected Hardy–Weinberg equilibrium, deficient heterozygote calls relative to expected Hardy–Weinberg equilibrium, low intensity, unexpected cluster positions, wide clusters, or low cluster separation (Grove et al., 2013). Once QC had been conducted in Genome Studio, the genotyping calls were exported from Genome Studio to PLINK format in the forward orientation with all samples and all variants included. The MapInfo (location) was updated for 29 markers that originally had no location (Supplementary Table 1), and the chromosome was updated for 121 markers from chromosome X to the pseudo autosomal region (Supplementary Table 2). Additional QC was performed in PLINK Version 1.07. Samples were first removed if they had a call rate <98% followed by markers that had a call rate <95%. Further samples were removed if they failed the following criteria: samples with identity by decent >18.75% or heterozygosity rate outside ±3 standard deviation of the mean, both determined using an linkage disequilibrium pruned version of the data set (indeppairwise 50 5 0.2) with only common autosomal variants (minor allele frequency [MAF] >0.1). Further common markers were removed if they had significant deviation (p-value <1.2E–6) from Hardy–Weinberg equilibrium in control samples (data not shown). The final NeuroX data set had 265,049 markers with an average sample call rate of 99.9%.
2.3. Samples harboring a known causative variant
A total of 1196 variations are documented in the PD online mutation database (http://www.molgen.vib-ua.be/PDMutDB/ accessed August 2013), AD&FTD online mutation database (AD&FTDMDB) (http://www.molgen.vib-ua.be/ADMutations/ accessed August 2013) (Cruts et al., 2012) and Human Prion Mutation Database (http://www.mad-cow.org/prion_point_mutations.html accessed November 2014) combined. These databases document variants across 16 genes known to cause familial forms of neurodegeneration (APP, C9orf72, CHMP2B, FUS, GRN, LRRK2, MAPT, PARK2, PARK7, PINK1, PRNP, PSEN1, PSEN2, SNCA, TARDBP, and VCP) and includes 3 genes that are linked to AD (APP, PSEN1 and PSEN2). The databases include variants that do not cause disease (not pathogenic), have unknown pathogenicity (pathogenic nature unclear), and are known to cause disease (known causative) (Supplementary Table 3). About 1075 of these database variants are SNPs, small insertions, or small deletions and therefore had the potential to be genotyped on the NeuroX.
The genomic position relative to the reference build GRCh37 (February 27, 2009), reference allele and alternative allele were successfully calculated for 265,828 markers (99%) on the NeuroX using an in-house script. A second in-house script was used to establish if any of these markers genotyped the 1075 database variants. It was established that 412 (38%) of these variants were genotyped on the NeuroX and included 38% of AD-related variants, 32% of frontotemporal dementia or amyotrophic lateral sclerosis–related variants, 47% of PD-related variants and 12% of prion-related variants. Of the 412 variants, 407 passed QC procedures and inspection of the cluster plots found they all clustered well.
The 407 database variants were filtered to retain only those labeled as causative and were also polymorphic in our sEOAD cohort, 4 variants fit this criterion. The sEOAD samples harboring these 4 variants were identified and their genotype verified with Sanger sequencing. As expected, these 4 variants were not located in APP, PSEN1, or PSEN2 as our sporadic cohort had been previously filtered to remove individuals harboring these variants.
2.4. Samples harboring a predicted pathogenic variant
It was established that 662 variants were genotyped on the NeuroX which passed QC and were located in one of the 16 genes linked to neurodegenerative diseases (Table 2), this list excluded the 407 known causative variants analyzed previously. The gene ID, European MAF from the 1000 Genomes Project (Genomes Project Consortium et al., 2012), polymorphism phenotyping (Polyphen) score, and Sorting Intolerant from Tolerant (SIFT) score were retrieved for all 662 variants using ENSEMBL’s variant effect predictor (McLaren et al., 2010), which was installed locally and run over the command line using reference genome build GRCh37, Polyphen HDIV database and predicting 1 consequence per variant.
Table 2.
Samples harboring a variant
| Marker | Position | Variant | Base change | MAF | Protein change |
Gene | Disease | Sample | Seq | Gender | AAO | APOE ε |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exm593516 | 6:162394349 | rs137853054 | aCg/aTg | 2.8e-04 | p.T240M | PARK2 | PD (R) | M177 (Het) | C | Female | 52 | 44 |
| NeuroX_PARK2_Gln34fs_del_AG | 6:162864411-2 | rs55777503 | cga.cAG.ggg/cga.cgg.ggt | 2.9e-04 | p.Q34fs | PARK2 | PD (R) | M099 (Het) | C | Female | 56 | 44 |
| NeuroX_PARK2_Gln34fs_del_A | 6:162864412 | rs748142049 | cga.cAg.ggg/cga.cgg.ggg | 8.2e-06 | p.Q34fs | PARK2 | PD (R) | M099 (Het) | U | Female | 56 | 44 |
| NeuroX_16:31196410 | 16:31196410 | - | gGc/gTc | U/K | p.G225V | FUS | ALS (R) | M215 (Het) | U | Female | 53 | 34 |
| Marker | Position | Variant | Base change | MAF | Protein change | Gene | Disease | Sample | Seq | Gender | AAO | APOE ε |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| exm1330962 | 17:44055753 | rs144397565 | gGc/gTc | 2.4e-05 | p.G107V | MAPT | FTD (D) | M820 (Het) | C | Male | 65 | 34 |
| exm1331018 | 17:44067341 | rs143956882 | tCc/tTc | 0.001 | p.S427F | MAPT | FTD (D) | M357 (Het) | C | Male | 53 | 33 |
| M143 (Het) | C | Female | 57 | 34 | ||||||||
| M697 (Het) | U | Female | 48 | 24 | ||||||||
| M245 (Het) | C | Female | 63 | 44 | ||||||||
| exm1331027 | 17:44068850 | rs143624519 | Gcc/Acc | 0.001 | p.A469T | MAPT | FTD (D) | M168 (Het) | C | Female | 58 | 33 |
| M172 (Het) | U | Female | 58 | 34 | ||||||||
| M382 (Het) | U | Female | 52 | 33 | ||||||||
| M691 (Het) | C | Male | 55 | 34 | ||||||||
| M699 (Het) | C | Male | 58 | 33 |
Samples identified by the NeuroX as harboring a known causative variant in 1 of the 13 non-AD familial genes (Top) or a predicted pathogenic variant in 1 of the 16 familial genes (Bottom). The name of the NeuroX marker (Marker) is followed by information about the variant genotyped by the marker, including the genomic position of the variant in reference genome build GRCh37 and format chromosome:base position (Position), and the reference ID of the variant according to dbSNP (Variant). The variant is given at nucleotide level (base change) on the sense strand, the minor allele frequency in the general population according to ExAC (MAF) and amino acid level (protein change). Also listed is the gene the variant resides in (Gene) and the disease most associated with the gene, the Mendelian pattern of inheritance is given in brackets (Disease). The sample(s) harboring the variant (Sample) is followed by the results of Sanger sequencing (Seq) and patient information including gender (Gender), age at onset (AAO) and APOE ε status (APOE ε).
Key: C, Sanger sequencing confirmed the genotype of this sample; D, dominant; FTD, frontotemporal dementia; R, recessive; U, Sanger sequencing did not confirm the genotype of this sample; U/K, unknown.
Variants were filtered to retain only those that were polymorphic in our sEOAD cohort, had a 1000 Genomes European MAF <0.01, and were predicted to be probably pathogenic; which is defined here as a “deleterious effect” by SIFT (≤0.05) or “probably damaging” by PolyPhen (≥0.909). Three variants fit this criterion. The sEOAD samples harboring these 3 variants were identified and their genotype verified with Sanger sequencing.
2.5. Sanger sequencing
Genomic DNA (gDNA) was amplified in a final volume of 15 µL using the following constituents and final concentrations: 2-ng/µL gDNA, 1-pM forward primer and 1-pM reverse primer, 1× Buffer (BioLabs), 0.2-mM deoxyribose nucleoside triphosphates (dNTPs, Thermo Scientific), 0.1-U/µL LongAmp Taq DNA polymerase (New England Biolabs), and molecular grade water to the required volume. The reaction was subjected to the following thermal conditions: initial denaturation step of 94 °C for 2 minutes, followed by 35 cycles of 94 °C for 30 seconds, 57 °C for 15 seconds, and 72 °C for 45 seconds, finished with a final extension step at 72 °C for 7 minutes. A reaction containing no gDNA was included as a negative control. The polymerase chain reaction (PCR) products were cleaned using ExoSAP-IT (Affymetrix). Primers used for sequencing were the same as those used for amplification (Eurogenomics). The primers used for p.T240M were forward 5′GCTCGTGTGGCAGAACAATA 3′ and reverse 5′ACACCCCACCTCTGACAAG 3′; the product was 202 bp (base pairs) long. The primers used for p.G255V were forward 5′ TGGCAATCAAGACCAGAGTG 3′ and reverse 5′ GATTCATGCAATCCTCCACA 3′; the product was 243 bp long. The primers used for p.Q34fs were forward 5′ TCAGGCATGAATGTCAGATTG 3′ and reverse 5′ CCTTCCAATT TCCTTGGTCA 3′; the product was 272 bp long. The primers used for p.G107V were forward 5′AGAGCTGAGGCACCTTGGTA 3′ and reverse 5′ ATGGCAGGCAATTCCAGTT 3′; the product was 235 bp long. The primers used for p.S427F were forward 5′ TCCACACGTTCC TCTGCTAA 3′ and reverse 5′ AGCAGCCTGGTTCTTTTCAA 3′; the product was 230 bp long. The primers used for p.A469T were forward 5′ GGCTGGTGTTGACTCTTGGT 3′ and reverse 5′ TCTTACCAGAGCTGGGTGGT 3′; the product was 206 bp long.
Amplicons were sequenced using the Sanger di-deoxy method in the forward and reverse directions. Cleaned PCR products were sequenced in a final volume of 10 µL using 4-µL PCR product and the following constituents: 0.5 pM forward/reverse primer, 1× BigDye Sequencing Buffer (Life Technologies), 0.25× BigDye Terminator Version 3.1 (Life Technologies), and molecular grade water to the required volume. The reaction was subjected to the following thermal conditions: 25 cycles of 96 °C for 30 seconds, 50 °C for 15 seconds, and finally 60 °C for 4 minutes. The reactions were cleaned using Performa DTR Gel filtration Cartridges (Edge Biosystems). The eluent was dried, and sequencing was performed on an ABI 3130 automated sequencer.
3. Results
sEOAD samples (n = 408) were genotyped on the NeuroX DNA microarray. These samples were screened for variants known to cause disease (known causative variants) of 13 genes linked to familial forms of neurodegeneration (non-AD familial genes) and variants predicted to be pathogenic (predicted pathogenic variants) in 16 genes linked to familial forms of neurodegeneration (familial genes).
3.1. Validated genotypes
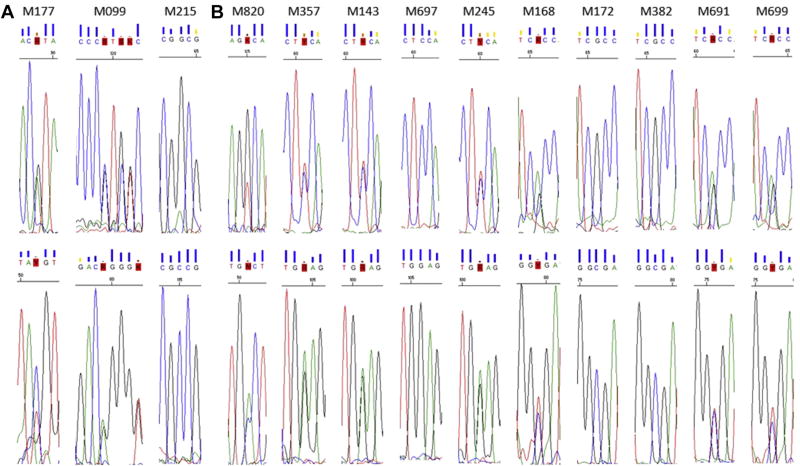
The NeuroX identified 3 sEOAD samples harboring a known causative variant in 2 of the 13 non-AD familial genes and 10 sEOAD samples harboring a predicted pathogenic variant in 1 of the 16 familial genes. All samples were clinically diagnosed as having probable AD. Sanger sequencing confirmed that 9 samples harbored a variant. Fig. 1 shows the sequence chromatograms for the 13 samples and Table 2 lists their genotypes.
Fig. 1.
Sequence chromatograms. Sequence chromatograms of all 13 individuals thought to harbor a pathogenic (A) or predicted pathogenic variant (B). Each individual was sequenced in forward (top row) and reverse (bottom row) orientations. Note that the forward orientation does not always correspond to the sense strand. The images were taken from Sequence Scanner (Applied Biosystems) with the variant at the centre and surrounded by 2 or 3 bases either side. Four individuals (M215, M697, M172, and M382) show wild-type sequence according to the chromatogram.
Sanger sequencing confirmed that 8 samples harbored the minor allele indicated by the NeuroX and 1 sample (M820) harbored an alternative minor allele. M820 appeared heterozygous for G>A base change (rs144397565, p.G170D), however sequencing confirmed it was heterozygous for G>T base change (rs144397565, p.G170V). Evidently this position is trimorphic, and as this marker uses the Infinium II probe design it was able to detect both minor alleles but was unable to differentiate between them.
3.2. Invalidated genotypes
Of the variants not confirmed by Sanger sequencing, 1 sample (M099) was identified as heterozygous for p.Q34fs (delA) in addition to the confirmed variant p.Q34fs (delAG) at the same position. Notably the probe sequence for both markers was identical. As both deletions are followed by the same nucleotide (G), the single nucleotide extension design meant this probe was capable of detecting both variants, in this instance both probes detected the delAG variant.
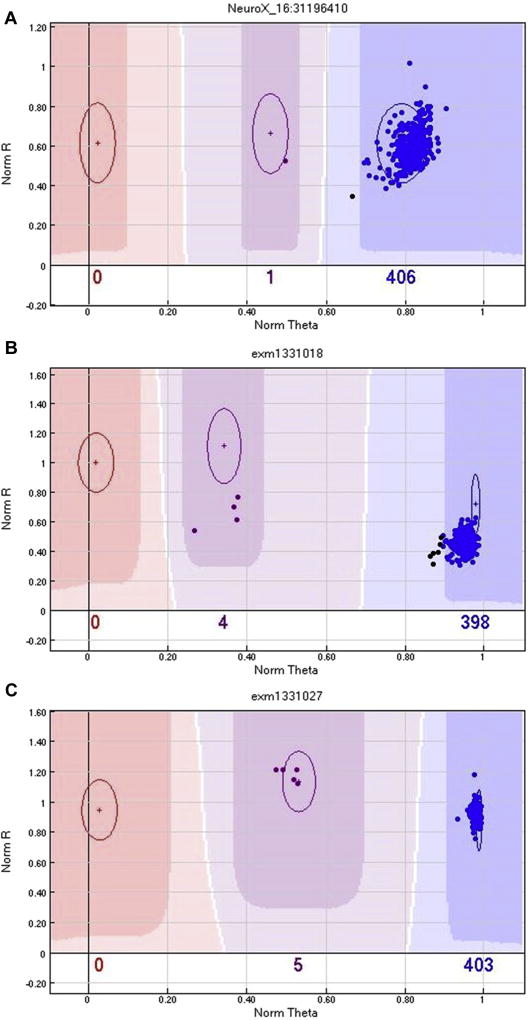
Four samples (M215, M697, M172, and M382) failed to verify with initial Sanger sequencing; the cluster plots were examined to establish call quality, which were found to be good (Fig. 2). Three of the four samples (M215, M697 and M382) were resequenced from the original DNA stock; all 3 gave the same result as originally obtained (unconfirmed), thereby obviating a sample “mix-up”. We were unable to resequence M172 as no additional DNA was available. It is interesting to note that variant p.G225V is located within a GGC repeat region, and this could be the reason for the discrepancy between the NeuroX genotype and sequencing result. Alternatively, the NeuroX genotypes could be correct for these samples, and the PCR reaction may have resulted in allele dropout (Blais et al., 2015); however, it would be advisable to use a different primer pair as this might permit detection of the other allele.
Fig. 2.
SNP cluster plot for genotypes unconfirmed with Sanger sequencing. SNP cluster graphs generated by Genome Studio for markers NeuroX_16:31196410 (p.G225V located in FUS) (A), exm1331018 (p.S427F located in MAPT) (B), and exm1331027 (p.A469T located in MAPT) (C). Each colored circle represents 1 individual, those colored red are homozygous mutant (TT), those colored purple are heterozygous (GT), those colored blue are homozygous wildtype (GG), and finally those colored black are not called. The plots show all 10 samples called as heterozygotes cluster well and do not explain why 4 samples failed to verify with Sanger sequencing (M215, M697, M172, and M382).
In addition to the above, 25 samples were identified as harboring the variant p.Q130fs (rs63750768, gaT.AGT/ga) in the GRN gene. Similar to the situation with M820 (as described in Section 3.1), Ghani et al. confirmed that this marker also genotypes a common alternative minor allele at the same position (rs25646, gaT/gaC) (Ghani et al., 2015). Consequently, this variant was discarded from further investigation.
4. Discussion
4.1. Known causative variants
Two sEOAD samples were confirmed heterozygous for a known causative variants; p.T240M (n = 1) and p.Q34fs (delAG, n = 1). Both of these variants are located in the gene PARK2. Known causative variants in PARK2 include point mutations and exon rearrangements/deletions/duplications, often as homozygote or compound heterozygotes in early-onset PD (EOPD).
Variant p.T240M has been seen in EOPD as a compound heterozygote with various exon deletions or duplications (Amboni et al., 2009; Deng et al., 2006; Periquet et al., 2003; Sironi et al., 2008), it has also been seen as a homozygote (Madegowda et al., 2005) and a heterozygote (Camargos et al., 2009). A compound heterozygote with an exon deletion has also been seen in 1 healthy individual (Deng et al., 2006). Variant p.Q34fs (delAG) has been seen in EOPD as a compound heterozygote with exon deletions or SNPs (Abbas et al., 1999; Guo et al., 2008, 2010; Hedrich et al., 2002; Illarioshkin et al., 2003; Koziorowski et al., 2010; Lesage et al., 2008; Lohmann et al., 2009; Scherfler et al., 2004), it has also been seen as a homozygote (Koziorowski et al., 2010; Scherfler et al., 2004) and a heterozygote (Brooks et al., 2009; Bruggemann et al., 2009). A compound heterozygote with an exon deletion has also been seen in LOPD (Lesage et al., 2008).
Previous findings would suggest that p.T240M and p.Q34fs (delAG) elicit risk for PD, in particular EOPD. Finding these variants in our sEOAD cohort would suggest that they could also elicit risk to sEOAD; however, we only found each of them in 1 sample (0.25% of our sEOAD cohort) as a heterozygote and we do not know if these individuals were compound heterozygotes. These variants could elicit risk to sEOAD; however, a large case-control association study would be needed to establish this.
Both of these patients (M117 and M099) had an APOE ε4ε4 status, and there was nothing unusual about their presentation or progress, which suggests that a misdiagnosis is unlikely. However, M099 had a mother who was said to have had motor neuron disease, so there was likely to have been physical signs in her, possibly consistent with a known causative variant in PARK2.
4.2. Predicted pathogenic variants
Seven sEOAD samples were confirmed heterozygous for a predicted pathogenic variant; p.G107V (n = 1), p.S427F (n = 3), or p.A469T (n = 2). All these variants are located in the gene MAPT and are named in reference to the longest tau transcript (tau-g). Most known causative variants in MAPT are SNPs with autosomal dominant inheritance and result in FTD.
Variant p.G107V was predicted to be probably damaging (1.00) by Polyphen and deleterious (0) by SIFT, it is located in exon 4 of MAPT, where no known causative variants have been documented. Variant p.S427F was predicted to be deleterious (0.02) by SIFT and probably damaging (0.99) by Polyphen, it is located in exon 4a, which is spliced out in the transcript htau40 and thus not present in the human brain (Liu and Gong, 2008; Pittman et al., 2006). It is unlikely this variant has a functional effect in the brain if the transcripts containing this variant are not present.
Variant p.A469T is also called p.A152T in htau40. It was predicted to be deleterious (0.05) by SIFT and benign (0.30) by Polyphen. This variant significantly increases the risk for both FTD (p-value = 0.0005, odds ratio = 3.0 [confidence interval: 1.6–5.6]) and AD (p-value = 0.004, odds ratio = 2.3 [confidence interval: 1.3–4.2]) when compared to controls (Coppola et al., 2012). In vitro site-directed mutagenesis of human tau complementary DNA showed the variant resulted in less efficient binding to microtubules and a pronounced increase in the formation of tau oligomers (Coppola et al., 2012). Furthermore, isogenic human iPSCs generated from fibroblasts saw the variant result in axonal degeneration and cell death (Fong et al., 2013). Whether heterozygous p.A469T in humans would cause the same effect is unknown. Notably, this variant is located in exon 7, the downstream residue (p.T153) is part of a Threonine-Proline motif that is phosphorylated during the cell cycle (Illenberger et al., 1998), and the upstream residue (p.I151) is seen to interact with microtubules using nuclear magnetic resonance (Mukrasch et al., 2009). Variant p.A469T could affect the functioning of the upstream or downstream residue, which might explain the experimental observations for this variant. This variant could elicit risk to sEOAD; however, a large case-control association study would be needed to establish this.
4.3. Considerations
Although DNA microarrays are cost efficient, they have several drawbacks as evident from this study. The results of Sanger sequencing found that a high proportion of variants failed to verify, this emphasizes the need to verify all genotypes called by DNA microarray technologies. The version of the NeuroX used in this study was only able to successfully genotype 182 of the 523 (35%) known causative variants documented in 3 online databases (Supplementary Table 3), since that time, the number of variants documented in the online databases has increased by 147, albeit not all of these will be important (causative), but highlights the additional issue of having to redesign chips as new personal variants come to light, which questions the cost effectiveness of DNA microarray technology for extremely rare variants. Nextgeneration sequencing (NGS) technology overcomes the limitations and drawbacks of DNA microarray technology, and the once debilitating cost of NGS has almost dissipated as the price has plummeted in recent years. There is no doubt that studies like this should be conducted; however, given the drawbacks of DNA microarray technology they would be better conducted using NGS technologies.
This study has made use of an in-silico approach to classify the pathogenicity of variants; however, in-silico predictions alone are insufficient to properly appraise a variant. Richards et al. have developed an approach that can help define the definition of “pathogenic” in the clinical and research setting with regard to Mendelian disorders (Richards et al., 2015), this approach makes use of additional types of data including population, functional, and segregation data. As our understanding of complex diseases increases, no doubt an approach incorporating several lines of data will be used to define pathogenic variants for non-Mendelian disorders such as sEOAD and LOAD.
5. Conclusions
We have screened sEOAD individuals for known causative and predicted pathogenic variants in 16 genes linked to neurodegenerative diseases. We have identified 9 sEOAD samples harboring a known causative variant or a predicted pathogenic variant in PARK2 and MAPT. These variants could elicit risk to sEOAD in addition to PD and FTD; however, further studies would be needed to establish this. This work highlights the need to screen sEOAD individuals for variants that are more classically attributed to other forms of neurodegeneration as there could be a degree of genetic overlap.
Supplementary Material
Acknowledgments
The University of Nottingham lab is funded by Alzheimer’s Research UK and the Big Lottery Fund (ART-BIG2009-1). Imelda S. Barber, PhD studentship is jointly funded by Alzheimer’s Research UK (ARUK-NCG2012B-1) and the School of Life Sciences at The University of Nottingham. Jose Bras and Rita Guerreiro’s Fellowships are funded by The Alzheimer’s Society.
Appendix
Details of ARUK consortium: Peter Passmore, David Craig, Janet Johnston, Bernadette McGuinness, and Stephen Todd (Centre for Public Health, School of Medicine, Queen’s University, Belfast, BT9 7BL, UK); Reinhard Heun (Royal Derby Hospital, Derby, DE22 3WQ, UK); Heike Kölsch (Department of Psychiatry, University of Bonn, Sigmund-Freud-Strasse 25, Bonn 53105, Germany); Patrick G. Kehoe (School of Clinical Sciences, John James Laboratories, University of Bristol, Bristol, BS16 1LE, UK); Emma R.L.C. Vardy (Ageing and Complex Medicine, Salford Royal NHS Foundation Trust, Stott Lane, Salford, M6 8HD, UK); Nigel M. Hooper, Stuart Pickering-Brown, Julie Snowden, Anna Richardson, Matthew Jones, David Neary, and Jennifer Harris (Institute of Brain, Behaviour and Mental Health, Faculty of Medical and Human Sciences, Oxford Road, University of Manchester, Manchester, M13 9PT, UK); Julie Snowden, Anna Richardson, Matthew Jones, David Neary, and Jennifer Harris (Cerebral Function Unit, Greater Manchester Neurosciences Centre, Salford Royal NHS Foundation Trust, Stott Lane, Salford, M6 8HD, UK); James Lowe (School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, NG7 2UH, UK); A. David Smith, Gordon Wilcock, and Donald Warden (OPTIMA, Nuffield Department of Clinical Neurosciences, Level 6, West Wing, John Radcliffe Hospital, University of Oxford, Oxford, OX3 9DU, UK); and Clive Holmes (Clinical and Experimental Science, University of Southampton, Southampton, SO17 1BJ, UK).
Footnotes
Disclosure statement
The authors have no conflicts of interest to disclose.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.neurobiolaging.2016.09.008.
References
- Abbas N, Lucking CB, Ricard S, Durr A, Bonifati V, De Michele G, Bouley S, Vaughan JR, Gasser T, Marconi R, Broussolle E, Brefel-Courbon C, Harhangi BS, Oostra AB, Fabrizio E, Bohme GA, Pradier L, Wood NW, Filla A, Meco G, Denefle P, Agid Y, Brice A French Parkinson’s Disease Genetics Study Group European Consortium Genetic Susceptibility Parkinson’s Disease. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. Hum. Mol. Genet. 1999;8:567–574. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- Amboni M, Pellecchia MT, Cozzolino A, Pieillo M, Vitale C, Barone P, Varrone A, Garavaglia B, Gambelli S, Federico A. Cerebellar and pyramidal dysfunctions, palpebral ptosis and weakness as presenting symptoms of PARK-2. Mov. Disord. 2009;24:303–305. doi: 10.1002/mds.22342. [DOI] [PubMed] [Google Scholar]
- Barber IS, Garcia-Cardenas JM, Sakdapanichkul C, Deacon C, Erazo GZ, Guerreiro R, Bras J, Hernandez D, Singleton A, Guetta-Baranes T, Braae A, Clement N, Patel T, Brookes K, Medway C, Chappell S, Mann DM, Morgan K ARUK Consortium. Screening exons 16 and 17 of the amyloid precursor protein gene in sporadic early-onset Alzheimer’s disease. Neurobiol. Aging. 2016;39:E1–E7. doi: 10.1016/j.neurobiolaging.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais J, Lavoie SB, Giroux S, Bussieres J, Lindsay C, Dionne J, Laroche M, Giguere Y, Rousseau F. Risk of misdiagnosis due to allele dropout and false-positive PCR artifacts in molecular diagnostics: analysis of 30,769 genotypes. J. Mol. Diagn. 2015;17:505–514. doi: 10.1016/j.jmoldx.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Bras J, Guerreiro R, Darwent L, Parkkinen L, Ansorge O, Escott-Price V, Hernandez DG, Nalls MA, Clark LN, Honig LS, Marder K, Van Der Flier WM, Lemstra A, Scheltens P, Rogaeva E, St George-Hyslop P, Londos E, Zetterberg H, Ortega-Cubero S, Pastor P, Ferman TJ, Graff-Radford NR, Ross OA, Barber I, Braae A, Brown K, Morgan K, Maetzler W, Berg D, Troakes C, Al-Sarraj S, Lashley T, Compta Y, Revesz T, Lees A, Cairns N, Halliday GM, Mann D, Pickering-Brown S, Dickson DW, Singleton A, Hardy J. Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum. Mol. Genet. 2014;23:6139–6146. doi: 10.1093/hmg/ddu334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J, Ding J, Simon-Sanchez J, Paisan-Ruiz C, Singleton AB, Scholz SW. Parkin and PINK1 mutations in early-onset Parkinson’s disease: comprehensive screening in publicly available cases and control. J. Med. Genet. 2009;46:375–381. doi: 10.1136/jmg.2008.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann N, Mitterer M, Lanthaler AJ, Djarmati A, Hagenah J, Wiegers K, Winkler S, Pawlack H, Lohnau T, Pramstaller PP, Klein C, Lohmann K. Frequency of heterozygous Parkin mutations in healthy subjects: need for careful prospective follow-up examination of mutation carriers. Parkinsonism Relat. Disord. 2009;15:425–429. doi: 10.1016/j.parkreldis.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Camargos ST, Dornas LO, Momeni P, Lees A, Hardy J, Singleton A, Cardoso F. Familial Parkinsonism and early onset Parkinson’s disease in a Brazilian movement disorders clinic: phenotypic characterization and frequency of SNCA, PRKN, PINK1, and LRRK2 mutations. Mov. Disord. 2009;24:662–666. doi: 10.1002/mds.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Khan QUA, Murray ME, Krishnan S, Aakre J, Pankratz VS, Thuy N, Ma L, Bisceglio G, Petersen RC, Younkin SG, Dickson DW, Boeve BF, Graff-Radford NR, Ertekin-Taner N. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurology. 2014;82:1455–1462. doi: 10.1212/WNL.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G, Chinnathambi S, Lee JJ, Dombroski BA, Baker MC, Soto-Ortolaza AI, Lee SE, Klein E, Huang AY, Sears R, Lane JR, Karydas AM, Kenet RO, Biernat J, Wang LS, Cotman CW, Decarli CS, Levey AI, Ringman JM, Mendez MF, Chui HC, Le Ber I, Brice A, Lupton MK, Preza E, Lovestone S, Powell J, Graff-Radford N, Petersen RC, Boeve BF, Lippa CF, Bigio EH, Mackenzie I, Finger E, Kertesz A, Caselli RJ, Gearing M, Juncos JL, Ghetti B, Spina S, Bordelon YM, Tourtellotte WW, Frosch MP, Vonsattel JP, Zarow C, Beach TG, Albin RL, Lieberman AP, Lee VM, Trojanowski JQ, Van Deerlin VM, Bird TD, Galasko DR, Masliah E, White CL, Troncoso JC, Hannequin D, Boxer AL, Geschwind MD, Kumar S, Mandelkow EM, Wszolek ZK, Uitti RJ, Dickson DW, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Alzheimer’s Disease Genetics Consortium. Ross OA, Rademakers R, Schellenberg GD, Miller BL, Mandelkow E, Geschwind DH. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum. Mol. Genet. 2012;21:3500–3512. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericakvance MA. Gene dose of apolipoprotein E type-4 allele and the risk of Alzheimer’s disease in late-onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Le WD, Hunter CB, Ondo WG, Guo Y, Xie WJ, Jankovic J. Heterogeneous phenotype in a family with compound heterozygous parkin gene mutations. Arch. Neurol. 2006;63:273–277. doi: 10.1001/archneur.63.2.273. [DOI] [PubMed] [Google Scholar]
- Fong H, Wang CZ, Knoferle J, Walker D, Balestra ME, Tong LM, Leung L, Ring KL, Seeley WW, Karydas A, Kshirsagar MA, Boxer AL, Kosik KS, Miller BL, Huang YD. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Reports. 2013;1:226–234. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani M, Lang AE, Zinman L, Nacmias B, Sorbi S, Bessi V, Tedde A, Tartaglia MC, Surace EI, Sato C, Moreno D, Xi Z, Hung R, Nalls MA, Singleton A, St George-Hyslop P, Rogaeva E. Mutation analysis of patients with neurodegenerative disorders using NeuroX array. Neurobiol. Aging. 2015;36:545.e9–545.e14. doi: 10.1016/j.neurobiolaging.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, Gudnason V, Harris TB, Kathiresan S, Kraaij R, Launer LJ, Levy D, Liu Y, Mosley T, Peloso GM, Psaty BM, Rich SS, Rivadeneira F, Siscovick DS, Smith AV, Uitterlinden A, van Duijn CM, Wilson JG, O’Donnell CJ, Rotter JI, Boerwinkle E. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JF, Xiao B, Liao B, Zhang XW, Nie LL, Zhang YH, Shen L, Jiang H, Xia K, Pan Q, Yan XX, Tang BS. Mutation analysis of Parkin, PINK1, DJ-1 and ATP13A2 genes in Chinese patients with autosomal recessive early-onset Parkinsonism. Mov. Disord. 2008;23:2074–2079. doi: 10.1002/mds.22156. [DOI] [PubMed] [Google Scholar]
- Guo JF, Zhang XW, Nie LL, Zhang HN, Liao B, Li J, Wang L, Yan XX, Tang BS. Mutation analysis of Parkin, PINK1 and DJ-1 genes in Chinese patients with sporadic early onset parkinsonism. J. Neurol. 2010;257:1170–1175. doi: 10.1007/s00415-010-5485-8. [DOI] [PubMed] [Google Scholar]
- Hedrich K, Marder K, Harris J, Kann M, Lynch T, Meija-Santana H, Pramstaller PP, Schwinger E, Bressman SB, Fahn S, Klein C. Evaluation of 50 probands with early-onset Parkinson’s disease for Parkin mutations. Neurology. 2002;58:1239–1246. doi: 10.1212/wnl.58.8.1239. [DOI] [PubMed] [Google Scholar]
- Illarioshkin SN, Periquet M, Rawal N, Lucking CB, Zagorovskaya TB, Slominsky PA, Miloserdova OV, Markova ED, Limborska SA, Ivanova-Smolenskaya IA, Brice A. Mutation analysis of the parkin gene in Russian families with autosomal recessive juvenile parkinsonism. Mov. Disord. 2003;18:914–919. doi: 10.1002/mds.10467. [DOI] [PubMed] [Google Scholar]
- Illenberger S, Zheng-Fischhofer QY, Preuss U, Stamer K, Baumann K, Trinczek B, Biernat J, Godemann R, Mandelkow EM, Mandelkow E. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: implications for Alzheimer’s disease. Mol. Biol. Cell. 1998;9:1495–1512. doi: 10.1091/mbc.9.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA. Neuropathology of dementia disorders. J. Alzheimers Dis. Parkinsonism. 2014;4:135. [Google Scholar]
- Koziorowski D, Hoffman-Zacharska D, Slawek J, Szirkowiec W, Janik P, Bal J, Friedman A. Low frequency of the PARK2 gene mutations in Polish patients with the early-onset form of Parkinson disease. Parkinsonism Relat. Disord. 2010;16:136–138. doi: 10.1016/j.parkreldis.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Lesage S, Lohmann E, Tison F, Durif F, Durr A, Brice A French Parkinson’s Disease Genetics Study Group. Rare heterozygous parkin variants in French early-onset Parkinson disease patients and controls. J. Med. Genet. 2008;45:43–46. doi: 10.1136/jmg.2007.051854. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann E, Thobois S, Lesage S, Broussolle E, du Montcel ST, Ribeiro MJ, Remy P, Pelissolo A, Dubois B, Mallet L, Pollak P, Agid Y, Brice A French Parkinson’s Disease Genetics Study Group. A multidisciplinary study of patients with early-onset PD with and without parkin mutations. Neurology. 2009;72:110–116. doi: 10.1212/01.wnl.0000327098.86861.d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madegowda RH, Kishore A, Anand A. Mutational screening of the parkin gene among South Indians with early onset Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry. 2005;76:1588–1590. doi: 10.1136/jnnp.2004.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PloS Biol. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Bras J, Hernandez DG, Keller MF, Majounie E, Renton AE, Saad M, Jansen I, Guerreiro R, Lubbe S, Plagnol V, Gibbs JR, Schulte C, Pankratz N, Sutherland M, Bertram L, Lill CM, DeStefano AL, Faroud T, Eriksson N, Tung JY, Edsall C, Nichols N, Brooks J, Arepalli S, Pliner H, Letson C, Heutink P, Martinez M, Gasser T, Traynor BJ, Wood N, Hardy J, Singleton AB International Parkinson’s Disease Genomics Consortium Parkinson’s Disease Meta-Analysis Consortium. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol. Aging. 2015;36:1605.e7–1605.e12. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M, Latouche M, Lohmann E, Rawal N, De Michele G, Ricard S, Teive H, Fraix V, Vidailhet M, Nicholl D, Barone P, Wood NW, Raskin S, Deleuze JF, Agid Y, Durr A, Brice A French Parkinson’s Disease Genetics Study Group European Consortium Genetic Susceptibility in Parkinson’s Disease. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain. 2003;126:1271–1278. doi: 10.1093/brain/awg136. [DOI] [PubMed] [Google Scholar]
- Pittman AM, Fung H-C, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Hum. Mol. Genet. 2006;15:R188–R195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, Goate A, Frommelt P, Ghetti B, Langbaum JBS, Lopera F, Martins R, Masters CL, Mayeux RP, McDade E, Moreno S, Reiman EM, Ringman JM, Salloway S, Schofield PR, Sperling R, Tariot PN, Xiong CJ, Morris JC, Bateman RJ Dominantly Inherited Alzheimer Network. Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology. 2014;83:253–260. doi: 10.1212/WNL.0000000000000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherfler C, Khan NL, Pavese N, Eunson L, Graham E, Lees AJ, Quinn NP, Wood NW, Brooks DJ, Piccini PP. Striatal and cortical pre- and post-synaptic dopaminergic dysfunction in sporadic parkin-linked parkinsonism. Brain. 2004;127:1332–1342. doi: 10.1093/brain/awh150. [DOI] [PubMed] [Google Scholar]
- Sironi F, Primignani P, Zini M, Tunesi S, Ruffmann C, Ricca S, Brambilla T, Antonini A, Tesei S, Canesi M, Zecchinelli A, Mariani C, Meucci N, Sacilotto G, Cilia R, Isaias IU, Garavaglia B, Ghezzi D, Travil M, Decarli A, Coviello DA, Pezzoli G, Goldwurm S. Parkin analysis in early onset Parkinson’s disease. Parkinsonism Relat. Disord. 2008;14:326–333. doi: 10.1016/j.parkreldis.2007.10.003. [DOI] [PubMed] [Google Scholar]
- WHO; Top Ten Leading Causes of Death by Country Income Group. Top Ten Causes of Death. World Health Organisation; Geneva: 2012. [Google Scholar]
- Wingo TS, Lah JJ, Levey AI, Cutler DJ. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch. Neurol. 2012;69:59–64. doi: 10.1001/archneurol.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.