Abstract
Bats have attracted attention in recent years as important reservoirs of viruses deadly to humans and other mammals. These infections are typically nonpathogenic in bats raising questions about innate immune differences that might exist between bats and other mammals. The APOBEC3 gene family encodes antiviral DNA cytosine deaminases with important roles in the suppression of diverse viruses and genomic parasites. Here, we characterize pteropid APOBEC3 genes and show that species within the genus Pteropus possess the largest and most diverse array of APOBEC3 genes identified in any mammal reported to date. Several bat APOBEC3 proteins are antiviral as demonstrated by restriction of retroviral infectivity using HIV-1 as a model, and recombinant A3Z1 subtypes possess strong DNA deaminase activity. These genes represent the first group of antiviral restriction factors identified in bats with extensive diversification relative to homologues in other mammals.
Keywords: restriction factors, bats, APOBEC3, evolution, antiviral immunity
Introduction
Bats are reservoirs of emerging viruses that are highly pathogenic to other mammals including humans. In recent years, viruses such as Ebola, Marburg, Hendra, Nipah, Sosuga, and severe acute respiratory syndrome coronavirus (SARS-CoV) have crossed species barriers from their natural hosts into humans (Calisher et al. 2006; Smith and Wang 2013; Amman et al. 2015; Hayman 2016). Intriguingly, experimental infections have demonstrated that specific bat species can be infected with Ebola, Marburg, SARS-CoV, Hendra, and Nipah viruses without the development of clinical symptoms (Swanepoel et al. 1996; Williamson et al. 1998; Middleton et al. 2007; Watanabe et al. 2010; Jones et al. 2015). This has led to efforts to uncover differences that might exist between the antiviral strategies of bats and other mammals (Baker et al. 2013; Zhang et al. 2013; Zhou et al. 2016).
Based upon studies of endogenous retroviruses, bats are an important source of mammalian retroviruses and many cross-species transmissions of retroviruses from bats to other mammals have been documented (Hayward et al. 2013; Cui et al. 2015). A number of antiviral restriction factors are known to be capable of inhibiting the normal replication of retroviruses; among the most well studied of these are the members of the APOBEC3 (A3) protein family. A3 proteins are capable of restricting the replication of several virus families including retroviruses, hepadnaviruses, and parvoviruses, in addition to retrotransposons which are genomic parasites (Chen et al. 2006; Muckenfuss et al. 2006; Renard et al. 2010). A3 proteins are DNA cytosine deaminases capable of hypermutating viral genomes and sterically hindering nascent DNA synthesis (Sheehy et al. 2002; Lecossier et al. 2003; Refsland and Harris 2013). All A3 proteins contain one or two zinc-coordinating cytosine deaminase domains (Z-domains) which are required for deaminase activity (Chiu and Greene 2008). A3 proteins are divided into three phylogenetic subgroups, Z1, Z2, and Z3, which are distinguished by the conservation of amino acid residues within the Z-domain (LaRue et al. 2009; Refsland and Harris 2013). We hypothesized that, given the prolonged history of coexistence between bats and their retroviruses, differences exist relative to other mammals in the antiviral genes responsible for retroviral restriction.
Pteropid bats are species within the family Pteropodidae, and are implicated as reservoirs of emerging human viral pathogens (Hayman 2016). Here, we report assemblies of the A3 loci in the closely related pteropids, Pteropus vampyrus and Pteropus alecto, which harbor Nipah and Hendra viruses, respectively (Halpin et al. 2000; Rahman et al. 2010). Several bat genomes (Fang et al. 2015) have been published in recent years; however, no available assembly has resolved the A3 gene locus. This failure is likely due to the presence of many repetitive sequences, generated by extensive gene duplication and recombination, which are known to hinder genome assembly (Zhang et al. 2013; Fang et al. 2015).
In this study, pteropid A3 loci were generated through the step-wise extension of cDNA-mapped gene scaffolds followed by remapping of sequence read archives, supported experimentally by an A3 expression analysis of bat spleen tissue. The functionality of P. alecto A3 subtypes were investigated through analysis of their ability to deaminate DNA, and restrict the infectivity of a model retrovirus. The role of A3-mediated restriction of ancient bat retroviruses was assessed using a hypermutation analysis of endogenous retroviruses within the genome of P. vampyrus.
Results
Pteropid Bats Express a Diverse Range of APOBEC3 Gene Products
Pteropid A3 gene expression was initially determined by searching for A3 homologues in the P. alecto transcriptome database through a tBLASTn analysis using the human A3 proteins A3A, A3C, and A3H, representing the three zinc-coordinating cytosine deaminase motif (Z-domain) subtypes A3Z1, A3Z2, and A3Z3, as search queries. A3 expression was subsequently assessed through analysis of cDNA (complementary DNA) generated from P. alecto spleen tissue, which harbors multiple immune cell types known to express A3 genes in humans (Refsland et al. 2010). This analysis revealed 20 distinct A3 mRNA transcripts derived from seven A3Z1 genes, five A3Z2 genes, and a single A3Z3 gene (supplementary table S1, Supplementary Material online). Pairwise comparison of these A3 transcripts (supplementary fig. S1, Supplementary Material online) reveal a wide range of sequence identities between pteropid A3 genes, indicating considerable diversity among homologs: A3Z1 genes range from 86% to 97% identity, whereas A3Z2 genes range from 54% to 99% identity.
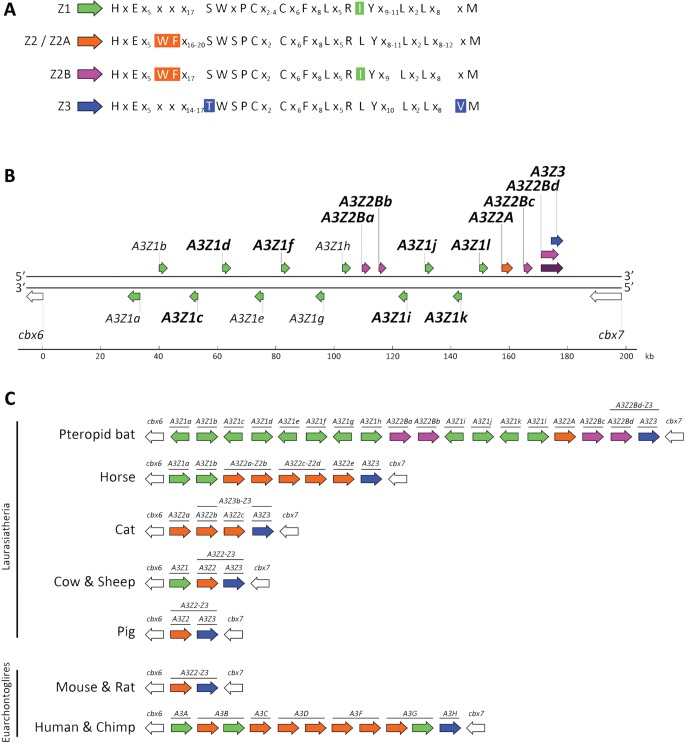
In addition to the expected A3Z1, A3Z2, and A3Z3 subtypes, several A3 amino acid sequences contained Z-domains distinct from the canonical domains as defined by a specific combination of key conserved residues (LaRue et al. 2009). This group, designated A3Z2B, carried both the A3Z2 domain-specific tryptophan-phenylalanine (WF) residues and the A3Z1 domain-specific arginine-adjacent isoleucine (RI) motif (fig. 1A). Of the 20 unique transcripts, 19 harbored a single Z-domain and one contained two Z-domains, an N-terminal A3Z2B domain paired with a C-terminal A3Z3 domain. These results indicate that pteropid bats express a more diverse repertoire of A3 mRNA than reported to date for other mammals (fig. 1C).
Fig. 1.
The pteropid bat A3 gene locus reveals the largest and most diverse known repertoire of A3 genes in mammals. (A) The amino acid sequences of the four Z-domain subtypes are depicted from the first to the last invariant amino acid residue. Invariant amino acid residues are represented by their letter, “x” indicates positions of variable amino acid identity, and numbers indicate the length of the variable amino acid sequence. For each subtype, the defining residue(s) are colored. (B) The A3 gene locus of Pteropus alecto is shown to scale. A3Z1, A3Z2A, A3Z2B, and A3Z3 gene coding regions are shown in green, orange, pink, and blue, respectively. The region encoding the A3Z2Bd-Z3 double-domain gene product is shown in maroon. A3 genes verified as being transcriptionally active are denoted in bold type. The CBX6 and CBX7 genes immediately upstream and downstream of the locus are shown in white. (C) Graphical schematic diagrams of sequenced mammalian A3 loci are depicted bound by the CBX6 and CBX7 genes, shown in white, positioned immediately upstream and downstream of the A3 genes.
Generation of the Pteropid APOBEC3 Gene Locus
The generation of the pteropid A3 loci was accomplished in a step-wise manner outlined in supplementary fig. S2, Supplementary Material online. The A3 gene locus in Eutherian mammals is located between the CBX6 and CBX7 genes (LaRue et al. 2008). BLASTn searches using the P. alecto A3 cDNA products as queries revealed that publicly accessible bat genomes of P. alecto and P. vampyrus did not contain assembled A3 gene loci located within the boundaries of CBX6 and CBX7. However, the bat A3 cDNA could be mapped, albeit across numerous discontiguous scaffolds, to the P. vampyrus genome accessible through the Ensembl database. The A3 locus contains many large, repetitive sequence elements that result in the failure of automated assembly. This problem was overcome by matching exons derived from sequenced cDNA, allowing the informed pairing of discontiguous scaffolds. This was not possible using the P. alecto genome as it did not contain sufficient assembled scaffolds to which A3 cDNA products could be mapped. It was reasonable to map P. alecto cDNA products against the P. vampyrus genome as these species are very closely related, with a recent divergence of ∼4.4 Ma and 97–99% genomic nucleotide sequence identity (Almeida et al. 2014). The P. vampyrus A3 locus was then generated through a step-wise extension of preexisting P. vampyrus gene scaffolds to join the gaps between exon-paired scaffolds, followed by remapping of the sequence read archives (SRA) to validate and generate the 190 kB P. vampyrus A3 locus. The P. alecto A3 locus was generated by mapping P. alecto reads against the P. vampyrus A3 locus and was found to be slightly shorter at approximately 188 kB as a result of multiple short 3–20 nt insertions and deletions. These indels occurred in the A3 loci since the divergence of P. alecto and P. vampyrus, likely as a result of nonhomologous recombination events, the frequency of which is known to be increased in regions of the genome that contain sections of repetitive DNA (Metzenberg et al. 1991).
Within the P. alecto A3 locus 18 gene coding regions were identified including 12 A3Z1, one prototypical A3Z2 (herein described as A3Z2A), four A3Z2B, and one A3Z3 (fig. 1B). The organization and number of the A3 genes was found to be identical between P. vampyrus and P. alecto. Neural network promoter prediction revealed promoter elements immediately upstream of each gene. Interferon-stimulated response element (ISRE) transcription factor binding sites for interferon regulatory factors and NF-κB were also identified in each promoter region (supplementary data S1, Supplementary Material online). These data indicate that each pteropid A3 gene is most likely capable of being transcribed and that their expression is potentially upregulated as part of the antiviral interferon response.
All P. alecto A3 cDNA sequences were successfully mapped against the assembled P. alecto loci, revealing various alternative splicing schemes that allow many of the A3 genes to express numerous transcripts (supplementary fig. S3 and table S1, Supplementary Material online). A comparison of the A3 loci of P. vampyrus and P. alecto against the A3 loci of other mammals (fig. 1C) reveals that pteropid bats possess the largest A3 locus yet reported, containing 18 putative A3 coding domains, 13 of which have been confirmed in this study as undergoing active transcription (fig. 1B).
Bats Possess a Novel Subtype of APOBEC3
Analysis of the phylogenetic relationships between the A3 proteins of P. alecto and other mammals (fig. 2) confirms the expected phylogenetic positions of the A3Z1, A3Z2A, and A3Z3 proteins, and reveals that the A3Z2B proteins, containing both Z1- and Z2-defining sequence motifs, phylogenetically cluster within the broader A3Z2 clade. Notably, pteropid A3Z2A and A3Z2B are positioned a greater sequence distance from each other than from the A3Z2 proteins of distantly related mammals, revealing that pteropid bats possess four phylogenetically distinct A3 Z-domain subtypes, two of which appear to be derivatives of the Z2 clade.
Fig. 2.
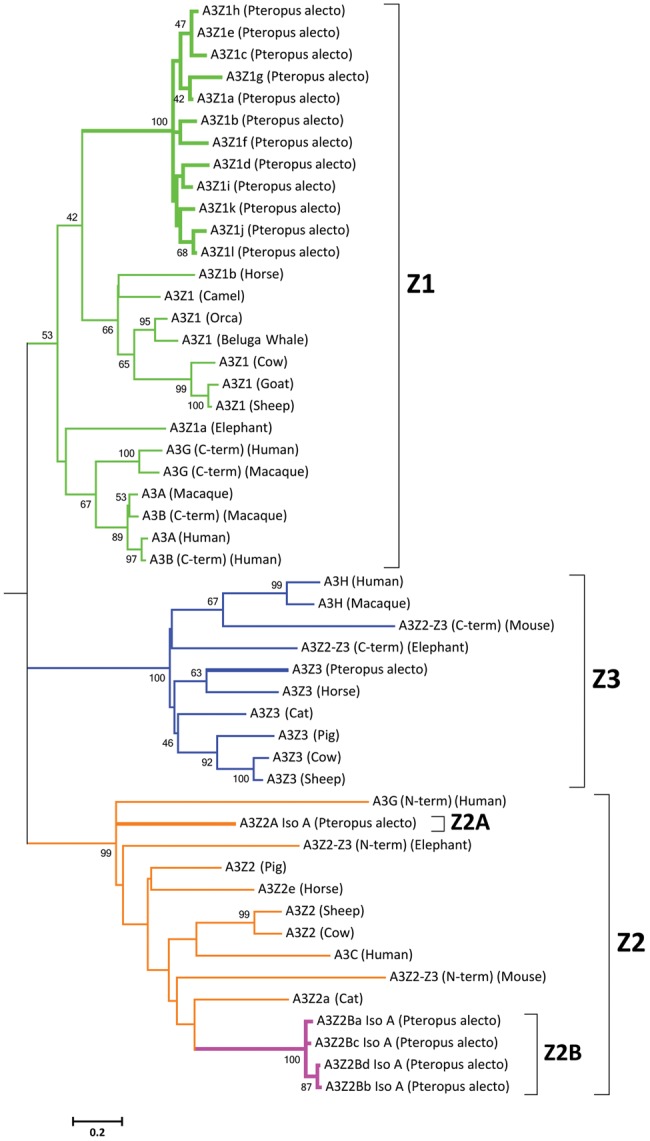
Mammalian APOBEC3 phylogeny. The phylogenetic relationships of mammalian A3 proteins were inferred using the maximum likelihood method and JTT + G model with 1,000 bootstrap replicates. A3 subgroups Z1, Z2, and Z3 are paralogs, which have expanded independently. Bootstrap values for nodes represented in >40% of trees are shown. A3Z1, A3Z2/Z2A, A3Z2B, and A3Z3 clades are shown in green, orange, pink, and blue, respectively. The scale represents the number of amino acid substitutions per site.
Bat APOBEC3 Proteins Are Potent DNA Mutators
We tested the activity of the bat A3 proteins for which expression was verified (supplementary table S1, Supplementary Material online) by assessing their ability to cause mutations in E. coli though a rifampicin mutagenesis assay, utilizing human A3B and A3G as positive controls because they are known to be expressed and enzymatically active in E. coli (Holden et al. 2008; Shi et al. 2015). Mutations in the rpoB gene increase with mutagenic stress mediated by A3 (Harris et al. 2002; Shi et al. 2015). Mutational frequencies above background levels were only observed for A3Z1 proteins, and we found that four bat A3Z1s showed a 10-fold or greater increase in mutation frequency (fig. 3A). In particular, A3Z1c isomer B showed over 1000-fold increase above empty vector and 100-fold above A3Z1c isomer A. To determine if the difference in mutation frequency correlated with differences in mutation signatures, we sequenced a PCR-amplified portion of the rpoB gene known to harbor hotspots for rifampicin resistance. Bat A3 mutation signatures were compared against human A3B and A3G (fig. 3B), which have dinucleotide sequence preferences for 5′-TC and 5′-CC, respectively. We found that bat A3Z1c isomer A was highly specific for causing mutation at a known 5′-TCG hotspot at nucleotide position 1585, which is also the preferred target site for A3B (Shi et al. 2015). In contrast, A3Z1c isomer B targeted sites at position 1565, 1585, and 1592, which have 5′-TCT, 5′-TCG, and 5′-TCC contexts, respectively. Decreased specificity may account for the high mutation frequency observed in the rifampicin mutagenesis assays. These results show that bat A3 proteins are functionally active and can mutate cytosines in a diverse range of dinucleotide contexts.
Fig. 3.
Functional analysis of bat A3 proteins. (A) A3 proteins were transformed in C43 (DE3) E. coli to measure mutational frequency in the rpoB gene. Each point represents an independent sample and bars represent median mutation frequency with numerical values above. (B) Mutation signatures in a portion of the rpoB gene by PCR amplification and Sanger sequencing. Position of the mutated nucleotide is underlined and in red with nucleotide changes below. Known hotspots targeted by related proteins are provided above. A1, APOBEC1; A3, APOBEC3; AID, Activation-induced deaminase.
Bat APOBEC3 Proteins Restrict HIV-1 Infectivity
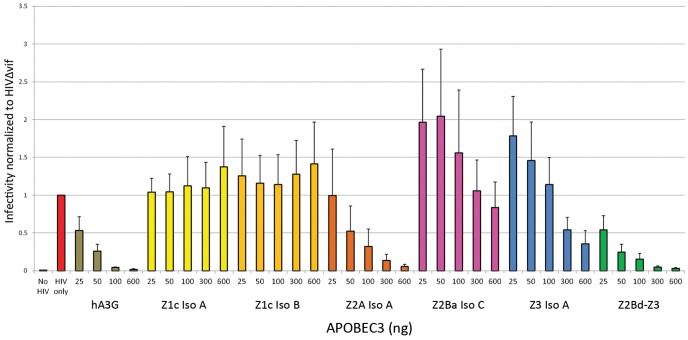
The antiviral activity of bat A3 proteins was determined by assessing the capacity of representatives of each bat A3 subtype to restrict HIV-1 infectivity in target cells. HIV-1 encodes the anti-A3 counter-measure, Vif, a protein that causes the proteolytic degradation of A3 through the recruitment of the cellular E3 ubiquitin ligase (Marin et al. 2003; Sheehy et al. 2003; Kane et al. 2015). HIV-1 stocks were generated by cotransfection of 293 T producer cells with bat A3 and an HIV-1 variant that does not express Vif. HIV-1 particles were harvested from the cell culture supernatant, and virion production was determined by quantifying virion-associated reverse transcriptase (RT) activity. HIV-1 infectivity was assessed by inoculating human TZM-bl target cells, which express luciferase in the presence of HIV-1 infection, with virus normalized to equivalent levels of virion-associated RT activity. Four bat A3 proteins demonstrated a dose-dependent inhibition of HIV-1 infectivity (fig. 4). The sole double Z-domain bat A3 protein, A3Z2Bd-Z3, produced the largest reduction in HIV-1 infectivity, which was comparable to inhibition by the human A3 protein, A3G. These data indicate that bat A3 proteins are functional restriction factors.
Fig. 4.
Antiviral activity of A3 proteins. Vif-deficient HIV-1 was generated in the presence of A3 proteins. Human 293 T cells were transfected with 400 ng of the HIVΔvif expression plasmid pIIIBΔvif and variable amounts (25–600 ng) of human or bat A3 expression plasmids. TZM-bl indicator cells were inoculated with 293 T cell culture supernatants normalized for viral particles by virion-associated RT activity. Infectivity relative to HIVΔvif alone was measured by quantifying luciferase activity in TZM-bl lysates. HIV, human immunodeficiency virus type 1; hA3G, human APOBEC3G; Z1c Iso A, Pteropus alecto APOBEC3Z1c isomer A; Z1c Iso B, Pteropus alecto APOBEC3Z1c isomer B; Z2A Iso A, Pteropus alecto APOBEC3Z2A isomer A; Z2Ba Iso C, P. alecto APOBEC3Z2Ba isomer C; Z3 Iso A, P. alecto APOBEC3Z3 isomer A; Z2Bd-Z3, P. alecto APOBEC3Z2Bd-Z3. Error bars represent the standard error of the mean from three independent assays.
Ancient Bat Retroviruses Were Hypermutated by APOBEC3
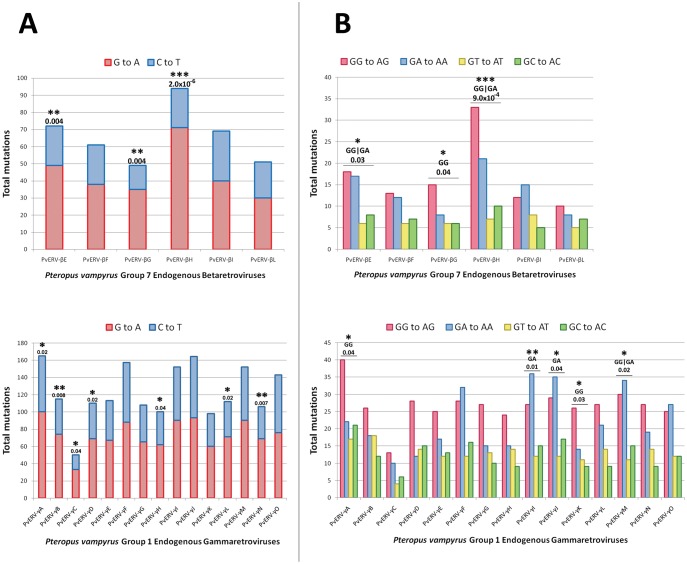
A3 activity results in C to U lesions in the negative (cDNA) strand, which lead to positive (genomic) strand G to A mutations in endogenous retroviruses (ERVs) within host genomes (Perez-Caballero et al. 2008). A3 proteins also exhibit local sequence-context bias in the nucleobases they deaminate. The presence of strand-biased, context-specific A3 mutagenic signatures in ERVs constitutes unambiguous evidence of a past encounter with one or more cellular A3 enzymes (Lee et al. 2008). To assess hypermutation of pteropid ERVs, we identified a group of closely related ERVs from each of the Betaretrovirus and Gammaretrovirus genera present in the genome of P. vampyrus. We interrogated the P. vampyrus genome since the P. alecto genome assembly does not contain sufficient contiguous endogenous retroviral sequences to conduct this analysis. A previously reported group of bat endogenous betaretroviruses, sub-classified as Group 7 betaretroviruses, was selected and a group of bat endogenous gammaretroviruses (herein described as Group 1 gammaretroviruses) was identified in the manner described previously (Hayward et al. 2013). Phylogenies were generated using the maximum likelihood method and an ancestral reconstruction was performed by extrapolation of the common ancestor of each group (see supplementary fig. S4, Supplementary Material online). Mutations in each ERV were identified by comparison against the group’s common ancestor. Strand-biased G to A hypermutation and context-specific GG and GA dinucleotide preferences were identified in both groups (fig. 5). These results are consistent with the hypothesis that ancient bat retroviruses were subjected to A3-mediated hypermutation, supporting the expected role of bat A3 proteins as antiviral restriction factors. The divergence times of the bat A3Z1 genes were estimated using a molecular clock dating analysis that indicates these genes arose through duplication events beginning approximately 27 Ma (supplementary fig. S5, Supplementary Material online).
Fig. 5.
Hypermutation analysis of endogenized retroviruses reveals that ancient bat retroviruses were subjected to restriction by A3 proteins. (A) Comparison of guanine to adenine (G to A) versus cytosine to thymidine (C to T) mutations on the coding (+) strand of endogenous retroviruses reveals strand-biased cytosine deamination. (B) Subsequent analysis of the dinucleotide context of the G to A mutations in the coding strand reveals that the GG to AG and GA to AA mutations typical of human A3G are the most common mutations in the endogenous retroviruses of bats. Statistical significance was calculated using Fisher’s Exact Test. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
This study was undertaken to address the question of differential antiviral gene evolution in bats, and more specifically, whether important differences exist in a major class of genes that control and restrict retroviruses and genomic retroelements. We demonstrated that bats possess 18 A3 genes, 13 of which are transcriptionally active in spleen tissue, with several proving active in a heterologous E. coli base mutation assay or an HIV-1 restriction assay. In addition to the expected A3Z1, A3Z2, and A3Z3 phylogenetic subtypes, we identified an additional division within the bat A3Z2 clade in which we designated A3Z2A as the prototypical A3Z2 subtype, and A3Z2B, as a subdivision that possesses a combination of invariant amino residues key to the amino acid character of both the A3Z1 and A3Z2 subtypes. Representative A3 loci have previously been delineated for every Eutherian order with the exceptions of Chiroptera (bats) and all orders within the clade Afrotheria (e.g., elephants, tenrecs, dugongs, and hyraxes) (Nakano et al. 2017). With the caveat that the A3 loci of the vast majority of mammalian species remain to be resolved, the pteropid bat species studied here are shown to possess the largest and most phylogenetically diverse antiviral A3 gene repertoire of any mammal reported to date.
Assessment of the functionality of bat A3 proteins through a rifampicin mutagenesis assay revealed several A3Z1 proteins as potent mediators of cytosine deamination with A3Z1 isomer B demonstrating a mutational frequency greater than human A3B and A3G. Although some A3Z1 proteins, and A3 proteins of other subtypes were not found to be catalytically active in this assay, numerous factors could account for inactivity within E. coli, such as protein misfolding or missing cellular cofactors and localization requirements. The capacity of bat A3 proteins to restrict retroviral infection was observed through the use of HIV-1 as a model target for A3 functionality. Four bat A3 proteins representing single and double Z-domain proteins of the Z2A, Z2B, and Z3 subtypes were observed to be capable of inhibiting HIV-1 infectivity in a dose-dependent manner. The bat A3Z1 proteins found to be mutagenic were not found to be capable of restricting HIV-1 infectivity. A possible explanation for this is that not all bat A3 proteins are capable of restricting this specific virus, as is the case for human A3 proteins such as A3B and A3C which inhibit SIV but not HIV, whereas A3F and A3G are potent inhibitors of HIV (Sheehy et al. 2003; Yu et al. 2004; Zheng et al. 2004). Although many endogenous retroviruses are present in bat genomes (Cui et al. 2012; Hayward et al. 2013), no exogenous bat retroviruses have yet been reported. To address our expectation that bat retroviruses could be subjected to restriction by bat A3 proteins we performed a hypermutation analysis of the endogenized retroviruses present in the genome of P. vampyrus and found evidence of strand-biased, context-dependent, cytosine deamination of both beta- and gammaretroviruses. These results implicate bat A3 proteins as the mediators of G to A hypermutation in bat retroviruses.
The large number of A3Z1 genes present in the pteropid A3 locus is surprising given the scarcity of this subtype among other mammals (LaRue et al. 2009). The presence of 12 A3Z1 genes in pteropid bats indicates an evolutionary advantage for pteropid bats in the maintenance and expansion of this subtype. The most comparable APOBEC3 protein in humans is A3A, the sole APOBEC3 single-domain Z1 protein. A3B and A3G are double-domain proteins that have a Z1 paired with a Z2A domain (fig. 1C). Endogenous A3A is cytoplasmically located (Land et al. 2013) but accesses the nucleus during telophase, the final stage of mitosis (Lackey et al. 2013). It has restrictive activity against retrotransposons (Chen et al. 2006; Koito and Ikeda 2013) and viruses including HIV-1, adeno-associated virus, and human papillomavirus (Chen et al. 2006; Berger et al. 2011; Warren et al. 2015). In humans, evidence suggests that A3Z1 domain-containing proteins may represent a double-edged sword, as potent inhibitors of genomic retrotransposons and a possible threat to the cell’s own genomic sequence integrity (LaRue et al. 2008; Roberts et al. 2013; Taylor et al. 2013; Burns et al. 2015; Green et al. 2016). One possible reason for the observed expansion of A3Z1 genes in bats may be related to the evolution of flight, which is hypothesized to be an important link to the physiological conditions allowing bats to be sources of high viral diversity (O’Shea et al. 2014). Increased metabolic capacity has been linked to the evolution of sustained flight in bats (Shen et al. 2010), and the by-products of oxidative metabolism include reactive oxygen species that are known to damage cellular DNA (Cooke et al. 2003). The most notable genetic adaptations involved in the development of flight in bats have been found to exist in the genes responsible for the detection and repair of cellular DNA damage (Zhang et al. 2013; Zhou et al. 2016). It is possible that enhanced tolerance to DNA damage would allow for the expansion of genes such as A3 whose products are known to cause such damage.
Many studies have indicated that A3 enzymes restrict endogenous transposable elements such as LINE-1 (long interspersed nuclear element 1) and Alu (Muckenfuss et al. 2006; Stenglein and Harris 2006; Kinomoto et al. 2007; Wissing et al. 2011). Interestingly, LINE-1 elements became extinct in the pteropid bat lineage approximately 24 Ma (Cantrell et al. 2008). Preliminary dating of the A3 gene expansions (supplementary fig. S5, Supplementary Material online) indicate that timing of the initial expansion of the bat A3Z1 subtype may coincide with this event and further investigation of a possible connection between pteropid A3 genes and the LINE-1 extinction may be warranted.
This study represents the first report of the differential evolution of an important antiviral gene family in bats relative to other mammals. The full account and explanation of the ability of bats to act as such effective hosts and transmitters of zoonotic viral pathogens will likely consist of many complex and interlinking factors beyond simple metrics such as gene numbers and amino acid sequence characteristics. Ongoing studies of this expanded gene family should further demystify the strategies and mechanisms utilized by bats that result in an impressive tolerance for viral infections.
Materials and Methods
APOBEC3 and Retroviral DNA Sequences
The accession numbers and genome locations of the mammalian APOBEC3 and retroviral genome sequences utilized in this study are listed in supplementary table S2, Supplementary Material online.
Generation of Bat Transcriptomes
Approval for the use of bat tissues was granted by the Commonwealth Scientific and Industrial Research Organization’s Australian Animal Health Laboratory Animal Ethics Committee (Protocol AEC1281). Transcriptome data sets of P. alecto were generated from pooled total RNA from mitogen-stimulated spleen, lymph node, white blood cells, and unstimulated bone-marrow and thymus from one pregnant female and one adult male, and the unstimulated thymus of a juvenile male as described previously (Papenfuss et al. 2012). The transcriptome of P. alecto is available at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/Traces/sra/; last accessed April 3, 2018) [SRA: SRP008674].
Transcriptome Analysis
To search for predictions of A3 gene expression within the bat transcriptomes, sequences of human A3 transcripts were obtained from Ensembl (http://www.ensembl.org/; last accessed April 3, 2018), specifically: A3A [Ensembl: ENST00000402255], A3C [Ensembl: ENST00000361441], and A3H [Ensembl: ENST00000348946]. The human A3 transcripts were translated into protein sequences using the CLC Genomics Workbench 8.0 (https://www.qiagenbioinformatics.com/; last accessed April 3, 2018). To identify transcripts of interest the tBLASTn function of CLC Genomics Workbench was used with the following parameters: BLOSUM62 matrix, word size = 3, E-values < 1 × 10−12, gap costs of existence 11, extension 1, with no filtering of regions of low complexity. Transcripts were confirmed as A3 homologues by initial identification of, in their translated protein sequences, the presence of the conserved Z-domain, Hx1Ex21–27S/TWSPCx2–4C (where “x” indicates a nonconserved position [Conticello et al. 2005; LaRue et al. 2009]), and subsequent identification of the A3 subtype-specific conserved motifs described in detail in LaRue et al. (2008, 2009).
cDNA Analysis
To confirm A3 gene product expression, polymerase chain reaction (PCR) amplification assays were performed using cDNA generated from P. alecto spleen tissue and various combinations of forward and reverse primers designed using the A3 transcript predictions identified in the transcriptome analysis. P. alecto trapping, tissue collection and RNA extraction was performed as reported previously (Cowled et al. 2011) except RNAlater (Ambion) preserved spleen from four male adult bats was pooled prior to tissue homogenization followed by total RNA extraction with the Qiagen RNeasy Mini kit with on-column removal of genomic DNA with DNase I. Total RNA was reverse transcribed into cDNA with the Qiagen Omniscript reverse transcriptase according to the manufacturer’s protocol except that the reaction contained 100 ng/μl total RNA, 1 μM oligo(dT)18 (Qiagen), and 10 μM random hexamers (Promega).
All PCR amplification assays were performed using the Roche FastStart High Fidelity PCR system (Cat # 04738292001) utilizing an annealing temperature gradient of 54–64°C in 2°C increments. Each reaction was made up to a total of 20 μl, containing 1 unit of polymerase, 2 ng of total cDNA, and 8 pmol of each primer. All other parameters for PCR amplification were performed according to the manufacturer’s recommended protocol. PCR reaction products were separated by agarose gel electrophoresis, using a 1% (w/v) agarose gel run at 120 V for 25 min. All DNA bands were excised from the gel and purified using the Qiagen QIAquick Gel Extraction kit (Cat # 28706) according to the manufacturer’s protocol. The gel extracted PCR products were cloned into the pCR2.1 plasmid vector using the Invitrogen TOPO TA Cloning kit (Cat # 450641) according to the manufacturer’s protocol and transformed into Escherichia coli TOP10 (Invitrogen). The nucleotide sequence of clones were determined by Sanger sequencing using the M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) sequencing primers.
APOBEC3 Gene Locus Assembly
To identify the gene scaffolds representing fragments of the A3 gene locus in the P. vampyrus genome, a BLASTn analysis was performed using the A3 gene products amplified in the cDNA analysis as queries. The P. vampyrus genome was downloaded from Ensembl and all BLASTn searches were performed locally using CLC Genomics Workbench with the following parameters: Match score 2, mismatch cost −3, word size = 5, E-values < 1 × 10−5, gap costs of existence 5, extension 2, with no filtering of regions of low complexity. To extend the sequence of each scaffold, 150 nt at both the 5′ and 3′ termini was used as the query sequence for a BLASTn search against the P. vampyrus Illumina whole genome sequencing (WGS) project SRA [SRA: SRP047390] using the NCBI BLASTn suite (http://blast.ncbi.nlm.nih.gov/Blast.cgi; last accessed April 3, 2018) with the following parameters: Program = megablast, maximum target sequences = 500, E-values < 1 × 10−20, word size = 28, maximum matches in a query range = 13, match score = 1, mismatch cost = 2, gap costs = linear, with no filtering or masking. All hits with a percentage identity of 97% or higher were downloaded and assembled using the CLC Genomics Workbench Assemble Sequences tool into a contiguous consensus sequence using the following parameters: Minimum aligned read length = 20, alignment stringency = high, conflicts = Vote (A, C, G, T). Consensus sequences were aligned with the original query to confirm that the assembly represented a terminally extended match of the original query sequence; they were subsequently used as a new query in an otherwise identical BLASTn search against the same SRA. This process was iteratively repeated until all scaffolds could be extended out far enough that their sequences overlapped with another scaffold. The extended and overlapping scaffolds were joined end to end to create a contiguous reference sequence construct representing the P. vampyrus A3 gene locus.
To validate the draft A3 locus reference sequence, all P. vampyrus WGS SRA were downloaded from NCBI and locally converted into paired FASTQ data sets using the NCBI SRA Toolkit 2.4.3 (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi; last accessed April 3, 2018). Each data set was subjected to a quality control check using the FastQC program 0.11.3 (Babraham Bioinformatics, Cambridge). Paired FASTQ data sets were imported into CLC Genomics Workbench using the Illumina import function and the CLC Genomics Workbench Next Generation Sequencing (NGS) Toolkit was used for all following tasks. All data sets were trimmed on the basis of quality using the default parameters. Data sets with short paired-read distances were assessed for the presence of overlapping pairs and these were merged where appropriate using the default parameters. The P. vampyrus data set included long paired reads with insert sizes of 2–20 kb which are helpful for the resolution of repetitive gene regions. Validation of the P. vampyrus A3 locus was performed by mapping all reads to the reference sequence construct using the following parameters: Linear gap cost, length fraction = 0.9, similarity fraction 0.9, with automatic detection of paired read distances. Other parameters were left in their default settings. Potential gene coding regions were identified by mapping P. alecto A3 cDNA sequences against the locus through a local BLASTn analysis using CLC Genomics Workbench with default settings. Promoter elements were predicted using the online Neural Network Promoter Prediction 2.2 tool (Reese 2001) (http://www.fruitfly.org/seq_tools/promoter.html; last accessed April 3, 2018) using the default parameters. ISRE were predicted using the online PROMO transcription factor binding site prediction tool (Farré et al. 2003) (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3; last accessed April 3, 2018) using the parameters: Species: Mammalia, Factors: Mammalia.
APOBEC3 Phylogenetic Analysis
The identities of the putative bat A3 gene products were confirmed by phylogenetic analysis. All reference sequences were downloaded from Ensembl (https://www.ensembl.org/; last accessed April 3, 2018) and aligned with P. alecto sequences using MUSCLE (Edgar 2004). Regions of low conservation and uncertain alignment were removed using the Gblocks program (Talavera and Castresana 2007). Phylogenetic relationships were inferred using the maximum likelihood (ML) method available in the MEGA6 program (Tamura et al. 2013), employing nearest-neighbor interchange and branch swapping, and the best-fit model of amino acid substitution was found to be JTT + G (Tamura et al. 2013). To determine the robustness of each node 1,000 bootstrap replications were incorporated.
Rifampicin Mutagenesis Assay
Bat A3 cDNAs were PCR amplified from parental vectors and subcloned into pET24(+) vectors (EMD Biosciences). Plasmids were transformed into calcium competent C43 (DE3) strain E. coli (gift from Dr. Do-Hyung Kim) and grown in LB plates with kanamycin. Single colonies were selected and grown in LB with kanamycin for 28 h to ensure stationary phase growth. Thereafter, cultures were spread on either rifampicin plates or serially diluted in M9 salt media and spread on kanamycin plates. Colony counts were obtained after 18 h. rpoB mutation frequency was obtained by dividing the number of viable colonies on rifampicin plates by colonies on kanamycin plates and correcting for the dilution factor. A portion of the rpoB gene was PCR amplified using primers: forward 5′-TTGGCGAAATGGCGGAAAACC and reverse 5′-CACCGACGGATACCACCTGCTG. PCR products were enzymatically purified using Exonuclease I and rSAP (NEB). Purified fragments were sequenced using the forward primer (GeneWiz, NJ).
Human Cell Cultures
Human embryonic kidney cells (293 T cells) and human epithelial cervical adenocarcinoma cells modified to express constitutively high levels of HIV-1 receptors and coreceptors (TZM-bl cells), were utilized in the inhibition of infectivity assay. The TZM-bl cells were obtained through the NIH AIDS Research and Reference Reagent Program (Wei et al. 2002). All cell cultures were maintained at 37°C, 5% CO2 in Dulbecco’s modified Eagle’s medium (Thermo) supplemented with heat-inactivated fetal bovine serum (100 ml/l; Invitrogen), glutamine (292 μg/ml; Invitrogen), and the antibiotics penicillin (100 units/ml; Invitrogen) and streptomycin (100 units/ml; Invitrogen).
A3 Retroviral Restriction Assay
The antiviral activity of bat A3 was assessed by examining the capacity to restrict HIV-1 infectivity. Bat A3 cDNAs were PCR amplified from parental vectors and subcloned into pcDNA3.1 mammalian expression vectors (Thermo Fisher, MA). 293 T cells were cotransfected with different quantities of bat A3 or human A3G (phu-A3GHA; [Mariani et al. 2003]) expression plasmids (25–600 ng) and 400 ng of pIIIBΔvif (Simon et al. 1995), which generates HIV-1 virions in the absence of Vif. Untransfected cells and cells transfected only with pIIIBΔvif were used as controls. Transfected cell cultures were incubated at 37°C, 5% CO2 for 48 h, and then virion-containing supernatants were collected and clarified by centrifugation at 200 × g for 5 min. Virus production was determined by quantifying virion-associated RT activity, as previously described (Wapling et al. 2005). TZM-bl cells were inoculated with RT normalized supernatant and incubated at 37°C, 5% CO2 for 48 h. HIV-1 infection of TZM-bl cells was assessed through quantitation of luciferase activity in cell lysates as previously described (Tyssen et al. 2010).
Hypermutation Analysis
The impact of cytosine deamination on the genomes of ancient bat retroviruses was assessed by hypermutation analysis (Lee et al. 2008). The genomes of all ERVs used in the analysis were extracted from the P. vampyrus genome. Multiple outgroups were tested to confirm that the position of the ancestral node remained consistent. For the betaretroviruses, we tested Jaagsiekte sheep retrovirus (JSRV) [NCBI: NC_001494], Squirrel monkey retrovirus (SMR) [NCBI: M23385], and the Mouse mammary tumor virus (MMTV) [NCBI: AF033807]; for the gammaretroviruses we tested the Moloney murine leukemia virus (MMLV) [NCBI: NC_001501], the Feline leukemia virus (FLV) [NCBI: AF052723], and the Gibbon ape leukemia virus (GaLV) [NCBI: M26927]. The most closely related betaretrovirus and gammaretrovirus to our ERV groups were JSRV and MMLV, respectively, and were employed as outgroups for the phylogenies. Sequence alignments and phylogenies were conducted as described for the APOBEC3 phylogenetic analysis, and the best-fit model of nucleotide substitution was found to be T92 + G for both groups of retroviruses. Ancestral sequences were computationally extrapolated using the MEGA6 program (Tamura et al. 2013) using the mutations accumulated in each ERV since integration by estimating the ancestral state of each node in the maximum likelihood phylogenetic tree. The state is chosen to be the one that maximizes the probability of the given sequence data. Mutational biases were then evaluated by analyzing an alignment of the ancestral sequence and the ERVs (supplementary data S2 and S3, Supplementary Material online) using the online Hypermut tool (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html; last accessed April 3, 2018) (Rose and Korber 2000) with context enforced on the ancestral reference sequence. The Hypermut tool incorporates Fisher’s exact test to determine statistical significance.
Molecular Clock Dating Analysis
To estimate the time of the expansion of A3Z1 genes in bat genome, molecular dating was performed using the Bayesian evolutionary method as implemented in BEAST v1.8.2 (Drummond et al. 2012). All reference nucleotide sequences were aligned using MUSCLE and adjusted manually. A recombination test was then performed using the RDP, GENECONV and MaxChi methods implemented in RDP3 (Martin et al. 2010). After removing recombinant sequences, all sequences were submitted to BEAST for estimation of the divergence time. Fossil node calibrations were taken from published literature (Kumar et al. 2017): human and elephant, 100–111 Ma; human and bat, 91–102 Ma; human and macaque, 27.61–31.28 Ma; horse and camel, 74–81 Ma; camel and sheep, 61.1–67.2 Ma; goat and sheep, 8.18–12.62 Ma; sheep and cow, 21.41–29.46 Ma; beluga whale and orca, 16.36–20.43 Ma. Bayesian phylogeny was inferred using the GTR + G substitution model, the Yule speciation tree prior and lognormal relaxed clock (Uncorrelated) prior. The BEAST processes were run for ten million generations until all relevant parameters converged [effective sample size (ESS)>200], with 10% of the Bayesian Markov chain Monte Carlo (MCMC) chains discarded as burn-in.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by Perpetual Trustees (AR/00255 to G.T. and L.-F.W.), the Australian Postgraduate Award and the Monash University Vice Chancellor’s Honours-PhD scholarship (to J.A.H.), the National Health and Medical Research Council Senior Research Fellowship (APP1117748 to G.T.), the National Research Foundation-Competitive Research Programme (NRF2012NRF-CRP001-056 to L.-F.W.), and the National Institute of Health and National Institute of Allergy and Infectious Diseases (R37AI064046 to R.S.H.). R.S.H. is an Investigator of the Howard Hughes Medical Institute and J.C. is supported by National Natural Science Foundation of China (31671324) and CAS Pioneer Hundred Talents Program. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program by the Burnet Institute. The pteropid A3 loci and cDNA sequences have been deposited in GenBank and are listed in supplementary table S2, Supplementary Material online.
References
- Almeida FC, Giannini NP, Simmons NB, Helgen KM.. 2014. Each flying fox on its own branch: a phylogenetic tree for Pteropus and related genera (Chiroptera: Pteropodidae). Mol Phylogen Evol. 77:83–95. [DOI] [PubMed] [Google Scholar]
- Amman BR, Albariño CG, Bird BH, Nyakarahuka L, Sealy TK, Balinandi S, Schuh AJ, Campbell SM, Ströher U, Jones MEB et al. , . 2015. A recently discovered pathogenic paramyxovirus, Sosuga virus, is present in Rousettus aegyptiacus fruit bats at multiple locations in Uganda. J Wildl Dis. 513: 774–779., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Schountz T, Wang LF.. 2013. Antiviral immune responses of bats: a review. Zoonoses Public Health 60:104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Durand S, Fargier G, Nguyen X-N, Cordeil S, Bouaziz S, Muriaux D, Darlix J-L, Cimarelli A.. 2011. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 79: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Leonard B, Harris RS.. 2015. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed J. 382: 102. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T.. 2006. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 193: 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA.. 2008. Loss of LINE-1 activity in the megabats. Genetics 1781: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD.. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 165: 480–485. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC.. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 26:317–353. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS.. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol Biol Evol. 222: 367–377. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J.. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 1710: 1195–1214. [DOI] [PubMed] [Google Scholar]
- Cowled C, Baker M, Tachedjian M, Zhou P, Bulach D, Wang L-F.. 2011. Molecular characterisation of Toll-like receptors in the black flying fox Pteropus alecto. Dev Comp Immunol. 351: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Tachedjian G, Tachedjian M, Holmes EC, Zhang S, Wang L-F.. 2012. Identification of diverse groups of endogenous gammaretroviruses in mega and microbats. J Gen Virol. 93(Pt_9): 2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Tachedjian G, Wang L-F.. 2015. Bats and rodents shape mammalian retroviral phylogeny. Sci Rep. 5:16561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 298: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Wang X, Mu S, Zhang S, Dong D.. 2015. BGD: a database of bat genomes. PLoS One 106: e0131296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, Messeguer X.. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 3113: 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AM, Landry S, Budagyan K, Avgousti DC, Shalhout S, Bhagwat AS, Weitzman MD.. 2016. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle 157: 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K, Young P, Field H, Mackenzie J.. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 81(Pt 8): 1927–1932. [DOI] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS.. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 105: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Hayman DT. 2016. Bats as viral reservoirs. Annu Rev Virol. 31: 77–99. [DOI] [PubMed] [Google Scholar]
- Hayward J, Tachedjian M, Cui J, Field H, Holmes E, Wang L-F, Tachedjian G.. 2013. Identification of diverse full-length endogenous betaretroviruses in megabats and microbats. Retrovirology 101: 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS.. 2008. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 4567218: 121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Schuh AJ, Amman BR, Sealy TK, Zaki SR, Nichol ST, Towner JS.. 2015. Experimental inoculation of Egyptian rousette bats (Rousettus aegyptiacus) with viruses of the ebolavirus and marburgvirus genera. Viruses 77: 3420–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JR, Stanley DJ, Hultquist JF, Johnson JR, Mietrach N, Binning JM, Jónsson SR, Barelier S, Newton BW, Johnson TL et al. , . 2015. Lineage-specific viral hijacking of non-canonical E3 ubiquitin ligase cofactors in the evolution of Vif anti-APOBEC3 activity. Cell Rep. 118: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K.. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 359: 2955–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A, Ikeda T.. 2013. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 347: 1812–1819. [DOI] [PubMed] [Google Scholar]
- Lackey L, Law EK, Brown WL, Harris RS.. 2013. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 125: 762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, Harris RS.. 2013. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem. 28824: 17253–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue R, Jonsson SR, Silverstein KAT, Lajoie M, Bertrand D, El-Mabrouk N, Hotzel I, Andresdottir V, Smith TPL, Harris RS.. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 91: 104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jónsson SR, Landau NR, Löchelt M.. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 832: 494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ.. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 3005622: 1112.. [DOI] [PubMed] [Google Scholar]
- Lee YN, Malim MH, Bieniasz PD.. 2008. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol 8217: 8762–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR.. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 1141: 21–31. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D.. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 911: 1398–1403. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P.. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 2619: 2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg A, Wurzer G, Huisman T, Smithies O.. 1991. Homology requirements for unequal crossing over in humans. Genetics 1281: 143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton DJ, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, Westbury HA, Halpin K, Daniels PW.. 2007. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J Comp Pathol. 1364: 266–272. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perković M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C.. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 28131: 22161–22172. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Aso H, Soper A, Yamada E, Moriwaki M, Juarez-Fernandez G, Koyanagi Y, Sato K.. 2017. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology 141: 31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea TJ, Cryan PM, Cunningham AA, Fooks AR, Hayman DT, Luis AD, Peel AJ, Plowright RK, Wood JL.. 2014. Bat flight and zoonotic viruses. Emerging Infect Dis. 205: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfuss AT, Baker ML, Feng Z-P, Tachedjian M, Crameri G, Cowled C, Ng J, Janardhana V, Field HE, Wang L-F.. 2012. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics 131: 261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Soll SJ, Bieniasz PD.. 2008. Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but Not TRIM5α proteins. PLoS Pathog. 410: e1000181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SA, Hassan SS, Olival KJ, Mohamed M, Chang L-Y, Hassan L, Saad NM, Shohaimi SA, Mamat ZC, Naim MS et al. , . 2010. Characterization of Nipah virus from naturally infected Pteropus vampyrus bats, Malaysia. Emerging Infect Dis. 1612: 1990., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 261: 51–56. [DOI] [PubMed] [Google Scholar]
- Refsland EW, Harris RS.. 2013. The APOBEC3 family of retroelement restriction factors In: Cullen BR, editor. Intrinsic immunity. Berlin Heidelberg: Springer; p. 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS.. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 3813: 4274–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard M, Henry M, Guétard D, Vartanian J-P, Wain-Hobson S.. 2010. APOBEC1 and APOBEC3 cytidine deaminases as restriction factors for hepadnaviral genomes in non-humans in vivo. J Mol Biol. 4003: 323–334. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G et al. , . 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 459: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT.. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 164: 400–401. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH.. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 4186898: 646.. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH.. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 911: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Shen Y-Y, Liang L, Zhu Z-H, Zhou W-P, Irwin DM, Zhang Y-P.. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci U S A. 10719: 8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H.. 2015. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J Biol Chem. 29047: 28120–28130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Southerling TE, Peterson JC, Meyer BE, Malim MH.. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J Virol. 697: 4166–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I, Wang L-F.. 2013. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 31: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS.. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 28125: 16837–16841. [DOI] [PubMed] [Google Scholar]
- Swanepoel R, Leman PA, Burt FJ, Zachariades NA, Braack LE, Ksiazek TG, Rollin PE, Zaki SR, Peters CJ.. 1996. Experimental inoculation of plants and animals with Ebola virus. Emerging Infect Dis. 24: 321.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 564: 564–577. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 3012: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS.. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2:e00534.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssen D, Henderson SA, Johnson A, Sterjovski J, Moore K, La J, Zanin M, Sonza S, Karellas P, Giannis MP et al. , . 2010. Structure activity relationship of dendrimer microbicides with dual action antiviral activity. PLoS One 58: e12309.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapling J, Moore KL, Sonza S, Mak J, Tachedjian G.. 2005. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J Virol. 7916: 10247–10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D.. 2015. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol. 891: 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Masangkay JS, Nagata N, Morikawa S, Mizutani T, Fukushi S, Alviola P, Omatsu T, Ueda N, Iha K et al. , . 2010. Bat coronaviruses and experimental infection of bats, the Philippines. Emerging Infect Dis. 168: 1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC.. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 466: 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MM, Hooper PT, Selleck PW, Gleeson LJ, Daniels PW, Westbury HA, Murray PK.. 1998. Transmission studies of Hendra virus (equine morbilli-virus) in fruit bats, horses and cats. Aust Vet J. 7612: 813–818. [DOI] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC.. 2011. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 28642: 36427–36437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, König R, Mariani R, Unutmaz D, Landau NR.. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 27951: 53379–53386. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W et al. , . 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 3396118: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y-H, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM.. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 7811: 6073–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JH, Mok L, Michalski WP.. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci U S A. 201518240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.