Abstract
Argonaute (Ago) proteins are encoded in all three domains of life and are responsible for the regulation of intracellular nucleic acid levels. Whereas some Ago variants are able to cleave target nucleic acids by their endonucleolytic activity, others only bind to their target nucleic acids while target cleavage is mediated by other effector proteins. Although all Ago proteins show a high degree of overall structural homology, the nature of the nucleic acid binding partners differs significantly. Recent structural and functional data have provided intriguing new insights into the mechanisms of archaeal and bacterial Ago variants demonstrating the mechanistic diversity within the prokaryotic Ago family with astonishing differences in nucleic acid selection and nuclease specificity. In this review, we provide an overview of the structural organisation of archaeal Ago variants and discuss the current understanding of their biological functions that differ significantly from their eukaryotic counterparts.
Keywords: Argonaute, DNA-guided DNA silencing, antiviral defense, archaea, bacteria, prokaryotic Argonaute
This review focuses on the mechanistic and structural diversity of the prokaryotic Argonaute protein family that constitute an additional prokaryotic defense system with high versatility, which is a consequence of the constant evolutionary pressure in the host-enemy arms race.
INTRODUCTION
Cellular live depends on the integrity of the genetic information stored in a cell. To ensure that the cellular blueprint encoded in the DNA is passed on correctly from one generation to the next, every cell is equipped with highly accurate DNA replication machineries. Additionally, defense mechanisms are in place that prevent foreign nucleic acids to infiltrate and expand in the cell. In eukaryotes, the nucleases Dicer and Argonaute 2 (hAgo2) are part of the RNA interference system that plays a role in antiviral defense (Ipsaro and Joshua-Tor 2015). Here, double-stranded RNAs (dsRNA) are recognized and degraded by the endonuclease Dicer. Dicer degradation products (termed small interfering RNAs, siRNAs) are loaded into hAgo2, which subsequently uses one of the RNA strands (termed guide strand) to identify complementary viral RNA targets via Watson-Crick base pairing. Ago catalyses the site-specific cleavage of the target RNA in case of full complementarity between RNA guide and target RNA. Besides its function in antiviral defense, hAgo2 plays a major role in post-transcriptional regulation mediated by endogenous small microRNAs (miRNA) and endogenous siRNAs (endo-siRNAs). Endo-siRNAs often arise from sense-antisense transcript hybrids and are suggested to be involved in the control of transposons (Piatek and Werner 2014). Similarly to the siRNA pathway, miRNAs and endo-siRNAs are processed by Dicer and loaded into hAgo2 that uses one of the strands as a guide to identify matching target mRNAs, which eventually leads to translational inhibition (Ipsaro and Joshua-Tor 2015) or transcript cleavage. Soon after the discovery of Ago, it became clear that Ago proteins are encoded in all three domains of life (Makarova et al. 2009; Swarts et al.2014a). The domain organisation of some bacterial and archaeal Ago variants is remarkably similar to their eukaryotic counterparts. An archaeal Ago variant from the hyperthermophilic organism Pyrococcus furiosus was also the first Ago for which a structure could be solved (Song et al.2004) followed by structures of the bacterial Ago from Aquifex aeolicus and Thermus thermophilus (Yuan et al.2005; Wang et al. 2008b). Prokaryotic Ago (pAgo) structures revealed many mechanistic details and enhanced our understanding of eukaryotic Ago (eAgo) action even before the structure of hAgo2 could be solved (Elkayam et al.2012; Schirle and MacRae 2012). The initial biochemical characterization of pAgos showed that these Ago variants also bind short nucleic acids and recognize target DNAs or RNAs leading to the nucleolytic cleavage of the target (Wang et al. 2009; Table 1). However, as Ago is only encoded in 10% of bacterial and in 30% of archaeal genomes (Makarova et al. 2009; Swarts et al. 2014a) and because Dicer homologs as well as other proteins associated with the RNAi pathways (e.g. the TAR RNA-binding protein TRBP or Zucchini) are absent from prokaryotic genomes, a universal prokaryotic silencing mechanism equivalent to eukaryotic RNAi seemed unlikely. Moreover, the widespread and versatile prokaryotic CRISPR-Cas (clustered regularly interspaced short palindromic repeats and associated Cas proteins), restriction modification systems as well as sugar-non-specific nucleases constitute powerful weapons against foreign nucleic acids (Horvath and Barrangou 2011; Hille and Charpentier 2016) and hence, the antiviral duties detected for eAgos seemed fulfilled by alternative molecular machineries in the prokaryotic world. The biological function of pAgos remained elusive for a long time, and only recent studies shed light on the functional role of Ago proteins in prokaryotes. Here, we will discuss the latest functional and physiological data that provide evidence for a role of pAgos in the defense system of prokaryotes. Intriguingly, while pAgos are mediating RNA-guided or DNA-guided DNA silencing and possibly RNA silencing in vivo, they can also act as a general nuclease in a guide-independent manner (Swarts et al.2017; Zander et al.2017). In line with the newly uncovered molecular mechanisms of pAgo action, novel structural studies unravelled the mechanistic basis for the different DNA silencing functions (Kaya et al.2016; Miyoshi et al.2016; Doxzen and Doudna 2017; Swarts et al.2017; Willkomm et al.2017). We will highlight the diversity in pAgo-mediated defense mechanisms, reflect on the evolutionary plasticity that led to the emergence of different Ago variants and shortly discuss the possibility of a synergistic link between Ago and CRISPR-Cas defense systems.
Table 1.
List of characterized prokaryotic Argonaute proteins and their guide specificity, 5΄-end specificity and silencing activity (n.d.—not determined).
| Argonaute variant | Guide | Preference (5΄-end of the guide) | Activity |
|---|---|---|---|
| Archaea | |||
| M. jannaschii (MjAGO) | DNA, (RNA) | 5΄-P-purine | DNA-guided DNA interference |
| A. fulgidus (AfAGO) | DNA, (RNA) | n.d. | ? |
| P. furiosus (PfAGO) | DNA | none | DNA-guided DNA interference |
| Bacteria | |||
| A. aeolicus (AaAGO) | DNA, (RNA) | n.d. | DNA-guided RNA interference |
| R. sphaeroides (RsAGO) | RNA, DNA | 5΄-P-U | RNA-guided DNA interference |
| T. thermophilus (TtAGO) | DNA | 5΄-P-C | DNA-guided RNA interference/DNA-guided DNA interference |
| M. piezophila (MpAGO) | RNA | 5΄-OH | RNA-guided DNA interference/RNA-guided RNA interference |
BIOLOGICAL FUNCTION OF BACTERIAL AND ARCHAEAL ARGONAUTE PROTEINS
By now, a number of bacterial and archaeal Ago variants have been characterized (Song et al.2004; Rivas et al.2005; Yuan et al.2005; Wang et al.2008a,b; Sheng et al.2014; Swarts et al.2014a,b, 2015a,b, 2017; Kaya et al.2016; Miyoshi et al.2016; Doxzen and Doudna 2017; Willkomm et al.2017). Depending on the organism, Ago co-purifies with short DNAs or RNAs. However, all characterized pAgos recognize DNA as target suggesting that all full-length pAgos are involved in DNA silencing. An exception might be the Argonaute variant from A. aeolicus for which only RNA cleavage was demonstrated in vitro so far (Yuan et al.2005). Recently, details regarding the guide biogenesis and silencing mechanisms have been elucidated (Swarts et al.2017; Zander et al.2017). These insights exemplify that pAgo variants, despite their high degree of structural conservation, are extremely variable in the molecular mechanisms that ultimately lead to silencing of foreign DNA.
Guide biogenesis and priming of Argonaute
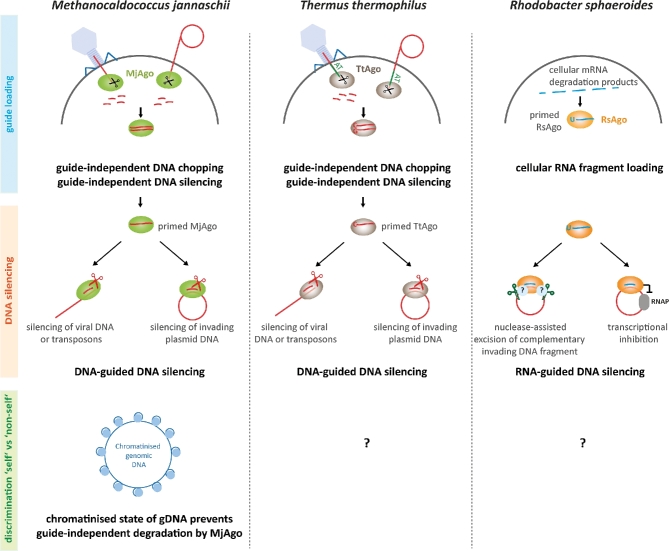
In the absence of a prokaryotic Dicer homolog, the production of short DNA or RNA guide strands that can be loaded into pAgo could not be assigned to a pre-processor nuclease. Recently, however, it could be shown that the archaeal Ago variant from Methanocaldococcus jannaschii (MjAgo) as well as a bacterial Ago from T. thermophilus (TtAgo) is able to process long dsDNA in a guide-independent manner, an activity termed DNA chopping (Swarts et al.2017; Zander et al.2017; Fig. 1). MjAgo was shown to fully degrade linear dsDNA fragments but also circular plasmid DNA in a non-specific fashion to produce short DNA strands suitable as DNA guides (Zander et al.2017). As MjAgo is derived from a hyperthermophilic organism, it seems likely that MjAgo can enter long dsDNAs at partly single-stranded sites formed regularly as a result of local DNA melting at high temperatures. Nucleolytic cleavage of one of the DNA strands leads to a free DNA end, which constitutes a starting point for stepwise degradation of the DNA yielding cleavage products of 8–17 nt in length. It has to be noted that MjAgo can also start cleavage from the 3΄-end and does not necessarily require a phosphate group for anchoring of the DNA guide in the MID domain (Zander et al.2017; unpublished data). Cleavage of a plasmid with MjAgo that was pre-loaded with previous degradation products of the very same plasmid proceeds significantly faster as compared to apo MjAgo suggesting that (i) the products of non-specific DNA digestion can be loaded into MjAgo and be used to recognize target DNA via Watson-Crick base pairing and (ii) guide-dependent cleavage of a DNA target is more efficient than guide-independent cleavage by MjAgo.
Figure 1.
Mechanisms of Argonaute-mediated DNA silencing in Archaea and Bacteria. Ago-mediated DNA interference pathways in prokaryotes exemplified for model systems from M. jannaschii (MjAgo), T. thermophilus (TtAGo), R. sphaeroides (RsAgo). MjAgo and TtAgo variants are able to degrade plasmid DNA or dsDNA in a guide-independent manner thereby creating short interfering DNAs that can be used for a subsequent sequence-specific guide-dependent degradation of DNAs. RsAgo recruits RNA guides from cellular transcript degradation products allowing RsAgo to target complementary foreign plasmid DNA for either direct nuclease-assisted cleavage of the target DNA or inhibition of transcription from the invading plasmid. How pAgo proteins differentiate between genomic and foreign DNA is not well studied but in M. jannaschii, the chromatinised genomic DNA is protected preventing guide-independent cleavage of the genomic DNA.
The bacterial TtAgo and archaeal Ago from P. furiosus (PfAgo) were shown to linearize plasmid DNA in the absence of a guide (Swarts et al. 2014b, 2015b). However, while MjAgo can fully degrade plasmid DNA via the chopping mechanism, it appears that TtAgo can only cleave long linear dsDNAs once starting from the respective 5΄-ends to generate short DNA guides. For PfAgo, only the linearization of plasmid DNA was described yet. Linearization of plasmid DNA by TtAgo requires an AT-rich site. In case of linear dsDNAs, cleavage only occurs if the DNA is GC-poor suggesting that TtAgo cleavage activity depends on a certain degree of DNA unwinding. AT-rich sequences are not a requirement for the guide-independent DNA cleavage by MjAgo. As observed for MjAgo, TtAgo-mediated cleavage of long dsDNAs does not result in products with a defined length but cleavage products cover a wide range of sizes from 8 to 26 nt. Usage of the cleavage products as functional guides was also demonstrated for TtAgo. While no experimental data are available yet, it seems likely that MjAgo and TtAgo bind dsDNAs created by the cleavage/chopping activity for guide acquisition and release one of the DNA strands after cleavage, a scenario comparable to the dsRNAs loading mechanism found in eAgos (Meister 2013). These data revealed that some members of the pAgo family fulfil a dual function as a non-specific guide-independent nuclease capable of guide generation and as guided, sequence-specific nuclease. Both nuclease modes mediate target DNA silencing.
In a cellular setting, however, the chopping of genomic DNA has to be prevented. Even though archaea and bacteria identify their own genomic DNA via the methylation pattern, which prevents cleavage by the cellular restriction endonucleases, DNA modification patterns do not influence MjAgo cleavage activity (Zander et al.2017). In M. jannaschii, the presence of histones protects the genomic DNA against Ago chopping activity (Zander et al.2017). How Ago distinguishes between ‘self’ and ‘non-self’ DNA in T. thermophilus is still unknown. However, while no bona fide histones are encoded in T. thermophilus, histone-like proteins like HU are present and might play a comparable protective role (Papageorgiou et al.2016). It seems feasible that either additional proteins regulate the chopping activity of pAgos or that replication might play a role in the enrichment of DNA guides derived from plasmid DNA. It is also possible that double-strand breaks which occur on stalled replication forks might serve as entry points for Ago. Many plasmids are efficiently replicated while bacterial and archaeal genomes often encode only one or just a few origins of replication leading to fewer double-strand breaks in the genome over time (Kelman and Kelman 2014). Consequently, free DNA ends are over-represented in replicating plasmids and linear DNA phage genomes rendering them more susceptible for Ago-mediated DNA degradation. However, it remains unclear how guides are generated in other prokaryotic organisms encoding Ago variants that can carry out guide-dependent cleavage of targets but for which no chopping activity was demonstrated yet (e.g. PfAgo, Marinitoga piezophila Ago (MpAgo)) (Swarts et al.2015b; Kaya et al.2016).
An alternative guide recruitment mechanism was identified for the bacterial Ago variant from Rhodobacter sphaeroides (RsAgo) (Olovnikov et al.2013). RsAgo binds an RNA guide in vitro to recognize an RNA or DNA target. In vivo studies demonstrated that RNA guide strands also associate with RsAgo in the cell. Loading of RsAgo with small RNAs and DNAs was even observed when RsAgo is heterologously expressed in Escherichia coli. The RNAs are mainly derived from the cellular transcriptome and RsAgo potentially recruits cellular RNA degradation products. Here, RsAgo selectively associates with RNA strands that carry a 5΄-U. Similarly, the Ago variant from MpAgo prefers RNA guides over DNA guides (Kaya et al.2016). However, the nucleic acids that associate with MpAgo in vivo have not been surveyed yet and consequently the source for its RNA guides is still unknown.
Ago-mediated DNA silencing
Which targets are silenced by pAgos? Sequencing of pAgo-associated nucleic acids revealed that sequences of mobile genetic elements like transposons and exogenous plasmid DNA accumulate in TtAgo and RsAgo suggesting that pAgos play a role in defense to degrade invasive genetic elements via a DNA silencing mechanism (Olovnikov et al.2013; Swarts et al.2014b; Fig. 1).
Based on a wide range of studies with TtAgo, the following scenario for DNA silencing in T. thermophilus can be drawn (Wang et al.2009; Sheng et al.2014; Swarts et al.2014b, 2017): after priming of TtAgo with a short DNA duplex, the passenger strand is cleaved and released from TtAgo. This allows the sequence-specific recognition of a target DNA (e.g. viral DNA, foreign plasmid DNA). In case of plasmid DNA, the target will be nicked once by TtAgo leading to the disintegration of the plasmid DNA. Alternatively, linear dsDNAs can be degraded by the previously described chopping mechanism. When exogenous plasmids are present in T. thermophilus, TtAgo reduces plasmid levels by a factor of four. As PfAgo makes use of DNA guides to cleave DNA targets and the presence of the ago gene in P. furiosus interferes with plasmid transformation (Swarts et al.2015b), it seems plausible that PfAgo employs a similar mechanism as observed for TtAgo to silence foreign DNAs. An analogous scenario could be imagined for MjAgo: loading of MjAgo with a guide allows the sequence-specific recognition of DNA targets that can be inactivated by target cleavage. However, unlike TtAgo, MjAgo does not nick plasmid DNA but guide-mediated cleavage would additionally allow guide-independent degradation of the plasmid DNA.
Degradation of target DNAs might yield new small interfering DNAs that can be loaded into Ago for another round of sequence-specific DNA silencing thereby enhancing the Ago-mediated defense mechanism. This process of secondary small interfering nucleic acid processing is slightly reminiscent of the ‘ping-pong’ cycle of the eukaryotic piRNA biogenesis pathway generating piRNAs that, in conjunction with P-element-induced wimpy testis (PIWI)-clade Ago proteins, represent a conserved defense mechanism in animal germ cells (Czech and Hannon 2016).
Interestingly, RsAgo is a catalytically inactive variant and hence, cleavage of a target is not possible. Nevertheless, in addition to guide RNAs, short complementary DNA fragments can be isolated from RsAgo (Olovnikov et al.2013). These DNAs are mainly derived from the exogenous RsAgo expression plasmid that was transformed into the cells. As RsAgo is encoded with a nuclease, it seems feasible that the nuclease carries out the DNA cleavage reaction. It has to be noted that cleavage seems to occur not opposite to nucleotide 10–11 of the guide (the canonical cleavage site for catalytic active Agos (Elbashir, Lendeckel and Tuschl 2001; Elbashir et al.2001)) but adjacent to the guide sequence resulting in a complementary DNA fragment with 3 nt overhangs. However, the co-encoded nuclease has not been biochemically characterized. Alternatively, RsAgo might silence foreign DNA by the repression of transcription on these DNAs. Here, RsAgo would represent a roadblock for the RNA polymerase on the DNA template.
Interplay of prokaryotic antiviral defense systems
Taken together, pAgo proteins represent an additional layer of the prokaryotic defense system that can act in a guide-independent and guide-dependent manner to silence DNA by nucleolytic cleavage. Due to the guide-independent nuclease activity of some pAgos, this defense system can act rapidly after invasion of the cell by foreign DNAs. For some prokaryotic organisms, it could be shown that the ago gene is constitutively expressed (Swarts et al. 2015a,b; Zander et al.2017), suggesting that Ago is used as a first line defense system allowing a fast reaction to combat invasive DNAs. In contrast to the CRISPR-Cas system, the pAgo-mediated defense has no memory as degradation fragments are not integrated into the genome. Even though this has not been demonstrated yet and phylogenetic patterns for both systems are largely overlapping (Makarova et al. 2009), Ago and CRISPR-Cas defense systems might interact with each other. Similar to Ago, the Cas1/2 complex processes invading DNA into short dsDNA fragments during the adaption phase of CRISPR-based defense (Jackson et al.2017). Possibly, degradation products can be handed over from Ago to Cas1/2 or vice versa. Interestingly, a small group of organisms encode its Ago within the CRISPR-Cas operon. For example, the ago genes from M. piezophila, Methanopyrus kandleri and Thermotoga profunda are encoded next to the cas1 and cas2 gene in the CRISPR-Cas locus (Kaya et al.2016). Here, an interplay between Ago and the CRISPR-Cas systems in the spacer acquisition phase seems likely but whether Ago interacts directly with Cas proteins is not known yet. Nevertheless, experimental data from the T. thermophilus system showed that in the presence of exogenous plasmids, Cas1 and Cas2 expression levels are elevated (Swarts et al.2015a). The stimulation of cas gene expression only occurs in the presence of Ago suggesting that Ago-mediated interference with plasmid DNA stimulates CRISPR adaptation. Moreover, some CRISPR-associated Cas proteins (e.g. Cas13a protein family) cleave RNAs non-specifically (East-Seletsky et al.2016; Liu et al.2017). These degradation products could potentially also serve as guides for pAgos.
STRUCTURES OF PROKARYOTIC ARGONAUTES REFLECT THE DIVERSITY OF TARGET RECOGNITION MECHANISMS
Ago proteins from all three domains of life are structurally highly conserved. However, the mechanistic differences found among pAgos arise from subtle structural changes and adaptation in the protein itself, which could only be discovered and understood with a new set of bacterial and archaeal Ago structures over the last 2 years (Sheng et al.2014; Kaya et al.2016; Miyoshi et al.2016; Doxzen and Doudna 2017; Swarts et al.2017; Willkomm et al.2017).
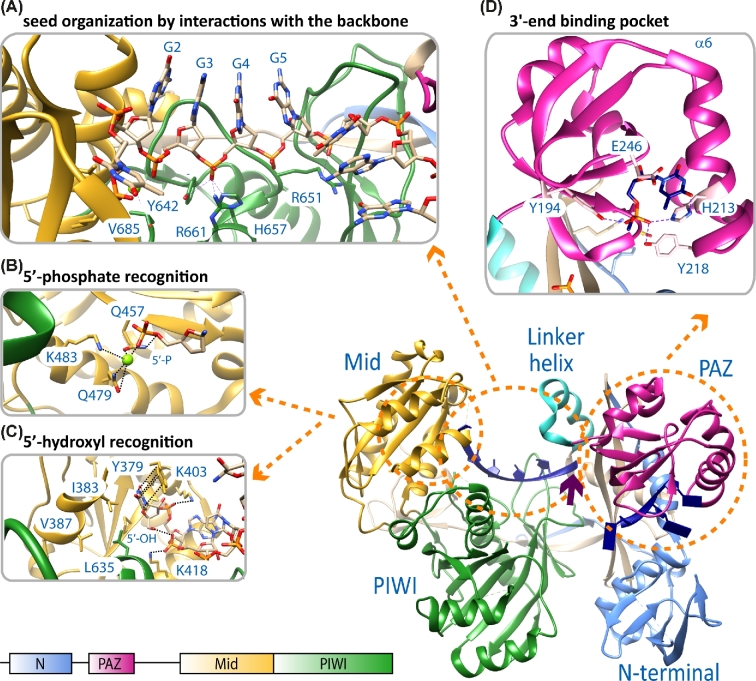
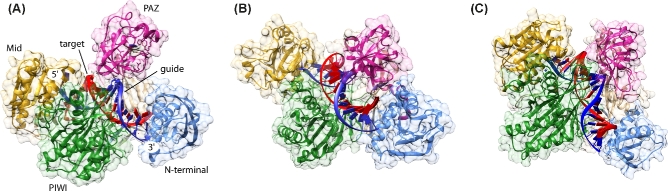
Full-length pAgos as well as eAgos are composed of four domains, which are organized in a bilobal fashion (Song et al.2004; Wang et al.2008b; Elkayam et al.2012; Nakanishi et al.2012; Schirle and MacRae 2012; Fig. 2). The N-terminal lobe consists of the N-terminal and the Piwi/Argonaute/Zwille (PAZ) domain and the C-terminal lobe contains the Mid and the PIWI domain that harbours the catalytic site of cleavage-active Agos (Fig. 2). pAgos are furthermore divided into two major groups termed long and short pAgos (Makarova et al. 2009; Swarts et al.2014a) (see section ‘Evolutionary aspects and diversity in the prokaryotic Argonaute world’). Whereas long pAgos contain all Ago-specific domains, short pAgos lack the PAZ domain, the N-terminal domain and consequently the L1 linker region. Therefore, the most conserved structural part of the pAgo clade is the C-terminal lobe comprising the L2 linker region, the Mid and the PIWI domain (Makarova et al. 2009). In contrast to eAgos, which are mostly restricted to use RNA as guide as well as target strands, pAgos tolerate a variety of RNA/DNA, DNA/RNA, RNA/RNA or DNA/DNA substrates (Willkomm et al.2015; Table 1).
Figure 2.
Structural features of Argonaute that influence guide binding. The main display shows the structural organization of the Argonaute protein from M. jannaschii in complex with a 21 nt guide DNA (blue) (pdb 5G5T). The domain organization of the protein is shown on the bottom left and the color code for the N-terminal (N, light blue), PAZ (magenta), Mid (yellow) and PIWI (green) domain is used throughout the figure. (A) The phosphate backbone of the nucleotides in the seed region of the guide interacts with residues of the Mid and PIWI domain. This way, the seed region is pre-organized to allow optimal stacking interactions for the recognition of matching target strands (pdb 3DLH, T. thermophilus Argonaute structure). In some Ago variants, the guide is kinked after the seed region. The kink (indicated by a purple arrow) is introduced by a helix in the linker region that connects the N-terminal and the C-terminal lobe. (B) The 5΄-nucleotide of the guide strand is buried in a binding pocket in the Mid domain, which is either lined with polar residues to stabilize a 5΄-phosphate (pdb 5G5T) or (C) with non-polar residues (pdb 5I4A) in case of a 5΄-hydroxyl bias of the Argonaute protein from M. piezophila. In both cases, some important residues stabilizing the guide's 5΄-end are highlighted. (D) In most crystal structures of Argonaute in complex with a guide, the 3΄-region of the guide is not resolved with the exception of the last three nucleotides. These nucleotides interact with the PAZ domain and the terminal 3΄-nucleotide is buried in a pocket formed by the PAZ domain. In case of Argonaute from M. jannaschii, a terminal 3΄-dT is preferred, although no base-specific interactions and only hydrogen bonding between the base and residues of Argonaute are observed (pdb 5G5T). However, the pocket is confined by the α6 helix.
The trajectory of a guide in the binary Ago-guide complex is well-defined. The guide strand can be divided into several functional sections, which fulfil different tasks during target recognition, binding and slicing (Wee et al.2012). The 5΄-end is anchored in the Mid domain binding pocket. The first eight bases of the guide counted from its 5΄-end comprise the seed region pre-organized for target recognition stabilized by the C-terminal lobe and the linker region between the two Ago lobes (Fig. 2A). The seed region is followed by the central part and the 3΄-end of the guide, which is attached to a binding pocket in the PAZ domain (Wang et al.2008a,b; Elkayam et al.2012; Schirle and MacRae 2012; Jung et al.2013; Zander et al.2014; Kaya et al.2016; Willkomm et al.2017). The first nucleotide at the 5΄-end of the guide, sometimes also called anchor nucleotide, is bent into a distinct binding pocket in the Mid-PIWI interface and therefore occluded from contacts to target nucleotides (Parker, Roe and Barford 2004; Ma et al.2005; Yuan et al.2005; Wang et al. 2008a; Elkayam et al.2012; Nakanishi et al.2012; Sasaki and Tomari 2012; Fig. 2A–C). Most of the Ago proteins described so far specifically recognize a phosphate group at the 5΄-end of the guide (Wang et al. 2008b; Frank, Sonenberg and Nagar 2010; Frank et al.2012; Miyoshi et al.2016; Willkomm et al.2017). The recognition of the phosphate group occurs in a highly conserved Mid binding pocket containing polar amino acids that are contacting the 5΄-phosphate either directly or via a coordinated metal ion to hold the guide strand in its position (Fig. 2B). However, a recently identified subfamily of pAgo proteins non-canonically selects 5΄-hydroxylated guide strands (Kaya et al.2016). The binding pocket of these Ago variants is composed of mainly hydrophobic amino acids that are stabilizing the 5΄-end of the guide with the 5΄-hydroxyl group being hydrogen-bonded to the phosphate group of the second base. The inverted first base is hold into place by π-stacking interactions with a tyrosine in the Mid domain (Fig. 2C). Moreover, a helix originating in the PIWI domain reduces the size of the Mid binding pocket, which additionally prevents the binding of bulky 5΄-phosphorylated guide strands. These different structural requirements might hint to different guide origins.
Another structural feature in the Mid domain binding pocket—the so-called nucleotide specificity loop (NSL)—is responsible for the recognition and selection of the first nucleotide at the 5΄-end of the guide in several Ago proteins (Frank, Sonenberg and Nagar 2010). In hAgo2, a bias towards guides with a 5΄-U has been observed and structural studies showed that specific contacts between residues of the specificity loop and uracil result in an increased affinity for a uridine (Frank, Sonenberg and Nagar 2010). Similar preferences for a specific 5΄-nucleotide have been also observed for archaeal MjAgo (preference for a 5΄-purine) and bacterial TtAgo (preference for a 5΄-dC) and RsAgo (preference for a 5΄-U) (Olovnikov et al.2013; Swarts et al.2014b; Willkomm et al.2017). Interestingly, the selection of a particular nucleotide at the 5΄-end of the guide strand is not always mediated by the NSL. A recent study showed that TtAgo preferentially selects target strands with a dG positioned at the 3΄-end of the target strand (3΄-G[t]) that is located opposite the 5΄-nucleotide of the guide. The 3΄-G[t] base is bound by a binding pocket formed by a residue on loop 2 and residues in the PIWI domain (Sheng et al.2014; Swarts et al.2017). Hence, the accumulation of 5΄-dC guides in TtAgo immunoprecipitations might also be a consequence of the preferred binding of the opposite target nucleotide. Interestingly, a similar binding pocket could be found in hAgo2 (Schirle, Sheu-Gruttadauria and MacRae 2014; Schirle et al.2015). Here, the binding of a 3΄-A[t] is mediated by a hydrogen-bond network in a binding pocket on the surface of hAgo2. Therefore, the concept of additional stabilizing interactions between a target and the Ago protein beyond guide-target complementarity might be found in other Ago proteins, too.
Whereas the 5΄-end stays bound during target binding, the 3΄-end of the guide is released from its binding pocket in the PAZ domain upon target binding caused by conformational changes especially in the N-lobe of the protein (Wang et al.2009; Parker 2010; Jung et al.2013; Sheng et al.2014; Zander et al.2014; Miyoshi et al.2016). There are no sequence-specific interactions between the 3΄-base and the PAZ binding pocket; however, in case of the archaeal MjAgo it was observed that the identity of the guide's 3΄-base influences the affinity of the Ago-guide complex (Willkomm et al.2017). Consequently, the efficiency of MjAgo-mediated target cleavage is also influenced by the base at the 3΄-end (Willkomm et al.2017). In case of MjAgo, the phosphate of a 3΄-dT nucleotide forms hydrogen bonds with residues Y194, H213 and Y218. Moreover, the α6 helix in the PAZ domain containing the residue H213 leads to a size reduction of the 3΄-end binding pocket (Fig. 2D). Apart from the adaptation of the binding pocket, the PAZ domain further stabilizes guide strands by its oligonucleotide/oligosaccharide (OB)-like fold (Lingel et al.2003; Song et al.2004; Hutvagner and Simard 2008).
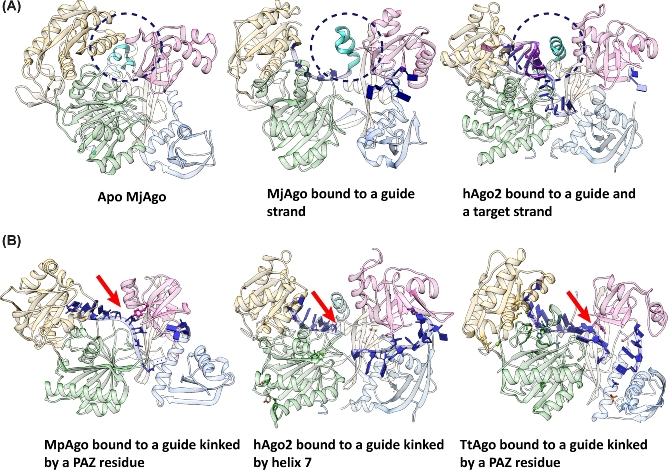
Nucleotides 2–8 of the guide strand counted from its 5΄-end are termed seed region. The bases of the seed region are solvent exposed in a helical conformation brought about by interactions between residues of the PIWI and Mid domain with the guide backbone. Therefore, the bases of the seed region can readily base pair with a matching target strand (Wang et al. 2008; Lambert, Gu and Zahler 2011; Elkayam et al.2012; Schirle and MacRae 2012; Swarts et al.2015b; Willkomm et al.2017; and Fig. 2A). The well-ordered seed region of the guide strand is followed by a significant kink introduced by a helix in the linker region (Fig. 3A) between the N-terminal lobe and the C-terminal lobe or residues of the PAZ domain (Fig. 3B). Kinking of the guide is a conserved feature found in all Ago-guide structures so far. The linker helix was proposed to act as a catalyst for seed-pairing accelerating target binding as well as dissociation (Klum et al.2017). It occludes the seed nucleotides beyond guide nucleotide 5 from base pairing with an incoming target strand and thereby creates a sub-seed region from guide nucleotide 2–5 (Klum et al.2017). On the one hand, upon recognition of a matching target strand, the linker helix moves and stabilizes base pairing by interactions with the minor groove of the guide-target duplex. On the other hand, guide-target interactions are weakened due to the kink in the guide strand. Therefore, the linker helix plays an important role in the binary as well as in the ternary complex (Klum et al.2017). This is also reflected by structural data that reveal that this helix is present in its full length only in the guide- and guide-target-bound complexes of Ago (Fig. 3A). A crystal structure of apo MjAgo reveals that the linker helix resembles a bi-partite helical motif in the absence of a guide (Willkomm et al.2017; Fig. 3A). If a guide is bound, the bi-partite motif shifts to the full-length linker helix that shapes the guide strand for efficient target binding (Schirle and MacRae 2012; Schirle, Sheu-Gruttadauria and MacRae 2014; Klum et al.2017; Willkomm et al.2017). Subsequent binding of a matching target strand requires a conformational change of approximately 4 Å to avoid a clash of the helix with target nucleotides that pair with the guide strand beyond guide nucleotide 5 in hAgo2 (Schirle et al.2014). Following extended base-pairing between guide and target, the linker helix interacts with the surface of the minor groove of the guide-target duplex and therefore stabilizes the interaction. Mismatches within the minor groove are disturbing interactions between the groove and the helix and therefore the release of non-matching targets is facilitated. In some cases, pAgo variants do not contain this linker helix. Here, the guide is kinked by residues from the PAZ domain (Wang et al. 2008b; Kaya et al.2016; Fig. 3B). It seems likely that the PAZ domain-induced kinking has a similar effect on Ago function as observed for the linker helix-induced kinking. Mutation of the kinking residues does not lead to reduced cleavage efficiency (Kaya et al.2016). Hence, kinking of the guide seems to be mainly necessary to avoid binding of targets with minimal complementarity.
Figure 3.
The guide strand is kinked to shape the guide for efficient target recognition. (A) Conformational changes of the linker helix upon binding of guide and target strand. In the Apo enzyme, the helix is present in a bi-partite motif, whereas in the binary and ternary complex the helix is found as an extended helix (seen in the structures of Argonaute from M. jannaschii), MjAgo, in its apo form (pdb: 5G5S) and in complex with a guide (pdb: 5G5T). Thereby, in the absence of the target, the guide is kinked and upon target binding the linker helix is stabilizing the guide-target duplex by interactions with the minor groove (hAgo2 (pdb: 4W5T)). Dotted circles indicat the position of the linker helix. (B) The sharp bend in the guide strand is either caused by residues that extend from the PAZ domain (e.g. M. piezophila [MpAgo] (pdb: 5I4A) and T. thermophilus Argonaute [TtAgo] (pdb: 3DLH) or by the linker helix (e.g. human Argonaute 2 [hAgo2] (pdb: 4W5N) and MjAgo (see A)). The red arrows indicate the position of the kink in the guide nucleic acid.
The 3΄-supplementary region located adjacent to the kink is not resolved in most Ago structures with the exception of the last three nucleotides of the guide (termed tail region) that can be clearly assigned in crystal structures (Wang et al.2008a,b; Elkayam et al.2012; Schirle and MacRae 2012; Jung et al.2013; Kaya et al.2016; Willkomm et al.2017). Information about the arrangement of the guide's supplementary region is only available for hAgo2. Here, it could be shown that these nucleotides with their bases facing inwards are forced through a narrow channel formed by the N-terminal and the PAZ domain (Schirle, Sheu-Gruttadauria and MacRae 2014). Because the 5΄- and 3΄-end of the guide is fixed in the Mid and the PAZ domain in all Agos, respectively, it can be deduced that the position of this part of the guide strand is similar for most Ago variants characterized so far.
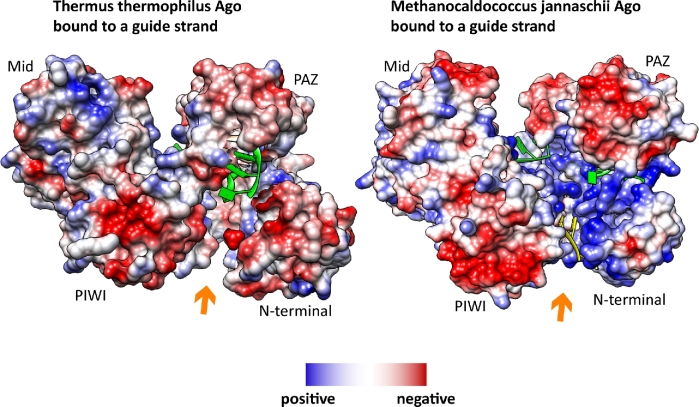
However, an additional possibility for the nucleic acid arrangement was found in case of MjAgo. An electron density representing nucleotides was found in between the N-terminal and the PIWI domain in the structure of the binary complex (Willkomm et al.2017). Interestingly, this region is exceptionally wide and very positively charged in MjAgo as compared to other Ago variants. This positively charged area seems to facilitate interactions with negatively charged nucleic acids (Fig. 4). Furthermore, this channel is important to enable efficient target cleavage for a subset of substrates (Willkomm et al.2017). This newly identified putative secondary nucleic acid binding channel adds to the complexity of mechanisms that enable pAgos to recognize and regulate target nucleic acids. Potentially, this binding channel allows MjAgo to load complex substrates like plasmid DNA or long dsDNA. Also in case of MpAgo the nucleic acid binding channel that is occupied by the guide strand is different from the canonical binding channel (Kaya et al.2016). In this case, the kink in the guide introduced by the PAZ domain is more pronounced than in other Ago variants (Fig. 3B).
Figure 4.
Unusual location of nucleotides in the Argonaute structure of M. jannaschii suggests the presence of a putative secondary nucleic acid binding channel. The crystal structure of M. jannaschii Argonaute (MjAgo) (pdb 5G5T) revealed electron density for nucleotides in a channel between the N-terminal and the PIWI domain (colored in yellow). The guide DNA in the canonical binding channel is indicated in green. Coulombic surface coloring shows that the channel in between the N-terminal and the PIWI domain (indicated by an orange arrow) is highly positively charged in case of MjAgo, which is not the case in e.g. the Ago protein from T. thermophilus (3DLH).
The mechanistic diversity of pAgos is also reflected by structural data of ternary complexes. Especially, the position and function of the N-terminal lobe shows a high degree of flexibility. In some cases, the N-terminal domain has the function of a wedge that blocks formation of base pairs between the guide and the target strand beyond guide nucleotide 16. Here, the guide is threaded through the N-PIWI tunnel and the target is located in between the PAZ and the N-terminal domain (Wang et al. 2009; Parker 2010; Kwak and Tomari 2012; Sheng et al.2014; Fig. 5A). In hAgo2, the N-terminal domain fulfils an active wedging function to unwind the passenger strand from the guide strand (Kwak and Tomari 2012). However, in some pAgos the N-terminal domain has a deviant function leading to different trajectories of the guide-target duplex (Fig. 5). For example, the N-terminal domain of MpAgo adopts a very unique position. Unlike in Agos with a wedge-like N-terminal domain base pairing in the 3΄-region of the guide-target duplex is not disturbed by the MpAgo N-terminal domain (Doxzen and Doudna 2017; Fig. 5B). To achieve this, the N-terminal domain is positioned closer to the PAZ domain than in case of the wedging N-terminal domain. Positively charged residues, which are positioned in a cleft at the PAZ-N-interface, stabilize the phosphate backbone of the target strand (Doxzen and Doudna 2017). These structural features allow for a straight guide-target duplex pathway within MpAgo (Fig. 5B). Similarly, in case of RsAgo the guide-target duplex is also not disrupted by the N-terminal domain. In order to maintain base pairing between guide and target, the N-terminal and the PIWI domain act cooperatively (Miyoshi et al.2016). However, the duplex bound by RsAgo does not adopt a completely straight conformation but is diverted by approximately 40° in comparison to the MpAgo-bound duplex (Doxzen and Doudna 2017). Doxzen and Doudna (2017) suggest that this bent duplex conformation is caused by a closer proximity of the N-terminal and the PIWI domain.
Figure 5.
The position of the N-terminal domain influences the trajectory of the guide-target duplex. (A) Thermus thermophilus Argonaute bound to a guide (blue) and a target strand (red) (pdb 4NCB). The 5΄- and the 3΄- end of the guide strand are indicated in the figure. Here, the N-terminal domain has a wedging function and disrupts base pairing beyond guide nucleotide 16. (B) Marinitoga piezophila Argonaute (MpAgo) bound to a guide (blue) and a target strand (red) (pdb 5UX0). The guide-target duplex occupies a nucleic acid binding channel across the N-terminal domain and maintains its base pairing even in the 3΄-region of the guide. The target backbone is stabilized by the residues in the interface between the N-terminal and the PAZ domain. (C) Rhodobacter sphaeroides Argonaute bound to a guide (blue) and a target strand (red) (pdb 5AWH). As observed for MpAgo, the N-terminal domain does not interrupt base pairing of the guide-target duplex. However, in contrast to MpAgo, the RsAgo-bound guide-target duplex is bent into the N-PIWI channel.
Taken together, the N-terminal domain can either disrupt or stabilize the duplex. Miyoshi et al. (2016) propose that this ambivalent function of the N-terminal domain directly correlates with the substrate usage of the respective Ago variant. For example, hAgo2 binds double-stranded substrates with subsequent release of the passenger strand. Here, the wedging function of the N-terminal domain is required. In contrast, RsAgo mainly recognizes unstructured transcripts that do not require unwinding. Miyoshi et al. (2016) furthermore suggest a higher stability of the guide-target duplex in case the N-terminal domain does not function as a wedge because complete base pairing between guide and target is possible. This would lead to an increased stability and consequently longer dwell times of the Ago-guide complex on the corresponding target strand, which suggests different mechanisms to regulate target nucleic acid levels.
EVOLUTIONARY ASPECTS AND DIVERSITY IN THE PROKARYOTIC ARGONAUTE WORLD
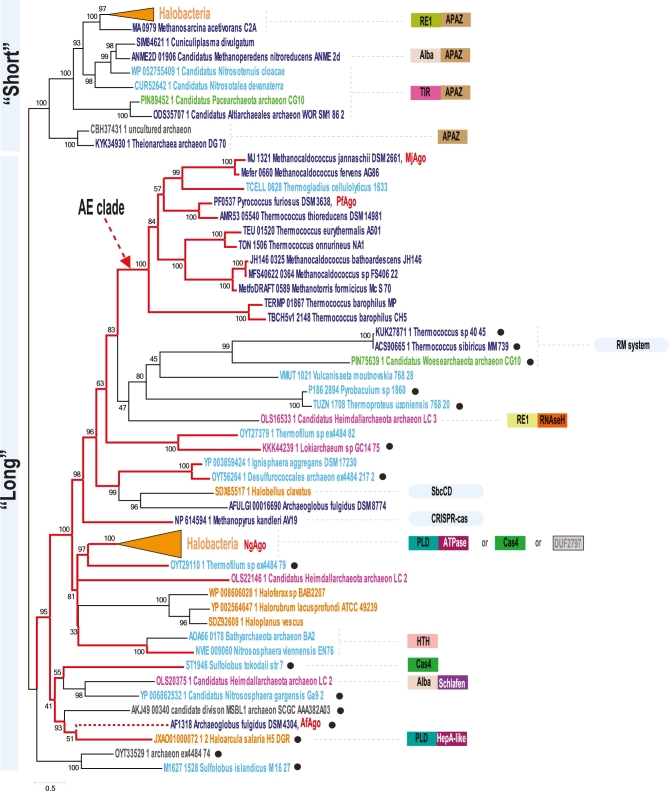
pAgos as any other defense system evolve under constant pressure of the proverbial ‘arms race’ with the targets against which this system is acting (Stern and Sorek 2011; Makarova, Wolf and Koonin 2013). This results in a high rate of protein divergence, domain rearrangement and frequent horizontal transfer (HGT) of the respective genes. Indeed, the detailed comparative genomic and phylogenetic analysis of pAgos published before revealed an evidence for both evolutionary processes (Swarts et al. 2014a). It has been shown that the distribution of pAgos is very patchy in genomes of both archaea and bacteria and the phylogenetic tree based on these protein sequences does not follow the respective species tree. This suggests a dominant role of HGT in the evolution of these families. Many pAgos have mutations of the catalytic residues suggesting that they are inactivated (unable to cleave nucleic acids) and many protein sequences are poorly conserved beyond Mid and PIWI domains suggesting their fast evolutionary rate and sub-functionalization. In this review, we focus primarily on archaeal pAgos, the group with many diverse representatives of this protein family. In agreement with previously published results phylogenetic analysis revealed two major clades of archaeal pAgos (Swarts et al. 2014a; Fig. 6). The first clade, ‘short’ pAgos, consists of proteins that have only Mid and PIWI domains and their PIWI domain is inactivated. These ‘short’ pAgos are always associated with the so-called APAZ domain (analog of PAZ), which is either fused to them or encoded separately. This domain is not structurally characterized yet. However, using sensitive methods of sequence similarity detection, such as HHpred (Soding, Biegert and Lupas 2005), a limited sequence similarity of the APAZ domain with N-terminal domain of Ago could be detected in agreement with predictions published previously (Burroughs, Ando and Aravind 2014). This suggests that the APAZ domain is rather not a PAZ-like domain but a homolog of the N-terminal pAgo domain. Most often an APAZ domain is fused to predicted nucleases, enzymes that could be involved in DNA modification or DNA-binding proteins comprising a two-component, guide-effector system (Fig. 6). The second clade, ‘long’ pAgos, is quite diverse, and it also includes some proteins with an inactivated PIWI domain and even proteins lacking the N-terminal region of Ago. Ironically, over a few last years the number of the pAgos lacking N-terminal domains, which belong to ‘long’ pAgo clade increased significantly (Fig. 6). This makes the description of this clade as ‘long pAgos’ rather misleading. Unlike bona fide ‘short’ pAgos, they are not associated with an APAZ domain, but their PIWI domain is also often inactivated. Considering that short pAgo sequences are not monophyletic, it appears that the loss of N-terminal domains and inactivation of PIWI domain are frequent events that occurred many times independently during the evolution of this family. Some of these short Agos are encoded in predicted operons or fused with other genes whose products can also play a role of effectors guided to the target by Agos, including predicted nucleases, DNA-binding proteins and even entire restriction-modification systems (Fig. 6).
Figure 6.
Phylogenetic tree of archaeal Argonaute proteins. Maximum-likelihood phylogenetic unrooted tree was built using the PhyML program (Guindon et al.2010) based on a multiple alignment, which was built by the MUSCLE program (Edgar 2004) for conserved blocks of MID and PIWI domains (211 informative positions) for 84 representative archaeal Argonaute proteins. To avoid redundancy, all archaeal proteins found using the PSI_BLAST program (Altschul et al.1997) in the NCBI non-redundant database (as of November 30, 2017) were clustered with 95% of amino acid identity and 90% length coverage, and a single representative from each cluster was taken for further analysis. The PhyML program was also used to compute bootstrap values indicated for all branches. Major archaea lineages are color coded as follows: orange—Halobacteria; dark blue—other Euryarchaea; light blue—Thaumarcharchaeota Aigarchaeota Crenarchaeota Korarchaeota (TACK) superphylum, green—Diapherotrites Parvarcheota Aenigmarchaeota Nanoarchaeota Nanohaloarchaea (DPANN) superphylum, purple—Asgard phylum. Collapsed branches corresponding to large groups of Halobacterial sequences are shown as triangles of the respective color. Species assignment for environmental archaea should be considered as tentative. Agos that are discussed in this manuscript are denoted by short names and colored red next to the full organism name. Red branches lead to the PIWI domains with preserved catalytic triad. The red dotted branch indicated that in this Ago variant a single canonical catalytic residue is substituted. The eAgo archaeal sister branch is indicated by the red arrow and denoted as AE-clade. Sequences in the ‘long’ pAgo clade lacking N-terminal domains are indicated by black circle next to the organism name. Genes or domains identified in the Ago neighborhoods are shown on the right side of the tree by colored boxes. Homologous domains are shown by boxes of the same color or pattern. Multigene defense systems encoded next to pAgo genes are shown by oval shapes. Abbreviations: RE1 and RE2 are predicted nucleases of restriction endonuclease superfamily; TIR—nucleotide-binding/processing domains of TIR family; Schlafen—predicted regulatory ATPase; APAZ—domain similar to the N-terminal domain of Ago, ‘analog of PAZ’ domain; Cas4—Cas4-like subfamily nuclease of restriction endonuclease superfamily; PLD—predicted nuclease of phospholipase D superfamily; Alba—chromatin binding protein Alba. Gray boxes indicate distinct families of uncharacterized proteins.
It has been shown that eAgos form a sister clade with pAgos from euryarchaeal thermophiles (Swarts et al. 2014a). Now we identified the first sequence from the anaerobic hyperthermophilic crenarchaeon, Thermogladius cellulolyticus, in this well-supported archaea-eukaryotic clade (AE-clade, Fig. 6). Several PIWI domain containing proteins have been also found in metagenomic assemblies of uncultured archaea, including those which were assigned to the Asgard group. Based on the analysis of phylogenetic markers Asgard archaea have been shown to be a sister group to eukaryotes (Zaremba-Niedzwiedzka et al.2017). Surprisingly, four Asgard pAgos do not form a monophyletic clade as it would be expected for related genomes and none of them groups with the AE-clade. Therefore, this analysis did not reveal any evidence that Argonautes were inherited by eukaryotes from the Asgard group. It appears that Asgard pAgos were acquired independently or even could be misassigned to the Asgard group. Despite the fact that all pAgos are structurally similar, amino acid signatures that determine their specificities to both guide and target nucleic acid were not identified. Moreover, only in eukaryotes the specificity to RNA guide and RNA target appears to be fixed. In pAgos, the guide/target specificity apparently could be shaped into any of the four possible specificity combinations. It is quite likely that the functional plasticity of the PIWI domain is an inherent feature of the ancient RNase H fold that is known to be able to interact with different single stranded nucleic acids (Ma et al.2008). This fold, in addition to PIWI domain containing proteins, includes a plethora of diverse nucleases, DDE transposases and RuvC-like domains of transposable elements and CRISPR-Cas system effectors as Cas9 and Cas12 (Nesmelova and Hackett 2010; Majorek et al.2014; Koonin, Makarova and Zhang 2017). This plasticity can be instrumental in the future to harness Agos for different biotechnological applications including genome editing.
CONCLUSIONS
Over the last years, an expanded set of pAgo variants has been characterized functionally as well as structurally. This opened our eyes for the diversity and plasticity of the pAgo clade including for example the unexpected ‘chopping activity’ of TtAgo and MjAgo. These and other pAgos (e.g. PfAgo) are handling up to three different nucleic acid strands (guide strand plus dsDNA). Hence, it would be of interest to understand the structural organisation of such ‘non-canonical’ Ago-substrate complexes. Additionally, so far only full-length pAgos have been characterized and we have no understanding of the functions and interactions of short pAgos. Organisms that encode short Agos seemingly have split up the protein into a separate Mid-PIWI protein and a protein that encompasses the N-terminal domain. Do these proteins form a functional complex? Do the co-encoded proteins (e.g. DNA binding and modification enzymes) form a complex with the APAZ domain and are these complexes dedicated to other functions? This question also arises for long pAgos that are co-encoded with Cas1 and Cas2 and for catalytically inactive Agos that are found in an operon with nucleases. Taken together, there is still much to discover in the bacterial and archaeal Ago world and these activities could be potentially harnessed to design tailor-made gene editing enzymes (Hegge, Swarts and van der Oost 2017). A recent example is the archaeal PfAgo, which can be used as programmable restriction endonuclease eliminating the need for specific restriction site sequences in the DNA of interest (Enghiad and Zhao 2017).
Acknowledgements
We gratefully acknowledge financial support by the Deutsche Forschungsgemeinschaft [DFG grant no. GR 3840/2-1 and SFB960-TP7 to DG] and by the intramural program of the US Department of Health and Human Services (to the National Library of Medicine) to KSM.
Conflict of interest. None declared.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, Aravind L. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip Rev RNA 2014;5:141–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. One loop to rule them all: the Ping-Pong cycle and piRNA-guided silencing. Trends Biochem Sci 2016;41:324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxzen KW, Doudna JA. DNA recognition by an RNA-guided bacterial Argonaute. PLoS One 2017;12:e0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016;538:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004;5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001;411:494–8. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001;15:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A et al. The structure of human argonaute-2 in complex with miR-20a. Cell 2012;150:100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enghiad B, Zhao H. Programmable DNA-guided artificial restriction enzymes. ACS Synth Biol 2017;6:752–7. [DOI] [PubMed] [Google Scholar]
- Frank F, Hauver J, Sonenberg N et al. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J 2012;31:3588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5΄-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010;465:818–22. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59:307–21. [DOI] [PubMed] [Google Scholar]
- Hegge JW, Swarts DC, van der Oost J. Prokaryotic Argonaute proteins: novel genome-editing tools? Nat Rev Microbiol 2018;16:5–11. [DOI] [PubMed] [Google Scholar]
- Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci 2016;371, 20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. Protection against Foreign DNA. In: Storz G, Hengge R. Bacterial Stress Responses, 2nd edn. American Society of Microbiology; 2011 [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22–32. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol 2015;22:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RD et al. CRISPR-Cas: Adapting to change. Science 2017;356, eaal5056. [DOI] [PubMed] [Google Scholar]
- Jung SR, Kim E, Hwang W et al. Dynamic anchoring of the 3΄-end of the guide strand controls the target dissociation of Argonaute-guide complex. J Am Chem Soc 2013;135:16865–71. [DOI] [PubMed] [Google Scholar]
- Kaya E, Doxzen KW, Knoll KR et al. A bacterial Argonaute with noncanonical guide RNA specificity. P Natl Acad Sci USA 2016;113:4057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman LM, Kelman Z. Archaeal DNA replication. Annu Rev Genet 2014;48:71–97. [DOI] [PubMed] [Google Scholar]
- Klum SM, Chandradoss SD, Schirle NT et al. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J 2018;37:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 2012;19:145–51. [DOI] [PubMed] [Google Scholar]
- Lambert NJ, Gu SG, Zahler AM. The conformation of microRNA seed regions in native microRNPs is prearranged for presentation to mRNA targets. Nucleic Acids Res 2011;39:4827–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E et al. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 2003;426:465–9. [DOI] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 2017;170:714–726.e10. [DOI] [PubMed] [Google Scholar]
- Ma BG, Chen L, Ji HF et al. Characters of very ancient proteins. Biochem Biophys Res Commun 2008;366:607–11. [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G et al. Structural basis for 5΄-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005;434:666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majorek KA, Dunin-Horkawicz S, Steczkiewicz K et al. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res 2014;42:4160–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 2013;41:4360–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013;14:447–59. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito K, Murakami R et al. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat Comms 2016;7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP et al. Structure of yeast Argonaute with guide RNA. Nature 2012;486:368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesmelova IV, Hackett PB. DDE transposases: structural similarity and diversity. Adv Drug Deliv Rev 2010;62:1187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov I, Chan K, Sachidanandam R et al. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell 2013;51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou AC, Adam PS, Stavros P et al. HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses. Extremophiles 2016;20:695–709. [DOI] [PubMed] [Google Scholar]
- Parker JS. How to slice: snapshots of Argonaute in action. Silence 2010;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J 2004;23:4727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek MJ, Werner A. Endogenous siRNAs: regulators of internal affairs. Biochm Soc Trans 2014;42:1174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 2005;12:340–9. [DOI] [PubMed] [Google Scholar]
- Sasaki HM, Tomari Y. The true core of RNA silencing revealed. Nat Struct Mol Biol 2012;19:657–60. [DOI] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science 2012;336:1037–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, Chandradoss SD et al. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. eLife 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science 2014;346:608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Zhao H, Wang J et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. P Natl Acad Sci USA 2014;111:652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005;33:W244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ et al. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004;305:1434–7. [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays 2011;33:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Hegge JW, Hinojo I et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res 2015b;43:5120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature 2014b;507:258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Koehorst JJ, Westra ER et al. Effects of Argonaute on gene expression in Thermus thermophilus. PLoS One 2015a;10:e0124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y et al. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 2014a;21:743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Szczepaniak M, Sheng G et al. Autonomous generation and loading of DNA guides by bacterial argonaute. Mol Cell 2017;65:985–998.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 2008a;456:921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 2009;461:754–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S et al. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008b;456:209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE et al. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 2012;151:1055–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willkomm S, Oellig CA, Zander A et al. Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein. Nat Microbiol 2017;2:17035. [DOI] [PubMed] [Google Scholar]
- Willkomm S, Zander A, Gust A et al. A prokaryotic twist on argonaute function. Life 2015;5:538–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 2005;19:405–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander A, Holzmeister P, Klose D et al. Single-molecule FRET supports the two-state model of Argonaute action. RNA Biol 2014;11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander A, Willkomm S, Ofer S et al. Guide-independent DNA cleavage by archaeal Argonaute from Methanocaldococcus jannaschii. Nat Microbiol 2017;2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017;541:353–8. [DOI] [PubMed] [Google Scholar]